SUMMARY
The enhanced disease resistance 1 (edr1) mutant of Arabidopsis confers enhanced resistance to bacterial and fungal pathogens. To better understand how edr1‐mediated resistance occurs, we performed transcriptome analyses on wild‐type and edr1 plants inoculated with the fungal pathogen Golovinomyces cichoracearum (powdery mildew). The expression of many known and putative defence‐associated genes was more rapidly induced, and to higher levels, in edr1 plants relative to the wild‐type. Many of the genes with elevated expression encoded WRKY transcription factors and there was enrichment for their binding sites in promoters of the genes upregulated in edr1. Confocal microscopy of transiently expressed EDR1 protein showed that a significant fraction of EDR1 was localized to the nucleus, suggesting that EDR1 could potentially interact with transcription factors in the nucleus. Analysis of gene ontology annotations revealed that genes associated with the endomembrane system, defence, reactive oxygen species (ROS) production and protein kinases were induced early in the edr1 mutant, and that elevated expression of the endomembrane system, defence and ROS‐related genes was maintained for at least 4 days after infection.
INTRODUCTION
Plants have evolved complex mechanisms to defend themselves against pathogens (Bent and Mackey, 2007). To identify the genes regulating plant defence responses, we have previously screened for Arabidopsis (Arabidopsis thaliana) mutants with enhanced resistance to virulent pathogens (Frye and Innes, 1998). The enhanced disease resistance 1 (edr1) mutant displays enhanced resistance to the fungus Golovinomyces cichoracearum, an obligate biotroph and causal agent of powdery mildew on Arabidopsis and many cucurbit species (Adam et al., 1999; Adam and Somerville, 1996). Although G. cichoracearum forms conidiophores (stalks of asexual spores) on the surface of susceptible leaves, resistance in edr1 is manifested as necrotic lesions at the site of infection and a reduction in conidiophores (Frye and Innes, 1998). In addition, edr1 mutants show greater callose deposition and form more papillae, and at an earlier time, than do wild‐type Col‐0 plants. EDR1 encodes a protein with a C‐terminal kinase domain and a putative N‐terminal regulatory domain (Frye et al., 2001). A recombinant protein containing the EDR1 kinase domain only is able to autophosphorylate and can phosphorylate the common kinase substrate myelin basic protein in vivo, demonstrating that EDR1 does, indeed, have kinase activity (Tang and Innes, 2002).
The enhanced resistance of the edr1 mutant is suppressed by mutations that reduce salicylic acid (SA) production (sid2, eds1 and pad4) or block SA perception (npr1/nim1) (Frye et al., 2001; Tang et al., 2005). Transgenic expression of NahG, which lowers endogenous SA levels, also eliminates the edr1‐mediated enhanced disease resistance phenotype (Frye et al., 2001). In contrast with the requirement for SA signalling in edr1‐mediated resistance, neither ethylene (ET) nor jasmonic acid (JA) appears to be necessary, as mutations in the ETHYLENE INSENSITIVE 2 (EIN2) or CORONATINE INSENSITIVE 1 (COI1) gene do not alter edr1‐mediated disease resistance (Frye et al., 2001).
In addition to regulating responses to pathogens, EDR1 also regulates responses to abiotic stresses, such as drought. When grown under drought conditions, the edr1 mutant is dwarfed and forms lesions, whereas growth is normal under optimal conditions (Tang et al., 2005). These phenotypes are suppressed by mutations in the SA signalling pathway (eds1, pad4 or npr1), indicating that the drought response is also dependent on SA and may share similarity with the pathogen response in edr1. In addition, the F‐box protein mutant, ore9, which shows delayed senescence in response to ET, restores wild‐type growth under drought conditions to the edr1 mutant, but does not abate the drought‐induced lesion phenotype (Tang et al., 2005).
EDR1 is most similar to the ET response regulator, CTR1, and four other proteins of unknown function in Arabidopsis. Despite this similarity, edr1 mutants have a normal triple response, unlike ctr1 mutants. However, when edr1 mutants are treated with ET, they senesce more rapidly than wild‐type Col‐0 (Frye et al., 2001). This response can be abolished by the presence of ein2, an ET signalling mutation, but it does not require SA responses (Tang et al., 2005). Taken together, the responses to pathogen, drought and ET in edr1 imply that EDR1 negatively regulates cell death in response to various stimuli.
A mechanism for CTR1‐mediated ET regulation proposes that two F‐box proteins, EBF1 and EBF2, target ET‐inducible transcription factors for proteasome‐mediated degradation (Guo and Ecker, 2003; Potuschak et al., 2003). This degradation is dependent on an active CTR1 protein and, in the absence of CTR1, the transcription factor EIN3 can accumulate. ET represses CTR1 activity, preventing the activity of EBF1/2, and this allows EIN3 to accumulate and activate ET responses. It is possible that EDR1 may negatively regulate cell death responses in a similar manner. Mutations in the F‐box protein ORE9 can block ET‐induced cell death in the edr1 mutant, as well as drought‐induced growth inhibition, suggesting that a repressor of these phenotypes accumulates in the ore9 mutant. However, not all edr1‐mediated responses can be blocked by ore9, indicating that ORE9 regulates only a subset of EDR1‐mediated responses.
All known edr1 mutant phenotypes can be suppressed by a specific missense mutation in the KEEP ON GOING (KEG) gene, which encodes an E3 ubiquitin ligase that is responsible for the degradation of the abscisic acid (ABA)‐inducible transcription factor ABI5. This result suggests that EDR1 may mediate cell death via a mechanism similar to the regulation of ET responses by CTR1, namely the targeting of transcription factors to the proteasome. Consistent with this model, quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analyses have revealed that some ABA‐inducible genes are expressed more highly in edr1 mutant plants, and this enhanced expression is abolished by the keg‐4 mutation (Wawrzynska et al., 2008).
Despite the extensive work performed on the edr1 mutant, there is still little information on how EDR1 negatively regulates cell death, particularly in response to G. cichoracearum. To investigate the control of cell death in the edr1 mutant, we performed microarray experiments to identify the genes whose regulation was affected by the edr1 mutation in the presence of powdery mildew. As expected, many of the genes upregulated in the edr1 mutant were defence response genes, indicating that EDR1 negatively regulates defence signalling pathways and that the removal of such repression in the edr1 mutant results in enhanced resistance. Significantly, the EDR1 protein was found to localize, at least part of the time, to the nucleus, suggesting that EDR1 may regulate directly the stability and/or activity of defence‐related transcription factors.
RESULTS
Identification of genes regulated by EDR1
Wild‐type Col‐0 and edr1 mutant plants were inoculated with G. cichoracearum and tissue was collected at 18, 36 and 96 h post‐inoculation (hpi). By 18 h, the fungus has germinated, penetrated the epidermal cells and begun to form haustoria (Fabro et al., 2008). By 36 h, infected cells have begun to form papillae and deposit callose. By 96 h, stalks of asexual spores (conidia) begin to form on wild‐type leaves, but very few formed on edr1 leaves; however, no cell death is observable in wild‐type or edr1 plants, even at 96 hpi, and visible powder has not begun to form (Frye and Innes, 1998). Tissue was also collected from plants immediately prior to inoculation as an uninfected control (0 h). High‐quality RNA was prepared from the collected tissue, including four biological replicates per genotype per time point, and analysed using Affymetrix ATH1 gene chips.
To identify genes that were negatively regulated by EDR1, we first selected genes that were upregulated by more than two‐fold in edr1 relative to wild‐type Col‐0 at any time point and that were determined to be significantly different (P≤ 0.05) using the Benjamini–Hochberg correction (Benjamini and Hochberg, 1995). This correction should reduce the false discovery rate to less than 5%. In addition, genes that were upregulated by more than two‐fold in edr1 or wild‐type Col‐0 after inoculation relative to uninoculated plants were selected. These datasets were then compared to identify genes that were upregulated in an edr1‐ and pathogen‐dependent manner. Genes whose expression was higher in edr1 than in Col‐0 at any time and was also higher in either Col‐0 or edr1, or both, after pathogen inoculation were selected (areas bounded by the yellow oval in Fig. 1; Table S1, see Supporting Information). This subset of genes contained 553 probe sets corresponding to 545 annotated genes. We refer to this subset as the edr1&pm‐upregulated gene set. It should be noted that, because of cost issues, we did not include an uninoculated control at each time point; thus, it is a formal possibility that some of the genes included in the powdery mildew‐induced gene set are upregulated as a result of circadian changes in gene expression instead of, or in addition to, powdery mildew infection. Nevertheless, all genes included in the edr1&pm‐upregulated gene set were more highly expressed in the edr1 mutant than in wild‐type Col‐0 during at least one time point.
Figure 1.
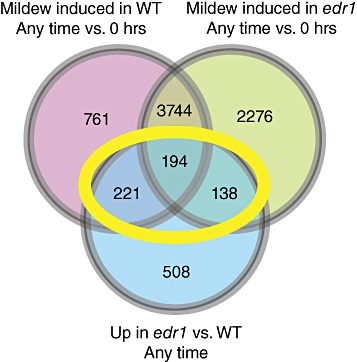
Venn diagram showing overlap between powdery mildew‐induced and enhanced disease resistance 1 (edr1)‐induced gene sets. The yellow oval indicates the set of edr1&pm‐upregulated genes used for the majority of the analyses. WT, wild‐type.
Upregulation of defence genes in the edr1 mutant
Many of the genes identified in the edr1&pm‐upregulated gene set (Table S1) are known to be involved in plant defence responses. For example, PBS3 and PAD4 are both required for SA accumulation (Glazebrook et al., 1996; Nobuta et al., 2007), and PR‐3, PR‐4, THI2.1, PDF1.4 and ATTI1 are associated with JA‐inducible defences. Commonly, SA and JA defences have antagonistic modes of action (Li et al., 2004), but may also have additive effects depending on hormone concentration and the type of pathogen encountered (Mur et al., 2006). PR‐3 and PR‐4 encode a chitinase and chitin‐binding protein with antifungal activity, respectively (Potter et al., 1993; Verburg and Huynh, 1991). THI2.1, PDF1.4 and ATTI1 are all induced by JA and may also have antifungal or antimicrobial activity (Epple et al., 1995; Silverstein et al., 2005). Other defence‐associated genes in the edr1&pm‐upregulated gene set include 12 genes with leucine‐rich repeat (LRR) domains, including three encoding a Toll/interleukin‐1 receptor (TIR) class nucleotide‐binding leucine‐rich repeat (NB‐LRR) disease resistance protein.
The edr1&pm‐upregulated gene set also contains five receptor‐like kinases (RLKs), including RLK5 and RLK6. RLK5 and RLK6 were identified in a search for genes that are regulated by pathogen‐inducible transcription factors and are known to be induced by SA treatment and by pathogens (Du and Chen, 2000). In addition to the RLKs identified, there are also eight putative LRR kinases. LRR kinases can act as receptors to transmit signalling information, often during defence responses (Shiu and Bleecker, 2001). An additional 11 kinases are present in the edr1&pm‐upregulated gene set, indicating that phosphorylation cascades are important for the defence mechanism induced by G. cichoracearum in edr1.
Significantly, the edr1&pm‐upregulated gene set contains at least 28 genes encoding transcription factors. The largest family of transcription factors in this gene set is the WRKY family, of which there are eight members, or 1.5% of the annotated genes in this gene set, compared with 61 of the 22 810 genes on the ATH1 chip (0.27%). There are at least 75 WRKYs in the Arabidopsis genome and many WRKYs have been implicated in controlling aspects of plant defence responses (Bhattarai et al., 2010; Eulgem et al., 2000). WRKY transcription factors have a conserved DNA‐binding domain, which contains a WRKY motif that is required for DNA binding (Ciolkowski et al., 2008). WRKYs also contain a zinc‐binding region in the DNA‐binding domain. WRKYs bind to the sequence (T)TGAC(C/T), known as the W‐box, in the promoter sequence of target genes (Ciolkowski et al., 2008).
A second class of transcription factors over‐represented in the edr1&pm‐upregulated gene set is the AP2/ERF family. There are seven AP2/ERFs present, comprising 1.28% of the genes, compared with 89 on the ATH1 chip, or 0.39% of the genes. AP2/ERF transcription factors were originally identified as genes that were induced in response to the hormone ET, but have since been shown to include genes that are induced in response to pathogen and during JA‐inducible defences (Gutterson and Reuber, 2004). Indeed, one AP2/ERF, ORA59, has been found to integrate JA‐ and ET‐mediated signalling pathways (Pre et al., 2008). AP2/ERF family transcription factors bind to the GCC‐box (GCCGCC) to activate transcription.
The edr1&pm‐upregulated gene set is also enriched for genes involved in reactive oxygen species (ROS) accumulation and turnover. Eleven genes annotated with putative peroxidase function were identified. Peroxidases act to oxidize other molecules through the use of H2O2 or O2, either as a way of preventing toxicity or to signal (Yoshida et al., 2003). One of the peroxidase genes identified in the edr1&pm‐upregulated gene set, ATP2a, has also been shown to be upregulated in response to wounding and may play a role in pathogen responses (Cheong et al., 2002).
Other genes that are present in the edr1&pm‐upregulated gene set include six genes that encode small heat shock proteins (sHSPs). sHSPs can act as molecular chaperones and have been identified in the regulation of responses to various stresses and developmental processes, including apoptosis (Basha et al., 2006). Genes encoding glutathione S‐transferase (GST) genes are also present. GSTs are involved in the regulation of the cellular redox state and are often induced during defence responses (Wagner et al., 2002).
Nine genes encoding flavin adenine dinucleotide (FAD)‐binding domain‐containing proteins were also identified as part of the edr1&pm‐upregulated gene set. These genes are closely related to a sunflower gene that encodes an antimicrobial protein with carbohydrate oxidase activity, Ha‐CHOX (Custers et al., 2004). Ha‐CHOX catalyses the production of H2O2 using glucose as a substrate. When Ha‐CHOX is overexpressed in tobacco, it confers greater resistance to Pectobacterium carotovorum. The nine FAD‐binding domain‐containing genes in the edr1&pm‐upregulated gene set are all members of the same family of proteins, sharing similarity across their entire length (Fig. S1, see Supporting Information).
To determine whether the FAD‐binding domain genes were induced by other pathogens, data from publicly available microarrays were analysed using the Genevestigator web‐based interface (https://www.genevestigator.ethz.ch/gv/index.jsp) (Hruz et al., 2008; Zimmermann et al., 2004). Using BiMax clustering, the available high‐quality arrays were analysed for conditions in which the FAD genes from our dataset were expressed in similar patterns (Fig. 2). The majority of these genes were induced by multiple pathogens, including fungi (e.g. Botrytis cinerea), oomycetes (Phytophthora infestans) and bacteria (Pseudomonas syringae). In addition, several of these genes were also induced by microbial‐associated molecular patterns (MAMPs) elf18, elf26 and chitin, and by some abiotic stresses, including osmotic and oxidative stresses. These results point to a role for this family of genes in controlling defence and stress responses, perhaps through the production of H2O2.
Figure 2.
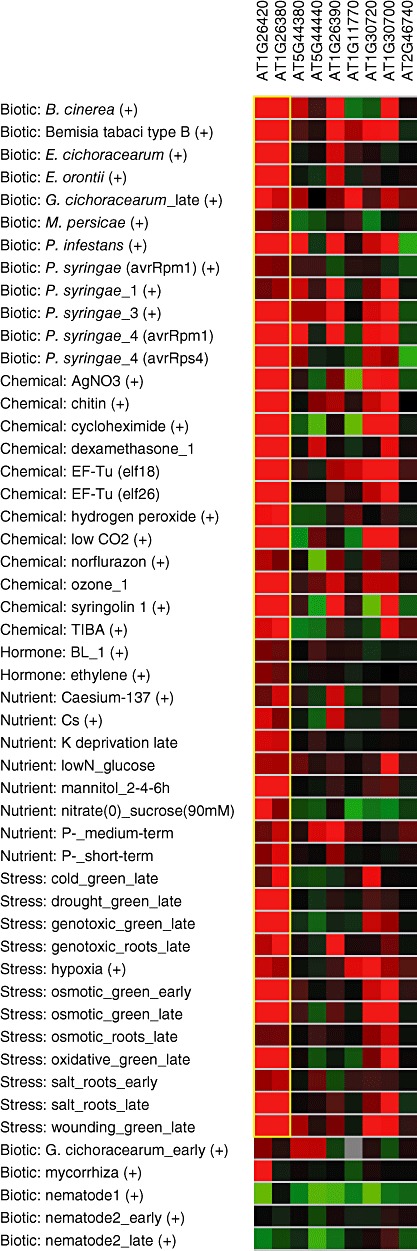
BiMax clustering of flavin adenine dinucleotide (FAD)‐binding domain‐containing genes. Red indicates induction and green indicates suppression, with brighter colours indicating a greater effect of the indicated treatment. The yellow box indicates the two genes with highly similar expression patterns over the indicated treatments.
Analysis of transcription factor motifs
To determine whether the edr1&pm‐upregulated gene set was enriched for genes that are induced by WRKY family transcription factors, a kilobase region of sequence upstream of the start codon was collected for all genes with AGI numbers in our dataset. These regions were then searched for W‐boxes to determine the frequency of this element. As a control, we analysed the equivalent upstream regions from all genes that remained unchanged (<1.155 fold up or down) in edr1 versus wild‐type, and also remained unchanged at all time points after infection in both edr1 and wild‐type plants (a total of 472 genes). Significantly, the upstream regions of genes from our dataset were enriched for W‐boxes, with 910 elements, or a frequency of 1.59 elements per gene, compared with the unchanged dataset, where there was a frequency of 0.79 elements per gene (Table 1). These data strongly suggest that WRKY‐regulated genes are upregulated in the edr1 mutant after pathogen treatment.
Table 1.
Frequencies of transcription factor binding sites in promoter regions of edr1&pm‐upregulated genes. A 1‐kb region 5′ of the start codon for each gene was analysed. This information was also calculated for genes that were unchanged in enhanced disease resistance 1 (edr1) mutants relative to Col‐0, and unchanged after pathogen infection in either genotype, and for all available upstream sequences in The Arabidopsis Information Resource (TAIR) (33 518 sequences).
| No. of elements (edr1&pm‐upregulated) | No. of elements (unchanged) | No. of elements (genome) | Frequency (edr1) | Frequency (unchanged) | Frequency (genome) | |
|---|---|---|---|---|---|---|
| W‐box (TTGACC/T) | 910 | 514 | 40 948 | 1.59 | 0.79 | 1.22 |
| GCC‐box (GCCGCC) | 52 | 60 | 3 528 | 0.091 | 0.127 | 0.11 |
| MYC2 (CACATG) | 267 | 204 | 16 556 | 0.47 | 0.43 | 0.49 |
| ARF (TGTCTC) | 270 | 284 | 17 690 | 0.47 | 0.60 | 0.53 |
| MYB2 (C/TAACG/TG) | 591 | 535 | 36 727 | 1.03 | 1.11 | 1.1 |
| GATA (T/AGATAG/A) | 1968 | 1404 | 107 927 | 3.43 | 2.98 | 3.2 |
| DREB (CCGAC) | 544 | 570 | 37 212 | 0.998 | 1.21 | 1.11 |
Using the same promoter scanning analysis as for the WRKY transcription factors, the number and frequency of GCC‐boxes in the promoter regions of genes from the edr1&pm‐upregulated gene set were calculated. There were 52 GCC‐boxes, a frequency of 0.091 per gene, compared with a frequency of 0.127 per gene in the unchanged dataset and 0.11 for all Arabidopsis genes (Table 1), indicating that the majority of the genes in the edr1&pm‐upregulated gene set were not regulated by AP2/ERF family transcription factors. Indeed, the lower than average frequency of GCC‐boxes suggests that the edr1&pm‐upregulated gene set is enriched in genes that lack AP2/ERF binding sites.
EDR1 localizes to the endoplasmic reticulum (ER) and the nucleus
The enrichment for WRKY transcription factors and genes containing their binding sites in the edr1&pm‐upregulated gene set suggests that EDR1 negatively regulates the activity of these transcription factors. To determine whether this could be occurring directly, we analysed the subcellular localization of full‐length EDR1 protein fused to super yellow fluorescent protein (sYFP2; Kremers et al., 2006) using confocal microscopy. This fusion protein was shown to be functional as EDR1‐sYFP expressed under the EDR1 native promoter was able to complement an edr1 Arabidopsis mutant in stable transgenic plants (Fig. S2, see Supporting Information). Unfortunately, we were unable to detect EDR1‐sYFP using confocal microscopy in these plants, probably because of the low level of expression from the native EDR1 promoter. To visualize EDR1‐sYFP, we thus transiently expressed it in Nicotiana benthamiana leaves using a dexamethasone‐inducible promoter. All transformed cells gave a similar pattern, displaying localization to internal membranes and to the nucleus (Fig. 3A). To confirm the nuclear localization, we co‐expressed EDR1‐sYFP with the nuclear protein GCN5‐mCHERRY (Bhat et al., 2004). GCN5‐mCHERRY fluorescence was confined to the nucleus and appeared to be excluded from the nucleolus (Fig. 3B). The nuclear portion of the EDR‐sYFP fluorescence co‐localized with GCN5. To determine whether the membrane localization was associated with ER, we co‐expressed EDR1‐sYFP with an ER marker consisting of the signal peptide of AtWAK2 (Arabidopsis thaliana wall‐associated kinase 2) at the N‐terminus of mCherry and the ER retention signal His–Asp–Glu–Leu at its C‐terminus (Nelson et al., 2007). Figure 3D–F shows that EDR1‐sYFP co‐localized with this ER marker outside of the nucleus. To assess whether the nuclear signal from EDR1‐sYFP could be a result of the degradation of EDR1‐sYFP, we performed immunoblot analyses using an anti‐green fluorescent protein (GFP) antibody (Fig. 3G). We observed a band of approximately 130 kDa, the expected size for intact EDR1‐sYFP, and a second band at approximately 70 kDa. Untagged EDR1 is readily cleaved, releasing a C‐terminal 50‐kDa fragment (without GFP) during extraction from either plant cells or from Escherichia coli (data not shown), despite the use of protease inhibitors in the extraction buffer. It is thus unclear whether this 70‐kDa protein is present in live cells, but, regardless, this EDR1‐YFP fragment should be too large to diffuse into the nucleus on its own. These observations indicate that a portion of the EDR1 pool accumulates in the nucleus.
Figure 3.
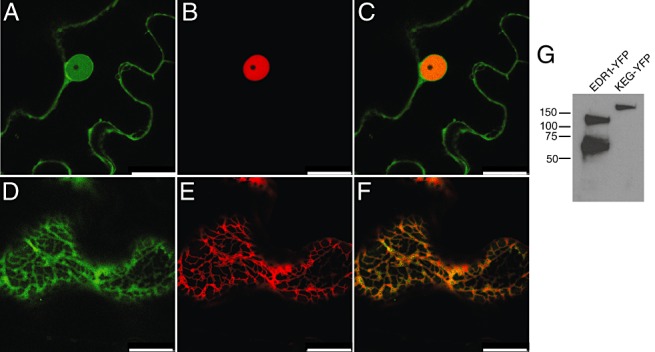
Subcellular localization of enhanced disease resistance 1 protein (EDR1). (A–C) EDR1‐sYFP and GCN5‐mCherry were transiently co‐expressed in Nicotiana benthamiana leaves and imaged using confocal laser scanning microscopy. (A) EDR1‐sYFP (a single optical section taken through the nucleus of an epidermal cell). (B) GCN5‐mCherry expressed in the same cell. (C) Overlay of (A) and (B). (D–F) EDR1‐sYFP and mCherry ER marker (see Experimental procedures) were transiently co‐expressed in N. benthamiana. (D) EDR1‐sYFP (a single optical section taken through the cell cortex of an epidermal cell). (E) mCherry‐HDEL. (F) Overlay of (D) and (E). (G) Immunoblot of EDR1‐sYFP extracted from N. benthamiana leaves. KEG‐sYFP is an unrelated yellow fluorescent protein (YFP) fusion protein included to show the specificity of the antibody. Scale bar, 25 µm.
Gene ontology analysis of genes in the edr1‐upregulated dataset
Gene ontology (GO) annotations have been assigned to nearly every gene in the Arabidopsis genome (Berardini et al., 2004). These annotations provide information about putative structure, function and cellular localization for the predicted protein products. By analysing the GO annotations of the edr1‐upregulated genes at each time point, it is possible to determine which categories of genes are enriched, and how these change temporally after infection with powdery mildew. For this analysis, genes whose expression was at least two‐fold higher in edr1 compared with wild‐type Col‐0 at any time point were selected (blue circle in Fig. 1). At 0 h, prior to inoculation with pathogen, the GO categories that were most significantly enriched (P≤ 0.0001) were ‘endomembrane system’, ‘cellulase activity’, ‘cell wall’, ‘external encapsulating structure’, ‘extracellular region’, ‘membrane’, ‘nutrient reservoir’, ‘response to heat’ and ‘apoplast’ (Table S2, see Supporting Information), suggesting that, in the absence of pathogen, the edr1 mutation primarily affects the expression of genes associated with secretion and the cell wall. Notably, the endomembrane system category remained highly enriched at 18, 36 and 96 hpi, suggesting that the secretory system may play an important role in edr1‐mediated defences. This is consistent with the ER localization of EDR1.
At 18 hpi, the number of categories that were enriched increased dramatically. Of the 25 GO categories enriched with P≤ 0.0001, the majority were associated with defence responses (e.g. ‘defence response’, ‘response to other organism’, ‘response to biotic stimulus’, ‘response to fungus’, ‘immune system process’, ‘response to chitin’, ‘innate immune response’, ‘systemic acquired resistance’, etc.). In addition, the ‘kinase activity’ category was highly enriched (P= 1.85 × 10−4). These observations suggest that the early response to G. cichoracearum in edr1 plants is a specific defence response regulated in part by kinase cascades.
At 36 hpi, many of the same categories were still enriched, with the notable exception of the kinase activity‐related categories (Table S2). In addition, at 36 hpi, the three new categories with the highest significance were ‘peroxidase activity’, ‘oxidoreductase activity’ and ‘antioxidant activity’. This implies that, by 36 hpi, edr1 plants produce significantly more ROS than do wild‐type plants. After 96 h, there was a reduction in the number of GO categories significantly enriched in edr1 plants because gene expression in wild‐type Col‐0 had caught up with the levels of expression in edr1 (Table S2). The GO analysis suggests that the response to G. cichoraceaum in edr1 mutant plants is primarily a more rapid and robust activation of defence genes, probably mediated by kinase signalling cascades that include increased ROS production.
BiMax clustering of edr1‐upregulated genes
Genes that are involved in defence pathways are induced in response to many different stimuli. To determine whether the edr1&pm‐upregulated gene set included genes that were also induced by other pathogens or stimuli, we used the BiMax clustering algorithm within the Genevestigator V3 web toolbox to analyse the expression of these genes across all Arabidopsis datasets involving biotic or abiotic stimulation, or comparing mutant plants with the wild‐type (Prelic et al., 2006). These analyses revealed a subset of genes from the edr1&pm‐upregulated gene set that were regulated in a similar manner in 10 different treatments (Fig. 4A). These 10 different experiments included infection with various virulent and avirulent pathogens, and responses to abiotic stresses such as ozone and wounding. Many of the genes that share regulation in these different categories are defence associated, including a flavin monooxygenase, a TIR‐NB‐LRR gene, two GSTs, a WRKY transcription factor and a cytochrome P450 gene that has been associated with defence responses (Fig. 4A). In addition, the FAD‐binding domain‐containing gene At1g30700, the gene most similar to Ha‐CHOX, was present in this BiMax cluster (Fig. 4A).
Figure 4.
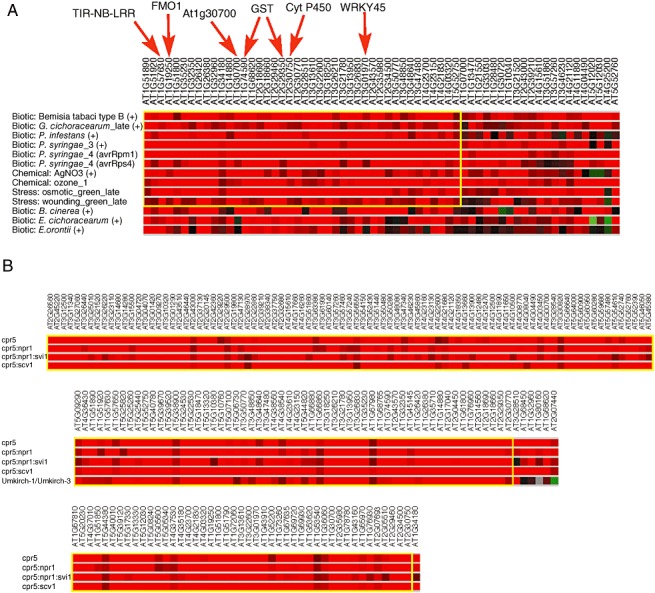
BiMax cluster analysis of edr1&pm‐upregulated genes. (A) Cluster analysis across the set of experiments housed within Genevestigator under the ‘stimulus’ category. (B) Cluster analysis across the set of experiments housed under the ‘mutant’ category. Only the largest clusters are shown, bounded by the yellow box in each.
BiMax clustering was also performed to compare gene regulation in different mutant backgrounds (Fig. 4B). Interestingly, the largest BiMax mutant cluster included the constitutive defence mutant cpr5 (Bowling et al., 1997). Over one‐third of the genes from the edr1&pm‐upregulated gene set were also upregulated in cpr5. This implies that enhanced resistance controlled by edr1 and cpr5 may be mediated by many of the same genes. CPR5 was identified in a screen for plants with enhanced resistance to pathogens. Unlike the edr1 mutant, however, the expression of defence genes, such as PR‐1, in the cpr5 mutant was high in the absence of pathogen.
DISCUSSION
The EDR1 kinase appears to negatively regulate cell death in response to pathogen infection, as well as in response to abiotic stress. Although it has been established that EDR1 is a functional kinase (Frye et al., 2001), we still have little information on how EDR1 regulates responses to pathogen infection. The transcriptome analyses described above revealed a set of genes that were upregulated in the edr1 mutant relative to wild‐type Arabidopsis after inoculation with G. cichoracearum. Of the 545 genes identified in this dataset, many are known to be involved in disease resistance from previous work. The presence of these genes indicates that the enhanced resistance to pathogen infection in edr1 is at least partly the result of the derepression of defence‐associated genes. This result supports the previous finding that edr1‐dependent enhanced resistance requires an intact SA signalling pathway (Frye et al., 2001), as many of these known genes are involved in SA pathways.
In addition, the genes that are induced in edr1 after pathogen inoculation encode for many signalling proteins, including putative NB‐LRR proteins and RLKs. The higher expression of these genes indicates that signalling and, possibly, the perception of pathogens are elevated in the edr1 mutant, and that one function of EDR1 may be to prevent unnecessary signalling. This may also serve to limit the perception of pathogen in the absence of immediate threat.
It should also be noted that, although the edr1 mutation enhances the expression of numerous defence and signalling genes following powdery mildew inoculation, this set of genes represents only a small percentage of the total number of genes induced by at least two‐fold following inoculation (Fig. 1). We identified nearly 4000 genes that were induced in both wild‐type and edr1 mutant plants during at least one time point following inoculation. The great majority of these (>95%) were not significantly affected by the edr1 mutation (i.e. they were induced similarly in wild‐type and edr1 plants), indicating that the edr1 mutation does not simply enhance the expression of all powdery mildew‐induced genes.
The number of genes identified as being induced by powdery mildew infection (4920 in wild‐type and 6352 in edr1) is larger than reported previously. For example, Zimmerli et al. (2004) examined gene expression in wild‐type Arabidopsis infected with the same strain of powdery mildew as used in this study and identified only 13 genes that were upregulated significantly at 24 hpi. More recently, Fabro et al. (2008) examined gene expression in wild‐type, npr1‐1 mutant and jar1‐1 mutant Arabidosis at 18 h following infection with the same powdery mildew strain and identified 117 induced genes. It is difficult to make direct comparisons between our study and those of Zimmerli et al. (2004) and Fabro et al. (2008), however, as their studies employed cDNA microarrays containing only about one‐half of the genes present on the ATH1 Affymetrix gene chip used in our study, two‐colour dye hybridization and different statistical tests. The reduced variability associated with Affymetrix chips relative to cDNA microarrays probably increased the sensitivity of our analyses. In addition, we sampled at later time points (36 and 96 h), allowing us to identify genes induced later in the infection process. Finally, many of the genes identified as upregulated in both edr1 and wild‐type plants at 18 and 36 h in our study may be under circadian regulation, which is one reason why genes that were not also upregulated in edr1 relative to wild‐type plants were excluded from our analyses (Fig. 1).
The analysis of GO annotations for the edr1‐upregulated gene set (blue circle in Fig. 1) revealed a distinct temporal pattern of gene induction during powdery mildew infection. At 18 hpi, numerous defence‐associated gene categories were highly enriched, including the ‘kinase’ category, whereas, at 36 hpi, the kinase category was no longer enriched, but several ROS‐related categories appeared. These results suggest that, immediately following pathogen inoculation, genes involved in signalling and defence responses are expressed more highly, and that, as the response continues, there is a shift from initial induction of signalling to a more sustained response, perhaps through the use of ROS as signalling molecules. By 96 h, most of the categories were no longer enriched, because gene expression in wild‐type plants had caught up.
To determine whether the genes in the edr1‐upregulated dataset were also regulated in response to other pathogens, we used the publicly available microarray data and the web‐based analysis tool Genevestigator V3 (Hruz et al., 2008; Zimmermann et al., 2004). Using the BiMax algorithm within Genevestigator (Prelic et al., 2006), we identified subsets of genes that were regulated in a similar manner in other microarray experiments (Fig. 4). Interestingly, many of the genes upregulated in edr1 were also induced in response to different pathogens as well as abiotic stress. That these genes are regulated by EDR1 suggests that, in the absence of pathogen, EDR1 serves to keep the transcription of these genes low or off, and, once a pathogen has been detected, EDR1 function is repressed, allowing for higher levels of transcription of pathogen‐inducible genes. Interestingly, a subset of edr1‐upregulated genes was also induced at greater levels during attack by the whitefly Bemisia tabaci. The induction of defence‐associated genes in response to B. tabaci may be caused by a wounding response, which is consistent with the observation that these same genes are also regulated in a similar manner in response to mechanical wounding.
BiMax clustering also revealed that many of the edr1‐upregulated genes were also more highly expressed in cpr5 mutants. Mutations in CPR5 cause constitutive expression of defence genes, such as PR genes and PDF1.2, elevated ROS in leaves and the formation of lesions that display the deposition of autofluorescent compounds (Bowling et al., 1997). Lesions induced on edr1 mutant leaves also show the deposition of autofluorescent compounds and elevated ROS (unpublished observations). In addition, like the edr1 mutant, cpr5 mutant plants display enhanced senescence (Jing et al., 2007; Yoshida et al., 2002). Interestingly, the majority of genes expressed in common between cpr5 and edr1 were not suppressed by the npr1 mutation in the cpr5 mutant (Fig. 4B). Previous work has demonstrated that edr1‐mediated disease resistance is dependent on NPR1 (Frye et al., 2001). It is possible that CPR5 functions downstream of NPR1, or that this subset of genes is not central to edr1‐mediated resistance. In addition, although these genes may be independent of NPR1 in a cpr5 background, they may still require NPR1 in the edr1 background. Further experiments are required to understand the signalling pathways controlling the expression of this subset in edr1.
The elevated levels of ROS and ROS‐associated gene expression in edr1 plants suggest that ROS may play a role in edr1 phenotypes, including enhanced sensitivity to drought (Tang et al., 2005). Recently, mutations in an EDR1‐like gene in rice, designated DSM1 (drought‐hypersensitive mutant1), have been shown to confer a similar enhanced sensitivity to drought (Ning et al., 2010). This drought sensitivity correlated with an increased sensitivity to oxidative stress and a reduction in the expression of two peroxidase genes and in peroxidase activity during drought stress. Similar to the edr1 mutant, transcriptome analysis of the dsm1 mutant revealed a large number of genes (678) whose expression was significantly upregulated during stress, suggesting that the dsm1 mutation may cause large disruptions to cellular homeostasis under stress conditions.
The set of edr1‐upregulated genes should include the genes directly responsible for the enhanced disease resistance phenotype of edr1 mutant plants. A particularly intriguing gene family identified in our dataset is the FAD‐binding domain family, which is related to the sunflower Ha‐CHOX gene. Ha‐CHOX was identified for its antimicrobial properties and was found to have carbohydrate oxidase activity, which is the ability to convert glucose into H2O2 (Custers et al., 2004). This class of protein may act to produce ROS that can act as either signalling molecules or as agents of cell death. ROS are produced in the cell during many different processes, including photosynthesis and defence responses (Apel and Hirt, 2004). During defence responses, ROS can be produced by a variety of proteins, including NADPH oxidases [also known as respiratory burst oxidase homologue (Rboh)] in the plasma membrane and peroxidases present in the apoplast (Allan and Fluhr, 1997; Torres et al., 2005; Vera‐Estrella et al., 1992). H2O2 production has been linked to programmed cell death (PCD) in response to pathogen (Dangl and Jones, 2001). However, overexpression of the Rboh protein AtRbohD limits PCD in response to P. syringae DC3000 and Botrytis cinerea (Torres et al., 2005). It appears that, in this case, ROS may act instead as a signalling molecule, delineating the area of infection. At the time of its discovery, Ha‐CHOX represented a new class of oxidase for the production of ROS with glucose as a substrate (Custers et al., 2004). This family of proteins represents another pathway for the production of ROS, and may contribute to the observed resistance of edr1 plants to powdery mildew infection.
Two classes of transcription factor were over‐represented significantly in the edr1‐upregulated dataset: WRKYs and AP2/ERFs. Both WRKY and AP2/ERF family transcription factors have been shown previously to be induced during defence responses and are known to induce defence‐related genes (Buttner and Singh, 1997; Eulgem and Somssich, 2007; Song et al., 2005). Consistent with the over‐representation of WRKY transcription factors, promoter scanning of the edr1‐upregulated genes showed them to be enriched for W‐boxes. Surprisingly, however, the same gene set had a lower than average frequency of AP2/ERF binding sites (GCC‐boxes), possibly indicating that the upregulated AP2/ERFs function as transcriptional suppressors.
WRKY transcription factors have long been associated with the control of defence gene induction. The WRKY‐box was originally identified in the promoters of PR genes from parsley (Rushton et al., 1996). Most WRKYs are transcriptional activators, but there is evidence that some can also act as repressors of transcription (Eulgem and Somssich, 2007). Two of the WRKY genes identified in the edr1‐upregulated dataset, WRKY38 and WRKY59, have also been shown to be induced by the overexpression of NPR1 and by treatment with the SA analogue benzothiadiazole S‐methylester, supporting a role for these two transcription factors in defence responses (Wang et al., 2006). Another WRKY present in the dataset, WRKY75, has been shown to be a regulator of phosphate uptake in roots and is induced under phosphate‐deficient conditions, indicating a role for this gene in nutritional stress responses (Devaiah et al., 2007).
AP2/ERF transcription factors were originally identified as proteins that modulated transcription in response to ET. In addition to proteins that are responsive to ET, ERF domain‐containing transcription factors can also be activated in response to pathogen, such as Pti4 from tomato (Chakravarthy, 2003), and in response to drought, such as DREB2A (Sakuma et al., 2006). Although none of the AP2/ERF genes identified in our dataset have a function yet ascribed, their similarity to other AP2/ERF transcription factors suggests that they may also be involved in defence‐ or drought‐related responses.
The promoters of several of the edr1‐upregulated transcription factors contain W‐boxes, indicating that they may be regulated by a positive feedback loop (data not shown), enabling a rapid response to even slightly elevated levels of these proteins. We hypothesize that EDR1 may function to regulate the level of these proteins by phosphorylation, which would then target them for proteasome‐mediated degradation. This model is supported by our localization studies, which showed that at least a fraction of the EDR1 protein was localized to the nucleus (Fig. 3), where it could interact with these transcription factors directly. A similar model has been proposed for the CTR1 kinase (Gagne et al., 2004; Guo and Ecker, 2003; Potuschak et al., 2003), which belongs to the same kinase subfamily as EDR1 (Frye et al., 2001). CTR1 regulates the level of the EIN3 transcription factor via direct or indirect phosphorylation of EIN3 on a specific threonine residue (T592; Yoo et al., 2008), which promotes its degradation by the proteasome (Gao et al., 2003). In this context, it is worth noting that, like EDR1, CTR1 has been localized to the ER, where it is associated with ET receptors (Gao et al., 2003); thus, if CTR1 phosphorylates transcription factors directly, it may also need to move between a membrane complex and the nucleus. Alternatively, it has been proposed that CTR1 may activate a mitogen‐activated protein kinase pathway which then leads to the phosphorylation of EIN3 on T592, but this remains to be shown (Yoo et al., 2008). A function for EDR1 and CTR1 in the nucleus is further supported by the finding that DSM1 from rice, which belongs to the same subfamily of kinases as EDR1 and CTR1, is primarily located in the nucleus (Ning et al., 2010).
Like CTR1, the majority of EDR1 protein appears to be associated with ER (Fig. 3). The significance of this localization is not yet clear, but is consistent with the GO analyses, which showed that genes associated with secretion and the endomembrane system are highly enriched in the edr1‐upregulated dataset (Table S2).
That EDR1 may regulate the levels of transcription factors by targeting them to the proteasome is supported by our previous finding that all edr1‐mediated phenotypes can be suppressed by a missense mutation in the KEG gene, which encodes a RING‐finger E3 ubiquitin ligase (Wawrzynska et al., 2008). Null mutations in KEG have been shown to cause elevated levels of the ABI5 transcription factor, a central regulator of ABA signalling during post‐germinative growth, and KEG and ABI5 can physically interact (Stone et al., 2006). These data suggest that KEG may ubiquitinate ABI5, targeting it for proteasome‐mediated degradation. ABI5 cannot be the only target of KEG, however, as an abi5 null mutation only partially suppresses a keg null mutation (Stone et al., 2006). We have proposed a model whereby EDR1 is responsible for the phosphorylation of at least a subset of transcription factors that are KEG substrates, and it is this phosphorylation that promotes the association with KEG (Wawrzynska et al., 2008). The transcription factors identified in the present study as being upregulated by the edr1 mutation represent candidates for testing this model.
EXPERIMENTAL PROCEDURES
Plant growth and inoculation conditions
Arabidopsis thaliana Col‐0 and edr1 seeds were sown on Metromix soil and placed at 4 °C for 3 days. Plants were then transferred to a growth room and grown under 9 h of daylight at a temperature of 23 °C. After 4 weeks, the plants were inoculated with G. cichoracearum using a settling tower approximately 1 m tall. Plants to be inoculated were placed at the bottom of the tower, which contained a Nytex mesh screen at the top. Four pad4 mutants with heavy powder growth were passed over the mesh 20 times each to transfer the spores to the plants below. The spores were allowed to settle for 30 min and the plants were transferred to growth chambers.
Tissue collection and RNA preparations
Tissue was collected for each time point (0, 18, 36 and 96 h) by harvesting four full rosettes per genotype per biological replicate. Four biological replicates were collected and placed in liquid nitrogen. Tissue was ground with a mortar and pestle and used for RNA preparations. High‐quality RNA was prepared using the Spectrum Plant Total RNA Kit (Sigma, St. Louis, MO, USA) and concentrated to >0.75 µg/µL with the RNEasy MinElute Cleanup Kit (Qiagen). RNA was then frozen in liquid nitrogen and shipped to the Center for Medical Genomics at the Indiana University School of Medicine, Indianapolis, IN, USA.
Transcriptome analyses
First‐strand cDNA synthesis, biotinylated cRNA synthesis, hybridization to Affymetrix ATH1 GeneChips® and chip scanning were carried out using the facilities of the Center for Medical Genomics at the Indiana University School of Medicine, Indianapolis, IN, USA. Data were processed using the Affymetrix MAS5 algorithm. Data analysis was carried out using ArrayAssist software (now sold under the name GeneSpring GX from Agilent Technologies, Santa Clara, CA, USA) and Genevestigator (https://www.genevestigator.ethz.ch/) (Hruz et al., 2008). Data were normalized using the GC‐RMA algorithm and log2‐transformed using ArrayAssist. Genes whose expression was at least two‐fold greater in edr1 than wild‐type Col‐0 for any time point with P≤ 0.05 using the asymptotic computation were selected. We also generated separate lists of genes that were induced at least two‐fold after inoculation with G. cichoracearum in wild‐type and the edr1 mutant. Each list of genes was then subjected to correction for multiple testing errors using the Benjamini–Hochberg method, and genes with a corrected P≤ 0.05 were selected, representing a false discovery rate of less than or equal to 5% (Benjamini and Hochberg, 1995). As a control for our promoter analyses, we also selected a set of genes unresponsive to either G. cichoracearum or the edr1 mutation (the unchanged dataset), defined as all genes whose fold change was less than 1.155 (up or down) in all comparisons. The 1.155‐fold change value was chosen in order to create a gene set of approximately the same size as the edr1‐upregulated gene set.
For GO analyses, we used the ArrayAssist program to identify GO terms that were significantly enriched (P≤ 0.05) in the set of genes upregulated in edr1 at each time point by at least two‐fold relative to the wild‐type (blue circle in Fig. 1). For bicluster analysis of the edr1&pm‐upregulated genes, we used the BiMax algorithm within the web‐based program Genevestigator V3 (Hruz et al., 2008; Prelic et al., 2006). Because the BiMax algorithm is limited to the analysis of 100 genes at a time, we divided our set of edr1&pm‐upregulated genes (yellow circle in Fig. 1) into five groups and analysed each group independently. Each group was subjected to BiMax cluster analysis with discretization set to 1.0 (Prelic et al., 2006). The ‘stimulus’ and ‘mutant’ microarray datasets within Genevestigator were analysed separately.
Promoter analyses
One kilobase regions upstream of the ATG start codon were collected for all the genes in the edr1‐upregulated dataset and the unchanged dataset using The Arabidopsis Information Resource (TAIR) bulk sequence retrieval tool (http://arabidopsis.org/tools/bulk/sequences/index.jsp). The promoter regions were scanned for six letter words using the TAIR motif analysis tool (http://arabidopsis.org/tools/bulk/motiffinder/index.jsp).
Construction of EDR1‐sYFP fusion proteins and subcellular marker proteins
To make translational fusions of EDR1 to sYFP2 (Kremers et al., 2006), a full‐length EDR1 cDNA without the stop codon and an sYFP2 cDNA with a stop codon were cloned into pDONR P1‐P4 and pDONR P4r‐P2 Gateway‐compatible vectors (Invitrogen, Carlsbad, CA, USA), respectively. The sequences of EDR1 and sYFP in the respective vectors were verified and the two pDONR vectors with EDR1 and sYFP were recombined into the pTA7002‐GW destination vector (Aoyama and Chua, 1997; McNellis et al., 1998), using multisite Gateway cloning technology from Invitrogen, to generate a dexamethasone‐inducible EDR1‐sYFP fusion protein construct. A similar cloning strategy was used to generate the dexamethasone‐inducible nuclear marker protein GCN5‐mCherry (Bhat et al., 2004). An ER marker was created by combining the signal peptide of AtWAK2 at the N‐terminus of mCherry and the ER retention signal His–Asp–Glu–Leu at its C‐terminus (Nelson et al., 2007). To generate an EDR1‐sYFP construct expressed under the native EDR1 promoter, approximately 1.5 kb of EDR1 5′ sequence was inserted into the binary vector pMDC32‐HPB in place of the 35S promoter in this Gateway‐compatible vector (Qi and Katagiri, 2009). EDR1‐sYFP was then recombined into the resulting vector as described above. A stop codon was included after the sYFP sequence; thus the HPB tag was not added.
Subcellular localization of EDR1
Fusion proteins were transiently expressed in leaves of N. benthamiana using agroinfiltration as described previously (Ade et al., 2007). For dexamethasone‐inducible constructs, leaves were imaged 24 h after the application of 50 µm dexamethasone. Intracellular fluorescence was observed by confocal laser scanning microscopy using a Leica SP5 AOBS inverted confocal microscope (Leica Microsystems, Bannockburn, IL, USA) equipped with argon ion (458‐, 476‐, 488‐, 496‐ and 514‐nm laser lines) and He‐Ne (561‐nm laser line) lasers and a Leica 63X NA1.2, HCX PL APO, water objective (Part# 506279). sYFP (excited by the 514‐nm argon laser) fluorescence was detected using the Leica AOBS system and a custom 522–545‐nm bandpass emission filter, whereas mCherry (excited using the 561‐nm He‐Ne laser) fluorescence was detected using the Leica AOBS system and a custom 595–620‐nm bandpass emission filter.
The integrity of the EDR1‐sYFP protein within N. benthamiana leaves was assessed by immunoblot analysis using rabbit polyclonal anti‐GFP antisera (Thermo Scientific, Waltham, MA, USA).
Complementation of the edr1 mutation with EDR1‐sYFP
The Arabidopsis edr1 mutant was transformed with the EDR1 native promoter EDR1‐sYFP construct described above using the floral dip transformation procedure (Clough and Bent, 1998). Plants containing the transgene were selected on agar plates using 30 µg/mL hygromycin. T2 generation plants were tested for complementation of drought‐induced senescence and lesion phenotypes of the edr1 mutant. Plants were grown in Metromix 360 in 4‐in plastic pots under 9 h of daylight for 3 weeks with watering as needed to keep the soil moist. At 3 weeks, watering was stopped. Ten days after cessation of watering, edr1 plants began to show yellow and brown lesions on the leaves and severe chlorosis on older leaves, whereas all leaves on wild‐type Col‐0 plants and edr1 plants transformed with EDR1‐sYFP remained green.
Data deposition
The raw and normalized gene expression data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) GEO expression database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE26679.
Supporting information
Fig. S1 Boxshade alignment of flavin adenine dinucleotide (FAD)‐binding domain proteins.
Fig. S2 Complementation of the Arabidopsis enhanced disease resistance 1 (edr1) mutation with EDR1‐sYFP.
Table S1 edr1&pm‐upregulated genes. All genes listed showed at least two‐fold higher expression in the enhanced disease resistance 1 (edr1) mutant compared with the wild‐type for at least one time point, and were induced at least two‐fold following inoculation by Golovinomyces cichoracearum in wild‐type and/or edr1 mutant plants.
Table S2 Gene ontology (GO) analysis. The enhanced disease resistance 1 (edr1)‐upregulated gene set was analysed for enrichment of GO terms at each time point. Only GO terms that were significantly enriched for at least one time point are listed. Expanded definitions of GO terms can be found at http://www.arabidopsis.org/servlets/Search?action=new_search&type=keyword
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Riyaz Bhat for valuable discussions and assistance with confocal microscopy and the Center for Genomics and Bioinformatics at Indiana University, Bloomington, IN, USA for assistance with gene expression analyses. We also thank the Indiana School of Medicine Center for Medical Genomics for Affymetrix GeneChip analyses and the Indiana University Light Microscopy Imaging Center for assistance with confocal microscopy. This work was supported by National Institutes of Health grant R01 GM063761 from the National Institute of General Medical Sciences to R.W.I. Part of this work was performed at the Indiana School of Medicine Center for Medical Genomics, which is supported in part by the Indiana Genomics Initiative at Indiana University (INGEN®, which is supported in part by the Lilly Endowment, Inc.).
Accession numbers: Table S1 (see Supporting Information).
REFERENCES
- Adam, L. and Somerville, S.C. (1996) Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana . Plant J. 9, 341–356. [DOI] [PubMed] [Google Scholar]
- Adam, L. , Ellwood, S. , Wilson, I. , Saenz, G. , Xiao, S. , Oliver, R.P. , Turner, J.G. and Somerville, S. (1999) Comparison of Erysiphe cichoracearum and E. cruciferarum and a survey of 360 Arabidopsis thaliana accessions for resistance to these two powdery mildew pathogens. Mol. Plant–Microbe Interact. 12, 1031–1043. [DOI] [PubMed] [Google Scholar]
- Ade, J. , DeYoung, B.J. , Golstein, C. and Innes, R.W. (2007) Indirect activation of a plant nucleotide binding site‐leucine‐rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 104, 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan, A.C. and Fluhr, R. (1997) Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9, 1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T. and Chua, N.H. (1997) A glucocorticoid‐mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Basha, E. , Friedrich, K.L. and Vierling, E. (2006) The N‐terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 281, 39943–39952. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 57, 289–300. [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Berardini, T.Z. , Mundodi, S. , Reiser, L. , Huala, E. , Garcia‐Hernandez, M. , Zhang, P. , Mueller, L.A. , Yoon, J. , Doyle, A. , Lander, G. , Moseyko, N. , Yoo, D. , Xu, I. , Zoeckler, B. , Montoya, M. , Miller, N. , Weems, D. and Rhee, S.Y. (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, R.A. , Borst, J.W. , Riehl, M. and Thompson, R.D. (2004) Interaction of maize Opaque‐2 and the transcriptional co‐activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol. Biol. 55, 239–252. [DOI] [PubMed] [Google Scholar]
- Bhattarai, K.K. , Atamian, H.S. , Kaloshian, I. and Eulgem, T. (2010) WRKY72‐type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene‐for‐gene resistance mediated by the tomato R gene Mi‐1. Plant J. 63, 229–240. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A. , Clarke, J.D. , Liu, Y. , Klessig, D.F. and Dong, X. (1997) The cpr5 mutant of Arabidopsis expresses both NPR1‐dependent and NPR1‐independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, M. and Singh, K.B. (1997) Arabidopsis thaliana ethylene‐responsive element binding protein (AtEBP), an ethylene‐inducible, GCC box DNA‐binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy, S. (2003) The tomato transcription factor Pti4 regulates defense‐related gene expression via GCC box and non‐GCC box cis elements. Plant Cell Online 15, 3033–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, Y.H. , Chang, H.S. , Gupta, R. , Wang, X. , Zhu, T. and Luan, S. (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolkowski, I. , Wanke, D. , Birkenbihl, R.P. and Somssich, I.E. (2008) Studies on DNA‐binding selectivity of WRKY transcription factors lend structural clues into WRKY‐domain function. Plant Mol. Biol. 68, 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Custers, J. , Harrison, S. , Sela‐Buurlage, M. , Van Deventer, E. , Lageweg, W. , Howe, P. , Van Der Meijs, P. , Ponstein, A. , Simons, B. , Melchers, L. and Stuiver, M. (2004) Isolation and characterisation of a class of carbohydrate oxidases from higher plants, with a role in active defence. Plant J. 39, 147–160. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Devaiah, B.N. , Karthikeyan, A.S. and Raghothama, K.G. (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L. and Chen, Z. (2000) Identification of genes encoding receptor‐like protein kinases as possible targets of pathogen‐ and salicylic acid‐induced WRKY DNA‐binding proteins in Arabidopsis. Plant J. 24, 837–847. [DOI] [PubMed] [Google Scholar]
- Epple, P. , Apel, K. and Bohlmann, H. (1995) An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis‐related proteins. Plant Physiol. 109, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T. and Somssich, I. (2007) Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Robatzek, S. and Somssich, I.E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Fabro, G. , Di Rienzo, J.A. , Voigt, C.A. , Savchenko, T. , Dehesh, K. , Somerville, S. and Alvarez, M.E. (2008) Genome‐wide expression profiling of Arabidopsis at the stage of Golovinomyces cichoracearum haustorium formation. Plant Physiol. 146, 1421–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, C.A. and Innes, R.W. (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, C.A. , Tang, D. and Innes, R.W. (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M. , Smalle, J. , Gingerich, D.J. , Walker, J.M. , Yoo, S.D. , Yanagisawa, S. and Vierstra, R.D. (2004) Arabidopsis EIN3‐binding F‐box 1 and 2 form ubiquitin‐protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z. , Chen, Y.F. , Randlett, M.D. , Zhao, X.C. , Findell, J.L. , Kieber, J.J. and Schaller, G.E. (2003) Localization of the Raf‐like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 278, 34725–34732. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. , Rogers, E.E. and Ausubel, F.M. (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H. and Ecker, J.R. (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)‐dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677. [DOI] [PubMed] [Google Scholar]
- Gutterson, N. and Reuber, T.L. (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7, 465–471. [DOI] [PubMed] [Google Scholar]
- Hruz, T. , Laule, O. , Szabo, G. , Wessendorp, F. , Bleuler, S. , Oertle, L. , Widmayer, P. , Gruissem, W. and Zimmermann, P. (2008) Genevestigator V3: a reference expression database for the meta‐analysis of transcriptomes. Adv. Bioinformatics 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, H.C. , Anderson, L. , Sturre, M.J. , Hille, J. and Dijkwel, P.P. (2007) Arabidopsis CPR5 is a senescence‐regulatory gene with pleiotropic functions as predicted by the evolutionary theory of senescence. J. Exp. Bot. 58, 3885–3894. [DOI] [PubMed] [Google Scholar]
- Kremers, G.J. , Goedhart, J. , van Munster, E.B. and Gadella, T.W., Jr (2006) Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry 45, 6570–6580. [DOI] [PubMed] [Google Scholar]
- Li, J. , Brader, G. and Palva, E.T. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. Plant Cell 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W. , Mudgett, M.B. , Li, K. , Aoyama, T. , Horvath, D. , Chua, N.H. and Staskawicz, B.J. (1998) Glucocorticoid‐inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 14, 247–257. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Atzorn, R. , Miersch, O. and Wasternack, C. (2006) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B.K. , Cai, X. and Nebenfuhr, A. (2007) A multicolored set of in vivo organelle markers for co‐localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ning, J. , Li, X. , Hicks, L.M. and Xiong, L. (2010) A Raf‐like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152, 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta, K. , Okrent, R.A. , Stoutemyer, M. , Rodibaugh, N. , Kempema, L. , Wildermuth, M.C. and Innes, R.W. (2007) The GH3 acyl adenylase family member PBS3 regulates salicylic acid‐dependent defense responses in Arabidopsis. Plant Physiol. 144, 1144–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, S. , Uknes, S. , Lawton, K. , Winter, A.M. , Chandler, D. , DiMaio, J. , Novitzky, R. , Ward, E. and Ryals, J. (1993) Regulation of a hevein‐like gene in Arabidopsis. Mol. Plant–Microbe Interact. 6, 680–685. [DOI] [PubMed] [Google Scholar]
- Potuschak, T. , Lechner, E. , Parmentier, Y. , Yanagisawa, S. , Grava, S. , Koncz, C. and Genschik, P. (2003) EIN3‐dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689. [DOI] [PubMed] [Google Scholar]
- Pre, M. , Atallah, M. , Champion, A. , De Vos, M. , Pieterse, C.M. and Memelink, J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelic, A. , Bleuler, S. , Zimmermann, P. , Wille, A. , Buhlmann, P. , Gruissem, W. , Hennig, L. , Thiele, L. and Zitzler, E. (2006) A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22, 1122–1129. [DOI] [PubMed] [Google Scholar]
- Qi, Y. and Katagiri, F. (2009) Purification of low‐abundance Arabidopsis plasma‐membrane protein complexes and identification of candidate components. Plant J. 57, 932–944. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J. , Torres, J.T. , Parniske, M. , Wernert, P. , Hahlbrock, K. and Somssich, I.E. (1996) Interaction of elicitor‐induced DNA‐binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15, 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Sakuma, Y. , Maruyama, K. , Osakabe, Y. , Qin, F. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought‐responsive gene expression. Plant Cell 18, 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H. and Bleecker, A.B. (2001) Plant receptor‐like kinase gene family: diversity, function, and signaling. Sci. STKE 2001, RE22. [DOI] [PubMed] [Google Scholar]
- Silverstein, K.A. , Graham, M.A. , Paape, T.D. and VandenBosch, K.A. (2005) Genome organization of more than 300 defensin‐like genes in Arabidopsis. Plant Physiol. 138, 600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C.P. , Agarwal, M. , Ohta, M. , Guo, Y. , Halfter, U. , Wang, P. and Zhu, J.K. (2005) Role of an Arabidopsis AP2/EREBP‐type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L. , Williams, L.A. , Farmer, L.M. , Vierstra, R.D. and Callis, J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, D. and Innes, R.W. (2002) Overexpression of a kinase‐deficient form of the EDR1 gene enhances powdery mildew resistance and ethylene‐induced senescence in Arabidopsis. Plant J. 32, 975–983. [DOI] [PubMed] [Google Scholar]
- Tang, D. , Christiansen, K.M. and Innes, R.W. (2005) Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in Arabidopsis by the EDR1 protein kinase. Plant Physiol. 138, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A. , Jones, J.D. and Dangl, J. (2005) Pathogen‐induced, NADPH oxidase‐derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana . Nat. Genet. 37, 1130–1134. [DOI] [PubMed] [Google Scholar]
- Vera‐Estrella, R. , Blumwald, E. and Higgins, V.J. (1992) Effect of specific elicitors of Cladosporium fulvum on tomato suspension cells: evidence for the involvement of active oxygen species. Plant Physiol. 99, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verburg, J.G. and Huynh, Q.K. (1991) Purification and characterization of an antifungal chitinase from Arabidopsis thaliana . Plant Physiol. 95, 450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, U. , Edwards, R. , Dixon, D.P. and Mauch, F. (2002) Probing the diversity of the Arabidopsis glutathione S‐transferase gene family. Plant Mol. Biol. 49, 515–532. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Amornsiripanitch, N. and Dong, X. (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. Plos. Pathog. 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynska, A. , Christiansen, K.M. , Lan, Y. , Rodibaugh, N.L. and Innes, R.W. (2008) Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol. 148, 1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.D. , Cho, Y.H. , Tena, G. , Xiong, Y. and Sheen, J. (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451, 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K. , Kaothien, P. , Matsui, T. , Kawaoka, A. and Shinmyo, A. (2003) Molecular biology and application of plant peroxidase genes. Appl. Microbiol. Biotechnol. 60, 665–670. [DOI] [PubMed] [Google Scholar]
- Yoshida, S. , Ito, M. , Nishida, I. and Watanabe, A. (2002) Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen‐defence responses in Arabidopsis thaliana . Plant J. 29, 427–437. [DOI] [PubMed] [Google Scholar]
- Zimmerli, L. , Stein, M. , Lipka, V. , Schulze‐Lefert, P. and Somerville, S. (2004) Host and non‐host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J. 40, 633–646. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P. , Hirsch‐Hoffmann, M. , Hennig, L. and Gruissem, W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Boxshade alignment of flavin adenine dinucleotide (FAD)‐binding domain proteins.
Fig. S2 Complementation of the Arabidopsis enhanced disease resistance 1 (edr1) mutation with EDR1‐sYFP.
Table S1 edr1&pm‐upregulated genes. All genes listed showed at least two‐fold higher expression in the enhanced disease resistance 1 (edr1) mutant compared with the wild‐type for at least one time point, and were induced at least two‐fold following inoculation by Golovinomyces cichoracearum in wild‐type and/or edr1 mutant plants.
Table S2 Gene ontology (GO) analysis. The enhanced disease resistance 1 (edr1)‐upregulated gene set was analysed for enrichment of GO terms at each time point. Only GO terms that were significantly enriched for at least one time point are listed. Expanded definitions of GO terms can be found at http://www.arabidopsis.org/servlets/Search?action=new_search&type=keyword
Supporting info item
Supporting info item
Supporting info item
Supporting info item


