Abstract
Background
Okadaic acid (OA), a toxin produced by several dinoflagellate species is responsible for frequent food poisonings associated to shellfish consumption. Although several studies have documented the OA effects on different processes such as cell transformation, apoptosis, DNA repair or embryogenesis, the molecular mechanistic basis for these and other effects is not completely understood and the number of controversial data on OA is increasing in the literature.
Results
In this study, we used suppression subtractive hybridization in SHSY5Y cells to identify genes that are differentially expressed after OA exposure for different times (3, 24 and 48 h). A total of 247 subtracted clones which shared high homology with known genes were isolated. Among these, 5 specific genes associated with cytoskeleton and neurotransmission processes (NEFM, TUBB, SEPT7, SYT4 and NPY) were selected to confirm their expression levels by real-time PCR. Significant down-regulation of these genes was obtained at the short term (3 and 24 h OA exposure), excepting for NEFM, but their expression was similar to the controls at 48 h.
Conclusions
From all the obtained genes, 114 genes were up-regulated and 133 were down-regulated. Based on the NCBI GenBank and Gene Ontology databases, most of these genes are involved in relevant cell functions such as metabolism, transport, translation, signal transduction and cell cycle. After quantitative PCR analysis, the observed underexpression of the selected genes could underlie the previously reported OA-induced cytoskeleton disruption, neurotransmission alterations and in vivo neurotoxic effects. The basal expression levels obtained at 48 h suggested that surviving cells were able to recover from OA-caused gene expression alterations.
Background
Okadaic acid (OA) is a marine toxin produced by several dinoflagellate species. It was firstly isolated from the black sponge Halichondria okadai [1] and is frequently found in several types of molluscs usual in the human diet as those from Mytilus or Ostrea genus. The ingestion of OA-contaminated shellfish results in a syndrome known as diarrhoeic shellfish poisoning (DSP) which is characterized by severe gastrointestinal symptoms including nauseas, vomit, diarrhoea and abdominal ache [2]. Although fatalities associated with DSP-contaminated shellfish have not been reported, this intoxication has become a serious problem for public health and for the economy of aquaculture industries in several parts of the world [3].
OA was found to be a very potent tumour promoter in two-stage carcinogenesis experiments in vivo involving mouse skin [4] or mucosa of the rat glandular stomach [5]. OA was also reported to induce different genotoxic, cytotoxic, and embryotoxic effects such as micronuclei [6-8], oxidative DNA damage [9,10], DNA strand breaks and alterations in DNA repair [11], mitotic spindle alterations [12,13], apoptosis [14,15], cell cycle disruptions [15,16], anomalies of the embryonic development [17] and teratogenicity [18]. Besides, despite the fact that DSP toxins are not classified as neurotoxins [19], some previous studies have already reported neurotoxic effects induced by OA including neuronal apoptosis and cytoskeleton alterations [20,21], deficits in spatial memory [22] and also cognitive deficits in rodents [23].
On the basis on these and other previous studies, OA represents other potential threats to human health besides DSP, even at concentrations within the nanomolar range. It is well-known that OA can inhibit specifically the serine/threonine protein phosphatases 1 (PP1) and 2A (PP2A) [24]; the number of physiological processes in which those phosphatases are involved is immense, including regulation of glycogen metabolism and coordination of the cell cycle and gene expression [25]. So this role of phosphatase inhibition by OA could explain most of the cell effects induced by this toxin [26]. However the number of controversial data in the literature continues increasing and further investigations on biochemical and molecular OA action mechanisms are required since the fact that non-phosphatase targets for OA are not known does not mean that they do not exist [3]. In fact, the existence of OA binding proteins other than phosphatases was demonstrated in several marine organisms [27,28].
In this study, a suppression subtractive hybridization (SSH) approach was used to identify genes differentially expressed in SHSY5Y cells in response to OA exposure at different times (3, 24 and 48 h). Sequences obtained by SSH were used to search for homology/identity to nucleotide and protein databases. Furthermore, differential expression patterns of 5 selected genes were also studied in OA-treated SHSY5Y cells at 3, 24 and 48 h by real-time PCR.
Methods
Cell culture and OA treatment
SHSY5Y cells (human neuroblastoma cell line) were obtained from the European Collection of Cell Cultures and cultured in nutrient mixture EMEM/F12 (1:1) medium with 1% non-essential aminoacids, 1% antibiotic and antimycotic solution and supplemented with 10% heat-inactivated foetal bovine serum, all from Invitrogen (Barcelona, Spain). The cells were incubated in a humidified atmosphere with 5% CO2 at 37°C. OA (CAS No. 78111-17-8) was purchased from Sigma-Aldrich Co. (Madrid, Spain) and dissolved in DMSO prior use.
Flasks with approximately 90% of confluence and 4 × 106 cells were chosen for the experiments. For the treatments, cells were incubated at 37°C for 3, 24 or 48 h in the presence of OA (100 nM) or the control dimethylsulfoxide (DMSO) at 1% of final volume.
Total RNA isolation and cDNA synthesis for SSH
After OA treatments, total RNA was isolated from SHSY5Y cells with TRIZOL® reagent (Invitrogen, Madrid, Spain) following the manufacturer's instructions, and then dissolved in nuclease-free water. RNA was quantified and quality checked using the NANODROP™ 1000 spectrophotometer (Thermo Scientific, Madrid, Spain). One microgram of total RNA from each sample was used as template to synthesize the first strand cDNA with the SMART™ PCR cDNA Synthesis Kit (Clontech, Madrid, Spain), a method for producing high quality cDNA from a low amount of starting material [29]. The double stranded cDNA was amplified with the same Kit according to the manufacturer's protocol.
Construction of subtracted cDNA libraries
SSH was carried out with the PCR-Select™ cDNA subtraction kit (Clontech, Madrid, Spain), as described by the manufacturer. Briefly, the double stranded cDNAs obtained from the step described above were digested with the restriction enzyme RsaI to obtain blunt-ends that are necessary for adaptor ligation. cDNA subtraction was carried out in two directions for the different exposure times. The forward subtracted libraries were made with the control cells cultured for 3, 24 or 48 h as the driver and OA-exposed cells (also cultured for 3, 24 or 48 h) as the tester. These forward reaction libraries were designed to produce clones that are overexpressed or up-regulated in OA-treated cells. The reverse libraries were made in the same way, but in this case the adapter ligated cDNA from OA-exposed cells were the driver and control cells were the tester. The reverse reaction library was designed to produce clones underexpressed or down-regulated in OA-treated cells. In either case the driver cDNA was added in excess during each hybridization to remove common cDNAs by hybrid selection and leaving overexpressed and novel tester cDNA to be recovered and cloned. The subtracted cDNA fragments were then inserted into yT&A® cloning vector, transformed into Escherichia coli ECOS707 (strain JM109) competent cells, and plated on LB agar plates containing 100 μg/ml ampicillin, 100 μl IPTG (100 mM) and 20 μl X-gal (50 mg/ml). The yT&A® cloning vector and the E. coli ECOS707 competent cells were purchased from Yeastern Biotech. Co., Ltd., (Taipei, Taiwan). From the six libraries, a total of 384 white recombinant colonies (corresponding to four 96-well plates) were picked.
Sequencing of the subtracted cDNA clones and bioinformatics analysis
Sequencing of all the cDNA clones from the six SSH libraries was carried out using the BigDye® Terminator v3.1 (Applied Biosystems) and an AB3730 sequencer (Applied Biosystems) at Secugen (Madrid, Spain). After excluding redundant and false-positive sequences, nucleic acid homology searches were performed against nucleotide databases at the National Center for Biotechnology Information (NCBI) using the Basic Local Alignment Search Tool (BLASTX and BLASTN) http://www.ncbi.nlm.nih.gov/BLAST to provide gene annotation. Homologies that showed identities over 60% and E-values of less than 1 × E-10 with more than 100 nucleotides were considered to be significant. The differentially expressed genes identified through expression analysis were classified according to the definition of Gene Ontology (GO) http://www.geneontology.org/ related to the aspects of biological and molecular function.
Differential screening of the subtracted libraries
With the aim of checking the level of background corresponding to common mRNAs in reverse and forward libraries we carried out a differential screening of subtracted libraries using the PCR select differentially screening kit (Clontech, Madrid, Spain), following the manufacturer's instructions. Briefly, PCR products from positive colonies were immobilized in nylon membranes and hybridized with forward- and reverse- probes. Those clones representing mRNAs truly differentially expressed should hybridize only with its corresponding forward-probe. Prior to hybridization forward- and reverse-probes were digested for removing adaptors. More than 90% of the clones tested resulted positive for the virtual Northern analysis (data not shown).
Simple gene set enrichment analysis
A simple gene set enrichment analysis was performed using FatiGO tool (Babelomics 4.2 suite, http://babelomics.bioinfo.cipf.es/). FatiGO takes two lists of genes and convert them into two lists of GO annotations. Then a Fisher's exact test for 2 × 2 contingency tables is used to check for significant over-representation of GO annotations in one of the sets with respect to the other one. Multiple test correction is applied as a measure of control for false positives. In our case, we conducted two single gene set enrichment analysis for KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways comparing our set genes from forward and reverse libraries with the rest of annotations in human genome (additional files 1 and 2).
Quantitative PCR
Five EST identified from SSH were chosen for their specific analysis with real-time PCR. First strand synthesis was performed on 100 ng of the same total RNA samples prepared for SSH from OA-treated and control SHSY5Y cells (for 3, 24 and 48 h) using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Oligonucleotide primers were designed based on the EST sequences determined for candidate differentially expressed genes using the web tool Universal ProbeLibrary (Roche) (Table 1). Quantitative PCR was run in triplicate using LightCycler® SYBR green I Master Kit (Roche) and LightCycler® 480 Real-time PCR Detection System (Roche). The PCR conditions were 95°C for 10 s, 60°C for 10 s, and 72°C for 5 s, for 45 cycles, and final extension of 5 min. A subsequent melting temperature curve of the amplicon was performed. Efficiency of target amplification was optimised prior to running samples for each of the five primer pairs by assaying four primer concentrations (200, 150, 100 and 50 nM). The number of amplification steps required to reach the threshold cycle number (Ct) was computed using LightCycler software 1.5.0 (Roche). Constant Ct values were observed at a 100 nM final primer concentration for each of the primer pairs. Ct values were calculated from the standard curve, entered into the qBasePlus software [30] and used to generate an input file for genNorm software v3.5 [31]. GenNorm determined the most stable reference genes out of the panel of candidate genes using expression stability analysis by pair wise correlations. Following the results of the genNorm, TPR, ACTB and NM23A genes were selected and run separately in all experiments under the same conditions. Normalised cDNA levels of each gene were calculated using qBasePlus [30] once the most stable reference genes were determined. The expression levels of each gene of the 3 h libraries were normalised against both TPR and ACTB, 24 h libraries genes were normalized against both ACTB and NM23A, and 48 h libraries genes were normalized against both TPR and NM23A.
Table 1.
Primers used in real-time PCR analysis
| Genes | Primers (5'-3') | Product (bp) |
|---|---|---|
| NEFM | TGCCGGCTACATAGAGAAGG | 62 |
| TCTCCGCCTCAATCTCCTTA | ||
| TUBB | CCCTCTGTGTAGTGGCCTTT | 68 |
| CCAGACAACTTTGTATTTGGTCA | ||
| SEPT7 | CACAATGTTCACCATTTTTCAAC | 82 |
| TCATTGAAGTTAATGGCAAAAGG | ||
| SYT4 | AAAGTTGTAAGGGGCCAATTC | 70 |
| ACCTCAGCTGCTACCACCAT | ||
| NPY | CGCTGCGACACTACATCAAC | 62 |
| CTCTGGGCTGGATCGTTTT | ||
| TPR | CCACCGAGCGAGGTGATA | 67 |
| AGAAGAAAGGCGAAGACCAGT | ||
| ACTB | AAGTCCCTTGCCATCCTAAAA | 91 |
| ATGCTATCACCTCCCCTGTG | ||
| NM23A | ACATCCATTTCCCCTCCTTC | 92 |
| AGCTTCCCTCCAAACTATGATG |
Statistical analysis
Experimental data were expressed as mean ± standard error. Statistical analyses between groups were carried out using Student's t-test and a P-value of < 0.05 was considered significant. Statistical analysis was performed using the SPSS for Windows statistical package (version 16.0).
Results and Discussion
The human SHSY5Y neuroblastoma cell line has been extensively used as a neuron model in many neurobiological, neurochemical, and neurotoxicological studies [32-37]. In the present study, we investigated the effects of OA, the main DSP toxin, on gene expression of SHSY5Y cells after 3, 24 and 48 h treatments.
Identification of genes with different transcript levels in OA-exposed SHSY5Y cells
For each exposure time 2 subtracted cDNA libraries (one forward and one reverse) were obtained. We isolated a total of 114 subtracted clones from the forward libraries (Table 2) and 133 from the reverse libraries (Table 3).
Table 2.
cDNAs selected from all 3 forward SHH libraries.
| Gene name | Gene Symbol | E-value | Genebank No. | Number of genes |
|---|---|---|---|---|
| 3 h | 24 | |||
| ATP synthase F0 subunit 6 | ATP6 | 2.00E-173 | XM_002345305 | 1 |
| 28S ribosomal RNA | 1.00E-36 | NR_003287 | 3* | |
| B-cell receptor-associated protein 31 | BCAP31 | 0.00E+00 | NM_005745 | 1 |
| calmodulin 2 | CALM2 | 0.00E+00 | NM_001743 | 1 |
| chromosome 6 open reading frame 125 | SGA-81M | 0.00E+00 | NM_032340 | 1 |
| coiled-coil domain containing 56 | CCDC56 | 0.00E+00 | NM_001040431 | 1 |
| ferritin | FTH1 | 2.00E-30 | NM_002032 | 2* |
| FSHD region gene 1 family, member B | FRG1B | 0.00E+00 | NR_003579 | 1 |
| H2A histone family, member Z | H2AFZ | 8.00E-153 | NM_002106 | 1 |
| H3 histone, family 3A | H3F3A | 1.00E-163 | NM_002107 | 1 |
| lysosomal protein transmembrane 4 alpha | LAPTM4A | 0.00E+00 | NM_014713 | 1 |
| mitochondrial ribosomal protein L42 | MRPL42 | 0.00E+00 | NM_172178 | 1 |
| fascin | FSCN1 | 3.00E-29 | AAH07539 | 1 |
| nerve growth factor receptor | TNFRSF16 | 6.00E-142 | NM_206915 | 1 |
| phosphoserine phosphatase | PSPH | 2.00E-139 | NM_004577 | 1 |
| regulator of G-protein signaling 5 | RGS5 | 0.00E+00 | NM_003617 | 1 |
| small nuclear ribonucleoprotein D3 | SNRPD3 | 0.00E+00 | NM_004175 | 1 |
| superoxide dismutase 1 | SOD1 | 0.00E+00 | NM_000454 | 1 |
| SWI/SNF related, matrix associated, actin dependent regulator of chromatin | SMARCE1 | 0.00E+00 | NM_003079 | 1 |
| ubiquitin C | UBC | 5.00E-39 | NM_021009 | 1 |
| Unknown genes | 1 | |||
| 24 h | 47 | |||
| calcyclin binding protein | CACYBP | 0.00E+00 | NM_014412 | 1 |
| 28S ribosomal RNA | 1.00E-36 | NR_003287 | 2* | |
| calmodulin 2 | CALM2 | 0.00E+00 | NM_001743 | 1 |
| casein kinase 1 | CSNK1A1 | 0.00E+00 | NM_001892 | 1 |
| chromosome 14 open reading frame 147 | SSSPTA | 9.00E-93 | NM_138288 | 1 |
| claudin domain containing 1 | CLDND1 | 0.00E+00 | NM_019895 | 1 |
| early growth response 1 | EGR1 | 0.00E+00 | NM_001964 | 1 |
| ferritin | FTL | 6.00E-161 | NM_000146 | 1 |
| glutamate-ammonia ligase (glutamine synthetase) | GLUL | 0.00E+00 | NM_002065 | 3* |
| glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 0.00E+00 | NM_002046 | 1 |
| GTP binding protein overexpressed in skeletal muscle | GEM | 0.00E+00 | NM_181702 | 2* |
| heat shock 70 kDa protein 8 | HSPA8 | 0.00E+00 | NM_006597 | 1 |
| heat shock protein 90 kDa alpha (cytosolic) | HSP90AA1 | 0.00E+00 | NM_005348 | 1 |
| integrator complex subunit 6 | INTS6 | 0.00E+00 | NM_001039937 | 1 |
| mab-21-like 1 | MAB21L1 | 0.00E+00 | NM_005584 | 1 |
| methionine adenosyltransferase II, beta | MAT2B | 9.00E-153 | NM_182796 | 1 |
| mitochondrial ribosomal protein L42 | MRPL42 | 0.00E+00 | NM_014050 | 1 |
| NADH dehydrogenase (ubiquinone) 1 | NDUFAB1 | 0.00E+00 | NM_005003 | 1 |
| neurofilament, medium polypeptide | NEFM | 0.00E+00 | NM_005382 | 1 |
| nudix-type motif 5 | NUDT5 | 0.00E+00 | NM_014142 | 1 |
| pituitary tumor-transforming 1 | PTTG1 | 0.00E+00 | NM_004219 | 1 |
| polo-like kinase 1 | PLK1 | 5.00E-103 | NM_005030 | 1 |
| ribosomal protein L15 | RPL15 | 0.00E+00 | NM_002948 | 1 |
| ribosomal protein L23a | RPL23A | 0.00E+00 | NM_000984 | 1 |
| RNA binding motif protein 7 | RBM7 | 3.00E-111 | NM_016090 | 1 |
| shisa homolog 2 | SHISA2 | 0.00E+00 | NM_001007538 | 1 |
| cytochrome b | CYTB | 0.00E+00 | XR_078322 | 1 |
| SNF2 histone linker PHD RING helicase | SHPRH | 0.00E+00 | NM_001042683 | 1 |
| TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor | TAF9 | 0.00E+00 | NM_003187 | 1 |
| Tax1 (human T-cell leukemia virus type I) binding protein 1 | TAX1BP1 | 0.00E+00 | NM_006024 | 1 |
| triosephosphate isomerase 1 | TPI1 | 0.00E+00 | NM_000365 | 2* |
| Unknown genes | 11 | |||
| 48 h | 43 | |||
| actin, beta | ACTB | 0.00E+00 | NM_001101 | 3 |
| adenylate kinase domain containing 1 | AKD1 | 0.00E+00 | NM_145025 | 1 |
| ADP-ribosylation factor-like 6 interacting protein 1 | ARL6IP1 | 0.00E+00 | NM_015161 | 1 |
| CD58 molecule | CD58 | 0.00E+00 | NR_026665 | 1 |
| clusterin | CLU | 0.00E+00 | NM_203339 | 1 |
| cyclin-dependent kinase inhibitor 1C | CDKN1C | 0.00E+00 | NM_000076 | 1 |
| cytochrome c oxidase subunit III | MT-CO3 | 6.00E-76 | XM_002342023 | 3* |
| family with sequence similarity 32 | FAM32A | 0.00E+00 | NM_014077 | 1 |
| ferritin | FTL | 0.00E+00 | NM_000146 | 1 |
| glutamate-ammonia ligase | GLUL | 0.00E+00 | NM_002065 | 1 |
| metallothionein 1X | MT1X | 2.00E-128 | NM_005952 | 1 |
| metastasis associated lung adenocarcinoma transcript | MALAT1 | 0.00E+00 | NR_002819 | 6* |
| microtubule-associated protein | MAPRE2 | 0.00E+00 | NM_014268 | 1 |
| M-phase phosphoprotein 8 | MPHOSPH8 | 0.00E+00 | NM_017520 | 1 |
| nudix -type motif 5 | NUDT5 | 0.00E+00 | NM_014142 | 1 |
| prostaglandin reductase 1 | PTGR1 | 0.00E+00 | NM_012212 | 1 |
| serine incorporator 3 | SERINC3 | 0.00E+00 | NM_198941 | 1 |
| cytochrome b | CYTB | 0.00E+00 | XR_078322 | 1 |
| solute carrier family 25 | SLC25A4 | 0.00E+00 | NM_001151 | 1 |
| S-phase kinase-associated protein 1 | SKP1 | 0.00E+00 | NM_170679 | 1 |
| TIMP metallopeptidase inhibitor 3 | TIMP3 | 3.00E-86 | NM_000362 | 1 |
| transcription elongation factor B (SIII) | TCEB1 | 3.00E-139 | NM_005648 | 1 |
| translocase of outer mitochondrial membrane 5 | TOMM5 | 0.00E+00 | NM_001001790 | 1 |
| tubulin, beta 2C | TUBB2C | 2.00E-120 | NM_006088 | 1 |
| tubulin, delta 1 | TUBD1 | 2.00E-81 | NM_016261 | 1 |
| tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 0.00E+00 | NM_007115 | 1 |
| tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | YWHAB | 8.00E-110 | NM_003404 | 1 |
| Unknown genes | 7 | |||
*Only one GenBank identity accession number and the highest Blastp E-value were given here because of the table capacity limitation.
Table 3.
cDNAs selected from all 3 reverse SHH libraries.
| Gene name | Gene Symbol | E-value | Genebank No. | Number of genes |
|---|---|---|---|---|
| 3 h | 35 | |||
| ferritin | FTL | 0.00E+00 | NM_000146 | 1 |
| phosphoglycerate kinase 1 | PGK1 | 0.00E+00 | NM_000291 | 1 |
| SMT3 suppressor of mif two 3 homolog 2 | SUMO2 | 5.00E-115 | NM_001005849 | 1 |
| ribosomal protein S14 | RPS14 | 2.00E-52 | NM_001025071 | 1 |
| ATPase | ATP6V0B | 1.00E-47 | NM_001039457 | 1 |
| tubulin, beta 2A | TUBB2A | 2.00E-27 | NM_001069 | 1 |
| actin, beta | ACTB | 0.00E+00 | NM_001101 | 1 |
| yippee-like 5 | YPEL5 | 3.00E-47 | NM_001127401 | 1 |
| translation elongation factor | EEF1A1 | 0.00E+00 | NM_001402 | 1 |
| early growth response 1 | EGR1 | 0.00E+00 | NM_001964 | 1 |
| glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 0.00E+00 | NM_002046 | 1 |
| proteasome 26S subunit, ATPase, 5 | PSMC5 | 0.00E+00 | NM_002805 | 1 |
| signal sequence receptor, alpha | SSR1 | 1.00E-130 | NM_003144 | 1 |
| protein disulfide isomerase family A, member 6 | PDIA6 | 0.00E+00 | NM_005742 | 1 |
| malate dehydrogenase | MDH1 | 0.00E+00 | NM_005917 | 2* |
| ubiquinol-cytochrome c reductase | UQCRH | 0.00E+00 | NM_006004 | 1 |
| translocase of inner mitochondrial membrane | TIMM17A | 0.00E+00 | NM_006335 | 1 |
| heat shock 70 kDa protein 8 | HSPA8 | 0.00E+00 | NM_006597 | 2* |
| peroxiredoxin 3 | PRDX3 | 0.00E+00 | NM_006793 | 1 |
| Sec61 beta subunit | SEC61B | 0.00E+00 | NM_006808 | 1 |
| CD24 molecule | CD24 | 0.00E+00 | NM_013230 | 1 |
| striatin, calmodulin binding protein 3 | STRN3 | 0.00E+00 | NM_014574 | 1 |
| nucleolar protein 11 | NOL11 | 6.00E-158 | NM_015462 | 1 |
| cleavage and polyadenylation factor subunit | PCF11 | 0.00E+00 | NM_015885 | 1 |
| zinc finger, CCHC domain | ZCCHC17 | 3.00E-138 | NM_016505 | 1 |
| homolog 2, suppressor of mek1 | SMEK2 | 0.00E+00 | NM_020463 | 1 |
| 5-azacytidine induced 2 | AZI2 | 2.00E-161 | NM_022461 | 1 |
| anaphase promoting complex subunit 13 | ANAPC13 | 2.00E-94 | NR_024401 | 1 |
| cytochrome c oxidase subunit II | MT-CO2 | 4.00E-150 | XR_078889 | 1 |
| NADH dehydrogenase subunit 4 | MT-ND4 | 7.00E-54 | ADG46850 | 1 |
| Unknown genes | 3 | |||
| 24 h | 44 | |||
| cholinergic receptor, nicotinic, alpha 3 | CHRNA3 | 0.00E+00 | NM_000743 | 1 |
| ribosomal protein L3 | RPL3 | 0.00E+00 | NM_000967 | 1 |
| ribosomal protein S6 | RPS6 | 0.00E+00 | NM_001010 | 1 |
| actin, beta | ACTB | 0.00E+00 | NM_001101 | 1 |
| casein kinase 2 | CSNK2B | 4.00E-176 | NM_001320 | 1 |
| eukaryotic translation elongation factor 1 alpha 1 | EEF1A1 | 0.00E+00 | NM_001402 | 1 |
| calmodulin 2 | CALM2 | 0.00E+00 | NM_001743 | 4* |
| septin 7 | SEPT7 | 0.00E+00 | NM_001788 | 1 |
| LIM domain only 1 (rhombotin 1) | LMO1 | 0.00E+00 | NM_002315 | 1 |
| proteasome beta type, 6 | PSMB6 | 0.00E+00 | NM_002798 | 1 |
| signal recognition particle 9 kDa | SRP9 | 0.00E+00 | NM_003133 | 1 |
| translocated promoter region | TPR | 0.00E+00 | NM_003292 | 1 |
| regulator of G-protein signaling 5 | RGS5 | 0.00E+00 | NM_003617 | 1 |
| NADH dehydrogenase Fe-S protein 2 | NDUFS2 | 0.00E+00 | NM_004550 | 1 |
| myeloid/lymphoid or mixed-lineage leukemia | MLLT11 | 4.00E-143 | NM_006818 | 1 |
| stathmin-like 2 | STMN2 | 0.00E+00 | NM_007029 | 1 |
| component of oligomeric golgi complex 4 | COG4 | 0.00E+00 | NM_015386 | 1 |
| RNA-binding region containing 3 | RNPC3 | 0.00E+00 | NM_017619 | 1 |
| zinc finger, matrin type 5 | ZMAT5 | 0.00E+00 | NM_019103 | 1 |
| synaptotagmin IV | SYT4 | 0.00E+00 | NM_020783 | 2* |
| non-metastatic cells 1, protein NM23A | NM23A | 0.00E+00 | NM_198175 | 1 |
| cytochrome c oxidase subunit III | MT-CO3 | 0.00E+00 | XM_002342023 | 4* |
| ATP synthase F0 subunit 6 | MT-ATP6 | 0.00E+00 | XM_002345305 | 4* |
| cytochrome c oxidase subunit II | MT-CO2 | 5.00E-149 | XR_078889 | 7* |
| NADH dehydrogenase subunit 4 | MT-ND4 | 0.00E+00 | XR_078993 | 1 |
| Unknown genes | 2 | |||
| 48 h | 54 | |||
| glutathione S-transferase | GSTP1 | 0.00E+00 | NM_000852 | 1 |
| neuropeptide Y | NPY | 0.00E+00 | NM_000905 | 1 |
| ribosomal protein L3 | RPL3 | 8.00E-120 | NM_000967 | 2* |
| ribosomal protein L4 | RPL4 | 0.00E+00 | NM_000968 | 1 |
| ribosomal protein L27a | RPL27A | 0.00E+00 | NM_000990 | 1 |
| ribosomal protein L31 | RPL31 | 1.00E-175 | NM_000993 | 1 |
| ribosomal protein S4 | RPS4X | 0.00E+00 | NM_001007 | 1 |
| chromosome 5 open reading frame 13 | C5orf13 | 3.00E-85 | NM_001142478 | 1 |
| adenylate kinase 2 | AKA2 | 7.00E-127 | NM_001625 | 1 |
| calmodulin 2 | CALM2 | 0.00E+00 | NM_001743 | 4* |
| translation initiation factor 4E | EIF4E | 0.00E+00 | NM_001968 | 1 |
| lactate dehydrogenase B | LDHB | 0.00E+00 | NM_002300 | 1 |
| NADH dehydrogenase (ubiquinone) 1 | NDUFC1 | 2.00E-100 | NM_002494 | 1 |
| signal recognition particle 9 kDa | SRP9 | 0.00E+00 | NM_003133 | 2* |
| regulator of G-protein signaling 5 | RGS5 | 2.00E-68 | NM_003617 | 4* |
| DNAJC25-GNG10 readthrough transcript | DNAJC25-GNG10 | 2.00E-166 | NM_004125 | 1 |
| guanine nucleotide binding protein | G protein | 0.00E+00 | NM_004126 | 1 |
| sperm associated antigen 7 | SPAG7 | 0.00E+00 | NM_004890 | 1 |
| heat shock protein 90 kDa | HSP90AA1 | 2.00E-166 | NM_005348 | 1 |
| chaperonin containing TCP1, subunit 3 | CCT3 | 0.00E+00 | NM_005998 | 1 |
| ATP synthase | ATP5L | 9.00E-140 | NM_006476 | 1 |
| mortality factor 4 like 1 | MORF4L1 | 5.00E-96 | NM_006791 | 1 |
| myeloid/lymphoid or mixed-lineage leukemia | MLLT11 | 0.00E+00 | NM_006818 | 1 |
| transmembrane emp24-like trafficking protein | TMED10 | 0.00E+00 | NM_006827 | 1 |
| dickkopf homolog 1 | DKK1 | 4.00E-70 | NM_012242 | 1 |
| signal peptidase complex subunit 1 | SPCS1 | 0.00E+00 | NM_014041 | 1 |
| mitochondrial ribosomal protein L42 | MRPL42 | 5.00E-142 | NM_014050 | 1 |
| HIG1 hypoxia inducible domain family | HIGD1A | 0.00E+00 | NM_014056 | 1 |
| signal peptidase complex subunit 2 | SPCS2 | 0.00E+00 | NM_014752 | 1 |
| mitochondrial ribosomal protein S7 | MRPS7 | 4.00E-156 | NM_015971 | 1 |
| splicing factor 3B | SF3B14 | 0.00E+00 | NM_016047 | 1 |
| hematological and neurological expressed 1 | HN1 | 5.00E-168 | NM_016185 | 1 |
| transmembrane protein 9 | TMEM9 | 0.00E+00 | NM_016456 | 1 |
| chromosome 20 open reading frame 3 | APMAP | 0.00E+00 | NM_020531 | 1 |
| synaptotagmin IV | SYT4 | 0.00E+00 | NM_020783 | 1 |
| ribosomal protein L41 | RPL41 | 0.00E+00 | NM_021104 | 1 |
| transmembrane protein 167A | TMEM167A | 0.00E+00 | NM_174909 | 1 |
| tubulin, beta | TUBB | 6.00E-101 | NM_178014 | 1 |
| THAP domain containing 5 | THAP5 | 6.00E-154 | NM_182529 | 1 |
| K(lysine) acetyltransferase 5 | KAT5 | 0.00E+00 | NM_182709 | 1 |
| gonadotropin-releasing hormone | GNRHR2 | 0.00E+00 | NR_002328 | 1 |
| cytochrome c oxidase subunit II | MT-CO2 | 1.00E-151 | XR_078889 | 1 |
| Unknown genes | 4 | |||
*Only one GenBank identity accession number and the highest Blastp E-value were given here because of the table capacity limitation.
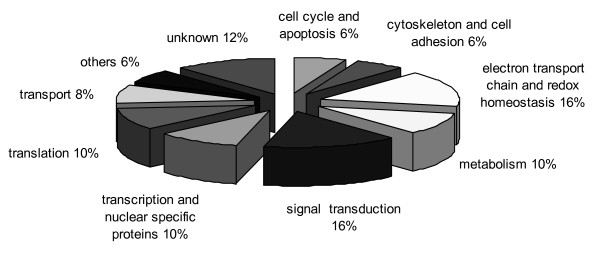
These characterized genes were associated with various functions including metabolism, signal transduction, and cytoskeleton and cell adhesion. The genes altered after the 3 h OA treatment were related to electron transport chain and redox homeostasis, signal transduction, metabolism, transcription, translation, cell cycle and apoptosis, and cytoskeleton and cell adhesion (Figure 1). Most of these genes are apparently involved in metabolism including electron transport chain and redox homeostasis. A few studies have previously reported the OA effects on the cell metabolism. Cable et al. [38] observed that OA affected the heme metabolism of human hepatic cell lines. Also, Shisheva and Shechter [39] showed that OA mimicked some of insulin bioeffects stimulating the glucose and lipid metabolism in rat adipocytes, and Tanti et al. [40] found that glycolysis was stimulated and glucose transport was increased after OA treatment in mouse skeletal muscle. More recently, another study showed that OA depressed the metabolic rate of rat hepatocytes and changed glucose uptake in these cells [41]. Related to electron transport chain, OA was previously found to induce alterations in mitochondrial membrane potential [42] and increased oxidative stress in the rat brain after intracerebroventricular injection [43], and in different cell types in vitro [10,44]. The altered expression levels in genes related to cell metabolism and electron transport chain found in this study could help to explain the effects described in all these works. Besides, 8% of the genes altered after the 3 h OA treatment were related to cellular transport processes. OA was previously found to interfere in the secretion of newly synthesized proteins and exocytosis in rats [45]; both effects could be related to the expression alterations found in the present study.
Figure 1.
Distribution by associated function of genes altered after 3 h OA treatment.
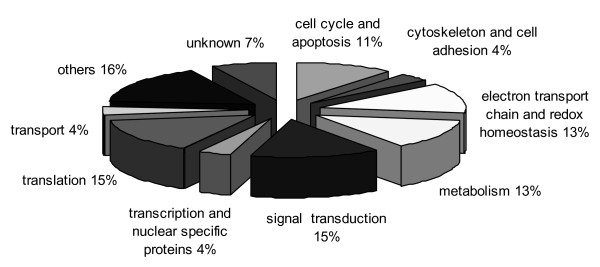
When cells were treated with OA for 24 h, the obtained genes were also categorized into different groups including translation, signal transduction, electron transport chain and redox homeostasis, metabolism, cell cycle and apoptosis, transcription and nuclear specific proteins, transport, and cytoskeleton and cell adhesion (Figure 2). Similar to the 3 h OA treatment, an important number of these genes are involved in metabolism including electron transport chain, but also a great percentage of genes related to translation were observed. The expression alterations found in the genes involved in processes of translation and transcription might be related to the previously reported OA-induced inhibition of protein synthesis [45,46].
Figure 2.
Distribution by associated function of genes altered after 24 h OA treatment.
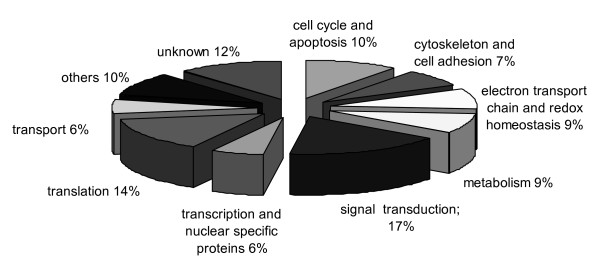
Among the genes altered after the 48 h OA treatment, most were related to signal transduction, translation, cell cycle and apoptosis, electron transport chain and redox homeostasis, metabolism, cytoskeleton and cell adhesion, transcription and nuclear specific proteins, and transport (Figure 3). Fewer genes related to metabolism and transcription were found altered at 48 h, but similarly to 24 h an important percentage of altered genes are involved in cell cycle and apoptosis. In another previous study, the gene expression alterations in mouse BALB/c3T3 cells after different OA treatment times (from 4 h to 24 days, 7.8 ng/ml) were evaluated by microarray analysis, and a total of 177 differentially expressed genes were identified [47]. The authors focused this study on the 31 genes found to be functionally involved in cell growth and/or maintenance, and observed that numerous genes associated with cell proliferation and cell cycle progression were down-regulated after OA treatment. Several genes related to apoptotic processes, some of them involved in the mitochondrial pathway of apoptosis, were also found to be altered. On the basis of their results, they concluded that multiple molecular pathways could be involved in OA-induced proliferation inhibition and apoptosis in these cells [47].
Figure 3.
Distribution by associated function of genes altered after 48 h OA treatment.
Two simple gene set enrichment analysis were performed using FatiGO tool to find which cellular KEGG pathways could be affected by OA exposure in SHSY5Y cells. The results obtained for the forward libraries revealed a total of 3 KEGG pathways altered: oocyte meiosis (hsa04114, adjusted p-value = 0.003), Parkinson's disease (hsa05012, adjusted p-value = 0.035), and cell cycle (hsa04110, adjusted p-value = 0.003). The genes corresponding to reverse libraries were significantly associated with KEGG pathways related to: glycolysis (hsa00010, adjusted p-value = 0.044), oxidative phosphorylation (hsa00190, adjusted p-value = 0.012), Vibrio cholerae infection (hsa05110, adjusted p-value = 0.044), pathogenic Escherichia coli infection (hsa05130, adjusted p-value = 0.044), Alzheimer's disease (hsa05010, adjusted p-value = 0.022), and ribosome (hsa03010, adjusted p-value < 0.001). Since most effects of OA come from binding to PP1 and PP2A, a possible explanation for the altered pathways could be the protein phosphatases inhibition induced by this toxin. In fact, inhibition of PP2A by OA has been previously demonstrated to increase tau phosphorylation, a pathological hallmark of Alzheimer's disease, in SHSY5Y cells [48].
Since OA was previously reported to induce several neurotoxic effects in mammalian cells [20-23] but the underlying mechanisms are still unknown, five specific genes associated with important neuronal structures and functions such as cytoskeleton and neurotransmission, were selected to confirm their expression levels in SHSY5Y cells by real-time PCR. Results obtained from these analyses are shown in Table 4.
Table 4.
Gene expression after real-time PCR.
| Control | OA (100 nM) | |
|---|---|---|
| 3 h treatment | ||
| NEFM | 0.7 ± 0.1 | 0.1 ± 0.0* |
| TUBB | 1.6 ± 0.4 | 1.0 ± 0.4 |
| SEPT7 | 1.7 ± 0.4 | 1.0 ± 0.1* |
| SYT4 | 1.4 ± 0.0 | 0.3 ± 0.0** |
| NPY | 0.6 ± 0.2 | 0.3 ± 0.0* |
| 24 h treatment | ||
| NEFM | 1.6 ± 0.0 | 5.6 ± 0.6* |
| TUBB | 1.5 ± 0.1 | 0.4 ± 0.0** |
| SEPT7 | 1.2 ± 0.1 | 0.9 ± 0.2 |
| SYT4 | 2.8 ± 0.1 | 0.7 ± 0.2* |
| NPY | 2.3 ± 0.3 | 1.3 ± 0.1* |
| 48 h treatment | ||
| NEFM | 1.3 ± 0.1 | 0.8 ± 0.3 |
| TUBB | 1.3 ± 0.1 | 1.5 ± 0.7 |
| SEPT7 | 0.6 ± 0.0 | 0.7 ± 0.0 |
| SYT4 | 0.9 ± 0.1 | 0.9 ± 0.0 |
| NPY | 1.1 ± 0.1 | 1.2 ± 0.1 |
*P < 0.05, **P < 0.01, significant difference with regard to the expression of reference gen.
NEFM, TUBB2A and SEPT7 expression: OA effects on neuronal cytoskeleton
The key role of cytoskeletal organization in several important neural processes such as neurite outgrowth [49], synaptogenesis [50], structural polarity and neuronal shape [51], axonal transport [52], and neurotransmitter release [53] has been characterized. Cell shape and structural polarity are lost in neurodegenerative diseases and neural aging [54,55].
OA was previously reported to induce several cytoskeleton alterations in different cell systems [56,57]. It has been hypothesized that these alterations could be due to different mechanisms that involve disruption of F-actin and ⁄or hyperphosphorylation and activation of kinases that stimulate tight junction disassembly, but the exact molecular mechanism has not been elucidated yet [3].
The cytoskeleton is made up of three kinds of protein filaments: actin filaments (also called microfilaments), intermediate filaments (IF) and microtubules, and other associated proteins. NEFM is the human gene which encodes for the neurofilament medium polypeptide. Neurofilaments (NEFs) are a neuron-specific type of intermediate filament proteins (10 nm in diameter) found at especially high concentrations along the axons, where they regulate axonal diameter. NEFs consist of three proteins according to their molecular weight: light neurofilament (NEFL), medium neurofilament (NEFM), and heavy neurofilament (NEFH) [58]. NEFM gene is often employed as a marker of neuronal differentiation [59]. NEF protein levels are correlative to neurite outgrowth, and its gene expression is dramatically altered in neurodegenerative diseases, including Parkinson's disease and Alzheimer's disease [60]. NEF protein levels have also been suggested as a potential biomarker in organophosphorous neurotoxicity [36]. Furthermore, neurite outgrowth can be promoted by nerve growth factor (NGF) via the regulation of NEF gene expression and NEF protein phosphorylation [61]. In the present study a statistically significant underexpression of this gene was found after 3 h OA treatment, but an overexpression was observed after 24 h, and no effects after 48 h, suggesting that OA deregulates NEFM expression at the short term (within at least the first 24 h), but then it stabilizes and return to control levels. It was previously described that tight coordination of the expression of neurofilament subunits is integral to the normal development and function of the nervous system. Imbalances in their expression are implicated in the induction of neurodegeneration in which formation of neurofilamentous aggregates is central to the pathology [62]. To our knowledge, no studies were reported before on the expression of this gene in human neuronal cells exposed to OA; nevertheless incubation of human fibroblasts [63] or rat brain tumour cells [64] with OA promoted the hyperphosphorylation of major intermediate filaments (IF) proteins, leading to the disassembly of IF networks, solubilisation of IF proteins, and disruption of desmosomes.
Microtubules are involved in many cellular functions, including mitosis, intracellular transport, determination of cell morphology, and differentiation [65]. In neurons, microtubules participate actively in the initial steps of neuronal polarization, the organization of intracellular compartments, the modelling of dendritic spines and the trafficking of cargo molecules to pre-, post- or extrasynaptic domains [66]. Tubulin, the subunit protein of microtubules, is a heterodimer, with both α- and β-tubulin isotypes, differing from each other in amino acid sequence [67]. In our study, the expression levels of TUBB2A, the gene encoding for the tubulin β chain 2A, were analyzed in OA-treated neuronal cells. Data obtained from the real-time PCR analysis showed that this gene is down-regulated in OA-treated SHSY5Y cells at 3 h and 24 h, significantly only in the second case, but at 48 h its expression levels were not different from the control. Microtubules were found to be altered after OA exposure in some previous studies mainly due to the hyperphosphorilation of tau, a microtubule-associated protein which promotes microtubule enlargement. Inhibition of PP2A activity by OA was suggested to produce the abnormal tau hyperphosphorylation in vivo after hippocampal injection in rats [68] and in vitro in metabolically competent brain rats [69], in mouse hippocampal HT22 cell line [70] and in human neuronal NT2N and SHSY5Y cells [48,71]. Besides, Yano et al. [56] found that OA induces reorganization of microtubules in human platelets via the phosphorylation of a microtubule-associated 90 kDa protein, and Benitez-King et al. [72] showed that OA produces cytoskeletal disorganization and microtubule disruption in N1E-115 neuroblastoma cells, as described in other neuronal cell culture models and in rat brain [20,73,74]. TUBB2A was characterized primarily as a neuronal β-tubulin isotype [75] and possess a high expression level in brain, peripheral nerves and muscles [76]. Tubulin isotype composition may be a determinant factor on microtubule functions. Therefore, changes in expression levels of tubulin subtypes would alter the microtubule dynamics. In this sense, Falconer et al. [77] demonstrated that TUBB2 is preferentially incorporated into stable microtubules during neuronal differentiation, and Hoffman and Cleveland [78] reported that the isotype TUBB2 is polymerized more efficiently than other isotypes. The higher expression observed in several types of tumours [79] and in cancer cells resistant to microtubule-binding drugs [80] could be related with the more stability of TUBB2A isotype. The underexpression of TUBB2A observed in this work might contribute to cytoskeletal disruption effects of OA in a similar way, since the major isotype in neuronal cells and the more would be incorporated in a lesser extent to microtubules of SHSY5Y OA-exposed cells.
Septins are an evolutionarily conserved family of cytoskeleton GTP-binding proteins [81]. They play putative roles in cytokinesis, cellular morphogenesis, polarity determination, vesicle trafficking and apoptosis [82-85]. Septins have been identified in all eukaryotic cells. Although yeast septins are better understood, the function of mammalian septins remains largely undefined [86]. SEPT7 is a member of the septin family which is abundantly expressed in the central nervous system [81], but its functional role has not been reported yet [87]. However, a previous study showed that SEPT7 is critical for spine morphogenesis and dendrite development during neuronal maturation [87] and other study confirmed that SEPT7 directly interacts with CENP-E via the C-terminal coiled-coil region. This SEPT7-CENP-E interaction is critical for a stable CENP-E localization to the kinetochore and for achieving chromosome alignment at the equator [88]. SEPT7 has been also related to oncogenesis. After investigating SEPT7 expression in a large number of human glioma tissue samples, Jia et al. [86] found that the expression of this gene was generally down-regulated in gliomas or even absent in some high-grade tumours. Furthermore, they showed that SEPT7 induces cell apoptosis through down-regulation of Bcl-2 and up-regulation of caspase-3, and increased cell apoptosis also contributes to the inhibitory effect of SEPT7 on glioma cell growth. Other studies showed that SEPT7 was much less expressed in brain tumours than in normal brain tissues [81,89] and that neuroblastoma patients with higher SEPT7 mRNA expression might have better prognosis [90]. In our study a significant decrease in the SEPT7 expression levels were found at 3 h, however no differences with regard to the control cells were observed after 24 or 48 h OA treatment.
Therefore, the results obtained from the genes related to cytoskeleton evaluated in this study suggest that the cytoskeleton disruption induced by OA described in previous works are due not only to the hyperphosphorylation of specific proteins caused by phosphatases inhibition, but also to short term alterations (mainly down-regulation) in the expression levels of relevant genes involved in the maintenance of the cell structure as the TUBB2A, NEFM or SEPT7 genes. The fact that no effects of OA were observed after 48 h treatment in any of these genes could be related to cells ability to recover and return to their normal expression levels. However, cell viability was enormously reduced after 48 h OA treatment (microscopic observations). Thus, the absence of gene expression alterations found at that time might also be due to the fact that those cells intensely altered by OA in their gene expression at 3 and 24 h treatments underwent apoptosis or necrosis, being absent at 48 h.
SYT4 and NPY expression: OA effects on synaptic neurotransmission
Synaptic neurotransmission is one of the most highly regulated of all vesicle trafficking events. Although many of the molecular components of synaptic vesicles, the presynaptic cytosol, and presynaptic plasma membrane have been identified, the mechanisms by which these components regulate stimulus evoked vesicle fusion and recycling are not completely understood yet. In this study, expression levels of two genes related to the neuronal signal transduction (SYT4 and NPY) after OA exposure were investigated.
The synaptotagmins (SYTs) are a family of proteins characterized by a short luminal NH2 terminus, one transmembrane region, and tandem C2A and C2B domains [91]. These synaptic proteins are also important in depolarization-induced, Ca2+-dependent fusion of the synaptic vesicles and presynaptic membrane [92]. Currently, it is thought that SYTs participate in the regulation of various steps during membrane fusion, primarily at the plasma membrane [93]. There are at least 17 SYTs isoforms that have the potential to act as modulators of membrane fusion events. SYT4 is particularly interesting since it has been found to be potentially involved in a wide variety of activities in the brain [94]; it is an immediate early gene that is up-regulated following neuronal depolarization [95] and maps to a region of human chromosome 18 associated with schizophrenia and bipolar disease [96]. Data obtained from real-time PCR showed that the SYT4 expression is inhibited by OA at 3 and 24 h exposure, but it recovers normal levels at 48 h treatment. In a previous study, loss of SYT4 results in a reduction of synaptic vesicles and a distortion of the Golgi structure in cultured hippocampal neurons [94]. Golgi disruptions were also found in rat pancreatic cells after OA exposure [45]. Besides, SYT4 affects a number of vesicle recycling properties in peptidergic nerve terminals in the posterior pituitary [97]. Interestingly, SYT4 also appears to play a role in the maturation of secretory granules in neuroendocrine cells [93], suggesting that it may also function in the movement of vesicles [91].
Neuropeptide Y (NPY) is a 36-amino acid peptide produced by neurons throughout the brain and by other secretor cells of the body. NPY has been associated with a number of physiological processes in the brain, including the regulation of energy balance, memory and learning, and epilepsy [98]. Similarly to SYT4, NPY expression levels after OA exposure were found to be down-regulated at 3 and 24 h, but expression levels similar to control were observed at 48 h. A deregulation of the hypothalamic NPY system has been proposed to be related to several pathological and pathophysiological states including cancer cachexia [99], hyperinsulinemia and hypercorticism [100], obesity and metabolic syndrome [101], and anorexia [102].
So far, no studies on SYT4 or NYP expression after OA exposure were reported, but several previous studies described neurotransmission alterations after OA exposure, and the down-regulation of genes involved in the synaptic processes found in this study could help to explain them. OA was found to inhibit mobilization of synaptic vesicles and depress Ca2+ release from sarcoplasmic reticulum in mouse neuromuscular junctions [103], to disrupt synaptic vesicle trafficking in goldfish bipolar cells [104], and to interfere with the formation of synaptic vesicle clusters in nerve terminals of frog neuromuscular junctions [105]. In vivo, OA significantly reduces electrically induced inhibitory non-adrenergic, non-cholinergic (NANC) neurotransmission responses in the rat gastric fundus, while leaving direct muscular effects of the inhibitory NANC neurotransmitters vasoactive intestinal peptide and nitric oxide unaffected, suggesting a neural site of action [106]. It was also reported that presynaptic clusters of synaptic vesicles at the frog neuromuscular junction can be disrupted by exposure to OA [105]. Furthermore, our data involving genes related to neurotransmission could also underlie the OA effects previously reported on the rodent nervous system in vivo such as hyperexcitation [107], spatial memory deficit and neurodegeneration [22] and cognitive deficits [48]. Similarly to the results obtained for the cytoskeleton genes expression, the expression levels of both SYT4 and NPY were highly depressed at 3 and 24 h OA exposure, but they went back to basal levels after 48 h, suggesting that surviving cells were able to recover from OA-induced gene expression alterations.
Conclusions
To elucidate the molecular mechanisms involved in the OA-induced neurotoxic effects, SSH was used in SHSY5Y cells to identify genes with altered expression level at designated treatment times in the promotion stage, including an early time point (3 h), a middle time point (24 h) and a late time point (48 h). A total of 247 known genes were found to be altered. At 3 h OA treatment genes altered are mainly involved in metabolism, including electron transport chain and transcription processes. At 24 and 48 h OA treatments, the percentage of genes related to translation, cell cycle and apoptosis increased. The percentage of genes related to signal transduction, cytoskeleton and metabolism was in general constant at the 3 treatment times.
The data obtained from SHH were confirmed by real-time PCR for 5 specific genes associated with neuronal cytoskeleton and neurotransmission: NEFM, TUBB2A, SEPT7, SYN4, and NPY. The expression levels of the three genes involved in cytoskeleton processes (NEFM, TUBB2A and SEPT7) were found to be altered at 3 and 24 h OA treatments. These alterations could help to explain the previously reported cytoskeleton modifications induced by OA including cell rounding, loss of stabilization of focal adhesions, loss of barrier properties, and loss of cell polarity [56,57,108-110]. The down-regulation observed at the short term (3 and 24 h) of the two genes participating in synaptic neurotransmission (SYT4 and NPY), might be the basis of several reported OA-induced neurotoxic effects [22,48,107]. No expression alterations were observed for any of the five studied genes at 48 h OA exposure, so surviving cells recovered their normal gene expression levels. In order to test whether current results are dependent on OA dose, similar experiments testing different OA concentrations are currently being carried out. Further investigations on the expression patterns of other relevant genes is required in order to completely understand the different effects induced by OA in these and other cells.
Authors' contributions
VV performed the cell culture, prepared SSH libraries, carried out the sequence analyses, did the molecular work and drafted the manuscript. JFT was involved in the conceptualization, design, and implementation of all experiments; helped with the SSH libraries and molecular work and revised the manuscript. EP helped with the experiment design and drafting and revision of the manuscript. JM, the first author's (VV) Ph.D. co-supervisor, was involved in the conceptualization, design, and revision of the manuscript. BL, the other first author's (VV) Ph.D. co-supervisor, was involved in the conceptualization, design and implementation of experiments, and took an active part in data interpretation and writing of this manuscript. All authors read and approved the final manuscript.
Supplementary Material
Single gene set enrichment analysis (KEGG pathways) of genes from forward libraries. Excel spreadsheet showing the results obtained from FatiGO analysis of the genes upregulated in SSH.
Single gene set enrichment analysis (KEGG pathways) of genes from reverse libraries. Excel spreadsheet showing the results obtained from FatiGO analysis of the genes downregulated in SSH.
Contributor Information
Vanessa Valdiglesias, Email: vvaldiglesias@udc.es.
Juan Fernández-Tajes, Email: jfernandezt@udc.es.
Eduardo Pásaro, Email: pspasaro@udc.es.
Josefina Méndez, Email: fina@udc.es.
Blanca Laffon, Email: blaffon@udc.es.
Acknowledgements
This work was funded by a grant from the Spanish Ministry of Science and Innovation (PSI2010-15115). V. Valdiglesias was supported by a fellowship from the University of A Coruña. Authors would like to thank the Genomics Service from INIBIC (Complejo Hospitalario Universitario A Coruña) for providing their facilities.
References
- Tachibana K, Scheurrer PJ, Tsukitani Y, Kikuchi H, Engen DV, Clardy J, Gopichand Y, Schimitz FJ. Okadaic acid, a cytotoxicity polyether from two marine sponges of genus Halichondria. J Am Chem Soc. 1981;103 [Google Scholar]
- Aune T, Yndestad M. In: Algal Toxins in Seafood and Drinking Water. Falconer IR, editor. London and New York: Academic Press; 1993. Diarrhetic shellfish poisoning; pp. 87–104. [Google Scholar]
- Vale C, Botana LM. Marine toxins and the cytoskeleton: okadaic acid and dinophysistoxins. FEBS J. 2008;275:6060–6066. doi: 10.1111/j.1742-4658.2008.06711.x. [DOI] [PubMed] [Google Scholar]
- Fujiki H, Suganuma M, Suguri H, Yoshizawa S, Takagi K, Uda N, Wakamatsu K, Yamada K, Murata M, Yasumoto T. et al. Diarrhetic shellfish toxin, dinophysistoxin-1, is a potent tumor promoter on mouse skin. Jpn J Cancer Res. 1988;79:1089–1093. doi: 10.1111/j.1349-7006.1988.tb01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma M, Fujiki H, Suguiri H, Yoshizwa S, Hirota M, Nakayasu M, Ojika M, Wakamatsu K, Yamada K, Sugimura T. Okadaic acid: an additional non-phorbol-12-tetrade-canoate-13-acetate type tumour promoter. Proc Natl Acad Sci USA. 1988;85:1768–1771. doi: 10.1073/pnas.85.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hegarat L, Puech L, Fessard V, Poul JM, Dragacci S. Aneugenic potential of okadaic acid revealed by the micronucleus assay combined with the FISH technique in CHO-K1 cells. Mutagenesis. 2003;18:293–298. doi: 10.1093/mutage/18.3.293. [DOI] [PubMed] [Google Scholar]
- Carvalho PS, Catian R, Moukha S, Matias WG, Creppy EE. Comparative study of domoic acid and okadaic acid induced-chromosomal abnormalities in the Caco-2 cell line. Int J Environ Res Public Health. 2006;3:4–10. doi: 10.3390/ijerph2006030001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdiglesias V, Laffon B, Pásaro E, Méndez J. Evaluation of okadaic acid-induced genotoxicity in human cells using the micronucleus test and γH2AX analysis. J Toxicol Environ Health A. 2011;74:980–992. doi: 10.1080/15287394.2011.582026. [DOI] [PubMed] [Google Scholar]
- Fessard V, Grosse Y, Pfohl-Leszkowicz A, Puiseux-Dao S. Okadaic acid treatment induces DNA adduct formation in BHK21 C13 fibroblasts and HESV keratinocytes. Mutat Res. 1996;361:133–141. doi: 10.1016/s0165-1161(96)90248-4. [DOI] [PubMed] [Google Scholar]
- Valdiglesias V, Laffon B, Pásaro E, Cemeli E, Anderson D, Méndez J. Induction of oxidative damage by the marine toxin okadaic acid depends on human cell type. Toxicon. 2011;57:882–888. doi: 10.1016/j.toxicon.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Valdiglesias V, Méndez J, Pásaro E, Cemeli E, Anderson D, Laffon B. Assessment of okadaic acid effects on cytotoxicity, DNA damage and DNA repair in human cells. Mutat Res. 2010;689:74–79. doi: 10.1016/j.mrfmmm.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Van Dolah FM, Ramsdell JS. Okadaic acid inhibits a protein phosphatase activity involved in formation of the mitotic spindle of GH4 rat pituitary cells. J Cell Physiol. 1992;152:190–198. doi: 10.1002/jcp.1041520124. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Paweletz N, Schroeter D. Effects of okadaic acid on mitotic HeLa cells. J Cell Sci. 1992;103:117–124. doi: 10.1242/jcs.103.1.117. [DOI] [PubMed] [Google Scholar]
- Rajesh D, Schell K, Verma AK. Ras mutation, irrespective of cell type and p53 status, determines a cell's destiny to undergo apoptosis by okadaic acid, an inhibitor of protein phosphatase 1 and 2A. Mol Pharmacol. 1999;56:515–525. doi: 10.1124/mol.56.3.515. [DOI] [PubMed] [Google Scholar]
- Valdiglesias V, Laffon B, Pásaro E, Méndez J. Okadaic acid induces morphological changes, apoptosis and cell cycle alterations in different human cell types. J Environ Monit. 2011;13:1831–1840. doi: 10.1039/c0em00771d. [DOI] [PubMed] [Google Scholar]
- Lerga A, Richard C, Delgado MD, Canelles M, Frade P, Cuadrado MA, Leon J. Apoptosis and mitotic arrest are two independent effects of the protein phosphatases inhibitor okadaic acid in K562 leukemia cells. Biochem Biophys Res Commun. 1999;260:256–264. doi: 10.1006/bbrc.1999.0852. [DOI] [PubMed] [Google Scholar]
- Casarini L, Franchini A, Malagoli D, Ottaviani E. Evaluation of the effects of the marine toxin okadaic acid by using FETAX assay. Toxicol Lett. 2007;169:145–151. doi: 10.1016/j.toxlet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Ehlers A, Stempin S, Al-Hamwi R, Lampen A. Embryotoxic effects of the marine biotoxin okadaic acid on murine embryonic stem cells. Toxicon. 2010;55:855–863. doi: 10.1016/j.toxicon.2009.12.008. [DOI] [PubMed] [Google Scholar]
- FAO (Food and Agriculture Organization) FAO Food and Nutritrion Paper 80. Rome: Food and Agriculture Organization of the United Nations; 2004. Marine Biotoxins. [Google Scholar]
- Arias C, Sharma N, Davies P, Shafit-Zagardo B. Okadaic acid induces early changes in microtubule-associated protein 2 and tau phosphorylation prior to neurodegeneration in cultured cortical neurons. J Neurochem. 1993;61:673–682. doi: 10.1111/j.1471-4159.1993.tb02172.x. [DOI] [PubMed] [Google Scholar]
- Nuydens R, de Jong M, Van Den Kieboom G, Heers C, Dispersyn G, Cornelissen F, Nuyens R, Borgers M, Geerts H. Okadaic acid-induced apoptosis in neuronal cells: evidence for an abortive mitotic attempt. J Neurochem. 1998;70:1124–1133. doi: 10.1046/j.1471-4159.1998.70031124.x. [DOI] [PubMed] [Google Scholar]
- He J, Yamada K, Zou LB, Nabeshima T. Spatial memory deficit and neurodegeneration induced by the direct injection of okadaic acid into the hippocampus in rats. J Neural Transm. 2001;108:1435–1443. doi: 10.1007/s007020100018. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Simpkins JW. Okadaic acid induces cognitive deficiency in rats. Society for Neuroscience. 2008. Abstract No. 556.4/BB25.
- Bialojan C, Takai A. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem J. 1988;256:283–290. doi: 10.1042/bj2560283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traoré A, Baudrimont I, Dano S, Sanni A, Larondelle Y, Schneider YJ, Creppy EE. Epigenetic properties of the diarrhetic marine toxin okadaic acid: inhibition of the gap junctional intercellular communication in a human intestine epithelial cell line. Arch Toxicol. 2003;77:657–662. doi: 10.1007/s00204-003-0460-0. [DOI] [PubMed] [Google Scholar]
- Sheppeck JEI, Gauss CM, Chamberlin AR. Inhibition of the Ser-Thr phosphatases PP1 and PP2A by naturally occurring toxins. Bioorg Med Chem. 1997;5:1739–1750. doi: 10.1016/S0968-0896(97)00146-6. [DOI] [PubMed] [Google Scholar]
- Schröder HC, Breter HJ, Fattorusso E, Ushijima H, Wiens M, Steffen R, Batel R, Müller WE. Okadaic acid, an apoptogenic toxin for symbiotic/parasitic annelids in the demosponge Suberites domuncula. Appl Environ Microbiol. 2006;72:4907–4916. doi: 10.1128/AEM.00228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Konoki K, Tachibana K. Isolation and characterization of okadaic acid binding proteins from the marine sponge Halichondria okadai. Biochemistry. 2007;46:11410–11420. doi: 10.1021/bi700490n. [DOI] [PubMed] [Google Scholar]
- Hillmann A, Dunne E, Kenny D. In: DNA and RNA Profiling in Human Blood: Methods and Protocols. Peter Bugert, editor. Vol. 496. 2009. cDNA Amplification by SMART-PCR and Suppression Subtractive Hybridization (SSH)-PCR; pp. 223–243. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by genomic averaging of multiple control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas M, Iglesias M, Llecha N, Comella JX. Extracellular-regulated kinases and phosphatidylinositol 3-kinase are involved in brain-derived neurotrophic factor-mediated survival and neuritogenesis of the neuroblastoma cell line SH-SY5Y. J Neurochem. 1999;73:1409–1421. doi: 10.1046/j.1471-4159.1999.0731409.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Santos C, Ferrer I, Reiriz J, Vinals F, Barrachina M, Ambrosio S. MPP+ increases alpha-synuclein expression and ERK/MAP-kinase phosphorylation in human neuroblastoma SH-SY5Y cells. Brain Res. 2002;935:32–39. doi: 10.1016/S0006-8993(02)02422-8. [DOI] [PubMed] [Google Scholar]
- Hasegawa T, Matsuzaki M, Takeda A, Kikuchi A, Furukawa K, Shibahara S, Itoyama Y. Increased dopamine and its metabolites in SH-SY5Y neuroblastoma cells that express tyrosinase. J Neurochem. 2003;87:470–475. doi: 10.1046/j.1471-4159.2003.02008.x. [DOI] [PubMed] [Google Scholar]
- Hong MS, Hong SJ, Barhoumi R, Burghardt RC, Donnelly KC, Wild JR, Venkatraj V, Tiffany-Castiglioni E. Neurotoxicity induced in differentiated SK-N-SH-SY5Y human neuroblastoma cells by organophosphorus compounds. Toxicol Appl Pharmacol. 2003;186:110–118. doi: 10.1016/S0041-008X(02)00016-9. [DOI] [PubMed] [Google Scholar]
- Cho T, Tiffany-Castiglioni E. Neurofilament 200 as an indicator of differences between mipafox and paraoxon sensitivity in SY5Y neuroblastoma cells. J Toxicol Environ Health A. 2004;67:987–1000. doi: 10.1080/15287390490447287. [DOI] [PubMed] [Google Scholar]
- Di Daniel E, Mudge AW, Maycox PR. Comparative analysis of the effects of four mood stabilizers in SH-SY5Y cells and in primary neurons. Bipolar Disord. 2005;7:33–41. doi: 10.1111/j.1399-5618.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- Cable EE, Kuhn BR, Isom HC. Effects of modulators of protein phosphorylation on heme metabolism in human hepatic cells: induction of delta-aminolevulinic synthase mRNA and protein by okadaic acid. DNA Cell Biol. 2002;21:323–332. doi: 10.1089/104454902753759735. [DOI] [PubMed] [Google Scholar]
- Shisheva A, Shechter Y. Effect of okadaic acid in rat adipocytes: differential stimulation of glucose and lipid metabolism and induction of refractoriness to insulin and vanadate. Endocrinology. 1991;129:2279–2288. doi: 10.1210/endo-129-5-2279. [DOI] [PubMed] [Google Scholar]
- Tanti JF, Grémeaux T, Van Obberghen E, Le Marchand-Brustel Y. Effects of okadaic acid, an inhibitor of protein phosphatases-1 and -2A, on glucose transport and metabolism in skeletal muscle. J Biol Chem. 1991;266:2099–2103. [PubMed] [Google Scholar]
- Espiña B, Louzao MC, Cagide E, Alfonso A, Vieytes MR, Yasumoto T, Botana LM. The methyl ester of okadaic acid is more potent than okadaic acid in disrupting the actin cytoskeleton and metabolism of primary cultured hepatocytes. Br J Pharmacol. 2010;159:337–344. doi: 10.1111/j.1476-5381.2009.00512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago J, Santaclara F, Vieites JM, Cabado AG. Collapse of mitochondrial membrane potential and caspases activation are early events in okadaic acid-treated Caco-2 cells. Toxicon. 2005;46:579–586. doi: 10.1016/j.toxicon.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Túnez JA, Druker-Colín R, Muñoz MC, Montilla P. Cytoprotection by melatonin, precursors and metabolites in an in vitro model of neurotoxicity induced by okadaic acid. Lett Durg Design Discov. 2005;2:316–321. doi: 10.2174/1570180054038396. [DOI] [Google Scholar]
- Ferrero-Gutiérrez A, Pérez-Gómez A, Novelli A, Fernández-Sánchez MT. Inhibition of protein phosphatases impairs the ability of astrocytes to detoxify hydrogen peroxide. Free Rad Biol Med. 2008;44:1806–1816. doi: 10.1016/j.freeradbiomed.2008.01.029. [DOI] [PubMed] [Google Scholar]
- Waschulewski IH, Kruse ML, Agricola B, Kern HF, Schmidt WE. Okadaic acid disrupts Golgi structure and impairs enzyme synthesis and secretion in the rat pancreas. Am J Physiol. 1996;270:G939–947. doi: 10.1152/ajpgi.1996.270.6.G939. [DOI] [PubMed] [Google Scholar]
- Matias WG, Bonini M, Creppy EE. Inhibition of protein synthesis in a cell-free system and Vero cells by okadaic acid, a diarrhetic shellfish toxin. J Toxicol Environ Health. 1996;48:309–317. doi: 10.1080/009841096161357. [DOI] [PubMed] [Google Scholar]
- Ao L, Liu JY, Gao LH, Liu SX, Yang MS, Huang MH, Cao J. Differential expression of genes associated with cell proliferation and apoptosis induced by okadaic acid during the transformation process of BALB/c 3T3 cells. Toxicol In Vitro. 2008;22:116–127. doi: 10.1016/j.tiv.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Simpkins JW. Okadaic acid induces tau phosphorylation in SH-SY5Y cells in an estrogen-preventable manner. Brain Res. 2010;1345:176–181. doi: 10.1016/j.brainres.2010.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Neurotransmitters in the regulation of neuronal cytoarchitecture. Brain Res Rev. 1988;13:179–212. doi: 10.1016/0165-0173(88)90020-3. [DOI] [PubMed] [Google Scholar]
- Vega IE, Hsu SC. The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J Neurosci. 2001;21:3839–3848. doi: 10.1523/JNEUROSCI.21-11-03839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid-Arregui A, De Hoop M, Dotti CG. Mechanism of neuronal polarity. Neurobiol Aging. 1995;16:239–243. doi: 10.1016/0197-4580(94)00190-C. [DOI] [PubMed] [Google Scholar]
- Reinsch SS, Mitchison TJ, Kirschner M. Microtubule polymer assembly and transport during axonal elongation. J Cell Biol. 1991;115:365–379. doi: 10.1083/jcb.115.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifaro JM, Vitale ML. Cytoskeleton dynamics during neurotransmitter release. Trends Neurosci. 1993;16:466–472. doi: 10.1016/0166-2236(93)90079-2. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Akiguchi I, Kameyama M, Mizuno N. Age-related changes of pyramidal cell basal dendrites in layers III and V of human motor cortex: a quantitative Golgi study. Acta Neuropathol (Berl.) 1985;65:281–284. doi: 10.1007/BF00687009. [DOI] [PubMed] [Google Scholar]
- Kowall NW, Kosik KS. Axonal disruption and aberrant localization of tau protein characterize the neuropil pathology of Alzheimer's disease. Ann Neurol. 1987;22:639–643. doi: 10.1002/ana.410220514. [DOI] [PubMed] [Google Scholar]
- Yano Y, Sakon M, Kambayashi J, Kawasaki T, Senda T, Tanaka K, Yamada F, Shibata N. Cytoskeletal reorganization of human platelets induced by the protein phosphatase 1/2 A inhibitors okadaic acid and calyculin A. Biochem J. 1995;307:439–449. doi: 10.1042/bj3070439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabado AG, Leira F, Vieytes MR, Vieites JM, Botana LM. Cytoskeletal disruption is the key factor that triggers apoptosis in okadaic acid-treated neuroblastoma cells. Arch Toxicol. 2004;78:74–85. doi: 10.1007/s00204-003-0505-4. [DOI] [PubMed] [Google Scholar]
- Qian Y, Zheng Y, Tiffany-Castiglioni E. Valproate reversibly reduces neurite outgrowth by human SY5Y neuroblastoma cells. Brain Res. 2009;1302:21–33. doi: 10.1016/j.brainres.2009.09.051. [DOI] [PubMed] [Google Scholar]
- Hofsli E, Wheeler TE, Langaas M, Lægreid A, Thommesen L. Identification of novel neuroendocrine-specific tumour genes. Br J Cancer. 2008;99:1330–1339. doi: 10.1038/sj.bjc.6604565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, MilleR CC. Neurofilaments and neurological disease. Bioessays. 2003;25:346–355. doi: 10.1002/bies.10251. [DOI] [PubMed] [Google Scholar]
- Lindenbaum MH, Carbonetto S, Grosveld F, Flavell D, Mushynski WE. Transcriptional and post-transcriptional effects of nerve growth factor on expression of the three neurofilament subunits in PC-12 cells. J Biol Chem. 1988;263:5662–5667. [PubMed] [Google Scholar]
- Thyagarajan M, Strong MJ, Szaro BJ. Post-transcriptional control of neurofilaments in development and disease. Experim Cell Res. 2007;313:2088–2097. doi: 10.1016/j.yexcr.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Yatsunami J, Fujiki H, Suganuma M, Yoshizawa S, Eriksson JE, Olson MO, Goldman RD. Vimentin is hyperphosphorylated in primary human fibroblasts treated with okadaic acid. Biochem Biophys Res Commun. 1991;177:1165–1170. doi: 10.1016/0006-291X(91)90662-Q. [DOI] [PubMed] [Google Scholar]
- Lee WC, Yu JS, Yang SD, Lai YK. Reversible hyperphosphorylation and reorganization of vimentin intermediate filaments by okadaic acid in 9L rat brain tumor cells. J Cell Biochem. 1992;49:378–393. doi: 10.1002/jcb.240490408. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Dynamics and mechanics of the microtubule plus end. Nature. 2003;422:753–758. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- Janke C, Kneusse M. Tubulin post-translational modifications: encoding functions on the neuronal microtubule cytoskeleton. Trends Neurosci. 2010;33:362–372. doi: 10.1016/j.tins.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Guo F, An T, Rein KS. The algal hepatotoxin okadaic acid is a substrate for human cytochromes CYP3A4 and CYP3A5. Toxicon. 2010;55:325–332. doi: 10.1016/j.toxicon.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YY, Liu H, Cong XB, Liu Z, Wang Q, Wang JZ, Zhu LQ. Acetyl-L-carnitine attenuates okadaic acid induced tau hyperphosphorylation and spatial memory impairment in rats. J Alzheimers Dis. 2010;19:735–746. doi: 10.3233/JAD-2010-1272. [DOI] [PubMed] [Google Scholar]
- Gong CX, Lidsky T, Wegiel J, Zuck L, Grundke-Iqbal I, Iqbal K. Phosphorylation of microtubule-associated protein tau is regulated by protein phosphatase 2A in mammalian brain. Implications for neurofibrillary degeneration in Alzheimer's disease. J Biol Chem. 2000;275:5535–5544. doi: 10.1074/jbc.275.8.5535. [DOI] [PubMed] [Google Scholar]
- Poppek D, Keck S, Ermak G, Jung T, Stolzing A, Ullrich O, Davies KJA, Grune T. Phosphorylation inhibits turnover of the tau protein by the proteasome: influence of RCAN1 and oxidative stress. Biochem J. 2006;400:511–520. doi: 10.1042/BJ20060463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick SE, Trojanowski JQ, Lee VM. Selective destruction of stable microtubules and axons by inhibitors of protein serine/threonine phosphatases in cultured human neurons. J Neurosci. 1997;17:5726–5737. doi: 10.1523/JNEUROSCI.17-15-05726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-King G, Túnez I, Bellon A, Ortíz GG, Antón-Tay F. Melatonin prevents cytoskeletal alterations and oxidative stress induced by okadaic acid in N1E-115 cells. Experim Neurol. 2003;182:151–159. doi: 10.1016/S0014-4886(03)00085-2. [DOI] [PubMed] [Google Scholar]
- Arendt T, Holzer M, Fruth R, Burckner MK, Gartner U. Paired helical filament-like phosphorylation of tau, deposition of beta/A4-amyloid and memory impairment in rat induced by chronic inhibition of phosphatase 1 and 2A. Neuroscience. 1995;69:691–698. doi: 10.1016/0306-4522(95)00347-L. [DOI] [PubMed] [Google Scholar]
- Kim D, Su J, Cotman CW. Sequence of neurodegeneration and accumulation of phosphorylated tau in cultured neurons after okadaic treatment. Brain Res. 1999;839:253–262. doi: 10.1016/S0006-8993(99)01724-2. [DOI] [PubMed] [Google Scholar]
- Sullivan KF, Cleveland DW. Identification of conserved isotype-defining variable region sequences for four vertebrate beta tubulin polypeptide classes. Proc Natl Acad Sci USA. 1986;83:4327–4331. doi: 10.1073/pnas.83.12.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsetos CD, Dráberová E, Legido A, Dumontet C, Dráber P. Tubulin targets in the pathobiology and therapy of glioblastoma multiforme. I. Class III beta-tubulin. J Cell Physiol. 2009;221:505–513. doi: 10.1002/jcp.21870. [DOI] [PubMed] [Google Scholar]
- Falconer MM, Echeverri CJ, Brown DL. Differential sorting of beta tubulin isoptypes into colchicine-stable microtubules during neuronal and muscle differentiation of embryonal carcinoma cells. Cell Motil Cytoskeleton. 1992;21:313–325. doi: 10.1002/cm.970210407. [DOI] [PubMed] [Google Scholar]
- Hoffman PN, Cleveland DW. Nrurofilament and tubulin expression recapitulates the development program during axonal regeneration: induction of a specific beta-tubulin isotype. Proc Natl Acad Sci USA. 1988;85:4530–4533. doi: 10.1073/pnas.85.12.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh IT, Luduena RF. The betaII isotype of tubulin is present in the cell nuclei of a variety of cancers. Cell Motil Cytoskleton. 2004;57:96–106. doi: 10.1002/cm.10157. [DOI] [PubMed] [Google Scholar]
- Estève MA, Carré m, Bourgarel-Rey V, Kruczynski A, Raspaglio G, Ferlini C, Braguer D. Bcl-2 down-regulation and tubulin subtype composition are involved in resistance of ovarian cancer cells to vinflunine. Mol Cancer Ther. 2005;5:2824–2833. doi: 10.1158/1535-7163.MCT-06-0277. [DOI] [PubMed] [Google Scholar]
- Hall PA, Jung K, Hillan KJ, Russell SE. Expression profilingthe human septin gene family. J Pathol. 2005;206:269–278. doi: 10.1002/path.1789. [DOI] [PubMed] [Google Scholar]
- Beites CL, Xie H, Bowser R, Trimble WS. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- Field CM, Kellogg D. Septins: cytoskeletal polymers or signalling GTPases? Trends Cell Biol. 1999;9:387–394. doi: 10.1016/S0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- Larisch S, Yi Y, Lotan R, Kerner H, Eimerl S, Tony Parks W, Gottfried Y, Birkey Reffey S, de Caestecker MP, Danielpour D, Book-Melamed N, Timberg R, Duckett CS. et al. A novel mitochondrial septin-like protein, ARTS, mediates apoptosis dependent on its P-loop motif. Nat Cell Biol. 2000;2:915–921. doi: 10.1038/35046566. [DOI] [PubMed] [Google Scholar]
- Kartmann B, Roth D. Novel roles for mammalian septins: from vesicle trafficking to oncogenesis. J Cell Sci. 2001;114:839–844. doi: 10.1242/jcs.114.5.839. [DOI] [PubMed] [Google Scholar]
- Jia ZF, Huang Q, Kang CS, Yang WD, Wang GX, Yu SZ, Jiang H, Pu PY. Overexpression of septin 7 suppresses glioma cell growth. J Neurooncol. 2010;98:329–340. doi: 10.1007/s11060-009-0092-1. [DOI] [PubMed] [Google Scholar]
- Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, Sheng M. Role of septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Wang F, Yan F, Yao PY, Du J, Gao X, Wang X, Wu Q, Ward T, Li J, Kioko S, Hu R, Xie W, Ding X, Yao X. Septin 7 Interacts with Centromere-associated Protein E and Is Required for Its Kinetochore Localization. J Biol Chem. 2008;283:18916–18925. doi: 10.1074/jbc.M710591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Jia ZF, Huang Q, Kang C, Wang GX, Zhang AL, Liu XZ, Zhou X, Xu P, Pu PY. Study on the anti-invasion effect of SEPT7 gene for U251MG glioma cell in vitro. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2008;25:262–267. [PubMed] [Google Scholar]
- Nagata T, Takahashi Y, Asai S, Ishii Y, Mugishima H, Suzuki T, Chin M, Harada K, Koshinaga S, Ishikawa K. The high level of hCDC10 gene expression in neuroblastoma may be associated with favorable characteristics of the tumor. J Surg Res. 2000;92:267–275. doi: 10.1006/jsre.2000.5918. [DOI] [PubMed] [Google Scholar]
- Bai J, Chapman ER. The C2 domains of synaptotagmin-partners in exocytosis. Trends Biochem Sci. 2004;29:143–151. doi: 10.1016/j.tibs.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Machado HB, Liu W, Vician LJ, Herschman HR. Synaptotagmin IV overexpression inhibits depolarization-induced exocytosis in PC12 cells. J Neurosci Res. 2004;76:334–341. doi: 10.1002/jnr.20072. [DOI] [PubMed] [Google Scholar]
- Ahras M, Otto GP, Tooze SA. Synaptotagmin IV is necessary for the maturation of secretory granules in PC12 cells. J Cell Biol. 2006;173:241–251. doi: 10.1083/jcb.200506163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur CP, Dean C, Pagratis M, Chapman ER, Stowell MHB. Loss of synaptotagmin iv results in a reduction in synaptic vesicles and a distortion of the golgi structure in cultured hippocampal neurons. Neuroscience. 2010;167:135–142. doi: 10.1016/j.neuroscience.2010.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vician L, Lim IK, Ferguson G, Tocco G, Baudry M, Herschman HR. Synaptotagmin IV is an immediate early gene induced by depolarization in PC12 cells and in brain. Proc Natl Acad Sci USA. 1995;92:2164–2168. doi: 10.1073/pnas.92.6.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson GD, Vician L, Herschman HR. Synaptotagmin IV: biochemistry, genetics, behavior, and possible links to human psychiatric disease. Mol Neurobiol. 2001;23:173–185. doi: 10.1385/MN:23:2-3:173. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Bhalla A, Dean C, Chapman ER, Jackson MB. Synaptotagmin IV: a multifunctional regulator of peptidergic nerve terminals. Nat Neurosci. 2009;12:163–171. doi: 10.1038/nn.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmers WF, El Bahh B. Neuropeptide Y and Epilepsy. Epilepsy Currents. 2003;3:53–58. doi: 10.1046/j.1535-7597.2003.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King PJ, Williams G. Role of ARC NPY neurons in energy balance. Drug News Perspect. 1998;11:402–410. doi: 10.1358/dnp.1998.11.7.659946. [DOI] [PubMed] [Google Scholar]
- Rohner-Jeanrenaud E, Jeanrenaud B. Central nervous system and body weight regulation. Ann Endocrinol. 1997;58:137–142. [PubMed] [Google Scholar]
- Kuo HW, Chou SY, Hu TW, Wu FY, Chen DJ. Urinary 8-hydroxy-2'-deoxyguanosine (8-OHdG) and genetic polymorphisms in breast cancer patients. Mutat Res. 2007;631:62–68. doi: 10.1016/j.mrgentox.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Kaye WH, Berrettini W, Gwirtsman H, George DT. Altered cerebrospinal fluid neuropeptide Y and peptide YY immunoreactivity in anorexia and bulimia nervosa. Arch Gen Psychiatry. 1990;47:548–56. doi: 10.1001/archpsyc.1990.01810180048008. [DOI] [PubMed] [Google Scholar]
- Hong SJ. Inhibition of mouse neuromuscular transmission and contractile function by okadaic acid and cantharidin. Br J Pharmacol. 2000;130:1211–1218. doi: 10.1038/sj.bjp.0703418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guatimosim C, Hull C, Von Gersdorff H, Prado MA. Okadaic acid disrupts synaptic vesicle trafficking in a ribbon-type synapse. J Neurochem. 2002;82:1047–1057. doi: 10.1046/j.1471-4159.2002.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz WJ, Henkel KW. Okadaic acid disrupts clusters of synaptic vesicles in frog motor nerve terminals. J Cell Biol. 1994;124:843–854. doi: 10.1083/jcb.124.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storr M, Folmer R, Kurjak M, Schusdziarra V, Allescher HD. Okadaic acid inhibits relaxant neural transmission in rat gastric fundus in vitro. Acta Physiol Scand. 2002;175:29–36. doi: 10.1046/j.1365-201X.2002.00959.x. [DOI] [PubMed] [Google Scholar]
- Arias C, Sharma N, Davies P, Shafit-Zagardo B. Okadaic acid induces early changes in microtubule-associated protein 2 and tau phosphorylation prior to neurodegeneration in cultured cortical neurons. J Neurochem. 1998;61:673–682. doi: 10.1111/j.1471-4159.1993.tb02172.x. [DOI] [PubMed] [Google Scholar]
- Tripuraneni J, Koutsouris A, Pestic L, De Lanerolle P, Hecht G. The toxin of diarrheic shellfish poisoning, okadaic acid, increases intestinal epithelial paracellular permeability. Gastroenterology. 1997;112:100–108. doi: 10.1016/S0016-5085(97)70224-5. [DOI] [PubMed] [Google Scholar]
- Niggli V, Djafarzadeh S, Keller H. Stimulusinduced selective association of actin-associated proteins (alpha-actinin) and protein kinase C isoforms with the cytoskeleton of human neutrophils. Exp Cell Res. 1999;250:558–568. doi: 10.1006/excr.1999.4548. [DOI] [PubMed] [Google Scholar]
- Romashko AA, Young MR. Protein phosphatase-2A maintains focal adhesion complexes in keratinocytes and the loss of this regulation in squamous cell carcinomas. Clin Exp Metastasis. 2004;21:371–379. doi: 10.1023/b:clin.0000046178.08043.f8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Single gene set enrichment analysis (KEGG pathways) of genes from forward libraries. Excel spreadsheet showing the results obtained from FatiGO analysis of the genes upregulated in SSH.
Single gene set enrichment analysis (KEGG pathways) of genes from reverse libraries. Excel spreadsheet showing the results obtained from FatiGO analysis of the genes downregulated in SSH.