Abstract
Background
GABA receptors are well known as the inhibitory receptors in the central nervous system and are also found in peripheral tissues. We have previously shown that GABA receptors are involved in lung development and fluid homeostasis. However, the microRNAs that regulate GABA receptors have not yet been identified.
Results
In this study, we used the online software, TargetScan and miRanda, to query the microRNAs that directly target GABA receptors and then selected some of them to verify experimentally using 3'-UTR reporter assays. Computational approaches predict many microRNA binding sites on the 3'-UTR of GABAA receptors, but not on GABAC receptors. 3'-UTR reporter assays only verified miR-181, miR-216, and miR-203 as the microRNAs that target GABA receptor α1-subunit among 10 microRNAs tested.
Conclusions
Our studies reinforce that microRNA target prediction needs to be verified experimentally. The identification of microRNAs that target GABA receptors provides a basis for further studies of post-transcriptional regulation of GABA receptors.
Background
GABA receptors are well known as the inhibitory receptors in the central nervous system [1,2]. However, GABA receptors are also found in several peripheral tissues [3-6]. The functions of GABA receptors in peripheral tissues are less studied. They may be involved in ion homeostasis [7], cell proliferation and differentiation [8], development [1], and hormone secretion [5,9].
We have initially identified GABA receptor π-subunit as a specific alveolar epithelial type II cell marker through DNA microarray analysis [10]. The expression pattern of the GABA receptor π-subunit is regulated by various culture conditions and is consistent with the type II cell phenotypes [11]. We have further identified 19 subunits of the ionotropic GABA receptors in alveolar epithelial cells [6]. Their expression is dynamically changed during lung development [12]. Functionally, GABA receptors play important roles in fluid homeostasis in the adult lung and fetal lung development [6,13].
GABA receptors can be classified into two major types: GABAA and GABAC as ligand-gated Cl- channels, and GABAB receptor as a metabotropic receptor coupled to a heterotrimeric G-protein. GABAA and GABAC receptors share a conserved structure that contains a long extracellular N-terminal region, 4 transmembrane domains (TM1-TM4), a large intracellular loop between TM3 and TM4, and a short extracellular C-terminus [1,2,14-16]. The N-terminal segment is responsible for ligand binding and subunit assembly. The TM2 domain forms the lining of the ion pore. The intracellular loop is the site for post-translational modifications and binding with other proteins. This loop harbors a number of consensus phosphorylation sites for protein kinase A and C (PKA and PKC) and tyrosine kinases [17].
MicroRNAs are small non-coding RNAs. They form a ribonucleoprotein complex, termed RISC that cleaves mRNA or represses protein translation. MicroRNAs regulate various biological processes [18,19]. Several microRNAs such as miR-17-92 cluster and miR-127 are involved in lung development [20-22]. MicoRNAs have also been implicated in many lung diseases including lung inflammation, Chronic Obstructive Pulmonary Disease, Asthma and Idiopathic Pulmonary Fibrosis [23-30]. Nevertheless, microRNAs that regulate GABA receptors have not yet been reported. In this study, we used online software, TargetScan (http://www.targetscan.org) [31] and mRanda (http://www.microrna.org) [32] to predict the microRNAs that possibly target to GABA receptors and then selected some of them to verify experimentally using 3'-UTR reporter assays. We found that miR-181, miR-23 and miR-216 target the GABA receptor α1-subunit.
Methods
Construction of microRNA expression vectors
Human microRNA expression vectors were constructed as previously described [22]. Mature microRNAs with the flanking sequences (~200 base pairs at each end) were PCR-amplified from human genomic DNA. The primers used for PCR amplification are listed in Table 1. The PCR products were inserted into a modified pLVX-Puro lenti-viral vector (Clontech) between CMV-driven enhanced green fluorescent protein (EGFP) and SV40 polyA terminal sequences.
Table 1.
Primers for miRNA expression vectors
| Forward | Reverse | |
|---|---|---|
| miR-15a | CCGCTCGAGTATTCTTTGTGTTTCCTAACCTAT | GCGGAATTCTCAATAATACTGAAAAGACTATC |
| miR-15b | CACCTCGAGCGGCCTGCAGAGATAATACTTC | GAGAATTCGTTGCTGTATCCCTGTCACACT |
| miR-16-1 | GGGCTCGAGAAACATAGATTTTTTTACATGC | CCCGAATTCAATTCCTCTAATGCTGCATAAG |
| miR-16-2 | CACCTCGAGCCTTAAAGTACTGTAGCAGCACAT | GAGAATTCGGTAAATCAAACACCAAGTGTACAG |
| miR-103-1 | CCGCTCGAGTACTTCCCAATCCATTTAAAGTAT | GCGGAATTCAAGTAGCAGATAATTCAAATTG |
| miR-103-2 | CGCCTCGAGTTCCAACAAATGTTTAATTACTTG | GGCGAATTCAAGTAGAGGAAGAGTGGAAGGT |
| miR-107 | CACCTCGAGGATACAATTACAACCCATGTC | GAGAATTCCTGTCTTTCTAATATACCTCAGTG |
| miR-137 | CACCTCGAGTTATGGATTTATGGTCCCGGTCAAG | GAGAATTCTCCAGTCCTGGTCACCAGAGGC^ |
| miR-146a | CACCTCGAGATCCACCCACATCAGCCTTC | GGTTGAAGACTGAATTCGAGTAGCAGCAGCAGCAAGAGAG |
| miR-146b | CACCTCGAGTCAGCTCCAAGCTCAGACCCTC | GGTTGGTCTCGAATTCAGCACTGAGGAAGGACCAGCAT |
| miR-181a-2 | CACCTCGAGTTCTCAGACATTCATTTGAGTC | CACGAATTCTCATCATGGACTGCTCCTTAC |
| miR-181b-1 | GTTCTCGAGTATGACTAAAGGTACTGTTGTTTC | GCTGGTCTCTAATTTGATACTGTACGTTTGATGGAC |
| miR-181c | CACCTCGAGCTGCACTGCTACATCTCCATCC | GAGAATTCACACCAGCTCTCCTCTCCAAAG |
| miR-181d | ATTCTCGAGCTCTCCTCCTCTCCCTCTTCATGCTC | ATAGAATTCAGATTGGGCCACTGCACTCCAGC |
| miR-203 | ATTCTCGAGGCGGGGCTGGGCTTGGCGGCTG | TTTGAATTCGCGCGCCCCCACCCAGCGGTTC |
| miR-216b | CCGCTCGAGTTTTCCTTCATTCTCTATGGAGAT | GGCGAATTCGATGCTAGTCTGGGAATCAAAC |
| miR-221 | CACCTCGAGCGACCTTCCTTCCATCCAGCTT | GAGAATTCTTTGACAGTTGAGGCAGGGAGAAG |
| miR-424 | CACCTCGAGCAGCTCCTGGAAATCAAATGGTG | GAGAATTCACTTACCCTGGCAGCGGAAACAATAC |
| miR-497 | CACCTCGAGATGGTCCGTCGCCTTCCAGTTG | GAGAATTCTGGGTGTGGGCAACAAAGACTC |
Construction of 3'-UTR reporter vectors
The full length 3'-UTR or the microRNA binding sites in the 3'-UTR of rat GABA receptors were PCR-amplified and inserted into the pRL-TK vector containing a Renilla luciferase (Promega). The primers used for PCR amplification are listed in Table 2.
Table 2.
Primers used for constructing 3'-UTR reporter vectors
| Names | 3'-UTR or binding Sites | Sequences |
|---|---|---|
| GABRA1-F-Spe I | 9-1962 | ggtactaGTCGTATTCTGTTGTTCAGTC |
| GABRA1-R-PspOMI | gttgggcccTCTATACATGAAATGTCCTTGG | |
| GABRG2-F | 2095-2102 (miR-103/107) | CTAGTATGGACTTTTACAATAAAAATGCTGCATTCTA |
| GABRG2-R | GGCCTAGAATGCAGCATTTTTATTGTAAAAGTCCATA | |
F: forward primers; R: reverse primers; microRNA binding sites are underlined
3'-UTR reporter assay
HEK 293T cells (2 × 104/well) were seeded in each well of a 96-well plate. After one day of culture, the cells were transfected with100 ng microRNA expression vector or control vector without miRNA insert, 2.5 ng 3'-UTR Renillia luciferase reporter vector and 15 ng pGL3 control vector (firefly luciferase reporter) using Lipofectamine. After a 2 day transfection, the cells were lyzed and dual luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). The Renilla luciferase activities were normalized with firefly luciferase activity. Data was expressed as a ratio to the control vector without miRNA insert.
Results and discussion
Both GABAA and GABAC receptors are ligand-gated Cl- channels. However GABAC receptors have very unique ligand binding characteristics in comparison with GABAA and GABAB receptors, including a high sensitivity to the physiological ligand, GABA, insensitivity to bicuculline, barbiturates, and bacofen, very weak de-sensitization, a smaller single-channel conductance, and a longer open time [14-16]. Eight different subunits of GABAA and GABAC receptors (α1-6, β1-3, γ1-3, δ, θ, ε, π, and ρ1-3) have been identified. The assembly of a heteropentamer, with at least one α-, one β-, and one γ-subunit, forms functional GABAA receptor channels. δ-, θ-, ε-, and π-subunits can substitute for the γ-subunit. However, GABAC receptors are exclusively composed of ρ subunits in the form of homo- or hetero-pentamers.
To identify the microRNA that may potentially regulate GABAA and GABAC receptors, we used the online computer software, TargetScan (v 5.2) to predict the binding sites of microRNAs on the 3'UTR of rat GABA receptors. We chose rat GABA receptors because we use rats as most of our animal or cell models. Since our microRNA expression vectors use human sequences, we only queried conserved microRNA target sites among mammals based on conserved 8 mer and 7 mer sites that match the seed sequence of a microRNA. The results are listed in Table 3. Among α-subunits, we found that α1 had most of the microRNA binding sites. There were 6 binding sites for 6 microRNAs on α1 3'-UTR. For other α-subunits, we found miR-128, miR-27ab, let-7/miR-98 for α6. We did not find any microRNAs for α2, α4, and α5. There was no information available for α3.
Table 3.
Predicted microRNAs targeting rat GABA receptor subunits by TargetScan and miRanda
| GABA receptor subunits | Entrez Gene symbol | Lengths of 3'-UTR in TargetScan (v5.2) | Conserved microRNAs targeting to GABA receptors predicted by TargetScan (v5.2) | Lengths of 3'-UTR in miRanda | Conserved microRNAs targeting to GABA receptors predicted by miRanda |
|---|---|---|---|---|---|
| α1 | GABRA1 | 2017 | miR-208/208ab, | 2018 | miR-129 (2), miR-130b, miR-136, miR-137, miR-148b-3p, |
| miR-499/499-5p, miR-181, | miR-150, miR-152, miR-181a (2) bc (3) d, miR-182, | ||||
| miR-216/216a, miR-137, | miR-186, miR-203 (2), miR-210, miR-216a, miR-26ab, | ||||
| miR-203 | miR-30acde, miR-30b-5p, miR-320, miR-340-5p, miR-361, | ||||
| miR-374, miR-375, miR-376c, miR-377, miR-384-5p, | |||||
| miR-410, miR-433, miR-488 (2), miR-539, miR-874 | |||||
| α2 | GABRA2 | 747 | 0 | NA | NA |
| α3 | GABRA3 | 0 | 0 | NA | NA |
| α4 | GABRA4 | 7478 | 0 | 88 | miR-186, miR-200bc, miR-203, miR-429, miR-495 |
| α5 | GABRA5 | 586 | 0 | 880 | miR-124, miR-132, miR-133ab, miR-195, miR-212, miR-223, |
| miR-30acde, miR-30b-5p, miR-322, miR-346, miR-376c, | |||||
| miR-378, miR-384-5p, miR-494 (2), miR-495, miR-539 | |||||
| α6 | GABRA6 | 809 | miR-128, miR-27ab, | NA | N/A |
| let7/miR-98 | |||||
| β1 | GABRB1 | 407 | miR-30a/30a-5p/30b/30b-5p/ | 428 | miR-103, miR-107, miR-128, miR-143, miR-148b-3p, |
| 30/384-5p (2), miR-103/107 | miR-152, miR-30a (2) c (2) d (2) e (2), miR-30b-5p (2), | ||||
| miR-384-5p (2), miR-411 | |||||
| β2 | GABRB2 | 5618 | miR-203,miR-135, miR-218, | 499 | miR-128, miR-199a-5p, miR-203, miR-33, miR-411, miR-485 |
| miR-21/590-5p, miR-10, | |||||
| miR-101, miR-19, miR-144, | |||||
| miR-9 (2), miR-455/455-5p, | |||||
| miR-33/33ab | |||||
| β3 | GABRB3 | 4060 | miR-26ab/1297, miR-204/211, | 508 | miR-122, miR-186, miR-199a-5p, miR-204, miR-210, miR-211, |
| miR-23ab, miR-27ab, miR-218 | miR-23ab, miR-26ab, miR-320, miR-324-5p, miR-329, | ||||
| miR-381, miR-539 | |||||
| γ1 | GABRG1 | 3428 | 0 | 240 | miR-203, miR-218, miR-379, miR-410, miR-455, miR-488 |
| γ2 | GABRG2 | 2106 | miR-150, miR-103/107 | NA | N/A |
| γ3 | GABRG3 | 96 | 0 | 114 | miR-15b, miR-16, miR-195, miR-26ab, miR-322, miR-497 |
| π | GABRP | 1178 | 0 | NA | N/A |
| δ | GABRD | 433 | 0 | 393 | miR-145, miR-19ab, miR-24, miR-328, miR-365 |
| ε | GABRE | 1485 | miR-22 | NA | N/A |
| θ | GABRQ | 80 | 0 | NA | N/A |
| ρ1 | GABRR1 | 464 | 0 | NA | N/A |
| ρ2 | GABRR2 | 113 | 0 | 191 | 0 |
| ρ3 | GABRR3 | N/A | N/A | 271 | miR-191 |
The conserved microRNAs targeting GABA receptors are predicted by Targetscan 5.2 http://www.targetscan.org/ and miRanda software (http://www.microrna.org). The number in parenthesis is the number of the binding sites and none means one binding site. N/A means no information is available in TargetScan or miRanda
For β-subunits, we found two binding sites for miR-30a/30a-5p/30b/30b-5p/30/384-5p and one binding site for miR-103/107. There were 15 binding sites for 15 microRNAs on β2 and 5 binding sites for 7 microRNAs on β3. For γ-subunits, we found two binding sites on γ2 and no binding sites on γ1 and γ3, There was only one binding site for ε and no binding sites on π-, δ-, θ-, ρ1- and ρ2-subunits. In general, the "common" subunits (α, β, and γ) had more miRNA target sites than the "rare" subunits (δ, θ, ε, π, and ρ). This is probably because these subunits had shorter 3'-UTRs, in particular for ρ-subunits.
We also used another software, miRanda to predict the microRNA that target to GABA receptors (Table 3). In general, miRanda predicted more microRNAs than TargetScan. There were some common microRNAs that were predicted by both software. For example, miR-137, miR-181, miR-203, and miR-216a for α1; miR-103, miR-107, miR-30, and miR-384-5p for β1; and miR-204, miR-211, miR-23, and miR-26 for β3.
We further utilized a recently developed software, miRWalk [33], to predict the miRNAs targeting 3'-UTR and open reading frame (ORF) of GABA receptors. The results are presented in Table 4. Obviously, this method is less stringent compared to TargetScan and miRanda, since the miRWalk query yielded 79 miRNAs for GABA receptor α1 subunit in comparison with only 6 by TargetScan and 29 by miRanda.
Table 4.
Predicted microRNAs targeting 5'-UTR, ORF and 3'-UTR region using miRWalk software
| GABA receptor subunits | Entrez Gene symbol | MicroRNAs targeting 5'-UTR | MicroRNAs targeting ORF | Numbers of microRNA targeting 3'-UTR with p-value < 0.05 |
|---|---|---|---|---|
| α1 | GABRA1 | miR-326, miR-28*, miR-29b-1*, | miR-341, miR-503, miR-150, miR-378 | 79 |
| miR-539, miR-542-5p, miR-147, | ||||
| miR-423, miR-598-5p | ||||
| α2 | GABRA2 | N/A | N/A | N/A |
| α3 | GABRA3 | miR-27b, miR-27a, miR-185, | miR-350, miR-431, miR-542-3p, miR-322, miR-323*, | 0 |
| miR-343, miR-346, miR-17-5p, | miR-140, miR-148b-3p, miR-29a*, miR-152, miR-497 | |||
| miR-93, miR-128, miR-143, | ||||
| miR-291a-5p, miR-20b-5p | ||||
| α4 | GABRA4 | N/A | N/A | N/A |
| α5 | GABRA5 | miR-345, miR-22, miR-451, | miR-24-1*, miR-24-2*, let-7d, miR-346, miR-153, | 37 |
| miR-541, miR-369 | miR-296, miR-376c, miR-466c | |||
| α6 | GABRA6 | 0 | miR-126*, miR-743b, miR-323*, miR-330*, miR-21*, | 0 |
| miR-153, miR-880 | ||||
| β1 | GABRB1 | miR-323, miR-219-2 | miR-140, miR-351, miR-324-5p, miR-325-3p, miR-7a*, | 26 |
| miR-10a-5p, miR-125a-5p, miR-125b-5p, miR-376b-5p, | ||||
| miR-384-3p | ||||
| β2 | GABRB2 | 0 | miR-20a*, miR-150, miR-297, miR-541 | 0 |
| β3 | GABRB3 | miR-188 | miR-300-5p, miR-350, miR-433, miR-881, miR-672, | 17 |
| γ1 | GABRG1 | miR-497, miR-322, miR-103-2, | miR-182, miR-216a, miR-483, miR-327, miR-338, | miR-126*, miR- |
| miR-103-1, miR-107 | miR-205, miR-296, miR-320, miR-880 | 379, miR-742, | ||
| miR-871 | ||||
| γ2 | GABRG2 | 0 | miR-182, miR-483, miR-382, miR-505 | 0 |
| γ3 | GABRG3 | miR-151*, miR-125a-3p | miR-142-3p, miR-298, miR-15b, miR-16, miR-28, | 14 |
| miR-34a, miR-195, miR-214, miR-290, miR-449a, | ||||
| miR-880, miR-708 | ||||
| π | GABRP | miR-28, miR-708 | miR-345-5p, miR-199a-3p, miR-873 | 0 |
| δ | GABRD | miR-210 | miR-322, miR-338, miR-193, miR-370, miR-497, miR-873 | 29 |
| ε | GABRE | 0 | miR-485, miR-484, miR-342-3p, miR-344-5p, miR-223, | 0 |
| miR-671, miR-322, miR-24, miR-139-3p, miR-199a-5p, | ||||
| miR-298, miR-483, miR-497, miR-743b, miR-672 | ||||
| θ | GABRQ | 0 | miR-350, miR-34c, miR-92a, miR-92a, miR-300-5p, | 0 |
| miR-92b, miR-7a*, miR-32 | ||||
| ρ1 | GABRR1 | miR-29b-1*, miR-26c | miR-338, let-7d, miR-204*, miR-421, miR-672, | 92 |
| miR-674-3p | ||||
| ρ2 | GABRR2 | miR-873, miR-134, miR-210, | miR-218*, let-7d, miR-34a, miR-204*, miR-421, | miR-191, |
| miR-207, miR-380, | miR-449a, miR-431, miR-381, miR-674-3p | miR-872* | ||
| ρ3 | GABRR3 | miR-350, miR-30c-1*, miR-30c- | miR-338, miR-341, miR-23a*, miR-143, miR-384-5p, | miR-191, miR- |
| 2*, miR-148b-3p, miR-152, | miR-324-3p, miR-30c, miR-30e, miR-30b-5p, miR-30d, | 344-1, miR-217, | ||
| miR-872*, miR-874, miR-672 | miR-30a, miR-204*, miR-539, miR-742, miR-873 | miR-883, miR-484 | ||
MicroRNAs which target 5'-UTR, open reading frame (ORF) and 3'-UTR of GABA receptors were predicted by miRWalk software http://www.ma.uni-heidelberg.de. N/A means that no information is available in miRWalk
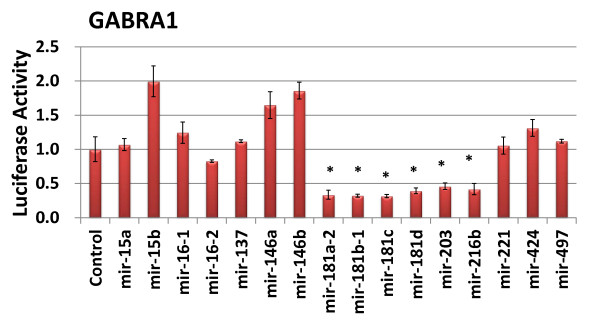
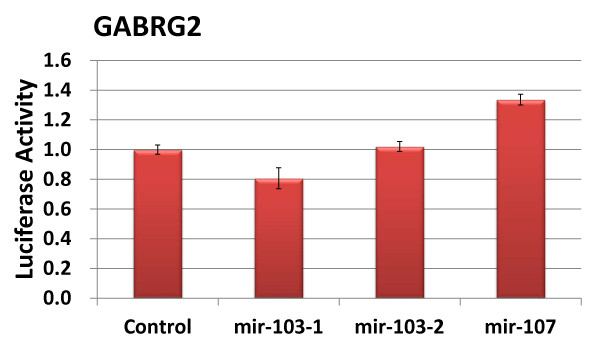
We selected two subunits, α1 and γ2 for experimental verification of the predictions. For α1-subunit, we constructed a 3'-UTR reporter vector, in which the 3'-UTR of α1-subunit was placed after a Renilla luciferase reporter gene (Table 2). For γ2-subunit, we cloned the predicted binding site of miR-103/107 into the downstream of a Renilla luciferase reporter gene. We then co-transfected a microRNA expression vector with the reporter into HEK293T cells to see whether the microRNA depressed the reporter activity. The firefly luciferase pGL3 vector was used for normalization. As shown in Figure 1, among the predicted microRNAs tested, miR-181, miR-216, and miR-203 inhibited the reporter activity. Four miR-181 isoforms, a-2, b-1, c and d-1 generated the same mature miR-181 and all of them depressed the reporter activity. All other microRNAs tested had no effects. The results suggest that miR-181, miR-216, and miR-203 are the micoRNAs that regulates GABA receptor α1-subunit. It is noted that miR-15, miR-16, miR-146, miR-221, miR-424 and miR-497 were predicated to target the GABA receptor α1-subunit by an earlier version (v.4.2) of TargetScan. Some of the miRNAs such as miR-181 are expressed in astrocytes naturally expressing GABA receptors [34]. For γ2-subunit, we did not find major effects of miR-103-1, miR-103-2, and miR-107 on the reporter activity (Figure 2).
Figure 1.
Effect of the predicted microRNAs on the 3'-UTR reporter activity of GABAA receptor α1-subunit. HEK 293T cells were transfected with the reporter and microRNA expression vectors and dual luciferase activities were assayed. The results were expressed as a ratio to the control microRNA vector. Data shown are means ± S.D. *P < 0.05 v.s. control. n = 3. Student t-Test.
Figure 2.
Effect of the predicted microRNAs on the binding site reporter activity of GABAA receptor γ2-subunit. HEK 293T cells were transfected with the reporter and microRNA expression vectors and dual luciferase activities were assayed. The results were expressed as a ratio to the control microRNA vector. Data shown are means ± S.D. n = 3.
It should be noted that we did not measure miRNA levels in the miRNA-overexpressed cells. Thus, there are possibilities that some of miRNAs may not be over-expressed in the experimental set-up; particularly for these miRNAs that had no effect on 3'-UTR reporter activity. However, the transfection efficiency is 90-100% under our experimental conditions based on the GFP reporter expression encoded in the same miRNA expression vector. Additionally, the effect of a miRNA on the luciferase activity does not necessarily mean that it was a direct effect on the binding of a miRNA to the 3'-UTR reporter construct. A miRNA could have indirect effects. The mutations of seed sequences in the miRNA binding sites are needed to exclude indirect effects. Further studies are also needed to see whether the overexpression of miR-181, miR-203, and miR-216 in a physiologically relevant cell type modifies GABA receptor expression, and whether these miRNAs are differentially regulated in diseased states.
It is also interesting to note that miR-15b and miR-146a/b actually increased the 3'-UTR reporter activity. It has been reported that miRNA increases translation [35]. However, it is also possible that this is a result of indirect effects.
We have previously shown that the activation of GABA receptors promotes fetal lung development [13]. The inhibition of miRNAs that target GABA receptors may increase receptor density and thus sensitivity of GABA receptors, which may benefit the development of therapy in treating diseases related to developmental anomalies.
Conclusions
In summary, computational approaches predict many microRNA binding sites on the 3'-UTR of GABAA receptors, but not on these of GABAC receptors. 3'-UTR reporter assays only verified miR-181, miR-216, and miR-203 as the microRNAs that target GABA receptor α1-subunit among 10 microRNAs tested. These studies reinforce that micoRNA target prediction needs to be verified experimentally. The identification of microRNAs that target to GABA receptors provides a basis for further studies of post-transcriptional regulation of GABA receptors.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CZ, TW and HM carried out experiments. CZ, CH and XX analyzed data and performed target predictions. LL conceived of the study, and participated in its design and coordination. LL and CZ drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Chunling Zhao, Email: chunlingzhao@sina.cn.
Chaoqun Huang, Email: Chaoqun.huang@okstate.edu.
Tingting Weng, Email: Tingting.Weng@uth.tmc.edu.
Xiao Xiao, Email: xiao.xiao@okstate.edu.
Hong Ma, Email: mh1985623@126.com.
Lin Liu, Email: lin.liu@okstate.edu.
Acknowledgements
We thank Ms. Tazia Cook for editorial assistance. This work was supported by the National Institutes of Health, HL-087884 and HL-095383 and OCAST, AR101-037.
References
- Ben Ari Y. Excitatory actions of GABA during development: the nature of the nurture. Nat Rev Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- Owens DF, Kriegstein AR. Is there more to GABA than synaptic inhibition? Nat Rev Neurosci. 2002;3:715–727. doi: 10.1038/nrn919. [DOI] [PubMed] [Google Scholar]
- Akinci MK, Schofield PR. Widespread expression of GABA(A) receptor subunits in peripheral tissues. Neurosci Res. 1999;35:145–153. doi: 10.1016/S0168-0102(99)00078-4. [DOI] [PubMed] [Google Scholar]
- Glassmeier G, Hopfner M, Buhr H, Lemmer K, Riecken EO, Stein H, Quabbe HJ, Rancso C, Wiedenmann B, Scherubl H. Expression of functional GABAA receptors in isolated human insulinoma cells. Ann N Y Acad Sci. 1998;859:241–248. doi: 10.1111/j.1749-6632.1998.tb11138.x. [DOI] [PubMed] [Google Scholar]
- Park HS, Park HJ. Effects of gamma-aminobutyric acid on secretagogue-induced exocrine secretion of isolated, perfused rat pancreas. Am J Physiol Gastrointest Liver Physiol. 2000;279:G677–G682. doi: 10.1152/ajpgi.2000.279.4.G677. [DOI] [PubMed] [Google Scholar]
- Jin N, Kolliputi N, Gou D, Weng T, Liu L. A novel function of ionotropic gamma-aminobutyric acid receptors involving alveolar fluid homeostasis. J Biol Chem. 2006;281:36012–36020. doi: 10.1074/jbc.M606895200. [DOI] [PubMed] [Google Scholar]
- Limmroth V, Lee WS, Moskowitz MA. GABAA-receptor-mediated effects of progesterone, its ring-A-reduced metabolites and synthetic neuroactive steroids on neurogenic oedema in the rat meninges. Br J Pharmacol. 1996;117:99–104. doi: 10.1111/j.1476-5381.1996.tb15160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mong JA, Nunez JL, McCarthy MM. GABA mediates steroid-induced astrocyte differentiation in the neonatal rat hypothalamus. J Neuroendocrinol. 2002;14:45–55. doi: 10.1046/j.1365-2826.2002.00737.x. [DOI] [PubMed] [Google Scholar]
- Mayerhofer A, Hohne-Zell B, Gamel-Didelon K, Jung H, Redecker P, Grube D, Urbanski HF, Gasnier B, Fritschy JM, Gratzl M. Gamma-aminobutyric acid (GABA): a para- and/or autocrine hormone in the pituitary. FASEB J. 2001;15:1089–1091. doi: 10.1096/fj.00-0546fje. [DOI] [PubMed] [Google Scholar]
- Chen Z, Jin N, Narasaraju T, Chen J, McFarland LR, Scott M, Liu L. Identification of two novel markers for alveolar epithelial type I and II cells. Biochem Biophys Res Commun. 2004;319:774–780. doi: 10.1016/j.bbrc.2004.05.048. [DOI] [PubMed] [Google Scholar]
- Jin N, Narasaraju T, Kolliputi N, Chen J, Liu L. Differential expression of GABA(A) receptor pi subunit in cultured rat alveolar epithelial cells. Cell Tissue Res. 2005;321:173–183. doi: 10.1007/s00441-005-1130-8. [DOI] [PubMed] [Google Scholar]
- Jin N, Guo Y, Sun P, Bell A, Chintagari NR, Bhaskaran M, Rains K, Baviskar P, Chen Z, Weng T, Liu L. Ionotropic GABA receptor expression in the lung during development. Gene Expr Patterns. 2008;8:397–403. doi: 10.1016/j.gep.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintagari NR, Jin N, Gao L, Wang Y, Xi D, Liu L. Role of GABA receptors in fetal lung development in rats. PLoS One. 2010;5:e14171. doi: 10.1371/journal.pone.0014171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormann J. The 'ABC' of GABA receptors. Trends Pharmacol Sci. 2000;21:16–19. doi: 10.1016/S0165-6147(99)01413-3. [DOI] [PubMed] [Google Scholar]
- Enz R, Cutting GR. Molecular composition of GABAC receptors. Vision Res. 1998;38:1431–1441. doi: 10.1016/S0042-6989(97)00277-0. [DOI] [PubMed] [Google Scholar]
- Zhang D, Pan ZH, Awobuluyi M, Lipton SA. Structure and function of GABA(C) receptors: a comparison of native versus recombinant receptors. Trends Pharmacol Sci. 2001;22:121–132. doi: 10.1016/S0165-6147(00)01625-4. [DOI] [PubMed] [Google Scholar]
- Moss SJ, Smart TG. Modulation of amino acid-gated ion channels by protein phosphorylation. Int Rev Neurobiol. 1996;39:1–52. doi: 10.1016/s0074-7742(08)60662-5. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W, Parnigotto PP, Warburton D. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De FS. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806–812. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc Natl Acad Sci USA. 2009;106:15819–15824. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Liu X, Nelson A, Nakanishi M, Kanaji N, Wang X, Kim M, Li Y, Sun J, Michalski J, Patil A, Basma H, Holz O, Magnussen H, Rennard SI. Reduced miR-146a increases prostaglandin Ein chronic obstructive pulmonary disease fibroblasts. Am J Respir Crit Care Med. 2010;182:1020–1029. doi: 10.1164/rccm.201001-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, Coarfa C, Zariff A, Milosavljevic A, Batts LM, Kheradmand F, Gunaratne PH, Corry DB. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. MIR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2010;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Anderson TA. MicroRNA: a new player in stem cells. J Cell Physiol. 2006;209:266–269. doi: 10.1002/jcp.20713. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk-Database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Ouyang YB, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2011. http://dx.doi.org/10.1016/j.mito.2011.09.001 [DOI] [PMC free article] [PubMed]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]