Abstract
Background
Serovars of the human pathogen Chlamydia trachomatis occupy one of three specific tissue niches. Genomic analyses indicate that the serovars have a phylogeny congruent with their pathobiology and have an average substitution rate of less than one nucleotide per kilobase. In contrast, the gene that determines serovar specificity, ompA, has a phylogenetic association that is not congruent with tissue tropism and has a degree of nucleotide variability much higher than other genomic loci. The ompA gene encodes the major surface-exposed antigenic determinant, and the observed nucleotide diversity at the ompA locus is thought to be due to recombination and host immune selection pressure. The possible contribution of a localized increase in mutation rate, however, has not been investigated.
Results
Nucleotide diversity and phylogenetic relationships of the five constant and four variable domains of the ompA gene, as well as several loci surrounding ompA, were examined for each serovar. The loci flanking the ompA gene demonstrated that nucleotide diversity increased monotonically as ompA is approached and that their gene trees are not congruent with either ompA or tissue tropism. The variable domains of the ompA gene had a very high level of non-synonymous change, which is expected as these regions encode the surface-exposed epitopes and are under positive selection. However, the synonymous changes are clustered in the variable regions compared to the constant domains; if hitchhiking were to account for the increase in synonymous changes, these substitutions should be more evenly distributed across the gene. Recombination also cannot entirely account for this increase as the phylogenetic relationships of the constant and variable domains are congruent with each other.
Conclusions
The high number of synonymous substitutions observed within the variable domains of ompA appears to be due to an increased mutation rate within this region of the genome, whereas the increase in nucleotide substitution rate and the lack of phylogenetic congruence in the regions flanking ompA are characteristic motifs of gene conversion. Together, the increased mutation rate in the ompA gene, in conjunction with gene conversion and positive selection, results in a high degree of variability that promotes host immune evasion.
Keywords: Chlamydia trachomatis, Evolution, MOMP, ompA, Phylogenetics
Background
Chlamydia trachomatis is an obligate intracellular bacterium that is the most frequently reported bacterial sexually transmitted disease in the United States [1], as well as the leading cause of infectious blindness worldwide [2]. Strains of C. trachomatis are differentiated into serovars based on the serospecificity of the major outer membrane protein (MOMP) that constitutes over 60% of all the surface-exposed proteins [3] and functions as a porin [4]. Serovars A-C primarily infect ocular tissue, serovars D-K typically infect urogenital tissue, and serovars L1-L3 infect lymphatic tissue. There appears to be a very high level of nucleotide identity across the genomes of the serovars [5,6] as even the most divergent genomes are over 99% identical in sequence [7]. Additionally, genomic data has demonstrated that the phylogenetic pattern of the serovars correlates with tissue tropism [8-10].
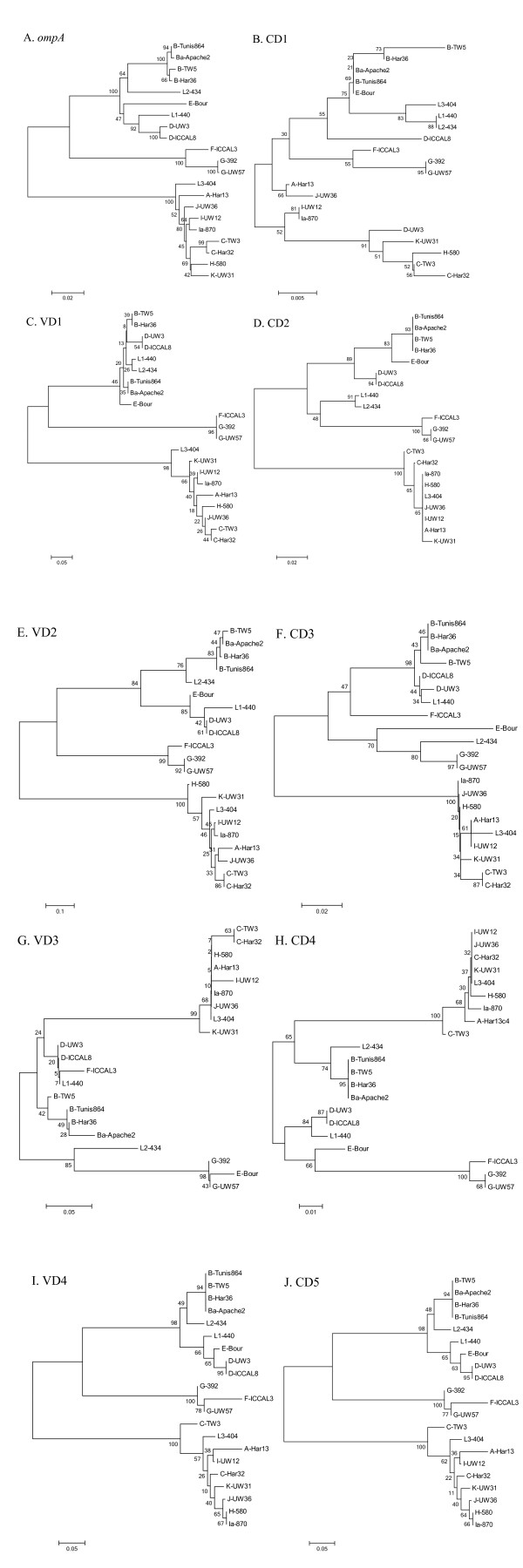
In contrast to the low level of nucleotide variation observed in most of the genome, the ompA gene that encodes MOMP is characterized by a very high level of variability as nearly 30% of the coding sequence exhibits polymorphisms [11]. For comparison, the porB gene in C. trachomatis also encodes a surface-exposed porin that elicits neutralizing antibodies [12,13], but it only varies at 1% of its nucleotide sites [8]. MOMP has four variable domains that encode the surface-exposed epitopes and contain the amino acid substitutions that define serospecificity; these are separated by five membrane-spanning constant domains that are considerably more conserved than the variable domains, but are nevertheless more polymorphic than the majority of the genes in the genome. Phylogenetic characterization of the ompA gene exhibits a pattern of strongly supported groupings that differ markedly from the tissue tropism [11,14]: serovars A, C, H, I, J, K, and L3 comprise one group; F and G a second; D, E, and L1 a third; and B and L2 a fourth (see Figure 1A).
Figure 1.
Phylogenetic analyses of the C. trachomatis ompA gene and individual constant and variable domains. Phylogenetic trees of the C. trachomatis ompA gene (A), as well as the five constant (CD1-5) and four variable (VD1-4) domains of the ompA gene (B-J), were constructed using the Maximum Composite Likelihood neighbor-joining method with pairwise deletions of alignment gaps. Results of 1000 bootstrap replicates are reported for each node.
The discordance between the ompA gene tree and trees from other C. trachomatis loci is considered to be due to intra-species recombination [8,9,15-17]. Recombination is thought to be rare in C. trachomatis due to the limited opportunity other bacteria, plasmids, or exogenous DNA have to enter a chlamydial inclusion [18], but genomic sequence data indicate that intra-species recombination has occurred in 8 genes in the genome, including ompA, and may have occurred in as many as 47 additional genes [17]. There is evidence that recombination has acted on portions of the ompA gene to create mosaic MOMP structures [19-22], as well as on the entire ompA gene in order to replace the MOMP of an isolate [8,9,16]. Further support that chlamydia has the means and mechanisms to acquire and stably incorporate exogenous DNA is the presence of a genomic island containing a tetracycline-resistance gene found in the chromosome of several swine isolates of Chlamydia suis [23]. In addition to recombination, diversity generated in MOMP can be attributed to the positive selection of non-synonymous substitutions that lead to beneficial amino acid replacements, especially in a gene that encodes an antigenic outer membrane protein.
It is currently unknown, however, if the increase in nucleotide diversity in ompA is solely due to recombination and positive selection. In order to establish if there are additional factors involved in the generation of ompA diversity, we examined the nucleotide and phylogenetic associations of the five constant and four variable domains of the ompA gene, as these regions are under different immune and functional constraints. We also evaluated the relationships of the loci surrounding the ompA gene. These results demonstrate that the synonymous nucleotide diversity generated in the ompA gene cannot be explained by recombination and positive selection alone, and indicate that this region is prone to increased mutations compared to other regions in the genome. The data also show that the increased nucleotide substitution rate and the lack of phylogenetic associations in the regions proximal to the ompA gene are characteristic motifs of gene conversion.
Results
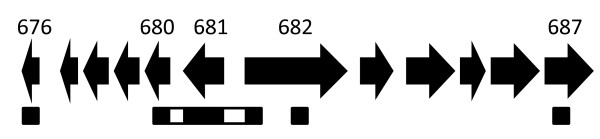
Twenty-one strains representing at least one of each serovar were used in this study (A/Har-13, B/Tunis 864, B/TW-5, B/Har-36, Ba/Apache 2, C/TW-3, C/Har-32, D/UW-3, D/IC-Cal 8, E/Bour, F/IC-Cal 3, G/UW-57, G/392, H/580, I/UW-12, Ia/870, J/UW-36, K/UW-31, L1/440, L2/434, L3/404). These are all lab-adapted strains that are common in the literature, and 18 of them were previously used to characterize regions throughout the genome [8]. These strains were sequenced at the following eight loci: CT676 (hypothetical gene), CT680 (rs2), CT680-1 (intergenic region), CT681 (ompA), CT681-2 (intergenic region), CT682 (5' end; pbpB), CT682 (middle; pbpB), and CT687 (yfhO2) (Figure 2).
Figure 2.
Schematic representation of the loci under investigation. The following eight regions were used in the study and are designated by their gene number and name: CT676 (hypothetical gene), CT680 (rs2), CT680-1 (intergenic region), CT681 (ompA), CT681-2 (intergenic region), CT682 (5' end; pbpB), CT682 (middle; pbpB), and CT687 (yfhO2). The arrows in the map represent the gene size and distance relative to each other; the boxes under the arrows represent the regions that were sequenced (black boxes are coding regions and open boxes are intergenic regions). Specific details on nucleotide region sizes and distances are in Table 1.
Sequence diversity in ompA and flanking regions
To assess the relationships of the ompA flanking regions, seven loci totaling ~2.7 kb within 10 kb of either side of the ompA gene were sequenced and analyzed. These included the entire contiguous non-coding regions on the 5' and 3' end of the ompA gene, as well as neighboring coding loci (Table 1). The rate of substitutions at both non-synonymous (dN) and synonymous sites (dS) in these genomic regions increase as they approach the ompA gene (Table 1). The ratio of transition to transversion events (ts/tv) in housekeeping loci outside of the ompA region of the genome is 10/1, but this rapidly decreases with proximity to ompA to a ratio of 1/1 (Table 1). Usually, the ts/tv ratio is biased towards transitions as these are biochemically simpler changes (purine to purine, and pyrimidine to pyrimidine), as was observed in the C. trachomatis housekeeping regions. A larger number of transversions, however, is indicative of a region more susceptible to mutation, though the ompA region may be biased due to saturation of nucleotide diversity [24]. Also, there are a number of insertion/deletion sites (indels) within the adjacent non-coding regions of ompA (10-12% of the nucleotide sites). Together, these data demonstrate that the percent of variable nucleotide sites and nucleotide diversity increase monotonically as ompA is approached.
Table 1.
Nucleotide analyses of the ompA gene and surrounding loci
| Loci | Gene | Distance from ompAA | #nt | #Δ nt sites | %Δ nt sites | dN | dS | dN/dS | ts/tv |
|---|---|---|---|---|---|---|---|---|---|
| 676 | CT676 | 4090-3803 | 347 | 16 | 4.61 | 0.003 | 0.065 | 0.05 | 8.6 |
| 680 | rs2 | 781-370 | 412 | 40 | 9.71 | 0.015 | 0.112 | 0.13 | 2.9 |
| 680-1 | 369-1 | 399 | 54 | 13.53 | ---- | ---- | ---- | 2.1 | |
| 681 | ompA | ---- | 1194 | 341 | 28.56 | 0.076 | 0.419 | 0.18 | 1.0 |
| 681-2 | 1-607 | 608 | 47 | 7.73 | ---- | ---- | ---- | 1.0 | |
| 682B | pbpB | 608-810 | 208 | 15 | 7.21 | 0.005 | 0.082 | 0.06 | 2.6 |
| 682C | pbpB | 1332-1772 | 442 | 17 | 3.85 | 0.004 | 0.030 | 0.13 | 3.8 |
| 687 | yfhO2 | 8873-9229 | 357 | 9 | 2.52 | 0.002 | 0.026 | 0.08 | ----D |
| HKE | 3096 | 23 | 0.74 | 0.001 | 0.002 | 0.61 | 10.0 | ||
ADistance based on serovar D/UW-3 genome
B5' end of gene
CMiddle of gene
DAll 9 polymorphisms were transitions
EHousekeeping gene data from Brunelle and Sensabaugh, 2006 [8]
Sequence diversity within ompA constant and variable domains
To compare the five constant domains within the ompA gene to the four interspersed variable domains [25], these nine regions were examined at the nucleotide level (Table 2). The five constant domains comprise a total of 927 nucleotide sites, 155 of which are variable (16.7%). Although the dN/dS ratio (0.04) is comparable to regions outside of ompA, the individual dN and dS value are each equal-to-greater than that observed in the adjacent loci. In contrast, 186 of the 267 (69.7%) total nucleotide sites in the four variable domains are variable, which is reflected in the elevated dN, dS, and dN/dS values compared to the constant domains. The considerably reduced rate of amino acid replacements in the constant regions implies they have been under a different level of selection; nonetheless, the constant domains have a high level of sequence diversity compared to regions outside of the ompA gene.
Table 2.
Nucleotide analyses of the constant (CD#) and variable (VD#) domains of the ompA gene
| # nt | #Δ nt sites | %Δ nt sites | dN | dS | dN/dS | ts/tv | |
|---|---|---|---|---|---|---|---|
| CD1 | 255 | 19 | 7.45 | 0.008 | 0.092 | 0.09 | 1.6 |
| CD2 | 165 | 43 | 26.06 | 0.024 | 0.927 | 0.03 | 2.7 |
| CD3 | 189 | 52 | 27.51 | 0.019 | 0.596 | 0.03 | 6.3 |
| CD4 | 150 | 26 | 17.33 | 0.014 | 0.421 | 0.03 | 2.4 |
| CD5 | 168 | 15 | 8.93 | 0.004 | 0.123 | 0.03 | 2.1 |
| CD1-5 | 927 | 155 | 16.72 | 0.013 | 0.314 | 0.04 | 2.0 |
| VD1 | 66 | 58 | 87.88 | 0.797 | 0.962 | 0.83 | 0.4 |
| VD2 | 66 | 51 | 77.27 | 0.640 | 0.983 | 0.65 | 0.9 |
| VD3 | 42 | 19 | 45.24 | 0.135 | 0.491 | 0.27 | 0.9 |
| VD4 | 93 | 58 | 62.37 | 0.281 | 0.648 | 0.43 | 1.1 |
| VD1-4 | 267 | 186 | 69.66 | 0.404 | 1.204 | 0.34 | 0.7 |
| ompA | 1194 | 341 | 28.56 | 0.076 | 0.419 | 0.18 | 1.0 |
Phylogenetic analysis of ompA and flanking regions
Individual phylogenetic trees were constructed for ompA (Figure 1A) and the seven regions flanking the ompA gene (Additional file 1) to assess if comparable or distinct associations existed between the ompA gene and the neighboring loci; differing phylogenetic topologies would be indicative of recombination. The phylogenetic trees were all different between the CT676 locus, the non-coding regions immediately adjacent to either side of ompA (CT680-1 and CT681-2), and the two different portions of the CT682 coding region. In addition, none of these regions were congruent with the phylogeny established for either ompA or that based on tissue tropism [8,9,16,26]. Interestingly, serovars L1-L3 grouped together in the trees from the flanking loci, but were separated only in the ompA tree. The lack of overall phylogenetic congruence observed in ompA and its neighboring regions can be ascribed to recombination.
Indels are informative events and can serve as valuable phylogenetic markers because there is a low probability that such events would occur in an identical region of two separate strains independently. Excluding single nucleotide indels, as well as those found in a repetitive stretch of a single nucleotide base (e.g. poly-T tracts) that could be susceptible to slip-strand mispairings, the indels in the flanking non-coding regions of ompA were examined. The non-coding region preceding the 5' end (CT681-2) had a 21 bp deletion in strains B/Har36, B/TW5, F/IC-CAL3. Since it was not found in B/Tunis 864 or Ba/Apache2, this indicates an association among these otherwise divergent strains not seen in any of the previous phylogenetic analyses. The 2, 3, 4, 6, and 9 bp indels in the non-coding region following the 3' end of ompA (CT680-1) were all congruent with the phylogenetic relationship of the region. In addition, there was a tandem 8 bp motif (TATTAGAA) in A/Har13 immediately adjacent to the stop codon of ompA. One 8 bp sequence is found in all the strains in this location, but the insertion in strain A/Har13 was apparently due to a duplication event. This region was sequenced from a different strain of the same serovar, A/Har1, and was found to have the same octamer repeat. This octamer occurs only 69 times in the C. trachomatis D/UW3 genome, but never in succession.
Phylogenetic analysis within ompA constant and variable domains
The nucleotide analysis above demonstrated that the variable and constant regions of the ompA gene are under different levels of selection (dN/dS = 0.34 and 0.04, respectively), which is in agreement with fact that each region is under different immunological and functional constraints. However, it is unknown if the disparity in synonymous nucleotide diversity between the variable and constant regions is due to recombination. The phylogenetic relationships among the nine regions were examined to determine if these regions evolved together or independently. If the phylogeny shows that they evolved together, then recombination was not a factor in generating the synonymous nucleotide diversity. Accordingly, trees were constructed from each of the nine variable and constant regions (Figure 1B-J). All of the trees were very similar to the ompA gene (Figure 1A), indicating the constant regions evolved in concert with the variable regions and independently of other regions in the genome. However, not all of the branching patterns in each region were identical; for example, strain F/IC-Cal3 did not group with strain G/UW-57 in the intermediate complex in constant domain 3 and variable domain 3 (Figure 1F, G). These differences coincide with the evidence of recombination with the ompA gene, and such heterogeneity would serve to explain some of the poor support (low bootstrap values) in an individual region. Because the phylogenies in all nine regions are highly similar, it implies that recombination did not shape the differences in synonymous nucleotide diversity between the variable and constant regions.
The synonymous and non-synonymous nucleotides in the ompA gene are under different selective constraints, and trees representing each class of sites from the ompA gene were constructed in order to compare their evolutionary histories (Additional file 2). These two trees were largely identical, indicating that the synonymous sites evolved in concert with the non-synonymous sites and are consistent with the ompA phylogeny.
Divergence rates from MoPn
The murine Chlamydia muridarum MoPn, of which there is one known isolate, is a closely related common ancestor of the human C. trachomatis isolates. By comparing the divergence rates in orthologous regions between the human C. trachomatis isolates and the C. muridarum isolate, differences in mutation rates for each locus can be assessed (Table 3). The amount of non-synonymous replacements (KA) between mouse and human isolates is higher in the ompA gene than other regions, but this is expected as positive selection is known to occur in the variable domains. The rate of synonymous change (KS) in ompA, as well as the overall nucleotide substitution rate (DXY), is comparable between mouse and human isolates (Table 3).
Table 3.
Divergence analysis of C. trachomatis human isolates and the murine C. muridarum MoPn isolate
| Locus | KA | KS | KA/KS | DXY |
|---|---|---|---|---|
| 676 | 0.056 | 1.461 | 0.04 | 0.12 |
| 680 | 0.064 | 0.694 | 0.09 | 0.16 |
| ompA | 0.112 | 0.981 | 0.11 | 0.24 |
| CD1-5A | 0.034 | 0.897 | 0.04 | 0.16 |
| VD1-4B | 0.569 | 1.396 | 0.41 | 0.72 |
| 682C | 0.032 | 0.645 | 0.05 | 0.14 |
| 682D | 0.019 | 1.170 | 0.02 | 0.16 |
| 687 | 0.094 | 1.070 | 0.09 | 0.24 |
| HKE | 0.052 | 0.915 | 0.06 | 0.19 |
AompA constant domains 1-5
BompA variable domains 1-4
C5' end of gene
DMiddle of gene
EHousekeeping gene data from Brunelle and Sensabaugh, 2006 [8]
Discussion
Examination of the ompA gene of C. trachomatis and its surrounding loci demonstrate an increase in nucleotide substitutions and differing phylogenetic histories compared to other regions of the genome. The increase in nucleotide substitution rates and the lack of any phylogenetic congruence in the regions flanking ompA are characteristic motifs of gene conversion. Gene conversion in bacteria is a non-reciprocal recombinatorial process in which one gene replaces another through strand invasion/displacement and repair via the recA-D proteins [27]. Such a process is common in bacteria; it occurs in at least 17 different genera [27], including Chlamydia [17,28,29]. Typically, gene conversion can serve one of two functions in the genome. First, it can promote diversification of antigenic outer membrane genes, as has been reported in species of Anaplasma [30], Bartonella [31], Borrelia [32,33], Mycoplasma [34], Neisseria [35,36], and Treponema [34]. Second, it can reduce nucleotide variation in functional genes by homogenizing homologs within a genome; there is evidence that all bacterial genomes with multiple-copy rDNA undergo gene conversion, which maintains the uniformity and the low-level of nucleotide diversity observed in the ribosomal gene sequences (concerted evolution) [37]. Both models, however, also lead to an increase in nucleotide diversity and indel events in the flanking regions [30,38,39]. This is what we observed in the ompA gene, supporting gene conversion of the ompA gene region.
Gene conversion alone does not explain the increase in polymorphisms within the ompA variable domains. An increase in the rate of amino acid replacements in the variable domains of the ompA gene compared to the constant domains is expected, as these regions are surface-exposed and are the main antigenic targets of the host immune system. However, the 3.8-fold increase in the rate of synonymous change in the variable domains relative to the constant domains is notable, especially considering that the constant domains contain more synonymous changes than most other genes in the genome. Though synonymous changes will hitchhike with the selection of favorable non-synonymous changes, this selection should occur for any polymorphism across the entire gene and not just in the variable domains. Nevertheless, in the ompA gene, the rate of synonymous change is clearly not evenly distributed. This disparity in the distribution of synonymous changes is not due to recombination of the variable domain because the phylogenetic trees for each constant and variable domain, in addition to the trees constructed from either synonymous or non-synonymous sites, are all congruent with each other and with the ompA gene. There appears to be some factor that increases the propensity of mutations in the variable regions (i.e. a mutational hotspot) that would explain the observations; a mutational hotspot has previously been demonstrated to occur in the trpA gene of C. trachomatis [40,41]. The high mutation rate observed in the synonymous sites in the variable domains compared to the constant domains appears to be evolutionarily conserved: the divergence analysis from the C. muridarum MoPn isolate indicates an equivalent level of synonymous change between the variable domains, the constant domains, and the neighboring genes.
It is not known what mechanism(s) promote the mutational increase in ompA and its neighboring loci relative to the rest of the genome. Codon bias causes a greater level of synonymous change in some genes as a way to increase transcriptional efficiency [42], but this would not occur in distinct segments of a single gene. Mutational bias associated with chromosomal location at the replication terminus [43] is also not a factor due to the fact that the ompA gene is only 60 kb from the origin of replication. One possible explanation is that a several-kilobase genomic region surrounding ompA has a unique secondary structure, which predisposes the region to increased mutations and decreased access to DNA repair machinery. While the whole region would be subject to increased mutations, more mutations could become fixed in the ompA gene if it had fewer functional constraints than the neighboring loci. In addition, the level of ompA expression may play a role: because ompA is one of the most highly expressed genes in C. trachomatis [44], it spends more time than other genes being single stranded and therefore may have a higher propensity of acquiring mutations in this state [45,46]. Mutations are more likely to occur in the non-template strand because it is single-stranded and not protected by the replication bubble, and is therefore more prone to DNA damage and deamination that can lead to nucleotide replacements, insertions, or deletions [47-50]. Coupled with the potential translesion activity of DNA polymerase I [51] and positive selection, these mutations in ompA could become permanent.
Conclusions
The discordant phylogenetic associations of the loci neighboring the ompA gene, along with the increased rate of nucleotide diversity in these regions, can be ascribed to recombination of the ompA gene via gene conversion. In addition, the high synonymous substitution rate within the variable domains of ompA appears to be the result of an unidentified influence that generates an increase in mutations within the gene or genomic region. Together, the increased mutation rate in the ompA gene acts in concert with positive selection and recombination to promote the high degree of variability observed in MOMP in order to evade immune detection.
Methods
Source of isolates
Nineteen serovars were originally obtained through Dr. J. Schachter, University of California, San Francisco (A/Har-1, A/Har-13, B/Tunis 864, B/Har-36, Ba/Apache 2, C/TW-3, C/Har-32, D/IC-Cal 8, E/Bour, F/IC-Cal 3, G/392, G/UW-57, H/580, Ia/870, J/UW-36, K/UW-31, L1/440, L2/434, L3/404). Three additional strains were provided by Dr. R. S. Stephens, University of California, Berkeley (B/TW-5, D/UW-3, I/UW-12).
DNA isolation from culture
DNA was isolated using the standard proteinase K digestion, phenol-chloroform-isoamyl extraction, and ethanol precipitation [52].
Genes of interest
The ompA gene sequences from these isolates are available in GenBank [GenBank: DQ064279-DQ064299] [8]. Primers were designed to amplify seven regions within 10 kb of either end of the ompA gene, and each PCR product was purified using ExoSAP-IT (GE Healthcare, Piscataway, NJ), verified on a 2% agarose gel, and sequenced on an ABI 377 sequencer using Big Dye 3 terminator chemistry (Applied Biosystems, Foster City, CA). Sequences were deposited in GenBank [GenBank: DQ063870-DQ063890; DQ063912-DQ064017; DQ064039- DQ064059]. Orthologous regions from the Mouse Pneumonitis (MoPn) biovar of Chlamydia muridarum strain Nigg were retrieved from the genome sequence [GenBank: AE002160] [53].
Alignment and analysis
The nucleotide sequences for each locus were aligned using ClustalX [54] employed in BioEdit http://www.mbio.ncsu.edu/BioEdit/bioedit.html. For coding regions, nucleotide sequences were translated to their amino acid sequences, aligned, and then translated back to their nucleotide sequences within the alignment. The software package MEGA4 [55] was used to calculate the following from the human isolates of C. trachomatis: the number of variable nucleotide sites in each locus; the number of non-synonymous substitutions per non-synonymous site (dN) and the number of synonymous substitutions per synonymous site (dS) using the Nei-Gojobori method and Jukes and Cantor correction with pairwise deletions for alignment gaps; and the ratio of transitions to transversions using pairwise deletion of alignment gaps (ts/tv). MEGA4 was also used to construct phylogenetic trees using the Maximum Composite Likelihood neighbor-joining method and pairwise deletions for alignment gaps. The only exception was the two trees built from either the synonymous or non-synonymous sites of ompA; for these analyses, the Nei-Gojobori neighbor-joining method was employed with the Jukes and Cantor correction and pairwise deletions for alignment gaps. All trees were supported by 1000 bootstrap replicates. The software package DnaSP v5 [56] was used to calculate the following divergence values with Jukes and Cantor correction between the human isolates of C. trachomatis and the MoPn isolate: non-synonymous substitutions per non-synonymous site (KA), synonymous substitutions per synonymous site (KS), and average number of substitutions per nucleotide site between populations (DXY).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BWB and GFS designed the study, BWB performed experiments, and BWB and GFS analyzed data and wrote the manuscript. Both authors read and approved the final manuscript.
Supplementary Material
Figure S1. Phylogenetic analyses of the C. trachomatis ompA gene flanking regions.
Figure S2. Phylogenetic analyses of the synonymous and non-synonymous sites of the ompA gene.
Contributor Information
Brian W Brunelle, Email: Brian.Brunelle@ars.usda.gov.
George F Sensabaugh, Email: sensaba@berkeley.edu.
Acknowledgements
We would like to thank Richard S. Stephens and Julius Schachter for the gift of the isolates, and Malcolm McGinnis for use of his sequencing facilities. We would also like to thank Heather Allen, John Bannantine, and Thaddeus Stanton for critical review of the manuscript.
This work was supported in part by a Faculty Bridging Grant to GFS. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
References
- CDC. Sexually Transmitted Disease Surveillance. Atlanta: US Department of Health and Human Services, Centers for Disease Control and Prevention. 2010.
- Mabey DC, Solomon AW, Foster A. Trachoma. Lancet. 2003;362(9379):223–229. doi: 10.1016/S0140-6736(03)13914-1. [DOI] [PubMed] [Google Scholar]
- Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31(3):1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavoil P, Ohlin A, Schachter J. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect Immun. 1984;44(2):479–485. doi: 10.1128/iai.44.2.479-485.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS. In: Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Stephens RS, editor. Washington, DC: American Society for Microbiology; 1999. Genomic autobiographies of Chlamydiae; pp. 6–26. [Google Scholar]
- Brunelle BW, Nicholson TL, Stephens RS. Microarray-based genomic surveying of gene polymorphisms in Chlamydia trachomatis. Genome Biol. 2004;5(6):R42. doi: 10.1186/gb-2004-5-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BM, Suchland RJ, Quinn KL, Davidson JR, Stamm WE, Rockey DD. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun. 2010;78(6):2544–2553. doi: 10.1128/IAI.01324-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle BW, Sensabaugh GF. The ompA gene in Chlamydia trachomatis differs in phylogeny and rate of evolution from other regions of the genome. Infect Immun. 2006;74(1):578–585. doi: 10.1128/IAI.74.1.578-585.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol. 2006;188(1):275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard DR, Toth GA, Batteiger BE. Polymorphic membrane protein H has evolved in parallel with the three disease-causing groups of Chlamydia trachomatis. Infect Immun. 2003;71(3):1200–1208. doi: 10.1128/IAI.71.3.1200-1208.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard DR, Boguslawski G, Jones RB. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect Immun. 1998;66(8):3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Stephens RS. Characterization and functional analysis of PorB, a Chlamydia porin and neutralizing target. Mol Microbiol. 2000;38(4):772–780. doi: 10.1046/j.1365-2958.2000.02167.x. [DOI] [PubMed] [Google Scholar]
- Kawa DE, Stephens RS. Antigenic topology of chlamydial PorB protein and identification of targets for immune neutralization of infectivity. J Immunol. 2002;168(10):5184–5191. doi: 10.4049/jimmunol.168.10.5184. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Peterson EM, de la Maza LM. Phylogenetic analysis of the outer-membrane-protein genes of Chlamydiae, and its implication for vaccine development. Mol Biol Evol. 1993;10(4):892–913. doi: 10.1093/oxfordjournals.molbev.a040048. [DOI] [PubMed] [Google Scholar]
- Feil E, Zhou J, Maynard Smith J, Spratt BG. A comparison of the nucleotide sequences of the adk and recA genes of pathogenic and commensal Neisseria species: evidence for extensive interspecies recombination within adk. J Mol Evol. 1996;43(6):631–640. doi: 10.1007/BF02202111. [DOI] [PubMed] [Google Scholar]
- Gomes JP, Bruno WJ, Nunes A, Santos N, Florindo C, Borrego MJ, Dean D. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 2007;17(1):50–60. doi: 10.1101/gr.5674706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Didelot X, Gandhi K, Dean D, Read TD. Interplay of recombination and selection in the genomes of Chlamydia trachomatis. Biol Direct. 2011;6:28. doi: 10.1186/1745-6150-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS. Chlamydial Infections, Proceedings of the Tenth International Symposium on Human Chlamydial Infections: 16-21 June 2002: 2002. Antalya, Turkey: International Chlamydia Symposium; 2002. Chlamydial evolution: a billion years and counting; pp. 3–16. [Google Scholar]
- Lampe MF, Suchland RJ, Stamm WE. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect Immun. 1993;61(1):213–219. doi: 10.1128/iai.61.1.213-219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham R, Yang C, Maclean I, Kimani J, Maitha G, Plummer F. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J Clin Invest. 1994;94(1):458–463. doi: 10.1172/JCI117347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman KL, Tavare S, Dean D. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J Bacteriol. 2001;183(20):5997–6008. doi: 10.1128/JB.183.20.5997-6008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman K, Black CM, Johnson RE, Stamm WE, Jones RB, Hook EW, Martin DH, Bolan G, Tavare S, Dean D. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J Bacteriol. 2004;186(8):2457–2465. doi: 10.1128/JB.186.8.2457-2465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan J, Rockey DD, Jones L, Andersen AA. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob Agents Chemother. 2004;48(10):3989–3995. doi: 10.1128/AAC.48.10.3989-3995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yoder AD. Estimation of the transition/transversion rate bias and species sampling. J Mol Evol. 1999;48(3):274–283. doi: 10.1007/PL00006470. [DOI] [PubMed] [Google Scholar]
- Stephens RS, Sanchez-Pescador R, Wagar EA, Inouye C, Urdea MS. Diversity of Chlamydia trachomatis major outer membrane protein genes. J Bacteriol. 1987;169(9):3879–3885. doi: 10.1128/jb.169.9.3879-3885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes A, Nogueira PJ, Borrego MJ, Gomes JP. Chlamydia trachomatis diversity viewed as a tissue-specific coevolutionary arms race. Genome Biol. 2008;9(10):R153. doi: 10.1186/gb-2008-9-10-r153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo G, Romero D. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol Rev. 2005;29(2):169–183. doi: 10.1016/j.femsre.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Somboonna N, Wan R, Ojcius DM, Pettengill MA, Joseph SJ, Chang A, Hsu R, Read TD, Dean D. Hypervirulent Chlamydia trachomatis clinical strain is a recombinant between lymphogranuloma venereum (L(2)) and D lineages. MBio. 2011;2(3):e00045–e00011. doi: 10.1128/mBio.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Makarova KS, Wolf YI, Koonin EV. Gene conversions in genes encoding outer-membrane proteins in H. pylori and C. pneumoniae. Trends Genet. 2001;17(1):7–10. doi: 10.1016/S0168-9525(00)02151-X. [DOI] [PubMed] [Google Scholar]
- Futse JE, Brayton KA, Knowles DP Jr, Palmer GH. Structural basis for segmental gene conversion in generation of Anaplasma marginale outer membrane protein variants. Mol Microbiol. 2005;57(1):212–221. doi: 10.1111/j.1365-2958.2005.04670.x. [DOI] [PubMed] [Google Scholar]
- Nystedt B, Frank AC, Thollesson M, Andersson SG. Diversifying selection and concerted evolution of a type IV secretion system in Bartonella. Mol Biol Evol. 2008;25(2):287–300. doi: 10.1093/molbev/msm252. [DOI] [PubMed] [Google Scholar]
- Rich SM, Sawyer SA, Barbour AG. Antigen polymorphism in Borrelia hermsii, a clonal pathogenic bacterium. Proc Natl Acad Sci USA. 2001;98(26):15038–15043. doi: 10.1073/pnas.071042098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66(8):3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noormohammadi AH, Markham PF, Kanci A, Whithear KG, Browning GF. A novel mechanism for control of antigenic variation in the haemagglutinin gene family of Mycoplasma synoviae. Mol Microbiol. 2000;35(4):911–923. doi: 10.1046/j.1365-2958.2000.01766.x. [DOI] [PubMed] [Google Scholar]
- Kline KA, Sechman EV, Skaar EP, Seifert HS. Recombination, repair and replication in the pathogenic Neisseriae: the 3 R's of molecular genetics of two human-specific bacterial pathogens. Mol Microbiol. 2003;50(1):3–13. doi: 10.1046/j.1365-2958.2003.03679.x. [DOI] [PubMed] [Google Scholar]
- Haas R, Meyer TF. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell. 1986;44(1):107–115. doi: 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- Liao D. Concerted evolution: molecular mechanism and biological implications. Am J Hum Genet. 1999;64(1):24–30. doi: 10.1086/302221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D. Gene conversion drives within genic sequences: concerted evolution of ribosomal RNA genes in bacteria and archaea. J Mol Evol. 2000;51(4):305–317. doi: 10.1007/s002390010093. [DOI] [PubMed] [Google Scholar]
- Santoyo G, Martinez-Salazar JM, Rodriguez C, Romero D. Gene conversion tracts associated with crossovers in Rhizobium etli. J Bacteriol. 2005;187(12):4116–4126. doi: 10.1128/JB.187.12.4116-4126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J Biol Chem. 2002;277(30):26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C. et al. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest. 2003;111(11):1757–1769. doi: 10.1172/JCI17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987;4(3):222–230. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H. Gene location and bacterial sequence divergence. Mol Biol Evol. 2002;19(8):1350–1358. doi: 10.1093/oxfordjournals.molbev.a004196. [DOI] [PubMed] [Google Scholar]
- Nunes A, Gomes JP, Mead S, Florindo C, Correia H, Borrego MJ, Dean D. Comparative expression profiling of the Chlamydia trachomatis pmp gene family for clinical and reference strains. PLoS One. 2007;2(9):e878. doi: 10.1371/journal.pone.0000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RE, Bergthorsson U, Ochman H. Transcription increases multiple spontaneous point mutations in Salmonella enterica. Nucleic Acids Res. 2003;31(15):4517–4522. doi: 10.1093/nar/gkg651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H. Origins of spontaneous mutations: specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu Rev Genet. 2002;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- Wright BE. A biochemical mechanism for nonrandom mutations and evolution. J Bacteriol. 2000;182(11):2993–3001. doi: 10.1128/JB.182.11.2993-3001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE. Stress-directed adaptive mutations and evolution. Mol Microbiol. 2004;52(3):643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21(3):195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beletskii A, Bhagwat AS. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci USA. 1996;93(24):13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hou J, Liu J. Chlamydial DNA polymerase I can bypass lesions in vitro. Biochem Biophys Res Commun. 2006;345(3):1083–1091. doi: 10.1016/j.bbrc.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Davis LG, Kuehl WM, Battey JF. In: Basic Methods in Molecular Biology. 2. Davis LG, Kuehl WM, Battey JF, editor. Norwalk, CT: Appleton and Lange; 1994. pp. 16–21. [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K. et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28(6):1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2(Unit 2 3) doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. pp. 1451–1452. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic analyses of the C. trachomatis ompA gene flanking regions.
Figure S2. Phylogenetic analyses of the synonymous and non-synonymous sites of the ompA gene.