Abstract
Both sorbitol and sucrose are imported into apple fruit from leaves. The metabolism of sorbitol and sucrose fuels fruit growth and development, and accumulation of sugars in fruit is central to the edible quality of apple. However, our understanding of the mechanisms controlling sugar metabolism and accumulation in apple remains quite limited. We identified members of various gene families encoding key enzymes or transporters involved in sugar metabolism and accumulation in apple fruit using homology searches and comparison of their expression patterns in different tissues, and analyzed the relationship of their transcripts with enzyme activities and sugar accumulation during fruit development. At the early stage of fruit development, the transcript levels of sorbitol dehydrogenase, cell wall invertase, neutral invertase, sucrose synthase, fructokinase and hexokinase are high, and the resulting high enzyme activities are responsible for the rapid utilization of the imported sorbitol and sucrose for fruit growth, with low levels of sugar accumulation. As the fruit continues to grow due to cell expansion, the transcript levels and activities of these enzymes are down-regulated, with concomitant accumulation of fructose and elevated transcript levels of tonoplast monosaccharide transporters (TMTs), MdTMT1 and MdTMT2; the excess carbon is converted into starch. At the late stage of fruit development, sucrose accumulation is enhanced, consistent with the elevated expression of sucrose-phosphate synthase (SPS), MdSPS5 and MdSPS6, and an increase in its total activity. Our data indicate that sugar metabolism and accumulation in apple fruit is developmentally regulated. This represents a comprehensive analysis of the genes involved in sugar metabolism and accumulation in apple, which will serve as a platform for further studies on the functions of these genes and subsequent manipulation of sugar metabolism and fruit quality traits related to carbohydrates.
Introduction
Carbohydrates provide energy and building blocks for plant growth and development. In addition, soluble sugars, including sucrose (Suc), glucose (Glc) [1], [2] and fructose (Fru) [3], [4], are known to act as signal molecules to regulate the expression of many key genes involved in plant metabolic processes and defense responses, consequently regulating plant growth and development [1], [5]. Carbohydrates are also central to quality and yield of crops. In fleshy fruits, the accumulation of soluble sugars during fruit development largely determines their sweetness at harvest.
Plants have evolved an elaborate system for sugar metabolism and accumulation in sink cells (Figure 1). Here, we name this system as Suc-Suc cycle (previously called ‘futile recycles’ [6]). In this system, once Suc is transported into sink cells (e.g. fruit, root or shoot tips), it is converted to Fru and Glc by neutral invertase (NINV, EC 3.2.1.26), or to Fru and UDP-glucose (UDPG) by sucrose synthase (SUSY, EC 2.4.1.13). The resulting Glc and Fru are then phosphorylated to glucose 6-phosphate (G6P) and fructose 6-phosphate (F6P) by hexokinase (HK, EC 2.7.1.1) and fructokinase (FK, EC 2.7.1.4). The interconversions between F6P, G6P, G1P, and UDPG are catalyzed by phosphoglucoisomerase (EC 5.3.1.9), phosphoglucomutase (EC 5.4.2.2) and UDP-glucose pyrophosphorylase (EC 2.7.7.9) in readily reversible reactions. The F6P produced in sugar metabolism enters glycolysis and the TCA cycle to generate energy and intermediates for other processes; G1P is used for starch synthesis; and both F6P and UDPG can be combined to re-synthesize Suc via sucrose phosphate synthase (SPS, EC 2.4.1.14) and sucrose-phosphate phosphatase (EC 3.1.3.24) [1]. Most of the Suc, Glc and Fru and other soluble sugars that have not been metabolized are transported into the vacuole by special transporter proteins located on the vacuole membrane. Once inside the vacuole, Suc can also be converted to Glc and Fru by vacuolar acid invertase (vAINV) [1]. This system operates in such a way that it not only allows carbon to be allocated into different pathways to satisfy sink growth and development, but also coordinates sugar metabolism and accumulation and maintains the balance in osmotic potential and turgor between cytosol and other subcellular compartments.
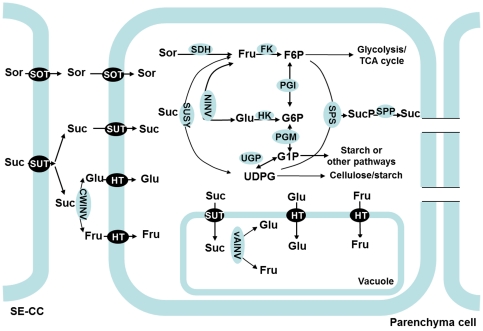
Figure 1. Sugar metabolism and accumulation in apple fruit [1], [14].
Both sorbitol (Sor) and sucrose (Suc) are unloaded to the cell wall space between sieve element-companion cell complex (SE-CC) and parenchyma cells in fruit [14]. Sor is taken up into parenchyma cells via sorbitol transporter (SOT). Suc is directly transported into parenchyma cells by plasma membrane-bound sucrose transporter (SUT), or converted to fructose (Fru) and glucose (Glc) in the cell wall space by cell wall invertase (CWINV), and then transported into the parenchyma cells by hexose transporter (HT). In the cytosol, Sor is converted to Fru by sorbitol dehydrogenase (SDH), while Suc can be converted to Fru and Glc by neutral invertase (NINV) or to Fru and UDP-glucose by sucrose synthase (SUSY). The resulting Glc and Fru can be phosphorylated to glucose 6-phsophate (G6P) and fructose 6-phosphate (F6P) by hexokinase (HK) and fructokinase (FK, specific for Fru). The conversions between F6P, G6P, G1P, and UDPG are catalyzed by phosphoglucoisomerase (PGI), phosphoglucomutase (PGM), and UDPG-pyrophosphorylase (UGP) in readily reversible reactions. The F6P produced in sugar metabolism enters glycolysis/TCA cycle to generate energy and intermediates for other processes. G1P is used for starch synthesis. UDPG can be used for cellulose synthesis or combined with F6P for re-synthesis of Suc via sucrose phosphate synthase (SPS) and sucrose-phosphatase (SPP). Most of the Fru, Glc and Suc that have not been metabolized are transported by special tonoplast transporters into vacuole for storage. Inside the vacuole, Suc can be also converted to Glc and Fru by vacuolar acid invertase (vAINV).
The concentration and distribution of sugars in plant cells are modulated by this Suc-Suc cycle system, which are affected by both internal and external factors, such as developmental processes [7]–[9], tissue types [10] and environmental conditions [5]. For example, rapid Suc accumulation in ripening fruit of melon (Cucumis melo) is attributed to higher SPS activity and lower invertase activity compared with young fruit, whereas activities of HK and FK are decreased [8]. In grape (Vitis vinifera) berries, the expression of two hexose transporter (HT) family genes, tonoplast monosaccharide transporter (TMT) and vacuolar glucose transporter (vGT) corresponds to massive accumulation of Glc and Flu in the vacuole [11]. The expression levels of hexokinases, OsHK5 and OsHK6, which function as Glc sensors, are up-regulated in rice (Oryza sativa) leaves by exogenous application of Glc and Fru [12]. However, our current understanding of the mechanisms controlling the homeostasis and accumulation of sugars in fleshy fruit still remains quite limited.
Apple (Malus domestica Borkh), a member of the Rosaceae family, is among the most important commercial fruit crops grown worldwide. Apple and other Rosaceae tree fruits synthesize sorbitol (Sor), in addition to Suc, in source leaves, and both Sor and Suc are translocated to and utilized in fruit, with sorbitol accounting for about 60–70% of the photosynthates produced in leaves and transported in the phloem [13]. After being unloaded from SE-CC complexes into the cell wall space in apple fruit [14], Sor is taken up into the cytosol of parenchyma cells by sorbitol transporter (SOT), and then converted to Fru by sorbitol dehydrogenase (SDH, EC 1.1.1.14) [15] (Figure 1); Suc is directly transported into parenchyma cells by sucrose transporters (SUT or SUC) located on the plasma membrane, or converted to Glc and Fru by cell wall invertase (CWINV) first, and then transported into parenchyma cells by hexose transporters [14], [16]. Compared with sink organs in model plants that import and metabolize sucrose alone (e.g. Arabidopsis, Solanum tuberosum, and Populus), apple is unique in the metabolism and accumulation of sugars: more than 80% of the total carbon flux goes through Fru(because almost all the sorbitol is converted to Fru and half of the sucrose is converted to Fru), and Fru accumulates to a much higher level than Glc in the fruit. Although there have been some reports on the accumulation of carbohydrates [9], [13], [17] and changes in the activities of related enzymes during apple fruit development [13], [14], [17], it remains unclear how sugar metabolism and accumulation is regulated at the gene expression level.
In this article, we identified members of the various gene families that encode key enzymes or transporters involved in sugar metabolism and accumulation using homology analysis based on Malus genome [18] and EST sequences and comparison of expression patterns in different tissues, and analyzed the relationship of their relative transcript abundance and activities of enzymes with sugar accumulation during apple fruit development.
Materials and Methods
Plant materials
Five-year-old ‘Greensleeves’ apple (M. domestica Borkh.) trees on M. 26 rootstocks were used in this study. All the trees were grown in 55-L plastic containers in a medium of 1 sand: 2 MetroMix 360 (v/v) (Scotts, Marysville, OH, USA) outdoors under natural conditions in Ithaca, NY, USA. They were trained as a spindle system and grown at a density of 1.5×3.5 m. The cropload of these trees was adjusted by hand thinning to 4 fruit per cm2 trunk cross-sectional area at 10 mm king fruit size. They were supplied with 15 mM N using Plantex® NPK (20–10–20) with micronutrients (Plantex Corp., Ontario, Canada) twice weekly during the growing season. Fungicides and pesticides were sprayed at regular intervals throughout the growing season to protect the plants from diseases and insects. At 40 (near the end of cell division), 74 (early stage of cell expansion), 108 (late stage of cell expansion), and 134 days (maturity) after bloom (DAB) [7], fruit samples were taken from the south side of the tree canopy between noon and 2:00PM under full sun exposure. On each sampling date, five replicates of fruit samples, with at least six fruit in each replicate from three trees, were harvested. The sampled fruit were immediately weighed, cut into small pieces after removing the core, and frozen in liquid nitrogen on site (It took about 2 min from harvest to frozen). To compare the expression patterns of related genes in source and sink tissues, we also obtained mature leaves, shoot tips at 40 DAB from the trees. All the frozen samples were stored at −80°C until use.
Identification of candidate genes
Candidate genes were identified by performing Blastp analysis against apple gene set (nucleic acid), in the Malus Genome Database from ‘Fondazione Edmund Mach Istituto Agrario San Michele All'Adige’, Italy (http://genomics.research.iasma.it/blast/blast.html) [18] using A. thaliana invertase, SUSY, HK, SPS, SUT, TMT and vGT sequences (obtained from The Arabidopsis Information Resource (http://arabidopsis.org/) as query (except for FK using Lycopersicon esculentum [19] as query), and an E-value of 1,00E-04 as threshold. The putative candidate gene sequences were retrieved from the Malus Genome Database: http://genomics.research.iasma.it/gb2/gbrowse/apple/. The corresponding sequences of candidate genes were then used for a BLAST search against the Malus EST database in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) to confirm that each predicted gene is expressed in Malus transcriptome while there is a high similarity EST sequence (score >300 bp, and identity >98%). Then, all ESTs sharing high similarity (>98%) with predicted genes were subjected to contig assembly (score >300 bp, and identity >98%; http://mobyle.pasteur.fr/cgi-bin/MobylePortal/). After similarity analysis between the predicted gene and its EST-constructed contig or EST, the divergent gene in splicing again underwent a Blastp analysis against all predictions in apple (nucleic acid) (http://genomics.research.iasma.it/blast/blast.html) using EST-constructed contig or EST sequence so that a concordant sequence with EST would be found in all predictions. Forty-one putative candidate genes involved in sugar metabolism in apple, including 3 CWINVs, 3 NINVs, 3 vAINVs (Table S1), 5 SUSYs (Table S2), 4 FKs (Table S3), 6 HKs (Table S4), 6 SPSs (Table S5), 5 SUTs, 5 TMTs and 2 vGTs (Table S6), were screened for expression analysis. Additionally, representative MdSOTs (MdSOT1, Genbank accession, AY237401, low Km; MdSOT2, AY237400, high Km [20]) and MdSDHs were also used for expression analysis. Although 17 predicted SDH homology genes were found in Malus genome [18], only MdSDH1 to MdSDH9 (SDH1, AY244806; SDH2, AY244807; SDH3, AY244809; SDH4, AY053504; SDH5, AY244811; SDH7, AY244813, SDH8; AY244812; SDH9, AY244810) had been systematically investigated as NAD-dependent sorbitol dehydrogenase [21], [22]. Since MdSDH2 shares high similarity of cDNA sequence with MdSDH3 to MdSDH9 [18], a pair of universal primers was designed for MdSDH2-SDH9 based on their conserved cDNA region.
Sequence similarities and phylogeny analyses
Sequence similarities were determined by performing Clustal V multiple alignments using Lasergene software (DNASTAR, USA). Phylogenetic analysis of Malus and A. thaliana or L. esculentum (Only for FK) amino acid sequences was performed using maximum likelihood (http://www.phylogeny.fr) [23]. For this, amino acid alignments were performed using the MUSCLE program [24], and maximum likelihood trees with 100 bootstrap replicates were constructed with the PHYML program [25] and the JTT amino acid substitution model. Phylogenic tree was visualized using Treedyn program [26]. Additionally, the subcellular localizations of candidate genes were predicted using the TargetP software (http://www.cbs.dtu.dk/services/TargetP; [27]) and WoLF PSORT version of PSORT II (http://wolfpsort.org/; [28]).
mRNA expression analysis
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to analyze expression of the genes involved in sugar metabolism and accumulation (Tables S1, S2, S3, S4, S5, 6). Total RNA was extracted from samples by the modified CTAB method [29], and DNase was used to clean out DNA before reverse-transcription. After analysis of sequence similarities, gene-specific primers (Table S7) were designed, using Primer5 software. Primer specificity was determined by RT-PCR and Melt Curve analysis. qRT-PCR was performed with a iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's protocol. The amplified PCR products were quantified by an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA), with iQ SYBR Green Supermix kit (Bio-Rad). Actin (CN938023) transcripts were used to standardize the different gene cDNA samples throughout the test. For all samples, five tubes of total RNA were extracted from five replicates, respectively, and then mixed in a tube used for reverse-transcription. qRT-PCR experiments were done with 3 technical replicates. The data were analyzed using the ddCT method in iQ5 2.0 standard optical system analysis software.
Assay of enzyme activities
SDH was extracted according to Park et al. [15] with some modifications. Each sample (0.50 g) was homogenized in 2 ml of 100 mM potassium phosphate (pH 7.8) buffer, containing 1 mM EDTA, 1 mM dithiothreitol (DTT), 1% BSA, 0.2% Triton X-100 and 1% (w/v) insoluble polyvinylpolypyrrolidone (PVPP). The homogenate was centrifuged at 16 000 g for 10 min at 2°C and 1 ml of the supernatant was desalted with a Sephadex G25 PD-10 column (Amersham BioSciences, Piscatway, NJ, USA) equilibrated with 125 mM Tris-HCl (pH 9.6). SDH activity was assayed in 1.0 ml reaction mixture containing 300 mM Sor, 1 mM NAD+, and 0.2 ml of the desalted extract in 100 mM Tris-HCl (pH 9.6), and NADH production was determined at 340 nm.
To extract CWINV, NINV, vAINV, SUSY, FK, HK, and SPS, 0.5 g sample was homogenized in 2 ml of 200 mM Hepes-KOH (pH 8.0) containing 5 mM MgCl2, 2 mM EDTA, 2.5 mM DTT, 2 mM Benzamidina, 0.1 mM leupeptin, 0,1% BSA, 2% glycerol, 1% Triton X-100) with 4% PVPP, similar to that used by Moscatello et al. [30]. The extract was centrifuged at 16 000 g for 20 min at 4°C, and immediately desalted in a Sephadex G25 PD-10 column, equilibrated with the extraction buffer at the concentration of 50 mM of Hepes-(KOH) (pH 7.4) but without Triton X-100 or DTT. For CWINV, the pellet was washed three times with the desalting buffer, and the protein on cell wall was solubilized by incubation in the extraction buffer with 1 M NaCl added at 4°C overnight. Then the extract was centrifuged and desalted as above. The desalted extract was used to assay soluble protein content and CWINV activity, while an aliquot of the desalted extract was boiled for 5 min to denature the enzyme as blank for each sample.
CWINV and vAINV were assayed for 60 min at 37°C, in an 200 µl assay mixture containing 100 mM phosphate-citrate buffer (pH 4.8), 100 mM sucrose, and 50 µl of the desalted extract or denatured extract (as blank). The assays were stopped by boiling for 3 min before adding 0.75 M Tris-HCl buffer (pH 8.5). The assay conditions for the NINV were the same except that the assay mixture contained Hepes-KOH (pH 7.2) as buffer. The amount of glucose produced from sucrose was determined by the enzyme-coupling method [10].
SUSY activity was determined according to Dancer et al. [31]. The enzyme extract (20 µl) was incubated at 27°C for 30 min in 100 µl final volume of assay medium containing 20 mM Hepes-KOH (pH 7.0), 100 mM sucrose and 4 mM UDP. The reaction was stopped by boiling in water for 3 min. Blanks contained the same assay mixture, but denatured extract was used. The UDPG content produced in the assay was measured spectrophotometrically following the reduction of NAD+ coupled to UDPG dehydrogenase activity in a reaction mixture (1.0 ml) containing 5 mM MgCl2, 2 mM NAD+ and 0.02 U UDPG dehydrogenase, and 100 µl of the reaction mixture for SUSY in 200 mM glycine (pH 8.9). The mixture was incubated at 27°C for 30 min. and NADH production was determined at 340 nm.
HK and FK activities were assayed by a continuous spectrophotometric assay as used by Renz and Stitt [32] with minor modifications. For HK, the assay mixture (0.5 mL) contained 50 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 2.5 mM ATP, 0.33 mM NAD+, 1 U of G6P dehydrogenase, 1 mM glucose and 25 µl of the desalted extract. For FK, one unit of phosphoglucoisomerase was also added and 0.4 mM Fru was used instead of Glc.
SPS was measured in a two-step assay, following the procedure of Stitt et al. [33]. The reaction mixture (200 µl total volumes) contained 50 mM Hepes-KOH (pH 7.4), 4 mM MgCl2, 1 mM EDTA, 4 mM F6P, 20 mM G6P, 3 mM UDPG and 100 µl of sample. The reaction was carried out at 27°C for 30 min and stopped by boiling in water for 3 min. Blanks were run, for each assay, by adding denatured extracts. After centrifugation for 1 min at 12 000 g, 75 µl of the reaction mixture was used for UDP measurement in a spectrophotometric assay in a final volume of 1.0 ml containing 50 mM Hepes-KOH (pH 7.0), 5 mM MgCl2, 0.3 mM NADH, 0.8 mM phosphoenolpyruvate, 14 U of lactate dehydrogenase, and 4 U of pyruvate kinase (to start the reaction).
Measurements of soluble sugars and starch
Soluble sugars and hexose phosphates were extracted and derivatized according to Wang et al. [34]. Briefly, 0.1 g sample was extracted in 1.4 ml 75% methanol with ribitol added as internal standard. After fractionation of non-polar metabolites into chloroform, 2 and 100 µl of the polar phase of each sample were taken and transferred into 2.0 ml Eppendorf vials for highly abundant metabolites (such as Sor, Suc, Glc, and Fru) and less abundant metabolites (such as G6P and F6P), respectively. They were dried under vacuum without heating and then derivatized with methoxyamine hydrochloride and N-methyl-N-trimethylsilyl-trifluoroacetamide sequentially [35]. After derivatization, metabolites were analyzed with an Agilent 7890A GC/5975C MS (Agilent Technology, Palo Alto, CA, USA) [34]. Metabolites were identified by comparing fragmentation patterns with those in a mass spectral library generated on our GC/MS system and an annotated quadrupole GC–MS spectral library downloaded from the Golm Metabolome Database (http://csbdb.mpimp-golm. mpg.de/csbdb/gmd/msri/gmd_msri.html) and quantified based on standard curves generated for each metabolite and internal standard.
The tissue residue after 75% methanol extraction for GC-MS analysis was re-extracted with 80% (v/v) ethanol at 80°C three times, and the pellet was retained for determination of starch. After digesting the residue with 30 U of amyloglucosidase (EC 3.2.1.3) at pH 4.5 overnight, starch was determined enzymatically as glucose equivalents [36].
Results
Candidate genes encoding key enzymes involved in sugar metabolism
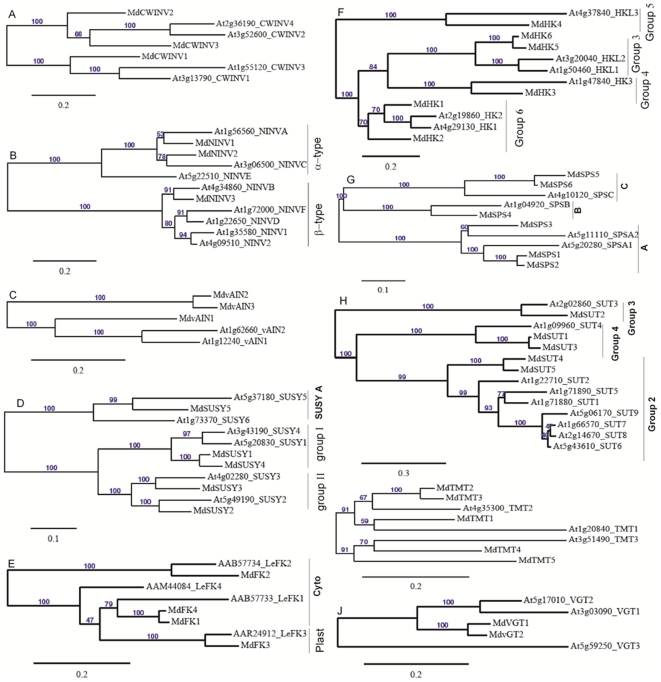
Blastp searches of the Malus Genome Database, using A. thaliana invertase, SUSY, HK, SPS, SUT, TMT and vGT sequences as query (except for FK using L. esculentum), allowed the identification of candidate genes in Malus, and these genes are expressed in Malus transcriptome based on their ESTs in Genbank. Nine genes encoding invertase were identified, including 3 CWINVs, 3 NINVs and 3 vAINVs. MdCWIN1 had high similarity with AtCWINV1 (At3g13790) and shared the same clade with AtCWINV1 and AtCWINV3 (At1g55120). Both MdCWINV2 and MdCWINV3 had high similarity and were in the same clade with AtCWIN2 (At3g52600) and AtCWINV4 (At2g36190) (Figure 2A). Both MdNINV1 and MdNINV2 shared high similarity of amino acid sequence, belonging to Arabidopsis ‘α-group’ with predicted mitochondrial or plastidic localization according to Nonis et al. [37], while MdNINV3 was in ‘β-group’ with predicted cytosolic localization (Figure 2B). MdvAINV1 was in the same clade with AtvAINV1 (At1g12240) and AtvAINV2 (At1g62660), whereas both MdvAINV2 and MdvAINV3 had high similarity and were in another clade (Figure 2C).
Figure 2. Maximum likelihood phylogeny of Malus genes encoding enzymes and transporters involved in sugar metabolism and accumulation with those from Arabidopsis or Lycopersicon esculentum.
The tree was produced using MUSCLE and PhyML with the JTT amino acid substitution model, a discrete gamma model with 4 categories and an estimated shape parameter of 1.0. Bootstrapping was performed with 100 replicates. A, cell wall invertase (CWINV); B, neutral invertases (NINV), α and β type NINV according to Nonis et al. [37]; C, vacuolar acid invertase (vAINV); D; sucrose synthase (SUSY), different types according to Bieniawska et al. [38]; E, fructokinase (FK), cytosolic and plastid fructokinases in tomato according to Granot [19]; F, hexokinase (HK), different groups according to Karve et al. [39]; G, Sucrose phosphate synthases (SPS), Arabidopsis types according to Lutfiyya et al. [40]; H, Sucrose transporter (SUT), different groups according to Braun & Slewinski [41]; I, tonoplast monosaccharide transporter (TMT); J, vacuolar glucose transporter (vGT).
Five SUSYs were identified in Malus and their predicted localization had possibilities in cytosol, mitochondria or plastids (data not shown). Both MdSUSY1 and MdSUSY4 shared high homology and were in the same group with AtSUSY1 (A5g20830) and AtSUSY4 (At3g43190) according to Bieniawska et al. [38]. MdSUSY2 and MdSUSY3 were in the same phylogeny group with AtSUSY2 (At5g49190) and AtSUSY3 (At4g02280), respectively. However, MdSUSY5 belonged to another Arabidopsis SUSY group: SUSY A [38] (Figure 2D).
Four orthologs of FK were found in Malus genome. MdFK1 had high similarity with cytosol-localized LeFK1 (AAB57733) and shared high similarity with MdFK4; MdFK2 belonged to another clade with cytosol-localized LeFK2 (AAB57734); MdFK3 encoded a predicted plastid-localized FK (data not shown) and had the highest similarity with LeFK3 (AAR24912) (Figure 2E).
Of the five ortholog of HK identified in Malus, both MdHK1 and MdHK2 had high homology with AtHK1 (At4g29130) and AtHK2 (At2g19860), respectively, and both belonged to HK ‘group 6’ according to Karve et al. [39]; MdHK3 was an ortholog gene of AtHK3 (At1g47840) and in the ‘group 4’ of Arabidopsis HK [39]; MdHK4 was in the same clade with AtHKL3 (At4g37840), which belonged to the ‘group 5’; MdHK5 and MdHK6 had high homology and were in the same clade with AtHKL1 (At1g50460) and AtHKL2 (At3g20040), both of which were in the ‘group 3’ [39] (Figure 2F).
Six MdSPSs identified in Malus covered 3 Arabidopsis SPS groups according to Lutfiyya et al. [40]. MdSPS1 to MdSPS3 belonged to the same group as Arabidopsis AtSPSA1 (At5g20280) and AtSPSA2 (At5g11110), and both MdSPS1 and MdSPS2 shared high similarity and had high homology with AtSPSA1, while MdSPS3 was homologous to AtSPSA2. MdSPS4 was an ortholog gene of AtSPSB (At1g04920) in another clade. Both MdSPS5 and MdSPS6 shared high similarity and were in the same group with AtSPSC (At4g10120) (Figure 2G).
Candidate genes encoding sugar transporters
Five ortholog genes of SUT were identified in the Malus genome. MdSUT1, the same as MdSUT1 (AY445915) reported by Fan et al. [16], shared high similarity with MdSUT3 and had high homology with AtSUT4, which belonged to SUC ‘group 4’ according to Braun & Slewinski [41]; MdSUT2 was in an independent clade with AtSUT3 (At2g02860) in the ‘group 3’; Both MdSUT4 and MdSUT5 showed high similarity and had high homology with AtSUT2 (At1g22710), which belonged to the ‘group 2’ [41] (Figure 2H).
Of the 5 ortholog genes of TMT identified in Malus, MdTMT1, MdTMT2 and MdTMT3 shared high similarity of amino acid sequence, and MdTMT1 had high homology with AtTMT1 (At1g20840) whereas both MdTMT2 and MdTMT3 had high homology with AtTMT2 (At4g35300). MdTMT4 showed high homology with AtTMT3 (At3g51490) and were in the same clade with MdTMT5 (Figure 2I).
Both MdvGT1 and MdvGT2 had high homology with AtvGT1 (At3g03090) and AtvGT2 (At5g17010), respectively, and were in the same clade, whereas they had low similarity with AtvGT3 (At5g59250) (Figure 2J).
Expression of genes in source and sink tissues
To determine tissue-specific expression levels of the candidate genes, qRT-PCR was used to analyze their mRNA relative expression abundance among mature leaves, shoot tips, young fruit (40 DAB) and mature fruit (135 DAB) (Table 1). MdSDH1 and MdSDH2-9 were expressed mainly in fruit, with the expression level of MdSDH2-9 being 80 times higher in young fruit than in mature leaves. MdSDH1 expression was higher in young fruit than in mature fruit, whereas the opposite was true for MdSDH2-9. MdCWINV1 expression was much lower in fruit than in mature leaves and shoot tips. Both MdCWINV2 and MdCWINV3 transcript levels were much lower in mature fruit than in shoot tips, with similar expression level of MdCWINV3 detected in shoot tips and young fruit. The expression levels of MdNINV1 and MdNINV2 were comparable among 4 different tissues, whereas MdNINV3 had much higher expression in fruit than in other tissue types. All 3 MdvAINVs (especially MdvAINV3) had lower transcript levels in mature fruit, with the expression level of MdvAINV1 being the highest in young fruit and the lowest in shoot tips. The expression levels of both MdSUSY1 and MdSUSY4 were lower in both young and mature fruit than in mature leaves, whereas those of both MdSUSY2 and MdSUSY3 were higher in young fruit than in mature leaves. MdSUSY5 expression was higher in young fruit than in mature leaves, with the highest expression detected in shoot tips (5 times more than in mature leaves). The expression levels of MdFKs were either similar (MdFK1, MdFK3 and MdFK4) or significantly higher (MdFK2) in shoot tips than in mature leaves; young fruit had comparable transcript abundance of MdFK2 and MdFK4, but lower expression levels of MdFK1 and MdFK3 than mature leaves; mature fruit had lower expression levels of MdFK2-4, but higher transcript abundance of MdFK1. The expression levels of MdHK1 were comparable among 4 tissue types, but MdHK2 and MdHK3 had much higher transcript abundance in young fruit than in mature leaves, especially MdHK3 (27.5 times higher). MdHK4 expression was highest in shoot tips, but lowest in mature fruit. In contrast, both MdHK5 and MdHK6 had the highest expression in mature fruit. The transcript levels of all 6 MdSPSs were much higher in mature fruit than in both mature leaves and shoot tips. The expression levels of MdSPS2-6 were higher in mature fruit than in young fruit, but the opposite was true for MdSPS1. In addition, both MdSPS5 and MdSPS6 showed the lowest expression levels in shoot tips (Table 1).
Table 1. Comparison of relative mRNA expression for genes encoding enzymes involved in sugar metabolism (including MdSDHs, MdCWINVs, MdNINVs, MdvAINVs, MdSUSYs, MdFKs, MdHKs, and MdSPSs) among mature leaves, shoot tips, young fruit (40 DAB) and mature fruit (135 DAB) of apple.
| MdSDH1 | MdSDH2-9 | MdCWINV1 | MdCWINV2 | MdCWINV3 | MdNINV1 | MdNINV2 | |
| Mature leaves | 1.00±0.33 | 1.00±0.25 | 1.00±0.18 | 1.00±0.26 | 1.00±0.23 | 1.00±0.19 | 1.00±0.18 |
| Shoot tips | 0.50±0.08 | 6.01±0.75 | 0.85±0.22 | 3.31±0.37 | 3.34±0.21 | 0.40±0.07 | 1.64±0.34 |
| Young fruit | 2.10±0.44 | 81.7±7.08 | n/d | 0.69±0.08 | 3.22±0.09 | 1.04±0.06 | 1.53±0.11 |
| Mature fruit | 17.6±2.01 | 49.2±6.83 | 0.03±0.01 | 0.06±0.01 | 0.02±0.01 | 0.60±0.13 | 0.73±0.12 |
Values are means of three technical replicates of the reverse transcribed RNA sample pooled from 5 biological replicates ± SD. n/d means no expression was detected.
qRT-PCR was performed with gene-specific primers, except that a pair of universal primers was designed from the conserved cDNA region of MdSDH2 to MdSDH9 for the expression of MdSDH2-9. For each sample, transcript levels were normalized with those of Actin, and the relative expression levels of each gene were obtained using the ddCT method while expression in mature leaves was designated as ‘1’.
Both MdSOT1 and MdSOT2 showed higher transcript levels in sink organs than in mature leaves, except that mature fruit had similar level of MdSOT2 as mature leaves (Table 2). Fruit had higher transcript levels of MdSUT1-4, but lower levels of MdSUT5 than mature leaves and shoot tips. The expression levels of MdTMT1 and MdTMT2 were higher in fruit than in shoot tips and mature leaves, with the highest expression detected in mature fruit; the transcript levels of MdTMT3, MdTMT4 and MdTMT5 were highest in young fruit, but shoot tips also had higher expression levels of MdTMT4 and MdTMT5 than mature leaves. The expression levels of both MdvGT1 and MdvGT2 were higher in fruit than in shoot tips and mature leaves, with the highest expression detected in young fruit (Table 2).
Table 2. Comparison of relative mRNA expression for genes encoding sorbitol transporter (SOT), sucrose transporter (SUT), tonoplast monosaccharide transporter (TMT), and vacuolar glucose transporter (vGT) among mature leaves, shoot tips, young fruit (40 DAB) and mature fruit (135 DAB) of apple.
| MdSOT1 | MdSOT2 | MdSUT1 | MdSUT2 | MdSUT3 | MdSUT4 | MdSUT5 | |
| Mature leaves | 1.00±0.24 | 1.00±0.13 | 1.00±0.15 | 1.00±0.08 | 1.00±0.11 | 1.00±0.18 | 1.00±0.09 |
| Shoot tips | 1.64±0.18 | 2.54±0.33 | 0.78±0.22 | 0.61±0.15 | 1.28±0.13 | 1.54±0.20 | 1.47±0.10 |
| Young fruit | 2.44±0.36 | 3.28±0.31 | 10.3±1.27 | 12.2±1.06 | 1.98±0.12 | 2.54±0.15 | 0.19±0.02 |
| Mature fruit | 3.07±0.53 | 1.02±0.04 | 4.50±1.46 | 2.95±0.13 | 2.58±0.43 | 1.90±0.32 | 0.48±0.04 |
Values are means of three technical replicates of the reverse transcribed RNA sample pooled from 5 biological replicates ± SD.
qRT-PCR was performed with gene-specific primers. For each sample, transcript levels were normalized with those of Actin, and the relative expression levels of each gene were obtained using the ddCT method while expression in mature leaves was designated as ‘1.’
Expression of genes during fruit development
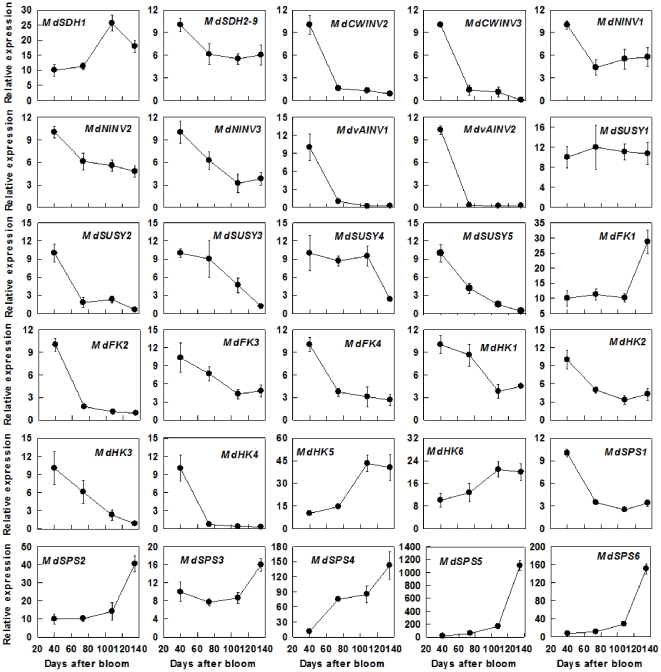
Transcript abundance of MdSDH1 increased with fruit development from 40 DAB to 108 DAB and then decreased slightly towards maturity, whereas the expression level of MdSDH2-9 showed a clear drop from 40 DAB to 74 DAB, and then remained unchanged to maturity (Figure 3). The expression levels of MdCWINV1 and MdvAINV3 were detected only in ripening fruit and 40-DAB-fruit, respectively (Data not shown). All of the transcript levels of MdCWINV2, MdCWINV3, MdNINV1, MdNINV2, MdvAINV1 and MdvAINV2 showed a dramatic decline from 40 DAB to 74 DAB, and then remained at low levels to maturity, whereas the transcript level of MdNINV3 showed a continuous decrease from 40 DAB to 108 DAB (Figure 3). During fruit development, the expression level decreased from 40 DAB to 74 DAB for MdSUSY2, from 74DAB to maturity for MdSUSY3, from 108 DAB to maturity for MdSUSY4, and throughout the fruit development for MdSUSY5, whereas no change in expression level was observed for MdSUSY1. Relative mRNA expression levels of MdFK1 remained unchanged until an obvious rise from 108 DAB to maturity. By contrast, MdFK2 expression had a large drop from 40 DAB to 74 DAB, and then maintained at a low level to maturity. However, both MdFK3 and MdFK4 expression showed decreasing trends throughout fruit development (Figure 3). The expression levels of MdHK1-4 decreased throughout fruit development, whereas the expression levels of both MdHK5 and MdHK6 increased towards fruit maturity. Except for a decrease in the transcript level observed for MdSPS1 during fruit development, all the other 5 MdSPSs showed increases in their expression levels with fruit development, particularly MdSPS5 and MdSPS6 (Figure 3).
Figure 3. Relative mRNA expression for genes encoding enzymes involved in sugar metabolism (including MdSDHs, MdCWINVs, MdNINVs, MdvAINVs, MdSUSYs, MdFKs, MdHKs, MdSPSs) during apple fruit development.
Quantitative RT-PCR was performed with gene-specific primers, except for MdSDH2-9 where a pair of universal primer was designed from the conserved cDNA region of MdSDH2 to MdSDH9. For each sample, transcript levels were normalized with those of Actin, and the relative expression levels of each gene were obtained using the ddCT method while expression in 40-DAB-fruit was designated as ‘10’. Values are means of three replicates of the reverse transcribed RNA sample pooled from 5 biological replicates ± SD.
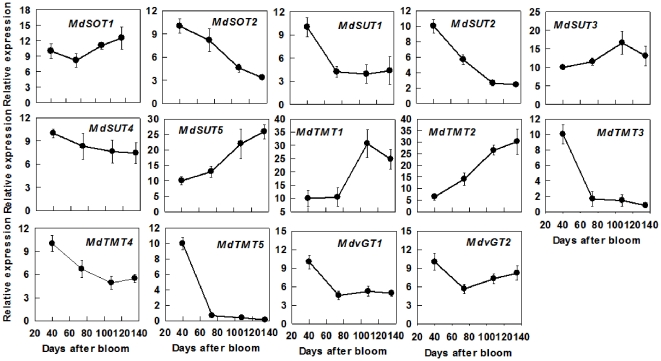
The transcript level of MdSOT1 showed a slight increase from 74 DAB to maturity, whereas that of MdSOT2 decreased throughout fruit development (Figure 4). The transcript level of MdSUT1 decreased from 40 DAB to 74 DAB and that of MdSUT2 decreased throughout fruit development; those of both MdSUT3 and MdSUT4 remained relatively stable; whereas that of MdSUT5 increased with fruit development. The transcript abundances of both MdTMT1 and MdTMT2 increased with fruit development, whereas those of MdTMT3, MdTMT4 and MdTMT5 decreased during fruit development. The expression levels of both MdvGT1 and MdvGT2 dropped from 40 DAB to 74 DAB and remained unchanged to maturity (Figure 4).
Figure 4. Relative mRNA expression for genes encoding sugar transporters (including MdSOTs, MdSUTs, MdTMTs and MdvGTs) during apple fruit development.
Quantitative RT-PCR was performed with gene-specific primers. For each sample, transcript levels were normalized with those of Actin, and the relative expression levels of each gene were obtained using the ddCT method while expression in 40-DAB-fruit was designated as ‘10’. Values are means of three technical replicates of the reverse transcribed RNA sample pooled from 5 biological replicates ± SD.
Activities of key enzymes in sugar metabolism during fruit development
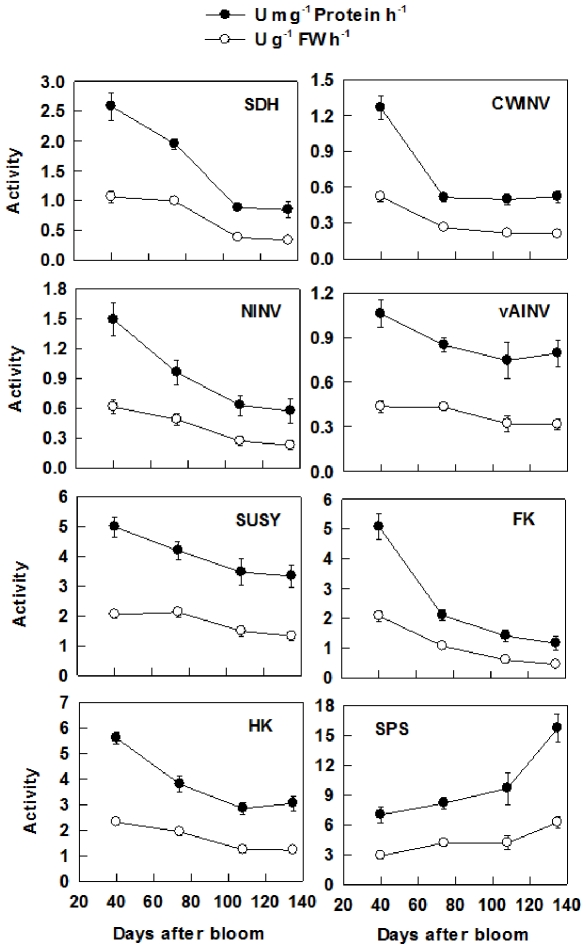
On a soluble protein basis, the activity of SDH decreased from 40 to 108 DAB, with the activity at 108 and 135 DAB being about 40% of that at 40 DAB (Figure 5), but there was no correlation between the activity of SDH and the expression level of SDH2-9 during fruit development (Table S8). The activity of CWINV decreased by 60% from 40 to 74 DAB and then remained unchanged to fruit maturity, which was correlated with the expression level of both CWINV2 and CWINV3 (Table S8). The activity of NINV decreased during fruit development (Figure 5), and was correlated with the expression level of both NINV2 and NINV3. The activity of vAINV decreased from 40 to 108 DAB and then remained unchanged to fruit maturity, which was correlated with the expression level of both vAINV1 and vAINV2. SUSY activity decreased with fruit development, and was correlated with the expression level of SUSY5. The activity of FK decreased with fruit development and was correlated with the expression level of FK2, FK3 and FK4. The activity of HK also decreased with fruit development, which was correlated with the expression level of HK2, HK3, and HK4. In contrast, the activity of SPS increased slightly from 40 to 108 DAB, and then increased by about 63% to fruit maturity, which was correlated with the expression level of SPS2, SPS5, and SPS6 (Table S8). When expressed on a fresh weight basis, the activities of all the enzymes showed similar trends as those on protein basis, but the degree of change was smaller (Figure 5).
Figure 5. Activities of key enzymes involved in sugar metabolism during apple fruit development.
SDH: Sorbitol dehydrogenase; CWINV: Cell wall invertase; NINV: Neutral invertase; vAINV: Vacuolar acid invertase; SUSY: Sucrose synthase; FK: Fructokinase; HK: Hexokinase; SPS: Sucrose-phosphate synthase. Values are means of five replicates ± SD.
Concentrations of soluble sugars and starch during fruit development
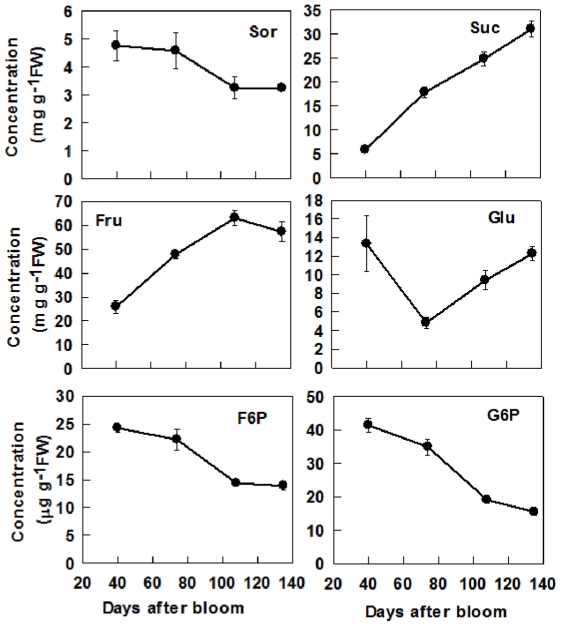
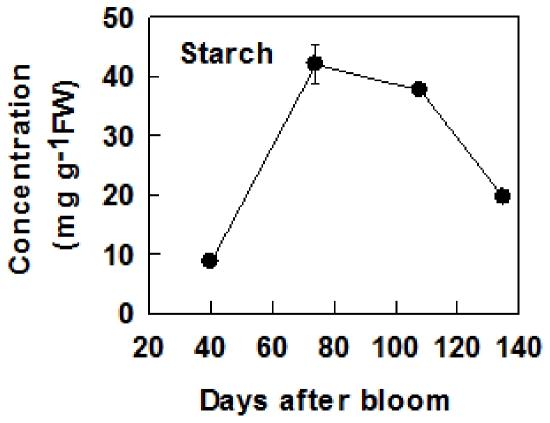
Sor concentration decreased with fruit development (Figure 6). Suc concentration increased nearly 5 times from 40 DAB to maturity (Figure 6). Fru concentration increased from 40 to 108 DAB, and dropped slightly at maturity, whereas Glc concentration decreased from 40 to 74 DAB, and then increased toward maturity. Both F6P and G6P decreased with fruit development (Figure 6). Of all the soluble sugars, Fru concentration was the highest in mature fruit, which was about 2 times that of Suc and 4 times that of Glc. Starch concentration increased rapidly from 40 to 74 DAB, was constant at the highest level from 74 to 108 DAB, and then decreased towards maturity (Figure 7).
Figure 6. Concentrations of sorbitol (Sor), sucrose (Suc), fructose (Fru), glucose (Glc), fructose 6-phosphate (F6P) and glucose 6-phosphate (G6P) during apple fruit development.
Values are means of five replicates ± SD.
Figure 7. Starch concentrations during apple fruit development.
Values are means of five replicates ± SD.
Discussion
Sugar metabolism and accumulation is developmentally regulated in apple
Our data clearly indicate that the metabolism of sugars in fruit is highly regulated by developmental processes. At the early stage of fruit development (40 DAB), high activities of SDH, NINV, SUSY, FK and HK found in this study and reported previously by Chourey and Berüter [17] match the high expression levels of their genes (Figures 3, 5). This makes rapid metabolism of the imported sugars possible to satisfy the requirement for energy and intermediates by cell division and growth at the early stage of fruit development. As a result, the concentrations of Fru, Suc and starch are low (Figure 6). As the requirement for energy and carbon skeletons decreases with fruit development, the expression levels and the activities of these enzymes decrease (Figures 3, 5), resulting in a smaller proportion of the imported sugars metabolized in the glycolysis/TCA cycle and subsequent amino acid and protein synthesis. Consequently, Fru and Suc accumulate in the vacuole [13] and starch synthesis is up-regulated to allow the storage of the extra imported sugars in the insoluble form in plastids [1]. At the late stage of fruit development, starch breaks down (Figure 6), and sucrose continues to accumulate, which elevates the total concentrations of soluble sugars, and the sweetness of the fruit to a maximum at maturity. From an evolutionary perspective, the maximum accumulation of soluble sugars at fruit maturity helps attract animals and human for seed dispersal.
Most Fru and Glc are stored in the central vacuole of parenchyma cells in apple, which occupies more than 80% of the cell's volume [13], [42]. Rapid accumulation of Fru during apple cell expansion results from the coordinated actions of three factors. First, abundant Fru is generated from sorbitol and sucrose. Although the transcripts and activities of SDH, invertase and SUSY decreased with fruit development (Figure 5), most sorbitol has been converted to Fru as indicated by its low concentration in fruit (Figure 6). Second, decreased expression and activity of FK (Figures 3, 5) indicates less Fru is metabolized with fruit development and more is available for accumulation. Finally, up-regulation of carrier proteins that are localized on the tonoplast membrane suggests active transport of Fru from cytosol into the vacuole is enhanced during fruit development. Although it remains unknown whether Fru transporters are present on the tonoplast membrane, vacuolar carrier proteins encoded by TMTs transport both Glc and Fru [43]. As reported in grape [11], 5 orthologs of TMT are present in the apple genome and all had much higher expression in fruit than in mature leaves and shoot tips (Table S6; Figure 4). The transcript levels of MdTMT1 and MdTMT2, both of which shared high amino acid similarity with AtTMT1 (Figure 2), showed similar developmental trends as Fru concentration (Figures 4, 6), suggesting both proteins may be involved in transporting Fru into the vacuole.
Glucose is primarily derived from the hydrolysis of the imported Suc via CWINV and NINV before significant starch breakdown occurs in fruit. Considering that phloem unloading of sorbitol and Suc involves an apoplastic step between SE-CC complexes and parenchyma cells in apple fruit [14], and sorbitol transporters are competitively inhibited by Glc and Fru [44], the hydrolysis of Suc by cell wall invertase is expected to operate only on a small scale to avoid excessive accumulation of sorbitol in the apoplast. Viewed this way, the majority of the unloaded Suc from SE-CC may be directly taken up into parenchyma cells by Suc transporters on the plasma membrane, and subsequently hydrolyzed via NINV. Decreases in the expression and activities of both CWINV and NINV (Figures 3, 5) indicate that the generation of Glc decreases with fruit development. At the same time, however, the decreased HK activity suggests that less Glc is metabolized as well. When starch breaks down rapidly towards maturity (Figure 7), Glc concentration increased only slightly (Figure 6). The developmental changes of Glc concentrations are consistent with the expression patterns of MdvGTs (Figure 4), and it has been confirmed that AtvGT1 is a specific Glc transporter located on the tonoplast membrane and has an important function in seed germination and flowering of Arabidopsis [45]. These results suggest that MdvGTs may play a role in transporting Glc into vacuole in apple fruit. In addition, the expression level of MdTMT3, which has high similarity with MdTMT2 (Figure 2), is consistent with high Glc concentration during early fruit development. Although AtTMT3 is hardly expressed at any stage of the plant's life cycle [43], the expression levels of MdTMT4 and MdTMT5, both of which belonged to the same clade as AtTMT3 (Figure 2), were much higher in sink tissues than in mature leaves, especially MdTMT5 in 40-DAB-fruit (Figure 4). Further work is needed to clarify their roles in sugar accumulation in apple fruit.
Besides hexoses, Suc also accumulates to a high concentration in the ripening apple fruit, although in young fruit a large proportion of Suc is in the apoplast and cytosol [42]. There are two sources of sucrose for accumulation: the imported sucrose that has not been metabolized and newly synthesized sucrose. The decreased transcript levels and activities of invertase and SUSY with fruit development (Figures 3, 5) are consistent with the accumulation pattern of Suc before rapid starch breakdown. The continued accumulation of Suc towards maturity is tightly associated with the rapid increase in the transcript levels and activities of SPS (Figures 3, 5). This suggests that newly synthesized Suc via SPS contributes significantly to the total Suc level at maturity. So, Suc accumulation in apple fruit approaching maturity is of SPS type according to the categorization of mechanisms controlling sugar accumulation in fruit by Yamaki [46], as in melon and strawberry [8], [47]. The glucose released from starch breakdown appears to contribute to Suc synthesis because Glc showed only a slight increase towards maturity (Figure 6). Although 5 orthologs of SUTs were identified in Malus genome (Figure 2), all are Suc/H+ symporters, which transport Suc into cytosol from apoplast or vacuole. So far, not a single Suc/H+ antiporter that transports Suc into vacuole has been identified in plants [41], [48]. However, it was reported recently that both TMT1 and TMT2 also transport Suc across the tonoplast membrane in Arabidopsis [49]. The expression patterns of both MdTMT1 and MdTMT2 (Figure 4) are in general agreement with that of Suc accumulation in fruit (Figure 6). Once Suc is inside the vacuole, it can be converted to Glc and Fru by vAINV. By changing the relative contribution of sugars, vAINV regulates the elongation of cotton fiber and Arabidopsis root in an osmotic dependent and independent manner, respectively [50]. However, the expression levels of 3 MdvAINVs were very low during apple fruit development except at 40 DAB, which is consistent with sustained Suc accumulation from cell expansion to fruit maturity (Figures 3, 5).
Differential expression patterns of multiple members of gene families for key enzymes involved in sugar metabolism suggest different roles in sugar metabolism
Each enzyme involved in Suc-Suc cycle is encoded by a family of genes that are differentially expressed in mature leaves, shoot tips and fruit. Because sugar metabolism and accumulation is tissue type-dependent, comparison of gene expression patterns, enzyme activities, and sugar metabolism and accumulation allows us to gain insights into the possible functions of these genes.
CWINV is typically considered as a sink-specific enzyme, and its activity is usually low in source leaves [51], [52]. However, all 3 MdCWINVs identified in apple had lower expression levels in fruit than in leaves, except for MdCWINV3 in 40 DAB-fruit (Table 1, Figure 3). Since the unloading of sucrose in shoot tips is symplastic [53], the high expression level of MdCWINV3 in both shoot tips and in young fruit suggests that this isoform of CWINV may play a role in cell division or cell growth [5], [54], and may not necessarily be related to apoplastic unloading of Suc in fruit [1]. The much higher expression level of MdCWINV2 in shoot tips than in fruit (Table 1) suggests that the role of this isoform in Suc unloading might be very small, if any.
The expression levels of MdNINV1 and MdNINV2, both of which belong to the α-group with a predicted mitochondrial and plastidic localization respectively (Figure 2), were comparable in different tissue types (Table 1). By contrast, MdNINV3, a predicted cytosolic NINV that has high homology with Arabidopsis cytosolic AtNINV genes: AtNINV1/AtNINV2 [55], had much higher transcript abundance in fruit than in leaves and shoot tips (Table 1, Figure 2), and its expression levels matched well with NINV activity during fruit development (Figure 3). These results suggest that MdNINV3 may play an important role in controlling Suc concentration in the cytosol of apple fruit.
In sinks where Suc is the only imported carbon, such as potato tubers [56], maize kernels [57] and kiwifruit fruit [30], SUSY activity is correlated with the sink strength of storage organs. In apple where Suc accounts for only about one third of the imported carbon, the expression levels of MdSUSY1 and MdSUSY4 were lower in fruit and shoot tips than in mature leaves, whereas the expression levels of other MdSUSYs in fruit were ∼2 times of those in mature leaves (except for that MdSUSY5 was much higher in shoot tips) (Table 1). Increased MdSUSY1 expression has been observed in the shoot tips of transgenic apple with decreased Sor supply but increased Suc supply from leaves, and both MdSUSY1 transcript level and SUSY activity were dramatically enhanced by Suc feeding [10]. MdSUSY1 had a relatively stable expression during fruit development, which is consistent with the finding of Janssen et al. [7], whereas MdSUSY4 expression showed a great drop towards fruit maturity. The expression pattern of neither MdSUSY1 nor MdSUSY4 is consistent with decreased SUSY activities during apple fruit development (Figures 3, 5). In contrast, the expression patterns of MdSUSY2, MdSUSY3 and MdSUSY5 showed general agreement with decreased SUSY activities during fruit development (Figures 3, 5). These results suggest that MdSUSY1 may play an important role in shoot tips whereas MdSUSY2, MdSUSY3 and MdSUSY5 may be largely responsible for the total SUSY activities in apple fruit.
Since more than 80% of the carbon flux goes through Fru in apple sink cells compared with only 50% in other plants where Suc is the only form of imported carbon, apple sinks that do not accumulate Fru significantly (both shoot tips and young fruit) are expected to have a stronger FK activity for Fru utilization than those that actively accumulate Fru (e.g. fruit during cell expansion). Consistent with this idea, the expression patterns of MdFKs were similar (MdFK1, MdFK3, and MdFK4) or significantly higher (MdFK2) in shoot tips than in mature leaves, and were higher (MdFK2-4) in young fruit than in mature fruit (Table 1, Figure 3). The higher expression of MdFK2 (ortholog of cytosol-localized LeFK2 [19]) in shoot tips than in young fruit (Table 1, Figure 2), with dramatic decline in the expression level in fruit at 74 DAB (Figure 3) suggests that it may play an important role in efficient utilization of Fru in both shoot tips and young fruit, and Fru accumulation during fruit cell expansion. The up-regulation of the expression of MdFK1 towards fruit maturity (Figure 3) appears to be consistent with phosphorylation of Fru for Suc synthesis.
Six HK genes were identified in apple in this study, which is similar to the number of HKs found in rice and Arabidopsis [12], [39]. HK has an important role in regulating carbon flow and energy status of the cell [57]. Both MdHK1 and MdHK2 are homologous to AtHK1 and AtHK2, respectively (Figure 2), and were predicted to target the secretory pathway while AtHK1 and AtHK2 are located on the outer mitochondrial membrane [39]. Transcript abundance of MdHK1 was comparable in different tissue types of apple, whereas MdHK2 had a higher expression level in fruit than in other tissues (Table 1). However, both expression levels decreased with fruit development (Figures 3, 6, 7), suggesting that both MdHK1 and MdHK2 may be more related to the modulation of glycolytic flux and energy status of the cell in young fruit [57]. MdHK3, as a predicted plastid localized protein (data not shown), is an ortholog gene of AtHK3 (a plastid enzyme, [39]), and much higher expression level of MdHK3 was observed during rapid starch accumulation (Figure 3). It remains unclear whether the protein encoded by MdHK3 plays a role in maintaining Glc concentration or controlling energy status in plastid, but it has been suggested that AtHK3 might only have a catalytic function [39]. Of the three HK genes in apple that are orthologs of the Arabidopsis AtHK1-3, MdHK4, an ortholog of AtHKL3, which is thought not to bind Glc [39], had a very low expression level in fruit after 74 DAB (Figure 3). MdHK5 and MdHK6 are in the same phylogeny clade with AtHKL1 and AtHKL2 (Figure 2), both of which were predicted to bind Glc with a relatively lower affinity than AtHK1 [39]. Karve and Moore [58] suggested that AtHKL1 functions as a negative regulator that limits plant growth under excessive Glc availability. Higher expression levels of MdHK5 and MdHK6 towards fruit maturity seem to imply that they may be related to fast utilization of the Glc released from starch breakdown.
The 6 MdSPSs identified in apple (Figure 2) cover all three main groups of plant SPSs [40]. MdSPS1-3 are in the same group as Arabidoipsis AtSPSA1 and AtSPSA2 (Figure 2). Both MdSPS1 and MdSPS2 share high homology of amino acid sequence, and are otholog genes of AtSPSA1, the main gene responsible for SPS activity in Arabidopsis leaves [59]. All MdSPS1-3 had higher expression in fruit than in mature leaves (Table 1). However, decreases in the expression of MdSPS1 with fruit development suggest that MdSPS1 might not be involved in Suc accumulation in apple fruit towards maturity. In contrast, higher expression levels of MdSPS2 and MdSPS3 towards fruit maturity are consistent with elevated SPS activity and fast accumulation of Suc during this period (Figures 3, 6). It has also been reported that the transcript level of CmSPS1, an ortholog gene of AtSPSA1 and MdSPS2 in melon, shows the same trend as SPS enzyme activity and Suc accumulation in melon fruit [8]. Although a mutant of AtSPSA2, an ortholog gene of MdSPS3, has no significant effect on SPS enzyme activity in Arabidopsis [59], the expression of MdSPS3 was also higher towards fruit maturity (Figure 3), which is similar to the result of a microarray analysis that EST sequence (EB123469) of MdSPS3 also showed up-regulation in mature apple fruit [7]. The transcript level of MdSPS4, an ortholog gene of AtSPSB whose mutant has no significant effect on SPS enzyme activity in Arabidopsis [59], was correlated with Suc concentration in apple (Figures 3, 6). Both MdSPS5 and MdSPS6 share high similarity and are in the same group with AtSPSC (Figure 2). Although a mutation in AtSPSC only caused a 13% decrease in SPS activity in Arabidopsis [59], the expression levels of both MdSPS5 and MdSPS6 were dramatically up-regulated in fruit towards maturity (Figure 3) whereas their expression levels were very low in shoot tips that do not accumulate significant amount of Suc (Table 1). This suggests that both MdSPS5 and MdSPS6 may play important roles in elevating SPS activity for Suc accumulation towards fruit maturity.
Significant correlations between the expression level of a member of a gene family and the total activity of its enzyme were taken as evidence for transcriptional regulation of enzyme activity during fruit development and were also used as a tool for identifying genes that play a role in controlling sugar metabolism in this study. However, it should be pointed out that the in vivo activity of many enzymes is also regulated at the posttranslational level, for example, SPS is regulated by protein phosphorylation and allosteric effects of G6P and inorganic phosphate [60]. The lack of a significant correlation between the expression level of members of MdSDHs and total SDH activity suggests that postranslational regulation might play a role in determining SDH activity.
Conclusions
Sugar metabolism in apple fruit is developmentally regulated to match the high requirements for energy and intermediates during the early stage of fruit development and sugar accumulation from the beginning of cell expansion to fruit maturity. At the early stage of fruit development, imported sorbitol and Suc are rapidly metabolized by high activities of SDH, invertase, SUSY, FK and HK to satisfy the requirement for energy and intermediates by cell division and growth. As fruit cell expansion continues, the requirement for energy and carbon skeleton decreases, and the activities of these enzymes are down-regulated. Decreased FK activity makes Fru available for accumulation in the vacuole while up-regulation of starch synthesis allows extra carbon stored in the insoluble form in plastids. At the late stage of fruit development, starch breaks down and up-regulation of sucrose synthesis via SPS contributes significantly to the continued Suc accumulation in the vacuole, elevating the total soluble sugars to a maximum at maturity. In addition, special transporters, e.g. proteins encoded by MdTMT1 and/or MdTMT2, may play important roles in controlling the accumulation of Fru and Suc in apple fruit. From an evolutionary perspective, accumulation of high concentrations of soluble sugars at maturity determines the sweetness of fruit, which serve as attractants for seed dispersal by animals, while low soluble sugars and high starch make immature fruit unsavory to protect seeds from animals or other herbivores.
This work represents a comprehensive analysis of genes involved in sugar metabolism and accumulation in apple. Comparison of the expression patterns of multigene families in mature leaves, shoot tips and fruit, and with those homologous genes functionally characterized in Arabidopsis and other plants allow us to gain insights into their functions in controlling sugar homeostasis and accumulation during apple fruit development. The genes identified in this study, such as MdFK1, MdFK2, MdNINV3, MdSPS5, MdSUT4, MdTMT1, MdTMT2 etc., and their expression data presented here will serve as a platform for further studies to understand sugar metabolism and accumulation in fruit and to manipulate sugar metabolism for improvement of fruit quality related to carbohydrates.
Supporting Information
Information of invertase genes identified in apple including cell wall invertase (CWINV), neutral invertase (NINV), and vacuole acid invertase (vAINV).
(DOC)
Information of sucrose synthase (SUSY) genes identified in apple.
(DOC)
Information of fructokinase (FK) genes identified in apple.
(DOC)
Information of hexokinase (HK) genes identified in apple.
(DOC)
Information of sucrose-phosphate synthase (SPS) genes identified in apple.
(DOC)
Information of sucrose transporter (SUT), tonoplast monosaccharide transporter (TMT) and vacuole glucose transporter (vGT) genes identified in apple.
(DOC)
Oligonucleotide sequences for primers used in this study.
(DOC)
Linear regression equations between enzyme activity (y, µmol mg−1 Protein h−1) and the relative expression level (x) of its genes, y = ax+b, during apple fruit development.
(DOC)
Acknowledgments
The authors would like to thank Richard Raba for maintaining the plants.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by Cornell University Agricultural Experiment Station. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 2.Mishra BS, Singh M, Aggrawal P, Laxmi A. Glucose and Auxin Signaling Interaction in Controlling Arabidopsis thaliana Seedlings Root Growth and Development. PLoS ONE. 2009;4:e4502. doi: 10.1371/journal.pone.0004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho Y-H, Yoo S-D. Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLoS Genetics. 2011;7:e1001263. doi: 10.1371/journal.pgen.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P, Wind JJ, Shia X, Zhanga H, Hanson J, et al. Fructose sensitivity is suppressed in Arabidopsis by the transcription factor ANAC089 lacking the membrane bound domain. Proc Natl Acad Sci USA. 2011;108:3436–3441. doi: 10.1073/pnas.1018665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan Y-L, Jin Y, Yang Y-J, Li G-J, Boyer JS. Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol Plant. 2010;3:942–955. doi: 10.1093/mp/ssq044. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen-Quoc B, Foyer CH. A role for ‘futile cycles involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot. 2001;52:881–889. doi: 10.1093/jexbot/52.358.881. [DOI] [PubMed] [Google Scholar]
- 7.Janssen BJ, Thodey K, Schaffer RJ, Alba R, Balakrishnan L, et al. Global gene expression analysis of apple fruit development from the floral bud to ripe fruit. BMC Plant Biol. 2008;8:16. doi: 10.1186/1471-2229-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai N, Cohen S, Portnoy V, Tzuri G, Harel-Beja R, et al. Metabolism of soluble sugars in developing melon fruit: a global transcriptional view of the metabolic transition to sucrose accumulation. Plant Mol Biol. 2011;76:1–18. doi: 10.1007/s11103-011-9757-1. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Li P, Cheng L. Developmental changes of carbohydrates, organic acids, amino acids, and phenolic compounds in ‘Honeycrisp’ apple flesh. Food Chem. 2010;123:1013–1018. [Google Scholar]
- 10.Zhou R, Cheng L, Dandekar AM. Down-regulation of sorbitol dehydrogenase and up-regulation of sucrose synthase in shoot tips of transgenic apple trees with decreased sorbitol synthesis. J Exp Bot. 2006;57:3647–3657. doi: 10.1093/jxb/erl112. [DOI] [PubMed] [Google Scholar]
- 11.Afoufa-Bastien D, Medici A, Jeauffre J, Coutos-Thevenot P, Lemoine R, et al. The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 2010;10:22. doi: 10.1186/1471-2229-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho J-I, Ryoo N, Eom J-S, Lee D-W, Kim H-B, et al. Role of the Rice Hexokinases OsHXK5 and OsHXK6 as Glucose Sensors. Plant Physiol. 2009;149:745–759. doi: 10.1104/pp.108.131227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaki S, Ino M. Alteration of cellular compartmentation and membrane permeability to sugars in immature and mature apple fruit. J Am Soc Hort Sci. 1992;117:951–954. [Google Scholar]
- 14.Zhang LY, Peng YB, Pelleschi ST, Fan Y, Lu YF, et al. Evidence for apoplasmic phloem unloading in developing apple fruit. Plant Physiol. 2004;135:574–586. doi: 10.1104/pp.103.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SW, Song KJ, Kim MY, Hwang JH, Shin YU, et al. Molecular cloning and characterization of four cDNAs encoding the isoforms of NAD-dependent sorbitol dehydrogenase from the Fuji apple. Plant Sci. 2002;162:513–519. [Google Scholar]
- 16.Fan RC, Peng CC, Xu YH, Wang XF, Li Y, et al. Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol. 2009;150:1880–1901. doi: 10.1104/pp.109.141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chourey PS, Berüter J. Sugar accumulation and changes in the activities of related enzymes during development of the apple fruit. J Plant Physiol. 1985;121:331–341. [Google Scholar]
- 18.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 19.Granot D. Role of tomato hexose kinases. Funct Plant Biol. 2007;34:567–570. doi: 10.1071/FP06207. [DOI] [PubMed] [Google Scholar]
- 20.Gao Z, Maurousset L, Lemoine R, Yoo SD, van Nocker S, et al. Sorbitol transporter expression in apple sink tissues: implications for fruit sugar accumulation and watercore development. J Am Soc Hort Sci. 2005;130:261–268. [Google Scholar]
- 21.Nosarzewski M, Clements AM, Downie AB, Archbold DD. Sorbitol dehydrogenase expression and activity during apple fruit set and early development. Physiol Plant. 2004;121:391–398. [Google Scholar]
- 22.Nosarzewski M, Archbold DD. Tissue-specific expression of sorbitol dehydrogenase in apple fruit during early development. J Exp Bot. 2007;58:1863–1872. doi: 10.1093/jxb/erm048. [DOI] [PubMed] [Google Scholar]
- 23.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucl Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 26.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 28.Horton P, Park KJ, Obayashi T, Nakai K. Protein subcellular localization prediction with WoLF PSORT. 2006. pp. 39–48. Proceedings of the 4th Annual Asia Pacific Bioinformatics Conference (APBC'06): 13–16 February 2006; Taipei, Taiwan.
- 29.Gasic K, Hernandez A, Korban SS. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep. 2004;22:437a–437g. [Google Scholar]
- 30.Moscatello S, Famiani F, Proietti S, Farinelli D, Battistelli A. Sucrose synthase dominates carbohydrate metabolism and relative growth rate in growing kiwifruit (Actinidia deliciosa, cv Hayward). Sci Hortic. 2011;128:197–205. [Google Scholar]
- 31.Dancer J, Hatzfeld WD, Stitt M. Cytosolic cycles regulate the turnover of sucrose in heterotrophic cell- suspension cultures of Chenopodium rubrum L. Planta. 1990;182:223–231. doi: 10.1007/BF00197115. [DOI] [PubMed] [Google Scholar]
- 32.Renz A, Stitt M. Substrate specificity and product inhibition of different forms of fructokinases and hexokinases in developing potato tubers. Planta. 1993;190:166–175. [Google Scholar]
- 33.Stitt M, Wilke I, Feil R, Heldt HW. Coarse control of sucrose-phosphate synthase in leaves: Alterations of the kinetic properties in response to the rate of photosynthesis and the accumulation of sucrose. Planta. 1988;174:217–230. doi: 10.1007/BF00394774. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Ma F, Cheng L. Metabolism of organic acids, nitrogen and amino acids in chlorotic leaves of ‘Honeycrisp’ apple (Malus domestica Borkh) with excessive accumulation of carbohydrates. Planta. 2010;232:511–522. doi: 10.1007/s00425-010-1194-x. [DOI] [PubMed] [Google Scholar]
- 35.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 36.Chen LS, Lin Q, Nose A. A comparative study on diurnal changes in metabolite levels in the leaves of three crassulacean acid metabolism (CAM) species, Ananas somosus, Kalanchoë daigremontiana and K. pinnata. J Exp Bot. 2002;53:341–350. doi: 10.1093/jexbot/53.367.341. [DOI] [PubMed] [Google Scholar]
- 37.Nonis A, Ruperti B, Pierasco A, Canaguier A, Adam-Blondon AF, et al. Neutral invertases in grapevine and comparative analysis with Arabidopsis, poplar and rice. Planta. 2008;229:129–142. doi: 10.1007/s00425-008-0815-0. [DOI] [PubMed] [Google Scholar]
- 38.Bieniawska Z, Paul Barratt DH, Garlick AP, Thole V, Kruger NJ, et al. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 2007;49:810–828. doi: 10.1111/j.1365-313X.2006.03011.x. [DOI] [PubMed] [Google Scholar]
- 39.Karve AA, Rauh B, Xia X, Kandasamy A, Meagher BR, et al. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta. 2008;228:411–425. doi: 10.1007/s00425-008-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lutfiyya LL, Xu N, D'Ordine RL, Morrell JA, Miller PW, et al. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J Plant Physiol. 2007;164:923–933. doi: 10.1016/j.jplph.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Braun DM, Slewinski TL. Genetic control of carbon partitioning in grasses: roles of sucrose transporters and tie-dyed loci in phloem loading. Plant Physiol. 2009;149:71–81. doi: 10.1104/pp.108.129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaki S. Isolation of vacuoles from immature apple fruit flesh and compartimentation of sugars, organic acids, phenolic compounds and amino acids. Plant Cell Physiol. 1984;25:151–166. [Google Scholar]
- 43.Wormit A, Trentmann O, Feifer I, Lohr C, Tjaden J, et al. Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell. 2006;18:3476–3490. doi: 10.1105/tpc.106.047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao ZF, Maurousset L, Lemoine R, Yoo SD, Van Nocker S, et al. Cloning, expression, and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiol. 2003;131:1566–1575. doi: 10.1104/pp.102.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aluri S, Büttner M. Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc Natl Acad Sci USA. 2007;104:2537–2542. doi: 10.1073/pnas.0610278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaki S. Physiology and metabolism of fruit development: biochemistry of sugar metabolism and compartmentation in fruits. Acta Hort (ISHS) 1995;398:109–120. [Google Scholar]
- 47.Hubbard NL, Pharr DM, Huber SC. Sucrose phosphate synthase and other sucrose metabolizing enzymes in fruits of various species. Physiol Plant. 1991;82:191–196. [Google Scholar]
- 48.Ayre BG. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol Plant. 2011;4:377–394. doi: 10.1093/mp/ssr014. [DOI] [PubMed] [Google Scholar]
- 49.Schulz A, Beyhl D, Marten I, Wormit A, Neuhaus HE, et al. Proton-driven sucrose symport and antiport is provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 2011;68:129–136. doi: 10.1111/j.1365-313X.2011.04672.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang L, Li XR, Lian H, Ni DA, He YK, et al. Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathway, respectively. Plant Physiol. 2010;154:744–756. doi: 10.1104/pp.110.162487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sturm A. Invertases primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999;121:1–7. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Coninck B, Le Roy K, Francis I, Clerens S, Vergauwen R, et al. Arabidopsis AtcwINV3 and 6 are not invertases but are fructan exohydrolases (FEHs) with different substrate specificities. Plant Cell Environ. 2005;28:432–443. [Google Scholar]
- 53.Patrick JW. Phloem unloading: Sieve element unloading and post-sieve element transport. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:191–222. doi: 10.1146/annurev.arplant.48.1.191. [DOI] [PubMed] [Google Scholar]
- 54.Tang GQ, Luscher M, Sturm A. Antisense repression and vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell. 1999;11:1–14. doi: 10.1105/tpc.11.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barratt DH, Derbyshire P, Findlay K, Pike M, Wellner N, et al. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA. 2009;106:13124–13129. doi: 10.1073/pnas.0900689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J. 1995;7:97–107. doi: 10.1046/j.1365-313x.1995.07010097.x. [DOI] [PubMed] [Google Scholar]
- 57.Claeyssen E, Rivoal J. Plant hexokinase isozymes: occurrence, properties, and functions. Phytochem. 2007;68:709–731. doi: 10.1016/j.phytochem.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Karve AA, Moore BD. Function of Arabidopsis hexokinase-like1 as a negative regulator of plant growth. J Exp Bot. 2009;60:4137–4149. doi: 10.1093/jxb/erp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun J, Zhang J, Larue CT, Huber SC. Decrease in leaf sucrose synthesis leads to increased leaf starch turnover and decreased RuBP regeneration-limited photosynthesis but not Rubisco-limited photosynthesis in Arabidopsis null mutants of SPSA1. Plant cell Environ. 2011;34:592–604. doi: 10.1111/j.1365-3040.2010.02265.x. [DOI] [PubMed] [Google Scholar]
- 60.Huber SC, Huber JL. Role and regulation of sucrose phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:431–444. doi: 10.1146/annurev.arplant.47.1.431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information of invertase genes identified in apple including cell wall invertase (CWINV), neutral invertase (NINV), and vacuole acid invertase (vAINV).
(DOC)
Information of sucrose synthase (SUSY) genes identified in apple.
(DOC)
Information of fructokinase (FK) genes identified in apple.
(DOC)
Information of hexokinase (HK) genes identified in apple.
(DOC)
Information of sucrose-phosphate synthase (SPS) genes identified in apple.
(DOC)
Information of sucrose transporter (SUT), tonoplast monosaccharide transporter (TMT) and vacuole glucose transporter (vGT) genes identified in apple.
(DOC)
Oligonucleotide sequences for primers used in this study.
(DOC)
Linear regression equations between enzyme activity (y, µmol mg−1 Protein h−1) and the relative expression level (x) of its genes, y = ax+b, during apple fruit development.
(DOC)