Abstract
Study Objectives:
To determine the effect of different genetic backgrounds on demographic and environmental interventions that affect sleep and evaluate variance of these measures; and to evaluate sleep and variance of sleep behaviors in 6 divergent laboratory strains of common origin.
Design:
Assessment of the effects of age, sex, mating status, food sources, and social experience using video analysis of sleep behavior in 2 different strains of Drosophila, white1118ex (w1118ex) and white Canton-S (wCS10). Sleep was also determined for 6 laboratory strains of Canton-S and 3 inbred lines. The variance of total sleep was determined for all groups and conditions.
Measurements and Results:
The circadian periods and the effects of age upon sleep were the same between w1118ex and wCS10 strains. However, the w1118ex and wCS10 strains demonstrated genotype-dependent differences in the effects upon sleep of sex, mating status, social experience, and being on different foods. Variance of total sleep was found to differ in a genotype dependent manner for interventions between the w1118ex and wCS10 strains. Six different laboratory Canton-S strains were found to have significantly different circadian periods (P < 0.001) and sleep phenotypes (P < 0.001). Three inbred lines showed reduced variance for sleep measurements.
Conclusions:
One must control environmental conditions in a rigorously consistent manner to ensure that sleep data may be compared between experiments. Genetic background has a significant impact upon changes in sleep behavior and variance of behavior due to demographic factors and environmental interventions. This represents an opportunity to discover new genes that modify sleep/wake behavior.
Citation:
Zimmerman JE; Chan MT; Jackson N; Maislin G; Pack AI. Genetic background has a major impact on differences in sleep resulting from environmental influences in Drosophila. SLEEP 2012;35(4):545-557.
Keywords: Genetic background, sleep measurement, drosophila, video analysis, variance
INTRODUCTION
While some demographic factors have been consistently reported to affect sleep in Drosophila, such as age1,2 and sex, 3–5 other factors such as the mating status of female flies, have been reported to either affect6 or not affect sleep.7 The different responses of mated females from the 2 different Canton-S strains used in these studies may be due to the accumulation of dissimilar genetic elements. However, this could also be because sleep was measured in these studies using the single infrared beam break system, the Drosophila Activity Monitoring System (DAMS). We have previously shown that video analysis is more accurate in determining sleep and wake than DAMS, which substantially overestimates sleep.8 We have also observed that the magnitude of these errors in estimation of sleep with the DAMS system is dependent on genotype.
In the study reported here, video analysis was used to compare the changes in sleep phenotype due to demographic and environmental factors that have been reported to affect sleep. We were particularly interested in determining the effect of different genetic backgrounds on demographic and environmental interventions that affect sleep. Therefore, we used two strains with different genetic backgrounds, white Canton-S and white1118ex.
We assessed the effects of age, sex, and mating status and the following environmental influences: (a) the type of food the flies were fed9,10; and (b) the social environment of the fly during the period before behavioral recording.7 We also determined, for each of these demographic and environmental interventions, the variance of total sleep in an effort to ascertain if particular interventions would reduce variance in both genotypes. Identification of an intervention that reduces variability in sleep quantity in multiple strains would be beneficial to the discovery of new genes and characterization of mutants. We also examined sleep behavior of divergent laboratory populations of the commonly used wild-type strain Canton-S to determine whether apparently identical strains from different laboratories had similar or dissimilar sleep and whether some of these strains demonstrated more variability in sleep/wake variables than others. We also assessed circadian rhythm in these divergent populations to determine whether the marked differences in sleep behavior we observed in these strains were also found in their circadian behavior. And finally we assessed sleep behavior and its variance in 3 highly inbred lines derived from a single wild-type population found in Raleigh, NC.11
Our results show that the response to environmental influences on sleep in flies differs between genotypes. Therefore, the response to particular environmental changes is genetically determined. Moreover, there are substantial differences in sleep amounts and other characteristics of sleep and wake in different Canton-S strains used in different laboratories, while only minor differences are found in the circadian period of activity in these strains. We propose that studies of sleep in Drosophila need to adopt strategies to minimize effects of genetic background.
METHODS
Drosophila Stocks and Culture Media
The Canton-S isogenized (CSiso) and w1118ex lines8 and white-eyed Canton-S (wCS10)12 line were described previously. The w1118ex strain was isogenized from a w1118 strain found in the Bloomington Stock Center (Indiana),13 while the wCS10 strain was created by crossing the same w1118 white allele into a CS strain for 10 generations.12 We chose these 2 w1118 strains because previous sleep studies have used strains containing the w1118 allele as an experimental control. 14–19 The other Canton-S lines used in these studies were gifts from Amita Sehgal (designated H), Daniel Marenda (designated DM), Amanda Crocker (designated QAC and EK), and Felice Elefant (designated FE). The isofemale inbred Raleigh lines (RAL-208, RAL-301, and RAL-303) were obtained from the Bloomington Stock Center and were described previously.11 We used 3 different food sources in this study: molasses media containing molasses, cornmeal, yeast and agar; dextrose media containing dextrose, cornmeal, yeast and agar; and sucrose media, which contains only sucrose and agar (Table S1 in supplement). The molasses and dextrose media were prepared by the Cell Center at the University of Pennsylvania Perelman School of Medicine.
Standard Behavioral Recording Procedure
Newly eclosed flies were collected under CO2 anesthesia, placed into vials with the appropriate food, and placed into an incubator on the same day. Flies were kept at 25°C on a 12 h light: dark cycle. Fly food vials were changed every 2–3 days. When flies were 9 days old, animals were removed from the incubator during the light period, anesthetized using CO2, and individually transferred into glass tubes with the appropriate food placed in one end and yarn in the other end to allow gas exchange. These individual tubes containing flies were then placed into trays of up to 28 tubes. Each tray was transferred to an incubator set at 25°C into the field of view of a Retiga 2000R digital camera (QImaging, Surrey, BC, Canada). Flies were acclimated overnight, and recordings were started at lights on (ZT0) when flies were 10 days old (see outline of experimental procedures in Table 1).
Table 1.
Experimental variables for each experimental group
| Experimental Groups | Food | Age | Social Experience | Mating Status | Sex |
|---|---|---|---|---|---|
| Male vs Female | Mo-Mo | 10 days | Grouped | Virgins | Males or Females |
| Young vs Old | Mo-Su | 8–10 days, 38 days (w1118ex), or 56 days (wCS10) | Grouped | Virgins | Females |
| Mated vs Virgin | Mo-Su | 10 days | Grouped | Virgins or Mated | Females |
| Molasses vs Dextrose | Mo-Mo or Dx-Dx | 10 days | Grouped | Virgins | Females |
| Switching to Sucrose | Mo-Su or Dx-Su | 10 days | Grouped | Virgins | Females |
| Group vs Isolation | Mo-Mo | 10 days | Grouped* or Isolated | Virgins | Females |
| Canton-S | Mo-Mo | 10 days | Grouped | Virgins | Females |
| Inbred Raleigh Strains | Mo-Mo | 10 days | Grouped | Virgins | Females |
The different variables we used for each of the experimental groups are summarized in this table. Mo-Mo = molasses-based food for entire study including behavioral recording, Mo-Su = flies kept on molasses-based food from eclosion and then switched to sucrose food for behavioral recording, Dx-Dx = dextrose-based food for entire study including behavioral recording, and Dx-Su = flies kept on dextrose-based food from eclosion and then switched to sucrose food for behavioral recording. Age = the flies' age in days at the start of the behavioral recording. Social Experience is the condition the flies were kept in before sleep recording, grouped = 8 to 10 flies, grouped* = 30 flies and isolated = 1 fly. Mating status is the condition of the fly when recorded. Sex is the sex of the flies used in the experiment.
Effects of Age
We selected ages for each strain which had < 50% of surviving flies based upon lifespan data: 38 days for w1118ex (40.5% survival) and 57 days for wCS10 (27.9% survival). Newly eclosed flies were placed in groups of 8 to 10 flies on molasses media, transferred into new vials containing molasses media every 2 to 3 days until day 36 (w1118ex) or day 55 (wCS10), when they were individually transferred into glass tubes containing sucrose media. Aged flies were then acclimated in the monitor tube overnight as described above for a single day and behavior recorded for 5 days starting at ZT0 on day 38 (w1118ex) or day 57 (wCS10). For this study, young controls were recorded starting at ZT 0 on day 8–10 for 5 days (Table 1).
Effects of Sex
To study the effects of sex, newly eclosed males and females were anesthetized using CO2, sorted by sex into groups of 8–10 flies and aged to day 9 as above on molasses media. Flies were then individually transferred into monitor tubes containing molasses media and recorded as described above for 5 days (Table 1).
Effects of Mating Status
We next studied the effect of mating in female flies. For both the virgin and mated groups, newly eclosed female flies were collected into groups of 10 and placed into a vial with molasses food, as described above. Flies in the virgin group were then recorded following the standard procedure as described above. For the mated group, one male was added to each vial of 10 females. All flies were then transferred to glass tubes containing sucrose media, followed by the standard procedure for recording at 10 days of age. After 3 days of recording, each female was removed from the glass tube and placed into a vial with molasses-based food to verify the fly had mated. The mated status of females was considered confirmed if larvae were observed in the vial after several days. The behavioral data of females in which the mated status could not be confirmed were eliminated from our analysis. Only 3 days of behavior were recorded in this aspect of our study to prevent the possibility of mated females sharing the monitor tube with actively moving larvae (Table 1).
Effects of Different Foods
Newly eclosed female flies were raised in groups of 10 with either the molasses or dextrose food (Table S1), using the standard behavioral protocol as described above. On day 9 when virgin females were anesthetized with CO2 and placed in individual tubes before behavioral recording, the tubes contained the same food females were raised upon, i.e., either molasses or dextrose food (Table 1).
Effects of Switching Foods to Sucrose
Newly eclosed females were raised in groups of 10 as above, but when transferred on day 9 the virgin flies were placed individually into tubes containing sucrose food, instead of the molasses or dextrose food upon which they had been raised from eclosion (Table 1).
Effects of Social Isolation versus Group-Raised Flies
We compared the effects of keeping flies in social isolation to being raised in groups. To do so, late stage pupae that had darkened wings were wetted using a small paintbrush and then gently removed with blunt forceps. The sex of each pupa was determined under a dissecting scope by determining the presence of sex combs. Pupae that lacked sex combs were sexed as females. Each female pupa was then placed in a 1.5 mL micro-centrifuge tube containing 150 μL molasses food on the bottom and a pierced lid that allowed for air exchange. Each of these tubes were placed so that they were physically and visually isolated from each other and kept at 25°C overnight. On the following day, each newly eclosed fly was anesthetized using CO2 and the sex was verified. For the isolated condition each individual female fly was transferred to a vial with molasses food. For flies kept in grouped conditions, newly eclosed female flies were combined into sets of 30 flies in vials containing molasses food. Upon reaching 9 days of age, flies from both conditions were transferred individually into monitor tubes containing molasses media for behavioral monitoring as described above (Table 1).
Behavior and Variance of Different Canton-S and Raleigh Inbred Lines
Newly eclosed females from the 6 Canton-S and 3 Raleigh inbred lines described above were collected into groups of 8–10 flies and raised upon molasses media for 9 days as described above. On day 9, flies were individually transferred into monitor tubes containing molasses media and their behavior recorded for 5 days beginning on day 10 (Table 1).
Determination of Circadian Period
Newly eclosed flies were collected and placed into vials of dextrose-based food in groups of ≥ 10 flies and kept at 25°C on a 12:12 L:D cycle. The vials were changed every 2–3 days. Flies were transferred into individual monitor tubes containing sucrose food during the light part of the L:D cycle at 6 days old and placed in DAMS monitors (Trikinetics, Waltham, MA), which were then transferred into an incubator set at 25°C on the same L:D cycle. The lights were disconnected when the flies were 8 days old, and the activity of the flies recorded using the DAMS monitor for an additional 7 days in D:D. The last 5 days of D:D data (ages 10 days to 14 days old) were used to calculate the period of activity using the Lomb-Scargle method20 in the Clocklab software package (Actimetrics, Wilmette, IL).
Statistical Analysis
Six variables describing sleep and wake were determined separately for daytime (ZT0 to ZT12) and nighttime (ZT12 to ZT0) for each group using our custom software8: total sleep, total wake, sleep bout number, wake bout number, median sleep bout duration, and median wake bout duration. The effects of specific interventions on sleep parameters within each fly strain was tested, using mixed effects linear regression, with day fixed effects and animal random effects. Mixed effects models are analogous to linear regression models for repeated measurements. The interventions examined in each of the fly strains were as follows: effects of age (young compared to old); effects of sex (male compared to female); effects of mating status (virgin compared to mated); and effects of social isolation (isolated compared to group-raised). For the food experiments, between food media group comparisons were conducted for the following food groups: molasses only (Mo-Mo), molasses switched to sucrose (Mo-Su), dextrose only (Dx-Dx), and dextrose switched to sucrose (Dx-Su). P-values reported for post hoc comparisons were Bonferroni adjusted for the following comparisons; Mo-Mo to Dx-Dx, Mo-Mo to Mo-Su, and Dx-Dx to Dx-Su. The sleep/wake variables in the 6 CS strains and 3 Raleigh inbred lines studied were also compared using the mixed effects linear regression with day fixed effects and fly random effect. No post hoc comparisons were made for the CS strains or the Raleigh inbred lines. We have chosen to show only the total sleep data in the figures for clarity. The remaining values are discussed in the text where appropriate, and all data can be found in the supplemental tables.
The variance of each sleep/wake variable was estimated for each intervention group by specifying models with unequal group variance. Significant differences in variance between genotypes and interventions were determined using a likelihood ratio test comparing models fitted with equal and unequal intervention group variances. The variance and significance of differences in variance for total sleep is presented as the sole measure of whether an intervention or demographic factor reduced or increased variance. Variance and the significance of differences in variance for other sleep and wake measures were determined but were found to lead to the same conclusion as total sleep alone and are not further described for sake of clarity.
As these parametric models assume normal distributions, the distribution of individual sleep/wake measures per group was critically examined for satisfaction of a normal distribution. Wake bout durations during the day exhibited an extreme right skew. However, this was considered somewhat artifactual, in that this resulted from a small number of flies having a few wake bouts of very long duration during the day. Therefore, the data for wake bout duration was log transformed to allow use of parametric modeling. In addition to log transformation of bout length values, in order to include data from flies with very few bouts, data from these flies were modified using a “Winsorising” technique.21 In brief, The Winsorising process simply replaced the largest observed value bout length (durations of > 360 min of wake) with the second largest value (durations of 360 min of wake). Analyses were conducted in SAS Version 9.2, The SAS Institute (Cary, NC).
We report our variance data as minutes squared, as the unit of variance is the average of the squared deviations of the variable. Variance was chosen rather than standard deviation because the standard deviation is an aggregate measure of the distribution of variance (i.e., standard deviation has no distribution of its own by definition), while the variance can be computed for each fly per day and has a distribution. These properties of variance make it ideal for testing variability, as variance is a direct measure of variability. We determined variance by subtracting the individual fly's sleep values from the mean of all flies' sleep measurements and squared it. The variance is reported as a single number, which is the average of all these squared deviations for total sleep for each genotype (wCS10, w1118ex, 6 Canton S, and 3 Raleigh strains) and each condition, i.e., male, female, old, young.
RESULTS
Circadian Period of the wCS10 and w1118ex Strains Are the Same
To evaluate whether the differences in the circadian clock of wCS10 and w1118ex strains contribute to observed differences in the sleep phenotypes, we determined the circadian period of activity for both males and females of these 2 strains. The periods for wCS10 and w1118ex males were not significantly different, 24.46 ± 0.26 h and 24.21 ± 0.58 h, respectively (P = 0.64, student t-test). The circadian period for females was also not significantly different (P = 0.21, student t-test) between the wCS10 and the w1118exstrains, 24.63 ± 0.72 h and 24.82 ± 0.42 h, respectively.
Sleep and Wake in Old Versus Young Flies of the w1118ex and wCS10 Genotypes
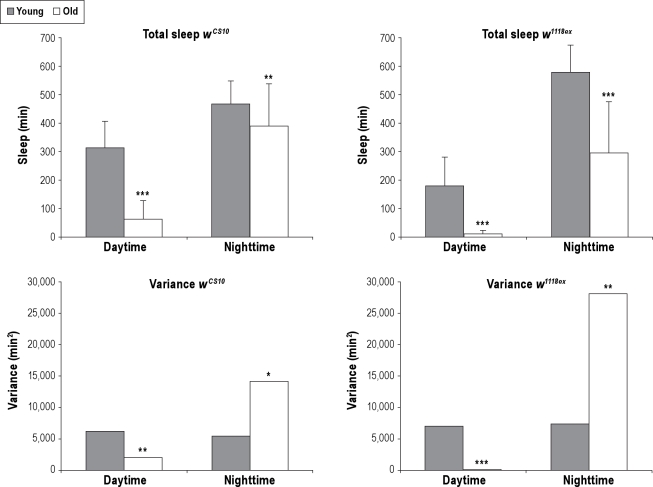
We have previously shown the effects of age on sleep as determined by video within the w1118ex strain.8 In the current study we repeated the examination of the effect of age on sleep measures in w1118ex as well as studied the effect of age in an additional wild-type strain, i.e., wCS10. We also determined the variance in total sleep measures of old and young flies in these 2 strains. We replicated our observation of significantly less total sleep for old flies than young flies both during the day (P < 0.001) and night (P < 0.001) in the w1118ex strain; we saw the same result for the wCS10 strain (P < 0.001 daytime and P = 0.003 nighttime; Figure 1). Older flies of both strains have long consolidated wake bouts with fewer shorter sleep bouts than young flies (Table S2). Although older flies of both strains have reduced nighttime sleep, age affects sleep architecture in each strain differently. Aged w1118ex flies do not have more bouts than young flies but have longer wake and shorter sleep bouts, whereas older wCS10 flies have more but significantly shorter sleep bouts (Table S2)
Figure 1.
Effects of age on total sleep and variance due to age. Age effects total sleep both in the daytime and nighttime and affects variance of total sleep in a time of day dependent manner. Upper Panels: The amount of total sleep for daytime and nighttime is shown for wCS10 and w1118ex strains. Average total sleep in minutes plus standard deviation is shown. Lower Panels: The amount of variance for daytime and nighttime is shown for wCS10 and w1118ex strains. Young animals = gray bar. Older animals = white bar. wCS10 young n = 30, wCS10 old n = 35. w1118ex young n = 29, w1118ex old = 33. Asterisks indicate significant differences between young and old groups; *P < 0.05, **P < 0.01, ***P < 0.001.
We next examined the effects of age of both strains upon the variance of total sleep. Age affected variance of daytime total sleep differently than nighttime total sleep in both of these strains (Figure 1). Younger flies had greater variance of total sleep during the day, while older flies had greater variance of total sleep at night. Therefore, for the wCS10 and w1118ex strains, although the effect of age upon total sleep was the same for both nighttime and daytime, the effect of age upon variance of total sleep was dependent upon time of day.
Effects of Sex on Sleep in the w1118ex and wCS10 Genotypes
As reported previously in other strains,3–5 males slept significantly more than females during the daytime in both the w1118ex and wCS10 strains (P < 0.001 for both) (Figure 2). This was due to males of both strains having a greater number and longer duration of sleep bouts than females (Table S3). During the nighttime, however, the 2 strains show different effects of sex upon sleep. As we have previously shown,8 w1118ex males slept significantly more at night than w1118ex females (P < 0.001). In contrast, wCS10 males slept significantly less at night than wCS10 females (P < 0.001; Figure 2). Males of both strains had fewer sleep bouts than females, but w1118ex males had significantly longer bouts, while wCS10 males had shorter sleep bouts than females of the same strain (Table S3)
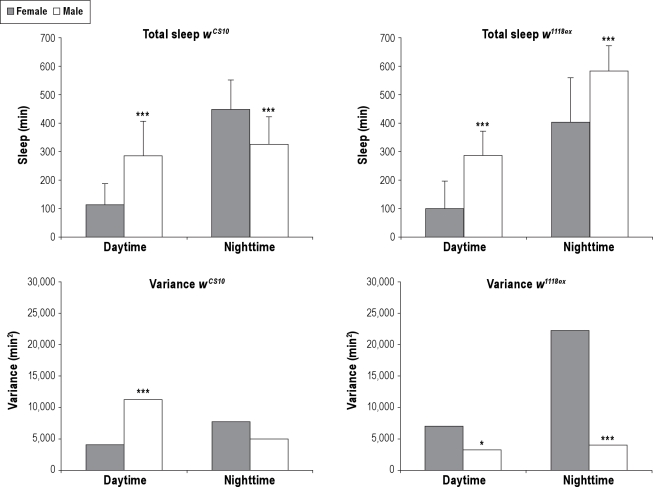
Figure 2.
Effect of sex on total sleep and variance due to sex. Sex affects total sleep and variance of total sleep in a strain dependent manner. Upper Panels: The amount of total sleep for daytime and nighttime is shown for wCS10 and w1118ex strains. Average total sleep in minutes plus standard deviation is shown. Lower Panels: The amount of variance for daytime and nighttime is shown for wCS10 and w1118ex strains. Females = gray bar. Males = white bar. wCS10 females n = 55, wCS10 males n = 48. w1118ex females n = 51, w1118ex males = 50. Asterisks indicate significant differences between male and female flies; *P < 0.05 ***P < 0.001.
Variance of total sleep also differed between sexes but the patterns of differences in variance were different between genotypes. For the wCS10 strain, female variance of daytime total sleep was significantly less than male variance, but nighttime total sleep variance was not significantly different (Figure 2). The opposite occurs in the w1118ex strain, as female w1118ex flies had significantly more variance than males in total sleep during both daytime and nighttime (Figure 2).
Thus, sex differences in sleep behaviors were very similar for sleep behavior between the 2 strains in the daytime, but total sleep time differed between sexes in a strain specific way during the nighttime (Figure 2). There were no common male or female sex differences in the variance of total sleep, given that the direction of the differences observed between males and females was strain specific (Figure 2).
Sleep in Mated Versus Virgin w1118ex and wCS10 Females
We also examined whether mating affected female sleep/wake behavior and the variance of these behaviors. The trends in daytime sleep behavior were in the same direction for mated females versus virgins in both w1118ex and wCS10 strains; i.e., less total sleep (Figure 3), fewer bouts, and longer wake bout durations (Table S3). However, only longer wake bout duration was significantly different for mated versus virgin females of the w1118ex strain (P = 0.006), whereas all of these behaviors were significantly different for wCS10 mated females (all P < 0.001; Table S3). During the nighttime, the only significant difference in sleep/wake parameters between mated and virgin females was that mated w1118ex females slept longer than virgins of the same strain (P = 0.021; Figure 3).
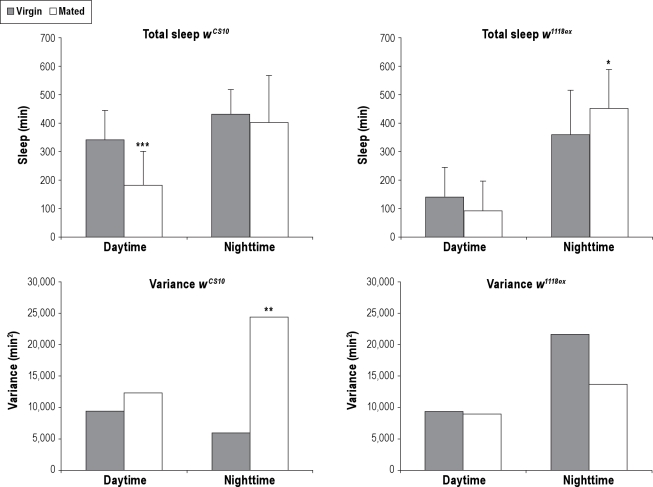
Figure 3.
Effects on total sleep and variance due to mating status in females. Mating status of females affects total sleep and variance of total sleep in a strain dependent manner. Upper Panels: The amount of total sleep for daytime and nighttime is shown for wCS10 and w1118ex strains. Average total sleep in minutes plus standard deviation is shown. Lower Panels: The amount of variance for daytime and nighttime is shown for wCS10 and w1118ex strains. Virgin females = gray bar. Mated females = white bar. wCS10 virgin females n = 20, wCS10 mated females n = 22. w1118ex virgin females n = 26, w1118ex mated females = 18. Asterisks indicate significant differences between virgin and mated females; *P < 0.05, **P < 0.01, ***P < 0.001.
The patterns of variance in total sleep between mated and virgin females were different between the 2 strains. Mated wCS10 females had greater variance in nighttime total sleep than virgin females (Figure 3). In contrast, mated females of the w1118ex strain had no significant differences in the variance of either daytime total sleep or nighttime total sleep than virgin flies (Figure 3).
Thus the effects of mating previous to behavioral recording were strain specific. As with age and sex, the pattern of differences in variance in total sleep were also strain specific
Effect of Food upon Sleep and Wake in w1118ex and wCS10
It has previously been reported that the composition of the food being fed during behavioral recording affects sleep behavior in female flies of a w1118 strain when 2% yeast extract is added to 5% sucrose without yeast.9 We examined the effects on sleep behavior and variance of sleep of 2 different foods, molasses- and dextrose-based (Table S1), in females of the wCS10and w1118ex genotypes. In addition, the effects upon sleep phenotype and variance of switching from either dextrose- or molasses-based food to 5% sucrose were examined.
Effects of Molasses versus Dextrose-Based Foods upon Sleep in wCS10 and w1118ex
Comparisons of specific food differences reveal that the 2 genotypes show a very similar behavioral response to being on dextrose versus molasses food during the daytime. Both wCS10and w1118ex females had increased total sleep (P < 0.001 and P = 0.036, respectively; Figure 4), increased sleep bout number (P = 0.004 and P = 0.016, respectively), and shorter wake bouts (P < 0.001 and P = 0.009, respectively) on dextrose media than on molasses media (Tables S5, S6). Both strains also had longer median sleep bout durations on dextrose versus molasses media (Table S5), but only the bout durations of wCS10females were significantly longer (P < 0.001; Table S6).
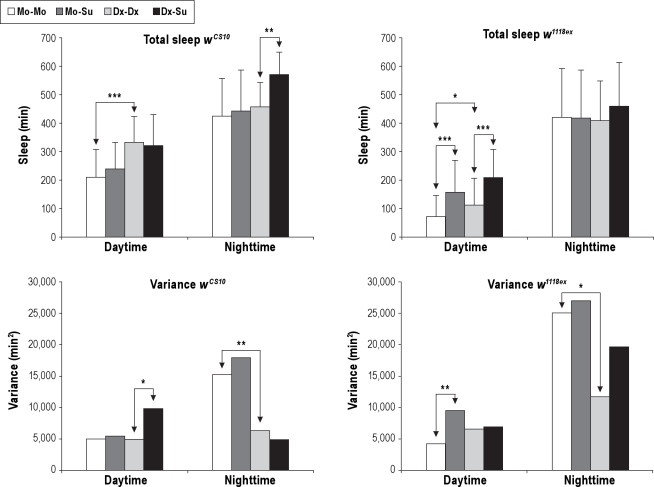
Figure 4.
Effects on total sleep and total sleep variance due to food. Different food regimens affect total sleep and variance of total sleep. Upper Panels: The amount of total sleep for daytime and nighttime is shown for wCS10 and w1118ex strains. Average total sleep in minutes plus standard deviation is shown. Lower Panels: The amount of variance for daytime and nighttime is shown for wCS10 and w1118ex strains. Animals kept on molasses for sleep recording (Mo-Mo) = white bar. Animals switched from molasses to sucrose for sleep recording (Mo-Su) = dark gray bar. Animals kept on dextrose for sleep recording (Dx-Dx) = light gray bar. Animals switched from dextrose to sucrose for sleep recording (Dx-Su) = black bar. For wCS10: Mo-Mo n = 52, Mo-Su n = 56, Dx-Dx n = 54 and Dx-Su n = 51. For w1118ex: Mo-Mo n = 56, Mo-Su n = 45, Dx-Dx n = 48 and Dx-Su n = 51. *P < 0.05, **P < 0.01, ***P < 0.001. Arrows indicate which groups are significantly different from each other.
During the nighttime, differences between the sleep phenotypes on the molasses and dextrose-based foods were dependent on strain. The wCS10females had fewer (P < 0.001) and longer duration sleep bouts (P = 0.001) on dextrose rather than on molasses, whereas in the nighttime, w1118ex females did not display significantly different behavior when on dextrose or molasses-based food (Tables S5, S6).
The effect of food upon variance of total sleep was affected by time of day in both strains. During the daytime on molasses- and dextrose-based foods no significant differences in the variance of total sleep were found in either genotype (Figure 4), whereas during the nighttime, flies kept on dextrose food had significantly decreased variance in total sleep compared to flies kept on molasses food in both the wCS10 (P < 0.01) and w1118ex strains (P < 0.05) (Figure 4).
Thus, the differences in sleep and wake of flies kept on 2 different food types were most profound on daytime sleep behavior for both strains, but neither strain showed a significant effect of the different foods upon variance of total daytime sleep. In contrast, while the nighttime total sleep behavior was not significantly different between foods, variance of total sleep on the dextrose-based food was decreased compared to flies kept upon molasses-based food for both the wCS10 and w1118ex strains.
Switching to Sucrose Food
Strain differences were also observed comparing the effects of flies being switched from either molasses- or dextrose-based food to sucrose-based food (Table S1) versus flies kept on the same food during acclimation and behavioral recording. In general, switching from either complex food to sucrose tended to consolidate sleep in these 2 strains (Table S5, S6); however, the time of day in which this happened was dependent on genotype. The wCS10 flies significantly consolidated nighttime sleep when switched from dextrose to sucrose (Tables S5, S6). The w1118ex strain showed significantly more total daytime sleep (Figure 4) and more consolidated daytime sleep (Tables S5, S6) when switched from either food to sucrose.
The effect upon variance of total sleep when changing to sucrose media from either molasses- or dextrose-based food also differed between the 2 genotypes. The wCS10strain showed a significant increase in daytime total sleep variance when switched from dextrose food to sucrose food during recording, while the w1118ex strain showed a significant increase in the variance of daytime total sleep when switched to sucrose from molasses-based food (Figure 4). Neither strain showed any significant changes in variance of total nighttime sleep when switched to sucrose from molasses- or dextrose-based foods.
Effects of Social Isolation upon Sleep Behavior and Variance of Sleep Behavior
Newly eclosed flies kept in isolation for 4 days before their behavior was recorded have been reported to have less daytime sleep and shorter daytime sleep bout durations than flies kept in groups of ≥ 4 flies.7 Flies kept in groups also had less variance in their total sleep across the day (Figure 1, Panel b).7 Therefore, we wanted to ascertain the effects of social isolation upon sleep/wake behavior and variance of these behaviors as determined by video analysis in the wCS10and w1118ex strains.
We observed the same loss of total daytime sleep with social isolation in both the w1118ex (P = 0.001) and wCS10(P < 0.001) strains, although to a lesser degree than reported previously (Figure 5). No significant differences in sleep bout duration between isolated and grouped flies were observed, but there was a significant decrease in the number of daytime bouts of sleep for isolated flies of the w1118ex (P = 0.022) and wCS10(P = 0.001) strains (Table S7). During the nighttime, there was a significant decrease in total sleep (P < 0.001; Figure 5), a decrease in median sleep bout duration (P < 0.001), and an increase in the median wake bout duration (P < 0.001) in isolated flies of the w1118ex strain (Table S7). However, no effects of social isolation upon nighttime sleep/wake parameters were observed in the wCS10strain.
Figure 5.
Effects on total sleep and variance of total sleep due to social isolation. Social isolation affects total sleep and variance of total sleep in a strain-dependent manner. Upper panels: The amount of total sleep for daytime and nighttime is shown for wCS10 and w1118ex strains. Average total sleep in minutes plus standard deviation is shown. Lower Panels: The amount of variance for daytime and nighttime is shown for wCS10 and w1118ex strains. Group-raised females = gray bar. Isolated females = white bar. wCS10 group-raised females n = 89, wCS10 isolated females n = 109. w1118ex group-raised females n = 46, w1118ex isolated females = 56. Asterisks indicate significant differences between socially isolated and group-raised flies; *P < 0.05, **P < 0.01, ***P < 0.001.
The effects of social isolation upon variance of sleep/wake parameters were strain dependent (Figure 5). For wCS10flies, the variance of both daytime and nighttime total sleep for group-raised flies was not significantly different from isolated flies, whereas the group-raised w1118ex flies had significantly greater variance in their daytime sleep than flies of the same strain raised in isolation.
CS Strains from Different Sources Have Very Different Sleep Behaviors
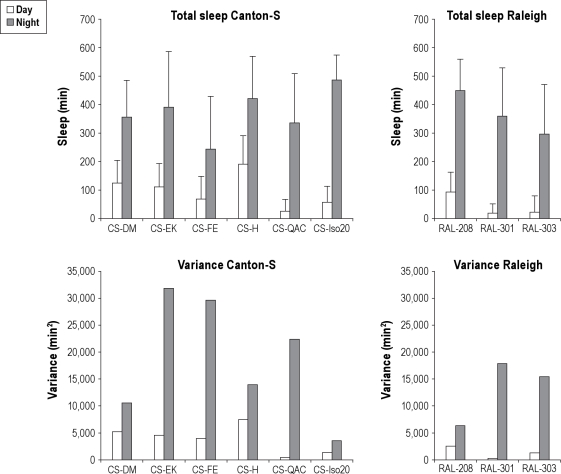
We next assessed differences in sleep from Canton-S strains originating from different laboratories. The Canton-S strain is a commonly used wild-type strain that was originally collected in the 1920s.22 We determined sleep amounts and compared this and the variance of sleep between flies of 6 different Canton-S strains—5 from other laboratories and an isogenized strain of Canton-S that we described previously.8 These studies were done using 10-day-old females raised in groups of 10 flies upon molasses-based food and kept on molasses food when transferred into their individual monitor tubes (see Table 1). There were highly significant differences between strains for all sleep and wake characteristics both during the daytime and nighttime (Table S8). There was a significant overall effect of genotype for each of the characteristics (P < 0.001 for all, except median nighttime sleep bouts, which was P = 0.003). Total nighttime sleep varied between these laboratory strains from a minimum of 246.4 ± 186.7 min in the FE strain to a maximum of 487.9 ± 88.5 min in the Iso20 strain (Figure 6).
Figure 6.
Total sleep and variance of CS and Raleigh lines. Total sleep and variance differ greatly between Canton-S strains from different laboratories (left panels) and three Raleigh inbred lines. Upper panels: The amount of total sleep in the daytime (day = white bars) and the nighttime (night = gray bars) for 6 Canton-S strains (left panels) and 3 Raleigh inbred lines (right panels). Average total sleep in minutes plus standard deviation is shown. Lower panels: The amount of variance for daytime (day = white bars) and the nighttime (night = gray bars) for 6 Canton-S strains and 3 Raleigh inbred lines. CS-DM n = 35, CS-EK n = 51, CS-FE n = 45, CS-H n = 51, CS-QAC n = 37, and CS-Iso20 N = 49. RAL-208 n = 64, RAL-301 n = 57, and RAL-303 n = 63. No post hoc comparisons were done between either the Canton-S or Raleigh inbred lines.
Circadian Period Differs Between Strains
We next assessed whether the strain differences in sleep were also found in another related behavior—the period of circadian rhythm (Table 2). The circadian periods were found to be significantly different between strains by ANOVA (P < 0.001). While the differences in period between strains were significant, the differences were not large (of the order of 4.3%); whereas the differences seen in sleep and wake behaviors are more extreme exceeding 2-fold for a number of parameters.
Table 2.
Circadian period of activity in the six Canton-S strains is different
| CS Strain | Activity Period (hours ± SD) | Post Hoc Comparisons |
|---|---|---|
| H | 25.07 ± 0.85 | a,c |
| DM | 25.55 ± 0.60 | a,b |
| QAC | 25.07 ± 0.25 | a,c |
| EK | 24.73 ± 0.74 | |
| FE | 25.31 ± 0.44 | a,b |
| Iso20 | 24.49 ± 0.42 |
The circadian period was calculated using the Lomb-Scargle method (Clocklab). The Canton-S lines were from Amita Sehgal (designated H), Daniel Marenda (designated DM), Amanda Crocker (designated QAC and EK) and Felice Elefant (designated FE) and our own laboratory (designated Iso20).The period of activity ± standard deviation is shown in hours. The significance of Tukey pair-wise post hoc comparisons are as follows: a = P < 0.005 versus Iso20; b = P < 0.005 versus EK; c = P < 0.05 versus DM.
Variance of Total Sleep Differs Greatly Between Canton-S Strains
The iso20 strain has the lowest variance of total sleep during the nighttime and second least variance in total sleep in the daytime (Figure 6). The generally lower variance seen in the iso20 strain may be due to the strain having been sib-mated for 20 generations previous to the study8; although this strain has been maintained as a population without selection or maintenance of isogeny since that time. Variance of daytime sleep was independent of nighttime variance of the same variable; for example, the QAC strain had the least variable daytime total sleep but the third highest amount of variance in nighttime total sleep (Figure 6).
Sleep Phenotypes and Variance of Three Isofemale Derived Inbred Lines
To further investigate the possibility that isogenization reduces variability of measures of sleep behavior, we recorded sleep and determined variance of sleep measures in 3 inbred lines, Ral-208, Ral-301, and Ral-303. These 3 lines are part of a collection of lines derived from individual fertilized females collected from the Raleigh, NC, area and then inbred for 20 generations.11 This is the same level of inbreeding that the CS-Iso20 strain underwent. These strains are likely to have a different genetic background than the Canton-S strains, which have been a laboratory strain for over 90 years; therefore, we did not analyze them in a single ANOVA. These strains showed very different sleep phenotypes, but tended to have reduced daytime variance in total sleep and middling variance of total nighttime sleep when compared to the Canton-S strains (Figure 6). These strains were inbred in 2003 and therefore predate the isogenization of the CS-Iso20 line, yet they have relatively less variance than the Canton-S, for which no inbreeding has been noted since the 1920s. Therefore, our data suggests that isogenization reduces variance of sleep measures and that this effect may persist for some time.
DISCUSSION
The factors of age,1,2 sex,4,5 and mating status in females6 have all been shown to affect sleep behavior in Drosophila. We have confirmed using video analysis the general changes in sleep behavior previously reported for these factors. However, which specific sleep behaviors and what time of day these effects are observed differ between the wCS10and w1118ex strains. While we did find significant differences in sleep phenotypes between these two strains, we did not find significant differences in their circadian periods. The wCS10strain was derived by crossing a w1118 strain for multiple generations into a Canton-S strain. It is possible that the wCS10and w1118ex strains have the chromosomal region immediately proximal to the white gene of their genomes in common, because they were both derived from an ancestral strain carrying the original white1118 allele isolated in 1984.23 However, the original w1118 allele, and therefore the w1118ex strain in these studies, was found as a spontaneous mutation in a stock derived from an entirely different wild-type Drosophila strain from Canton-S, Oregon-R.23,24 Therefore, differences in sleep/wake behavior observed between the wCS10and w1118ex strains are most likely because these strains have different genetic backgrounds.
Both Food and Social Experience Prior to Behavioral Recording Profoundly Affect Sleep Phenotype
In this study both wCS10and w1118ex strains raised on two different foods had significantly different sleep/wake behaviors, although the effects varied between genotype (Figure 4, Tables S5, S6). Adding 2% yeast to sucrose food (same formula as this study, see Table S1) has been reported to lead to increases in total sleep (only in the daytime), increases in sleep bout number, and decreases in wake bout duration relative to females of a w1118 strain kept on sucrose food without added yeast.9 Likewise, increasing the yeast concentration fivefold while keeping the sucrose concentration constant is reported to increase daytime sleep with no effect on nighttime sleep.10 It has been proposed that the addition of yeast affects sleep behavior through alteration of ratios of dietary amino acids.9 The differences we observed between the wCS10 and w1118ex strains is unlikely to be due to differences in yeast content of the food, since the molasses and dextrose foods do not significantly differ in yeast amounts, 3.2% and 2.9%, respectively. Molasses is a mixture of compounds containing a small amount of protein,25 and the molasses recipe used in this study also has a higher concentration of cornmeal, which also contains amino acids26 than the dextrose-based food (Table S1). Therefore, it is possible that differences in the amino acid content of the two foods we studied lead to alterations in sleep behavior. We conclude that the different genetic backgrounds of the wCS10 and w1118ex strains determine the strain-dependent manner in which the two foods affect sleep and wake.
Another environmental factor that affects sleep is the degree of social isolation during early adulthood. Ganguly-Fitzgerald et al.7 observed that flies kept in groups of at least four flies for four days before behavioral recording, slept significantly longer than flies raised in isolation from the pupae stage. In our studies social isolation also profoundly reduced the daytime total sleep of both strains we examined (Figure 5). We also observed a decrease in the nighttime sleep of isolated w1118ex females but not in wCS10; i.e., the effect of social isolation on nighttime sleep is dependent on genotype. We did not observe a significant decrease in daytime sleep bout duration as previously reported,7 but we did find a reduction in the number of sleep bouts (Table S7). The lack of a significant decrease in daytime sleep bout duration may be because of differences in genetic backgrounds of the CS strain used in the previous study versus the wCS10 and w1118ex strains used in this study. Also video analysis estimates daytime sleep more accurately than DAMS, which we have shown to overestimate sleep bout duration.8
The effect on sleep and wake of social experience and the type of food the flies are exposed to indicates that these and other environmental conditions need to be controlled in a rigorously consistent manner to ensure that sleep data may be compared between experiments and between laboratories.
Dextrose-Based Food Reduced Variance in Nighttime Sleep Measures
This study also ascertained the impact of demographic and environmental factors on variance of sleep behavior of two white-eyed strains of Drosophila melanogaster. Any intervention that reduces variance consistently will be of value because reduced variance in sleep measures enables investigators to more readily detect the effects of genetic mutations or experimental interventions upon sleep. In all cases, however, the variance observed in daytime behavior did not predict the variance observed in the same behavior in the nighttime. Variance of total sleep was affected by sex and mating status of females in a strain-dependent manner. The amount of variance observed due to the environmental parameters of food source and extent of social interactions during early adult stage were also found to be strain dependent. However, both the wCS10 and w1118ex strains kept on dextrose food had less variance in nighttime total sleep than when kept on molasses-based food. This was also true for other nighttime sleep/wake parameters (data not shown). Variance in daytime sleep was not significantly affected by either molasses or dextrose-based foods (Figure 4). Therefore, we conclude that the dextrose-based food generally reduces variance of nighttime sleep behavior.
Canton-S Strains from Different Laboratories Are Not Equal
A somewhat surprising result was the very large differences in all aspects of sleep/wake behavior between different Canton-S strains kept in different laboratories. The original wild type CS strain was collected in Canton, Ohio, by Calvin Bridges in the 1920s. This strain has been kept for many generations separately in different laboratories since that time. We examined the sleep and wake behavior of six CS lines which came from different laboratories, one of which had recently been isogenized for twenty generations.8 Every sleep and wake behavior examined in this study was significantly different among the six CS lines, during both the daytime and the nighttime (Table S8). This demonstrates that for determining sleep, one laboratory line of CS is not the equivalent of another CS line from a different source. The impact of this non-equivalency can be seen in recent studies of the effect of aging upon sleep phenotype in Drosophila. We have found in this study that age has a consistent effect upon sleep and wake between w1118ex and wCS10 strains. Older flies of both strains slept less and had shorter sleep bout durations than young flies (Figure 1, Table S2). This phenotype is very similar to the aging phenotype seen in a previous study in two CS-strains1 and a more recent study which examined sleep phenotype across the lifespan in a CS strain.27 These findings are in contrast to another study, which observed that total sleep amounts increased and sleep bout duration did not change with age.28 In this study all of the mutant and wild-type sibs were crossed in to a single Canton-S strain.28 The authors attributed the discrepancy between their study and the previous study to differences in genetic background and rearing conditions.28 We agree that genetic background differences are the likely explanation for the different results in these studies.
Our findings of large differences in sleep between CS strains from different laboratories suggest that these strains have accumulated differences in their genetic backgrounds over time. There are a number of mechanisms which could account for these differences, which are not mutually exclusive. Over time, different strains maintained in different laboratories can end up with different alleles for multiple genes that would lead to significant effects upon sleep and wake behavior, through either mutation events (which would generate new alleles) or the exclusion of specific alleles through genetic drift. Another mechanism of genetic variation, copy number variation, has recently been shown to be more common than initially believed and conserved across numerous species, including Drosophila.29–31 Therefore, copy number variation could also account for differences in genetic background between Canton-S strains. It is also possible that epigenetic modifications which have become fixed and heritable would lead to the differences we observe in sleep behavior between CS-strains.32 Recently, it has been shown that stress-induced epigenetic modifications have been passed down for several generations from a single event and that repeated events increased the number of generations these modifications persisted in the populations.33
There are also differences in the circadian period for these six CS strains, although the maximum difference in period seen between these six strains is only 4.3% (Table 2), whereas the maximum difference seen between strains for daytime total sleep is greater than 600%, and nighttime sleep maximum difference is approximately twofold. Therefore, although the circadian periods for these strains were different from one another, the magnitude of the difference was small compared to the large differences seen in sleep/wake behaviors. The magnitude of sleep behavior differences versus the magnitude of differences in circadian period likely reflects that sleep may be determined by more genes than circadian period. Evidence for as many as 1,659 genes determining sleep in Drosophila was found in a quantitative trait loci analysis of 40 inbred lines derived from individual female flies, all collected from Raleigh, NC.34
Strategies to Address Background Issues
This effect of genetic background is not unique to sleep, as evidenced by the recent controversy about whether sirtuins affect longevity in Drosophila.35,36 Results from one laboratory may not be replicated in another laboratory using a seemingly identical strain. It is conceivable that a particular mutation could have distinctive effects upon sleep when crossed into different strains of Canton-S because of different sets of modifier genes. This background issue is not unique to Drosophila since it also occurs in mice.37 Investigators who routinely used mouse mutants convened to discuss the issue and developed recommendations to address genetic background issues.38 One strategy in Drosophila to avoid this issue is to conduct forward genetic screens looking for mutants with extreme alterations in sleep that are likely to be less sensitive to modifier genes. This strategy has led to the identification of mutants with a loss of function of the Shaker K channel39 and the gene sleepless,40 both of which have profound reductions in sleep as well as other neurological abnormalities. But evaluating only extreme phenotypes limits the identification of many of the genes altering sleep.
An additional solution to this issue is to demonstrate that a particular environmental change or altered expression of a gene produces the same phenotype in at least two unrelated strains; this would strengthen the case for the effect of specific mutations or genetic interventions (such as overexpression or knockdown) as not being dependent on genetic background. Crossing all mutations or genetic constructs into the same well characterized wild-type strain is also a method to reduce genetic background within an experiment. For behavioral studies using knockdown or overexpression, the lines containing the expression and driver constructs (UAS and GAL4, for example) should ideally have as similar a genetic background as practical. This entails a minimum of 8 backcrossings into a common background strain.35 Our data also suggest that isogenization may reduce the amount of variance in sleep. This needs to be confirmed in other studies. Use of an inbred line for backcrossing could reduce the background issue for mutations and/or genetic constructs of interest while reducing variability in behavioral measures as well. Lastly the development of the site-specific phiC31 integrase system in the fly41–43 allows the opportunity to create gene expression lines (containing constructs for overexpression or RNAi), which will all have the same genetic background without backcrossing and additionally prevent variability due to random insertion of the construct in the genome.
In one sense this major effect of genetic background represents a challenge to studying sleep in Drosophila. An alternative view is that differences in the amount of sleep/wake in different strains found in different laboratories are not a problem but an opportunity. As the cost of genome sequencing decreases due to new technologies, identification of modifier genes in these divergent strains from different laboratories becomes feasible, which will likely lead to discoveries of new genes that modify sleep/wake behavior.
CONCLUSION
Our data demonstrate that relatively minor differences in environmental conditions have major impacts upon sleep behavior. Therefore, investigators must control environmental conditions in a rigorously consistent manner to ensure that sleep data may be compared between experiments and laboratories. Genetic background also has a significant impact upon changes in sleep behavior in response to environmental and demographic factors. There are a number of strategies which can be implemented to address the effect of genetic background, such as: demonstrating that the effects of mutations or interventions are the same in multiple unrelated backgrounds, crossing all constructs and mutations for a minimum of 8 generations into the same strain, and using site specific integration, such as the phiC31 system, to ensure constructs are in an identical genetic background. As new sequencing technologies are developed the differences in sleep/wake behavior observed in different backgrounds becomes an opportunity to discover new genes involved in sleep and wake regulation. An important first study would be to discover the contribution of genetic background to the differences observed by different laboratories of the effect of age upon sleep.
DISCLOSURE STATEMENT
This was not an industry supported study. Funds for the endowment of Allan I. Pack's professorship (John Miclot Professorship) are provided by the Phillips/Respironics Foundation. Mr. Maislin is an employee of Biomedical Statistical Consulting. The other authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Aging (AG-17628 to Dr. Pack) and the National Institute of Neurological Disorders and Stroke (NS-055821 to Dr. Zimmerman). Work for this study was performed at Center for Sleep & Circadian Neurobiology, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA.
REFERENCES
- 1.Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci U S A. 2006;103:13843–7. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 3.Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–39. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 4.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 5.Andretic R, Shaw PJ. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol. 2005;393:759–72. doi: 10.1016/S0076-6879(05)93040-1. [DOI] [PubMed] [Google Scholar]
- 6.Isaac RE, Li C, Leedale AE, Shirras AD. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc Biol Sci. 2010;277:65–70. doi: 10.1098/rspb.2009.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganguly-Fitzgerald I, Donlea J, Shaw PJ. Waking experience affects sleep need in Drosophila. Science. 2006;313:1775–81. doi: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman JE, Raizen DM, Maycock MH, Maislin G, Pack AI. A video method to study Drosophila sleep. Sleep. 2008;31:1587–98. doi: 10.1093/sleep/31.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catterson JH, Knowles-Barley S, James K, Heck MM, Harmar AJ, Hartley PS. Dietary modulation of Drosophila sleep-wake behaviour. PLoS ONE. 2010;5:e12062. doi: 10.1371/journal.pone.0012062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broughton SJ, Slack C, Alic N, et al. DILP-producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging Cell. 2010;9:336–46. doi: 10.1111/j.1474-9726.2010.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan KW, Carbone MA, Yamamoto A, Morgan TJ, Mackay TF. Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biol. 2007;8:R172. doi: 10.1186/gb-2007-8-8-r172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urizar NL, Yang Z, Edenberg HJ, Davis RL. Drosophila homer is required in a small set of neurons including the ellipsoid body for normal ethanol sensitivity and tolerance. J Neurosci. 2007;27:4541–51. doi: 10.1523/JNEUROSCI.0305-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoskins RA, Phan AC, Naeemuddin M, et al. Single nucleotide polymorphism markers for genetic mapping in Drosophila melanogaster. Genome Res. 2001;11:1100–13. doi: 10.1101/gr.178001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABA(A) receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–9. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–7. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 16.Seugnet L, Suzuki Y, Vine L, Gottschalk L, Shaw PJ. D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol. 2008;18:1110–7. doi: 10.1016/j.cub.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bushey D, Tononi G, Cirelli C. The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci. 2009;29:1948–61. doi: 10.1523/JNEUROSCI.4830-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chugh DK, Weaver TE, Dinges DF. Neurobehavioral consequences of arousals. Sleep. 1996;19:S198–201. doi: 10.1093/sleep/19.suppl_10.s198. [DOI] [PubMed] [Google Scholar]
- 19.Crocker A, Sehgal A. Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. J Neurosci. 2008;28:9377–85. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Dongen HP, Olofsen E, VanHartevelt JH, Kruyt EW. Searching for biological rhythms: peak detection in the periodogram of unequally spaced data. J Biol Rhythms. 1999;14:617–20. doi: 10.1177/074873099129000984. [DOI] [PubMed] [Google Scholar]
- 21.Hasings C, Mosteller F, Tukey JW, Winsor CP. Low moments for small samples: a comparative study of order statistics. Ann Math Stat. 1947;18:413–26. [Google Scholar]
- 22.Linnen C, Tatar M, Promislow D. Cultural artifacts: a comparison of senescence in natural, lab-adapted and artificially selected lines of Drosophila melanogaster. Evol Ecol Res. 2001;3:877–88. [Google Scholar]
- 23.Hazelrigg T, Levis R, Rubin GM. Transformation of white locus DNA in drosophila: dosage compensation, zeste interaction, and position effects. Cell. 1984;36:469–81. doi: 10.1016/0092-8674(84)90240-x. [DOI] [PubMed] [Google Scholar]
- 24.Bingham PM. The regulation of white locus expression: a dominant mutant allele at the white locus of Drosophila melanogaster. Genetics. 1980;95:341–53. doi: 10.1093/genetics/95.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Q, Li X, Yan M, et al. High-level production of heterologous proteins using untreated cane molasses and corn steep liquor in Escherichia coli medium. Appl Microbiol Biotechnol. 2010;87:517–25. doi: 10.1007/s00253-010-2536-0. [DOI] [PubMed] [Google Scholar]
- 26.Gianazza E, Viglienghi V, Righetti PG, Salamini F, Soave C. Amino acid composition of zein molecular components. Phytochemistry. 1977;16:315–7. [Google Scholar]
- 27.Haselton AT, Halpern R, Vinson R, Klein R. Effects of nutrition on lifelong sleep and activity patterns in Drosophila melanogaster (Diptera: Drosophilidae) Ann Entomol Soc Am. 2011;104:749–60. [Google Scholar]
- 28.Bushey D, Hughes KA, Tononi G, Cirelli C. Sleep, aging, and lifespan in Drosophila. BMC Neurosci. 2010;11:56. doi: 10.1186/1471-2202-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:19920–5. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–31. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 31.Schrider DR, Hahn MW. Gene copy-number polymorphism in nature. Proc Biol Sci. 2010;277:3213–21. doi: 10.1098/rspb.2010.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33:70–4. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 33.Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–61. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 34.Harbison ST, Carbone MA, Ayroles JF, Stone EA, Lyman RF, Mackay TF. Co-regulated transcriptional networks contribute to natural genetic variation in Drosophila sleep. Nat Genet. 2009;41:371–5. doi: 10.1038/ng.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burnett C, Valentini S, Cabreiro F, et al. Absence of effects of Sir2 overexpression on lifespan in C.elegans and Drosophila. Nature. 2011;477:482–5. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer JH, Morris SN, Chang C, Flatt T, Wood JG, Helfand SL. dSir2 and Dmp53 interact to mediate aspects of CR-dependent lifespan extension in D.melanogaster. Aging. 2009;1:38–48. doi: 10.18632/aging.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doetschman T. Influence of genetic background on genetically engineered mouse phenotypes. Methods Mol Biol. 2009;530:423–33. doi: 10.1007/978-1-59745-471-1_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva AJ, Simpson EM, Takahashi JS, et al. Mutant mice and neuroscience: recommendations concerning genetic background. Banbury Conference on genetic background in mice. Neuron. 1997;19:755–9. doi: 10.1016/s0896-6273(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 39.Cirelli C, Bushey D, Hill S, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–92. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 40.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–6. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–77. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–82. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D.melanogaster. Science. 2006;314:1747–51. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]