Abstract
Type 2 diabetes mellitus is characterized by insulin resistance and failure of pancreatic β-cells producing insulin. Autophagy plays a crucial role in cellular homeostasis through degradation and recycling of organelles such as mitochondria or endoplasmic reticulum (ER). Here we discussed the role of β-cell autophagy in development of diabetes, based on our own studies using mice with β-cell-specific deletion of Atg7 (autophagy-related 7), an important autophagy gene, and studies by others. β-cell-specific Atg7-null mice showed reduction in β-cell mass and pancreatic insulin content. Insulin secretory function ex vivo was also impaired, which might be related to organelle dysfunction associated with autophagy deficiency. As a result, β-cell-specific Atg7-null mice showed hypoinsulinemia and hyperglycemia. However, diabetes never developed in those mice. Obesity and/or lipid are physiological ER stresses that can precipitate β-cell dysfunction. Our recent studies showed that β-cell-specific Atg7-null mice, when bred with ob/ob mice, indeed become diabetic. Thus, autophagy deficiency in β-cells could be a precipitating factor in the progression from obesity to diabetes due to inappropriate response to obesity-induced ER stress.
Keywords: autophagy, diabetes mellitus, endoplasmic reticulum stress, insulin-secreting cells, unfolded protein response
Introduction
Two major types of cell death are apoptosis and necrosis, however, recently another type of cell death, autophagy, was defined. Autophagy is also called as type 2 programmed cell death, in comparison with apoptosis, programmed cell death type 1 (Boya et al., 2005). Although morphologic description of autophagy had been reported decades ago (Ashford and Porter, 1962), its physiologic role has been unknown. However, elucidation of the genes of autophagy using yeast system revolutionized this field and opened a new era of molecular autophagy (Nakatogawa et al., 2009). Mammalian homologues were also identified, and the roles of mammalian autophagy genes in diverse physiological and pathological systems are being intensely investigated. Recent investigations employing techniques of molecular autophagy showed that autophagy protects cells both constitutively and during cellular stress or starvation (Lum et al., 2005). Furthermore, dysregulated autophagy is being implicated in diverse pathological processes such as neurodegenerative diseases, cancer, and infection (Levine and Kroemer, 2008).
Autophagy (derived from the Greek meaning "self eating") exists in all eukaryotic cells and is evolutionarily conserved from yeast to humans (Levine and Klionsky, 2004). Autophagy is a dynamic process involving the rearrangement of subcellular membranes (referred as an autophagosome) to sequester cytoplasm and organelles for delivery to lysosomes, where the sequestered material is degraded and recycled (Klionsky and Emr, 2000). Autophagy is basically cell-protective; however, it may also promote cell death through excessive degradation of cellular constituents, depending on the cellular and environmental context (Levine and Yuan, 2005). Among the three types of cell death, the roles of apoptosis and necrosis in type 1 or type 2 diabetes have been extensively studied (Kaneto et al., 1995; Kim et al., 1999, 2007a, 2007b). However, the role of autophagy in diabetes or body metabolism has been far from clear, but recent observations suggest that autophagy may be an important factor in the development and prevention of diabetes.
Role of autophagy in pancreatic β-cell physiology
It has been reported that ubiquitinated-protein aggregates were formed in pancreatic β-cells during oxidative stress associated with hyperglycemia and were regulated by autophagy (Kaniuk et al., 2007). Because accumulation of protein aggragates may damage cells, such data suggested that autophagy may contribute to the regulation of β-cell survival and death, but direct relationship between autophagy and β-cell death was not demonstrated. To address this issue, a mouse model whose β-cells were deficient in autophagic activity would be valuable. By crossing autophagy-related 7 (Atg7)-floxed mice (Atg7F/F) (Komatsu et al., 2005) with RIP-Cre mice, we and others generated β-cell-specific Atg7-knockout mice (Atg7Δβ-cell). Atg7 is an E1-like gene that is essential for the formation and completion of autophagosomes (Komatsu et al., 2005). Atg7Δβ-cell mice showed significant hyperglycemia, glucose intolerance and hypoinsulinemia (Ebato et al., 2008; Jung et al., 2008). Morphologic analysis showed decreased β-cell mass, along with increased β-cell death and reduced β-cell proliferation. Due to the reduced β-cell mass, pancreatic insulin content was also reduced.
When pancreatic islets were isolated from the Atg7Δβ-cell mice and challenged with glucose to investigate glucose-stimulated insulin release ex vivo, both the basal and stimulated insulin secretion were significantly attenuated in β-cells of the Atg7Δβ-cell mice compared to control mice. In addition, when glucose-induced changes of cytosolic Ca2+ concentrations ([Ca2+]c) were measured in viable islet cells, they were significantly impaired in islets of Atg7Δβ-cell mice compared to control islets (Jung et al., 2008).
While soluble short-lived ubiquitinated proteins are removed through proteasomal pathway, insoluble or large long-lived heavily ubiquitinated proteins are preferentially cleared through autophagy pathway (Kirkin et al., 2009). Consistently, Immunohistochemistry of autophagy-deficient β-cells showed a number of large ubiquitinated inclusion bodies inside the cells. In addition, p62 which is a polyubiquitin-binding protein and also a specific substrate of autophagy itself (Komatsu et al., 2007), also accumulated in autophagy-deficient β-cells. Confocal immunofluorescent microscopy revealed the ubiquitin and p62 were co-localized in the same inclusion bodies (Jung et al., 2008). Hence, large inclusion bodies were formed in autophagy-deficient β-cells that comprise ubiquitin, p62 and degenerated proteins. Pathophysiological roles of inclusion body formation in wild-type cells or autophagy-deficient cells including β-cells await further investigations (Komatsu et al., 2007).
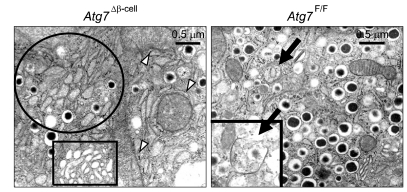
Analysis of the ultrastructural changes using electron microscopy revealed that some β-cells of Atg7Δβ-cell mice contained remarkably reduced numbers of insulin granules compared to wild-type β-cells. Further magnification showed swelling of mitochondria and cisternal distension of rough endoplasmic reticulum (ER) and Golgi complex even in apparently normal-looking β-cells of Atg7Δβ-cell mice at lower magnifications (Jung et al., 2008) (Figure 1).
Figure 1.
Electron microscopic analysis of the pancreas at 20 weeks of age. Mitochondria (white arrowheads), rough ER (circles) and Golgi complex (rectangles). Those in β-cells of Atg7Δβ-cell mice were swollen and distended compared to those of control Atg7F/F mice. A classical autophagosome, an organelle surrounded by double membrane structure was seen in wild-type β-cells (black arrow) (Reproduced from the original paper in Cell Metabolism 8:318-325, 2008).
These morphologic findings implied the existence of mitochondrial dysfunction and ER stress, which could lead to β-cell death and malfunction. We speculate that the decreased function and mass of β-cells of Atg7Δβ-cell mice might be due to mitochondrial dysfunction and/or ER stress, because mitochondria and ER are main cellular organelles rejuvenated by autophagy and critical for β-cell function and survival (Silva et al., 2000; Back et al., 2009). These results suggest that autophagy is necessary to maintain structure, mass and organelle function of β-cells. While these results suggest an important role for autophagy in β-cell homeostasis, the role for autophagy in diabetes has not been fully elucidated. Because ER and autophagy are closely related to each other, and ER stress is important in the pathogenesis of diabetes, β-cell autophagy may be related with ER stress response, also called unfolded protein response (UPR), which is a cellular response to ER stress induced by accumulation of unfolded proteins in ER lumen. The following sections will deal with relationships between autophagy, UPR, ER stress, and diabetogenesis.
ER and autophagy
Evidences that ER is involved in autophagic process are as follows: First, ER is one of the most important sources of isolation membrane, the starting structure of autophagosomes. Previous delicate 3D electron tomography and immunolabelling on thin slice sections showed that protein disulfide isomerase (PDI), an ER-resident protein, exists in both sides of membranes that sandwiched isolation membrane, suggesting that isolation membrane is forming autophagosome cradle stacked between two ER membranes (Hamasaki and Yoshimor, 2010). This structure is called omegasomes, and double FYVE-containing protein 1 (DFCP-1) has been used as a marker molecule of omegasomes (Matsunaga et al., 2010). In such a way, aggregated or terminally misfolded proteins in ER lumen may be able to be cleared by autophagic process as follows. Usually misfolded or excessive unfolded proteins are cleared through ubiquitination and proteasomal degradation in the cytoplasm in a process called ER-associated protein degradation (ERAD) (Smith et al., 2011). Thus, misfolded protein should be unfolded again and be transported across ER membrane back to cytoplasm in a process called retrograde translocation. However, terminally misfolded protein cannot be unfolded for retrograde translocation and ERAD. Hence, terminally misfolded protein or insoluble protein aggregates cannot be easily processed by ERAD, and autophagy may be the only way to remove such protein aggregates. For example, terminally misfolded proteins in the ER lumen may be able to move into autophagosomes stacked between two ER membranes as described above without traversing ER membrane or retrograde translocation. Second, ER is one of the important target organelles of autophagy in a process called "ER-phagy" or "reticulophagy" (Bernales et al., 2007). Third, we have also observed distention of ER in autophagy-deficient β-cells (Jung et al., 2008). Finally, the close relationship between ER and autophagy was demonstrated by recent reports showing activation of autophagic process by ER stress, which was mediated by induction of Atg12 expression, JNK activation or mTOR inhibition (Ogata et al., 2006; Yorimitsu et al., 2006; Kouroku et al., 2007). The significance of such findings is not clear, but may be related to the increased demand for autophagic removal of misfolded proteins in ER lumen in the process of ER stress or accumulation of unfolded proteins.
ER stress and diabetes
Two most important pathgenetic mechanisms of type 2 diabetes are insulin resistance and β-cell failure (DeFronzo et al., 1979). Numerous theories have been proposed to explain insulin resistance and β-cell failure. Several theories proposed different mechanisms for insulin resistance and β-cell failure. However, some theories suggested the same pathogenetic mechanism could explain both insulin resistance and β-cell failure, which is simple and more attractive, from the point of view of Occam's razor. ER stress theory is one of such a unitarian theory. ER stress by lipid injury or cytokines and subsequent JNK activation have been implicated as a cause of insulin resistance (Ozcan et al., 2004, 2006), although effector molecules or lipid metabolites causing direct ER stress has not been identified. ER stress has also been reported to be important in β-cell failure. Pancreatic β-cells are constantly exposed to a great demand for insulin production and, thus, unusually heavy load for UPR. Hence, control of ER stress or UPR is particularly important in β-cells. Furthermore, β-cell response not only to high glucose but also to low glucose may take a form of ER stress response in β-cells (Back et al., 2009). High glucose induces increased insulin synthesis which will lead to an increased amount of unfolded protein and activation of PERK, while low glucose leads to decreased synthesis of insulin through eIF2α phosphorylation (Gomez et al., 2008). Therefore, pancreatic β-cells are particularly susceptible to ER stress, which is aggravated by obesity, insulin resistance, lipids and other physiologic or pathologic stimuli (Scheuner et al., 2005). ER stress or subsequent JNK activation can lead to decreased insulin production, reduced β-cell mass or even β-cell death (Kaneto et al., 2002; Solinas et al., 2006). Thus, meticulous and continuous control of UPR is needed in β-cells to ensure appropriate insulin release according to ambient glucose level, and failure of such an exquisite control would lead to dysregulated glucose homeostasis and β-cell failure or death.
Compromised UPR machinery in autophagy-deficient β-cells
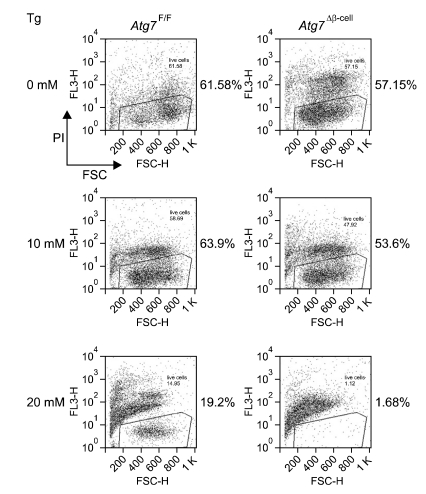
Because of the aforementioned close relationship between UPR and ER stress and that between diabetes and ER stress, we studied the expression of UPR genes in autophagy-deficient β-cells. In fact, this is the reverse question of aforementioned ER stress-induced autophagy induction. In other word, it is well known that ER stress induces autophagy, then what is the effect of autophagy on ER stress or UPR? In our experiment to address these questions, we unexpectedly observed significantly decreased expression of almost all UPR genes in autophagy-deficient β-cells (Quan et al., 2011), which was contrary to our expectation that UPR gene expression would be upregulated due to ER stress which was suspected on the basis of ER distention in autophagy-deficient β-cells. Then, we realized that UPR is a response to ER stress, but not ER stress itself. While investigators usually suspect the existence of ER stress when they observe ER stress response, the relationship between ER stress and ER stress response is not always linear. For instance, appropriate UPR is an adaptive response to ER stress, and deficient UPR in the face of ER stress could be a real danger to cells (Merksamer et al., 2008). Thus, we hypothesized that autophagy is important in the maintenance of appropriate UPR machinery and autophagy deficiency could lead to impaired UPR response and vulnerability toward ER stress. In our experiment to prove our hypothesis, we observed that autophagy-deficient β-cells are more prone to cell death when they are treated with ER stressors such as thapsigargin in vitro (Figure 2).
Figure 2.
Viability of autophagy-deficient islet β-cells from Atg7Δβ-cell mice after treatment with thapsigargine (Tg), a classical ER stressor. Viability presented in this figure as the number on each panel was measured by FACS analysis after staining with propidium iodide. Autophagy-deficient β-cells were more susceptible to thapsigargin-induced cell death, probably because of insufficient UPR gene expression.
The mechanism of the compromised UPR gene expression in autophagy-deficient β-cells is not clear. We observed impaired expression of noncatalytic regulatory subunits of PI3K, p85 and p85β (Quan et al., 2011) that bind to X-box binding protein 1 (XBP-1) in an insulin-dependent manner and are important in the basal and stimulated expression of diverse UPR genes (Park et al., 2010; Winnay et al., 2010). Thus, deficient p85 and p85β expression might be a cause of compromised UPR in autophagy-deficient β-cells.
Lipid and ER stress
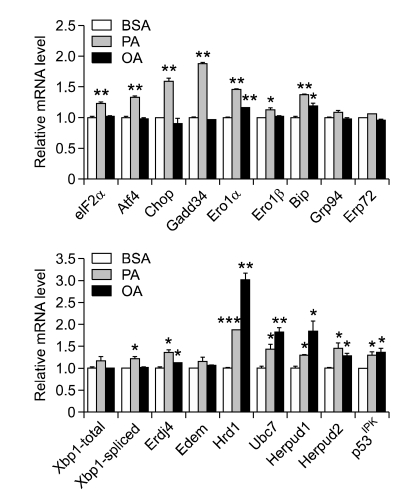
While thapsigargin is a classical pharmacological ER stressor (Li et al., 1993), lipid is a physiological ER stressor that is relevant to the pathogenesis of diabetes and metabolic syndrome (Cunha et al., 2008). As an in vivo counterpart, obesity is a well-known inducer of ER stress, while the effector molecules or lipid metabolites directly causing ER stress is not yet clearly demonstrated. The mechanism of ER stress by lipid injury or obesity might entail altered ER luminal homeostasis by lipid overload, changes of lipid composition of ER membrane, or sequestration of ER machinery to accommodate lipid droplets. When we studied the impact of lipids on ER in our system, we confirmed that lipids induce expression of diverse UPR genes both in insulinoma cells and primary islet cells, showing that lipids are ER stressors and increase demand for UPR by lipids in those cells (Figure 3). Induction of UPR gene expression by lipid was significantly lower in autophagy-deficient β-cells compared to wild-type β-cells, suggesting that demand for UPR is increased by lipid injury and the increased demand for UPR is unmet in autophagy-deficient β-cells. After confirming that lipid injury is an ER stressor, we investigated whether autophagy-deficient β-cells with compromised URP machinery is more susceptible to treatment with lipids in vitro. As hypothesized, we found that autophagy-deficient β-cells were more susceptible to injury by high doses of palmitic acid, the most abundant saturated fatty acid in vivo (Quan et al., 2011). These results show that autophagy-deficient β-cells with compromised UPR machinery are more vulnerable to physiological ER stressors as well as pharmacological ER stressors.
Figure 3.
Induction of UPR gene expression in insulinoma cells by palmitic acid (PA) or oleic acid (OA). Increased expression of UPR genes by free fatty acids suggests that lipids are physiological ER stressors and demand for UPR is increased by lipids. ***P < 0.001, **P < 0.01, *P < 0.05 VS BSA. BSA, bovine serum albumin; PA, palmitic acid; OA, oleic acid.
Role of β-cell autophagy in diabetes and UPR of β-cells in vivo
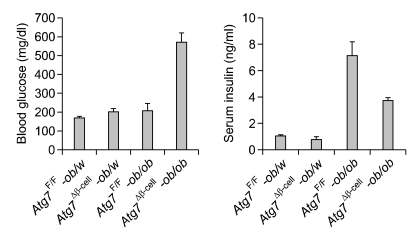
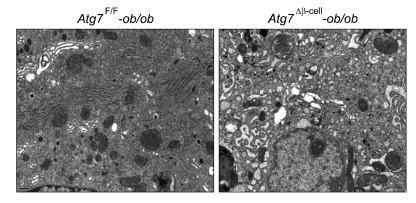
The functional significance of the increased susceptibility of autophagy-deficient β-cells to ER stressors in vitro should be tested in vivo. Animal models for this purpose were generated by breeding mice with β-cell-specific Atg7-deficiency (Atg7Δβ-cell mice) with ob/ob mice. It has been reported that obesity imposes ER stress in β-cells in vivo (Scheuner et al., 2005). Increased UPR gene expression of β-cells of Atg7-wild type (Atg7F/F)-ob/ob mice was confirmed, which is consistent with previous reports (Scheuner et al., 2005) and suggests increased demand for UPR by obesity in vivo. Intriguingly, Atg7Δβ-cell-ob/ob mice developed severe diabetes, while littermate Atg7Δβ-cell-ob/w mice or Atg7F/F-ob/ob mice developed only mild hyperglycemia (Quan et al., 2011). Random blood glucose level was very high in Atg7Δβ-cell-ob/ob mice, while it was only mildly increased in littermate Atg7Δβ-cell-ob/w or Atg7F/F-ob/ob mice (Figure 4). These results suggest that autophagy-deficient β-cells are susceptible to ER stress imposed by obesity in vivo probably due to compromised UPR machinery. This hypothesis was supported by increased β-cell apoptosis and marked accumulation of reactive oxygen species (ROS) in pancreatic islets, and also severely impaired β-cell function of Atg7Δβ-cell-ob/ob mice (Quan et al., 2011). Consistently, fasting serum insulin level was significantly reduced in Atg7Δβ-cell-ob/ob mice compared to Atg7F/F-ob/ob mice that showed high serum insulin level in compensation for obesity-induced insulin resistance (Figure 4). Electron microscopy demonstrated severe ER distention in β-cells of Atg7Δβ-cell-ob/ob mice, in comparison with minimal and moderate ER distention in Atg7F/F-ob/ob and Atg7Δβ-cell-ob/w mice, respectively (Figure 5). These results are in line with a previous report that autophagy-deficient β-cells showed defects in compensatory increase of β-cells mass in response to high-fat diet (Ebato et al., 2008). However, diabetes was not observed in those mice fed high-fat diet.
Figure 4.
Metabolic profile of Atg7Δβ-cell-ob/ob mice and their control mice. Random blood glucose level (left) and fasting serum insulin level (right) in each type of male mice at 12 weeks of age.
Figure 5.
Electron microscopic analysis showed that ER in pancreatic islets of Atg7Δβ-cell-ob/ob mice was severely distended (right), while only minimal distention was observed in islets of Atg7F/F-ob/ob mice (left). ER in islets of Atg7Δβ-cell-ob/w mice showed moderate distention (data not shown), similar to that in Atg7Δβ-cell mice (See Figure 1).
In summary, β-cell autophagy is necessary for appropriate UPR machinery constitutively and in response to ER stress such as lipid injury. Lipid or obesity appears to increase demand for UPR, which was unmet in autophagy-deficient β-cells of Atg7Δβ-cell-ob/ob mice. Observation that overt diabetes developed in Atg7Δβ-cell-ob/ob mice but not in Atg7Δβ-cell mice or ob/ob mice, suggest the possibility that autophagy deficiency in pancreatic β-cells due to genetic predisposition or other causes such as aging could be a factor in the progression from obesity to diabetes.
Lipid and autophagy
Now we understand that autophagy is a protective mechanism against ER stress imposed by obesity and its deficiency leads to compromised UPR in response to ER stress. Then, we next asked the reverse question. What is the effect of lipid injury on autophagy level or activity? This is not a simple question, and consensus has not been reached regarding the effect of lipid on autophagy level or autophagic activity. Some papers showed increased autophagy level by lipid, while others reported otherwise. Different levels of autophagy depending on the stages or ages of the experimental animals have also been suggested (Ebato et al., 2008; Singh et al., 2009; Koga et al., 2010; Shibata et al., 2010; Yang et al., 2010). Such inconsistencies are partly due to technical difficulties associated with assay of in vivo autophagy level and autophagic activity (Klionsky et al., 2008). Particularly, differentiation of autophagy level and autophagic activity is difficult in vivo, as explained below. Thus, we employed GFP-LC3+ mice that express GFP fused to LC3 globally and could provide reliable results regarding in vivo autophagic activity (Hosokawa et al., 2006; Klionsky et al., 2008). In those mice, GFP puncta represent autophagosomes and, thus, direct visualization of autophagy level and quantitation of autophagosome number are possible in vivo (Mizushima et al., 2004). To determine the changes in autophagy level by obesity in vivo, we crossed ob/w mice to GFP-LC3+ mice to finally derive GFP-LC3+-ob/ob mice. The number of GFP puncta reflecting autophagy level was apparently increased in pancreatic islets of GFP-LC3+-ob/ob mice compared to GFP-LC3+-ob/w mice (Quan et al., 2011), suggesting that autophagy level is increased by obesity in vivo. However, the number of LC3 puncta and autophagosomes could be apparently increased by the blockade of autophagic process at the lysosomal step due to clamp effect, rather than true increase in autophagic activity. Thus, we studied cleavage of GFP in GFP-LC3+ mice which reflects autophagic degradation or cleavage of substrates in lysosomes and, thus, autophagic activity (Hosokawa et al., 2006). Indeed, Western blotting demonstrated that GFP cleavage has occurred in islets of GFP-LC3+-ob/ob mice which was not observed in those of GFP-LC3+-ob/w mice (Quan et al., 2011), suggesting that lysosomal steps of autophagic process had occurred and autophagic activity was indeed increased in pancreatic islets of ob/ob mice. Therefore, our data show that both autophagy level and autophagic activity are increased by lipid or obesity in multiple tissues in vivo. However, the level of p62 level, a specific substrate of autophagy (Komatsu et al., 2007), was elevated in islets of GFP-LC3+-ob/ob mice compared to GFP-LC3+-ob/w mice, which is apparently inconsistent with increased autophagic activity (Klionsky et al., 2008). To resolve this inconsistency, we performed a proteolysis assay that represents the eventual degradation of autophagic protein substrates (Klionsky et al., 2008). Lysosomal degradation of long-lived proteins measured by counting release of incorporated C14-leucine was significantly inhibited by palmitic acid or oleic acid (P < 0.01-0.001) (Quan et al., 2011), suggesting that while lipids enhance apparent autophagic activity to eliminate lipids through 'lipophagy' (Singh et al., 2009), lipid overload decreases proteolysis, probably due to shunting or sequestration of the autophagic machinery toward lipophagy. This phenomenon could be a 'trade-off' between removal of lipid and proteolysis, which may be occurring in recent era due to excessive energy supply. Thus, the relationship between lipid and autophagy is complex and may be dependent on cell types, experimental conditions or assay methods. Further exquisite studies employing diverse advanced technology will answer the exact relationship between lipid and UPR and its significance in pathophysiological contexts.
Autophagy and Insulin resistance
In addition to β-cell dysfunction, dysregulated autophagy may possibly be involved in insulin resistance as well, because ER stress has been implicated not only in β-cell dysfunction but also in insulin resistance (Ozcan et al., 2004). Because autophagy deficiency could lead to abnormal ER stress or ER stress response, impaired autophagy may affect insulin resistance. A recent paper reported that autophagy is deficient in the liver of obese mice and overexpression of Atg7 restored insulin sensitivity together with alleviated expression of ER stress markers (Yang et al., 2010). However, it is still controversial whether ER stress is a cause of insulin resistance or simply associated with insulin resistance. Recently, autophagy was reported to participate in the down-regulation of insulin receptors and ER stress-mediated insulin resistance as an adaptive process in ER stress (Zhou et al., 2009). Conversely, insulin resistance has been reported to suppress autophagy (Liu et al., 2009). Although these findings suggest that autophagy may be related to insulin resistance, direct causal relationship between them would require further investigation.
Conclusion and future perspective
While obesity is a well-known risk factor for diabetes, obesity and diabetes are not the same. Not all human subjects with obesity or experimental animals with obesity develop diabetes. Our results suggest the possibility that deficiency of β-cell autophagy could be a factor in the progression from obesity to diabetes.
Because of the elucidation of the machinery of molecular autophagy and its relevance to the maintenance of functions of organelles such as ER, autophagy is emerging as a new target of intense researches and drug discovery aiming at diseases that are caused by or associated with ER dysfunction such as diabetes. If pathogenetic role of dysregulated autophagy in the development of natural human diabetes can be confirmed, a new class of drugs will be possible based on a new principle.
Acknowledgements
This study was supported by the Bio R&D Program (2008-04090) and the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Korea (A080967). Lee M-S is the recipient of the Global Research Laboratory Grant of the National Research Foundation of Korea (K21004000003-10A0500-00310) and a grant from the 21C Frontier Functional Proteomics Project of the Korean Ministry of Science & Technology (FPR08B1-210).
Abbreviations
- Atg7
autophagy-related 7
- DFCP-1
double FYVE-containing protein 1
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- PDI
protein disulfide isomerase
- ROS
reactive oxygen species
- UPR
unfolded protein response
- XBP-1
X-box binding protein 1
References
- 1.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back SH, Scheuner D, Han J, Song B, Ribick M, Wang J, Gildersleeve RD, Pennathur S, Kaufman RJ. Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 2009;10:13–26. doi: 10.1016/j.cmet.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 4.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunha DA, Hekerman P, Ladrière L, Bazarra-Castro A, Ortis F, Wakeham MC, Moore F, Rasschaert J, Cardozo AK, Bellomo E, et al. Initiation and execution of lipotoxic ER stress in pancreatic beta-cells. J Cell Sci. 2008;121:2308–2318. doi: 10.1242/jcs.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol. 1979;237:E214–E223. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 7.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Gomez E, Powell ML, Bevington A, Herbert TP. A decrease in cellular energy status stimulates PERK-dependent eIF2α phosphorylation and regulates protein synthesis in pancreatic beta-cells. Biochem J. 2008;410:485–493. doi: 10.1042/BJ20071367. [DOI] [PubMed] [Google Scholar]
- 9.Hamasaki M, Yoshimor T. Where do they come from? Insights from autophagosome formation? FEBS Letters. 2010;584:1296–1301. doi: 10.1016/j.febslet.2010.02.061. [DOI] [PubMed] [Google Scholar]
- 10.Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Letters. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Jung HS, Chung KW, Kim JW, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, et al. Loss of autophagy diminishes pancreatic b-cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Kaneto H, Fujii J, Seo HG, Suzuki K, Matsuoka T, Nakamura M, Tatsumi H, Yamasaki Y, Kamada T, Taniguchi N. Apoptotic cell death triggered by nitric oxide in pancreatic b-cells. Diabetes. 1995;44:733–738. doi: 10.2337/diab.44.7.733. [DOI] [PubMed] [Google Scholar]
- 13.Kaneto H, Xu G, Fujii N, Kim S, Bonner-Weir S, Weir GC. Involvement of c-Jun N-terminal kinase in oxidative stress-mediated suppression of insulin gene expression. J Biol Chem. 2002;277:30010–30018. doi: 10.1074/jbc.M202066200. [DOI] [PubMed] [Google Scholar]
- 14.Kaniuk NA, Kiraly M, Bates H, Vranic M, Volchuk A, Brumell JH. Ubiquitinated-protein aggregates form in pancreatic beta-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes. 2007;56:930–939. doi: 10.2337/db06-1160. [DOI] [PubMed] [Google Scholar]
- 15.Kim HS, Han MS, Chung KW, Kim S, Kim E, Kim MJ, Jang E, Lee HA, Youn J, Akira S, et al. Toll like receptor 2 senses beta-cell death and contributes to the initiation of autoimmune diabetes. Immunity. 2007a;27:321–333. doi: 10.1016/j.immuni.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Kim S, Millet I, Kim HS, Kim JY, Han MS, Lee MK, Kim KW, Sherwin RS, Karin M, Lee MS. NF-kappa B prevents beta cell death and autoimmune diabetes in NOD mice. Proc Natl Acad Sci U S A. 2007b;104:1913–1918. doi: 10.1073/pnas.0610690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YH, Kim S, Kim KA, Yagita H, Kayagaki N, Kim KW, Lee MS. Apoptosis of pancreatic b-cells detected in accelerated diabetes of NOD mice:no role of Fas-Fas ligand interaction in autoimmune diabetes. Eur J Immunol. 1999;29:455–465. doi: 10.1002/(SICI)1521-4141(199902)29:02<455::AID-IMMU455>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 18.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress(PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 25.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 26.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li WW, Alexander SA, Cao X, Lee AS. Transactivation of the grp78 promoter by Ca2+ depletion. J Biol Chem. 1993;268:12003–12009. [PubMed] [Google Scholar]
- 29.Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulinresistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 31.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 35.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 37.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SW, Zhou Y, Lee J, Lu A, Sun C, Chung J, Ueki K, Ozcan U. The regulatory subunits of PI3K, p85a and p85b, interact with XBP-1 and increase its nuclear translocation. Nat Med. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quan W, Hur KY, Lim Y, Oh SH, Lee JC, Kim HC, Kim GH, Kim SH, Kim HL, Lee MK, et al. Autophagy deficiency in beta cells leads to compromised unfolded protein response and progression from obesity to diabetes in mice. Diabetologia. 2011;55:392–403. doi: 10.1007/s00125-011-2350-y. [DOI] [PubMed] [Google Scholar]
- 40.Scheuner D, Vander Mierde D, Song B, Flamez D, Creemers JW, Tsukamoto K, Ribick M, Schuit FC, Kaufman RJ. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 41.Shibata M, Yoshimura K, Tamura H, Ueno T, Nishimura T, Inoue T, Sasaki M, Koike M, Arai H, Kominami E, et al. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem Biophys Res Com. 2010;393:274–279. doi: 10.1016/j.bbrc.2010.01.121. [DOI] [PubMed] [Google Scholar]
- 42.Silva JP, Köhler M, Graff C, Oldfors A, Magnuson MA, Berggren PO, Larsson NG. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet. 2000;26:336–340. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- 43.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solinas G, Naugler W, Galimi F, Lee MS, Karin M. Saturated fatty acids inhibit induction of insulin gene transcription by JNK-mediated phosphorylation of insulin-receptor substrates. Proc Natl Acad Sci USA. 2006;103:16454–16459. doi: 10.1073/pnas.0607626103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winnay JN, Boucher J, Mori MA, Ueki K, Kahn CR. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat Med. 2010;16:438–445. doi: 10.1038/nm.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou L, Zhang J, Fang Q, Liu M, Liu X, Jia W, Dong LQ, Liu F. Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol Pharmacol. 2009;76:596–603. doi: 10.1124/mol.109.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]