Abstract
Heparin is a linear sulfated polysaccharide widely used in medicine because of its anticoagulant properties. The various sulfation and/or acetylation patterns on heparin impart different degrees of conformational change around the glycosidic bonds and subsequently alter its function as an anticoagulant, anticancer, or antiviral drug. Characterization of these structures is important for eventual elucidation of its function but presents itself as an analytical challenge due to the inherent heterogeneity of the carbohydrates. Heparin octasaccharide structural isomers of various sulfation patterns were investigated using ion mobility mass spectrometry (IMMS). In addition to distinguishing the isomers, we report the preparation and tandem mass spectrometry analysis for multiple sulfated or acetylated oligosaccharides. Herein, our data indicate that heparin octasaccharide isomers were separated based on their structural conformations in the ion mobility cell. Subsequent to this separation, isomers were further distinguished using product ions resulting from tandem mass spectrometry. Overall, IMMS analysis was used to successfully characterize and separate individual isomers and subsequently measure their conformations.
Introduction
Heparin, commonly used in clinical settings as an anticoagulant, is a member of the glycosaminoglycan (GAG) family of anionic polysaccharides and is typically found in the granules of mast cells and granulated cells of organs1. It is also highly sulfated, non-branched, and a heterogeneous mixture consisting of basic repeating disaccharide units of hexuronic acid linked (a1→4) to D-glucosamine 1. Sulfation commonly occurs at the 2,O- position of the hexuronic acid and 6,O- and/or N-position of glucosmine while sulfation on the 3,O- position of glucosamine rarely occur2–4. Biologically, heparin interacts with numerous proteins such as proteases, growth factors, lipid binding proteins, pathogen proteins, and adhesion proteins. It regulates physiological and/or pathological functions such as inflammation, morphogenesis, and angiogenesis5. Heparin is composed of over 90% L-iduronic acid, which exists in equilibrium between the chair (1C4) and the skew boat (2S0) conformations 6.
These flexible conformations of L-iduronic acid are heavily influenced by adjacent sulfated residues and thus influence its ability to bind certain protein motifs and/or binding pockets as exemplified in both heparin hexasaccharide/fibroblast growth factor-2 and pentasaccharide/antithrombin complexes 7, 8. Additionally, various sulfated substitution patterns of heparin affect the conformation around the glycosidic linkages9. Thus, by gaining a deeper understanding of the three dimensional structure of heparin and its various conformations as result of different sulfation patterns, we may further discern the importance heparin undertakes in numerous biological niches, particularly in its ability to bind and recruit chemokines.10, 11
Inherently, structure analysis of heparin is difficult not only due to its size and sequence variety (~100KDa), but because of the net negative charge resulting from the sulfate groups. A more common method for analyzing heparin is to depolymerize it into manageable smaller oligosaccharides through enzymatic and/or chemical reactions12, 13. Smaller oligosaccharides are then separated through characterization techniques including LC, capillary electrophoresis (CE), NMR, circular dichroism, FTIR, and small angle scattering14–16. These methods can provide structural information on heparin such as the conformations surrounding the uronic acid residue or the glycosidic bonds. In addition, mass spectrometry, either stand-alone or coupled with solution separation techniques such as HPLC or CE, can both quantify and identify heparin disaccharides of various sulfation/acetylation patterns16, 17. However, the characterization of heparin isomers still presents a challenge; although identical in elemental composition, isomers of varying sulfation patterns may present with vastly different structures that are difficult to distinguish by classic mass spectrometric methods.
More recently, ion mobility mass spectrometry (IMMS) has emerged as a viable method for separating small molecules and proteins in the gas phase based on measurement of their arrival time distributions (ATD) and their relative collision cross sections (CCS)18–20. This burgeoning new area of mass spectrometry has been used in the structural characterization of oligonucleotides21, 22, glycans23, and peptides24–26, to cite just a few, and it provides positional and structural isomer separation. In the case of heparin, IMMS measurements were able to distinguish L-iduronic acid from its C-5 epimer, D-glucoronic acid of heparan sulfate hexasaccharide27. Herein, we describe a method that employs nano-electrospray coupled with quadrupole-traveling wave ion mobility time of flight mass spectrometry to separate heparin octasaccharide isomers with subsequent analysis and detection of the different conformations stemming from varying sulfation patterns. We also report on the preparation of these GAG isomers with site specific sulfation or acetylation patterns.
We specifically focused our study on heparin octasaccharides as they have been previously shown to have a significant biological impact. Heparin octasaccharides have been shown to interact with antithrombin, cytokine fibroblast growth factor-2, and chemokines as these are closely intertwined with the biological processes of anticoagulation, angiogenesis, and inflammation, respectively11, 28, 29. Herein, we illustrate the ability of IMMS to separate heparin octasaccharide isomers according to their sulfation or acetylation patterns. Based on these observations, our data indicate that not only does IMMS allow for separation of heparin octasaccharide isomers but these experiments can also provide extensive structure information about heparin and may to lead to a better understanding of heparin binding.
EXPERIMENTAL SECTION
Materials
Heparin octasaccharide was purchased from V-labs, INC (Covington, LA). N-methylpyrrolidinone, trimethylamine-sulfur trioxide, sodium carbonate, dimethylsulfoxide, and acetic anhydride were purchased from Sigma-Aldrich Corp. (St. Louis, MO). Oligonucleotides TTTTTTT (T7), CCCCCCC (C7), and ATATAT ((AT)3) were purchased from Invitrogen (Carlsbad, CA). Heparinase I, II, and III from Flavobacterium heparinum were purchased from Seikagaku Corp. (East Falmouth, MA). PD-10 column was purchased from GE healthcare (Piscataway, NJ). Disaccharide standards for compositional analysis were purchased from Calbiochem (La jolla, CA). The IonPac AS7 anion exchange column was purchased from Dionex (Sunnyvale, CA). AG-5W Resin was purchased from Bio-Rad (Hercules, CA). All solvent used were purchased from Fisher Scientific (Fair Lawn, NJ).
Preparation of desulfated or N-acetlyated heparin octasaccharides
We prepared various heparin octasaccharide libraries of different sulfation patterns using similar methods 30, 31. To prepare the de-2,O sulfated heparin octasaccharide library, 500μg of a fully sulfated heparin octasaccharide library was dissolved in 500μl of 0.2M NaOH. The alkaline solution was frozen and then lyophilized. The lyophilized yellow powder was dissolved in 500μl of Milli-Q water and adjusted to pH 7.0 with 10% acetic acid. The solution was then desalted using a PD-10 column with subsequent elution with Milli-Q water. A final sample volume of 100μl was attained using a speed-vac. To prepare the de-6,O sulfated heparin octasaccharide library, 250μl of 2mg/ml fully sulfated heparin octasaccharide library was first applied to a Dowex 50W (X-8, H+, 20–50 mesh) spin column followed by 500μl of pyridine prior to elution and subsequent flash freezing and lyophilization. This pyridinium-heparin octasaccharide powder was then dissolved in 500μl of N-methlypyrrolidinone:water (90/10;v/v) and incubated for 3 hours at 90°C. After cooling to room temperature, the solution was applied to a PD-10 column, eluted with water, flash frozen, and then lyophilized. The white solid powder was then dissolved in water followed by addition of 1mg each of trimethylamine-sulfur trioxide and sodium carbonate. The Reaction proceeded for 24 hours at 55°C followed by desalting using a PD-10 column. A final volume of 100μl was obtained using a speed-vac. To prepare the de-N-sulfated octasaccharide library, pyridinium-heparin octasaccharide powder was dissolved in 500μl of dimethyl sulfoxide:water (95/5; V/v) at 50°C for 1.5hrs, desalted with a PD-10 column, and brought to a final volume of 100 μl via speed-vac. To generate the N-acetylated heparin octasaccharide library, De-N- sulfated heparin octasaccharide library was lyophilized with subsequent addition of both 500μl of 0.25M sodium phosphate buffer (pH=7.5) and 50μl of acetic anhydride at 4°C. Total pH was adjusted to 7.0 with 0.4M NaOH. A final volume of 100μl was attained from a PD-10 column using a speed-vac.
Strong anion exchange chromatography analysis
Both chemically modified heparin octasaccharide and dodecasulfated heparin octasaccharide libraries were separated by strong anion exchange chromatography (SAX)27, 32. Briefly, SAX analysis was performed on a Waters Delta 600 system (Waters Corp., Milford, Ma) coupled to a UV-VIS spectrophotometer set at 232nm. Each of the aforementioned libraries synthesized previously was individually loaded onto an Ion PAC AS7 column (4.00×250mm) with flow rate of 1ml/min in solution A with 4M NaCl (pH 3.7) and solution B H2O (pH 3.7). The Gradient was run as follows: 0–12% solution A for 15minutes and 12–70% A for 45minutes, total gradient time is one hour. Eluted peaks were manually collected. Fractions were then desalted on a PD-10 column. Concentrations were determined by Nano-UV-VIS spectrometry with ε=5500M−1cm−1 in 30mM HCl at 232nm. Samples were applied to a spin column of NH4+ resin (AG 50W X-2, 100–200 mesh) converted from H+ resin using 1M ammonium acetate.
Disaccharide compositional analysis
Quantitative disaccharide compositional analysis was performed to validate the separated heparin octasaccharides17, 33. Briefly, 0.1μg of each of the fractionated heparin octasaccharides was dissolved in 20mM ammonium acetate containing 2mM calcium acetate (pH=7). Fractions were completely digested using 2mU each of heparinase I, II, and III at 37 °C for 15hrs. The reaction was quenched by adding 10mM ammonium hydroxide dissolved in MeOH/H2O (1/1; V/V). Addition of 5μM of the internal standard I-P (ΔUA2S-GlcNCOEt6S) was prepared for quantification. Approximately, 2ul of each digested sample was directly infused into an LTQ mass spectrometer equipped with an electrospray ionization (ESI) source (Thermo Finnigan, San Jose, CA). Both precursor masses and their subsequent tandem mass spectra were used to identify and quantify 12 diagnostic ions. Heparin derived from bovine lung was used as a control to validate our procedure34. Measurements were obtained in triplicate.
Ion mobility mass spectrometry (IM-MS) and calibration curve for collision cross section measurements
Ion mobility mass spectrometry analysis was performed on the Synapt G2 HDMS system (Waters Corp., Milford, MA) with nano-ESI. The samples were analyzed in negative ion mode with a capillary voltage at 0.6kV, source temperature at 40°C, and calibration of TOF mass analyzer adjusted to a mass accuracy within 3ppm from 50 to 1200m/z. The Synapt G2 parameters were optimized as follows: sample cone voltage at 5V, cone voltage at 1V, trap collision energy at 1V, and transfer collision energy at 0V to avoid unnecessary desulfation and fragmentation of heparin octasaccharides. For optimal ion mobility separation, the traveling wave velocity and pulse height were set at 300m/s and 15V, respectively. Both ion mobility and helium cells were kept at 3.45mbar of nitrogen and 142mbar of helium, respectively. Three independent measurement were acquired for each sample and were processed with Masslynx(V4.1) software. To obtain collision cross sections (CCS) of heparin octasaccharides, a calibration curve was constructed using a mixture of 20μM T7, C7 and (AT)3 in MeOH:H2O (1/1; V/V) containing 10mM ammonium hydroxide solution as previously described32. CCS calculations were performed according to previously described protocols 35. Absolute collision cross sections (CCS) of oligonucleotides were determined using conventional IMS-MS from the laboratory of Michael T. Bowers at the University of California, Santa Barbara. Corrected ATDs of three standard oligonucleotides for each charge state were plotted against the corrected CCS. After a calibration curve was constructed using a linear fit, the CCSs were determined using the mathematical formula derived from the calibration curve and then corrected for charge state and reduced mass, as indicated in the equation below:
where Y is the corrected CCS of oligonucleotides and X is the corrected ATDs of oligonucleotides.
Results and discussion
Preparation, separation, and purification of heparin octasaccharide libraries
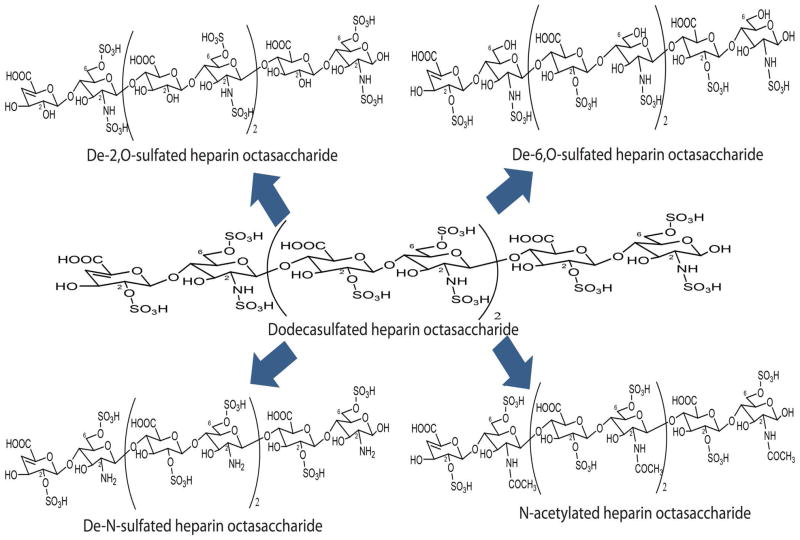
The structures of specific de-sulfated or N-acetylated heparin octasaccharides were prepared using different chemical reactions (Figure 1). Chemically modified heparin octasaccharide libraries were subsequently separated by strong anion exchange chromatography (SAX) based on their charge and structure36. Each fractioned peak was identified using Ion mobility mass spectrometry and our heparin/HS Oligosaccharide Sequencing tool (HOST program) 33. Dodecasulfated octasaccharide library, an initial reactant, contained mostly dodecasulfated species (71.34min retention times) along with minor by-products of sulfated species at various positions. Upon individual chromatographic separation of four distinct heparin octasaccharide libraries via HPLC, numerous compounds of varying sulfation and/or acetylation levels were revealed. (FigureS-1 Supporting information). Additionally, there are several 8 sulfated species for both de-2,O and de-6,O sulfated heparin octasaccharides present in the chromatogram possibly indicative of impurities and/or the presence of by-products from the chemical reactions. Specifically, we optimized the preparation of de-6, O sulfated species to a 3 hour reaction to avoid unnecessary desulfation at any position due to an excessive reaction with subsequent degradation of backbond of octasaccharide. In lieu of dialysis, which is a commonly used procedure, we used size exclusion SPE. Although size exclusion SPE was effective at lowering the concentration of sodium chloride solution, sulfated and acetylated groups of heparin octasaccharide were strongly bound to alkali and alkaline metal ions based on polyelectrolyte theory 37, 38. Cation exchange (NH4+) spin columns were then used to remove metal ion coordination to the heparin octasaccharides.
Figure 1.
Heparin octasaccharide structures. The number of carbon atom is indicated at each structure.
Disaccharide compositional analysis
We next used compositional analysis on each heparin octasaccharide in order to validate their sulfation and/or acetylation patterns17, 34. De-N- sulfated (31.99mim), de-6,O-sulfated (29.36min)and de-2,O sulfated (39.17min) heparin octasaccharides, each containing eight sulfated groups, were digested using Heparinases I,II, and II. Digested samples were observed as isomers at m/z of 247 with z=2−. Subsequent MS/MS analysis allowed the isomers to be distinguished from each other based on the identification of diagnostic ions17 (Table S-1 Supporting information). De-N-sulfated, de-6,O-sulfated, de-2,O sulfated and N-acetylated species were comprised of 99±0.12% I-H(ΔUA2S-GlcN-6S), 99±0.16% III-S (ΔUA2S-GlcNS), 99±0.35% II-S(ΔUA-GlcNS6S), and 99±0.39% I-A (ΔUA2S-GlcNAc-6S), respectively.
Mass spectrometry and ion mobility analysis of heparin octasaccharides
Collected fractions were subjected to ESI-MS (Figure S-2 Supporting information). Heparin octasaccharides were observed under various charge state distributions with 4− and 5− as the predominant charge state. Sodium adducts were also observed due to strong electrostatic interactions occurring between either sulfated groups or the negative charges that result from proton loss.
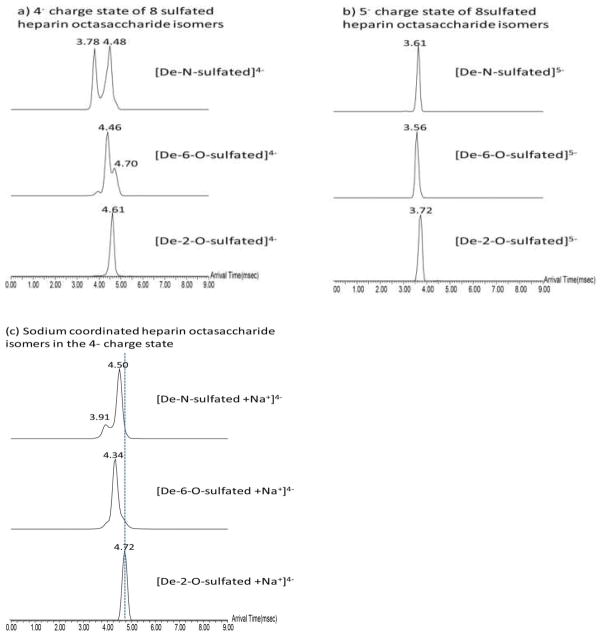
We next investigated the use of ion mobility mass spectrometry (IMMS) to discriminate the isomeric heparin octasaccharide isomers of differing sulfation patterns. Arrival time distributions (ATDs) of the three octasaccharide isomers each retaining 8 sulfates were obtained at both the 4− and 5− charge states (Figure 2). The arrival time distribution for de-2,O-sulfated octasaccharide was 4.61±0.03 msec at the 4− charge state. For the de-6,O-sulfated species, two arrival times could be measured at the 4− charge state with the predominant arrival time at 4.46±0.03 msec. Similarly for the de-N- sulfated octasaccharide, two ion populations were observed at the 4− charge state with arrival times at 3.78±0.03 msec and 4.48±0.03 msec. The presence of two conformations may emanate from sulfate removal at the N-position of glucosamine resulting in a more flexible L-iduronic acid. Observation of the different conformation changes of iduronic acid with desulfation of N-glucosamine have been previously observed 39. The peak resolution measured for the two ion populations of de-N-sulfated species was measured at 1.3 (baseline resolution is 1.5)40. The fact that we were able to resolve two ion populations within the ion mobility cell at this resolution suggests the presence of two different conformations having the same sulfation pattern. For the 5− charge state, the arrival time distributions for de-2,O-suflated, de-6,O-sulfated, and de-N- sulfated octasaccharides were 3.72±0.03 msec, 3.56±0.03 msec, and 3.61±0.03 msec, respectively. One conformation observed at the 5− charge state resulting from charge-charge repulsion may interrupt the flexible conformation changes of the glycosidic bond as compared to those at the 4− charge state. These different conformations allude to the significant impact that charge state contributes to gas phase structures and their subsequent measurement.
Figure 2.
The arrival time distributions for eight sulfated heparin octasaccharide isomers (a) in the 4− charge state and (b) in the 5− charge state and (c) for sodium ion coordinated eight sulfated heparin octasaccharide isomers in the 4− charge state. The arrival times are from three individual measurements and standard deviation is ±0.03msec
Contrary to de-N- and de-6,O-sulfated octasaccharides, each of which had two ion populations, only one ion population was observed for the de-2,O sulfated species. Previous studies using NMR have shown that the non-sulfated iduronic acid of heparin had a predominant chair 1C4 conformation39, 41–43. Our finding suggests that the absence of sulfation at the 2-O position of L –iduronic acid yields a single conformation. However, when sulfated, the conformer can adopt either a chair 1C4 and/or a skew 2S0 conformations 44. The existence of the sulfated L-iduronic acid in both the de-N- and de-6,O- sulfated octasaccharide may explain the presence of two ion populations that we observed while only one ion population was observed when no sulfation was present on the L-iduronic acid of the de-2,O-sulfated species.
Lastly, in order to further increase the efficacy of ion mobility at segregating isomers from one another, we investigated the effect of sodium adduction on these isomeric heparin octasaccharides (Figure 2(c)). Although the arrival times of sodium ion coordinated isomers were similar to those of sodium free isomers, the ion mobility measurements made were sufficient to individually distinguish the isomers from one another. Arrival time distributions for octasaccharides coordinated with one sodium ion could be more easily distinguished from each other than their sodium free counterparts. The bound sodium ions may cause the formation of a more stable conformation in the gas phase thus reducing the freedom of the different conformations that can be formed when little to no sodium is present 39, 42 In addition, the ion population of the sodium coordinated De-N-sulfated heparin octasaccharide was predominantly observed at 4.50±0.03 msec, compared to that of the sodium free species. This observation could explain that two De-N-sulfated species conformations seem to collapse to one upon sodium binding. Thus, measurements made of isomers with a counter ion such as sodium, may induce one stable conformation thereby allowing for their more efficient separation via ion mobility mass spectrometry
Collision cross section measurements of heparin octasaccharides
From each of the measured arrival time distributions, we next calculated the corresponding collision cross sections (CCS) of the heparin octasaccharides at the 4− and 5− charge states using the oligonucleotide calibration linear curve (Table 1). CCSs of octasaccharides at the 5− charge state were larger than those of species at the 4− charge state most likely due to the presence of charge-charge repulsion and therefore gaining elongation of the structures. Comparison of the most compact structures of the three 8 sulfated isomers at the 4− charge state showed the smallest CCS was that of the de-N-sulfated isomer measured at 338.8 Å2. The largest de-2,O sulfated oligosaccharide was measured at 379.3 Å2 and correlated to a 40.5 Å2 CCS difference, while the de-6,O species had a 371.5 Å2 collision cross section for the 4− charge state. Observation of these different conformations resulting from varying sulfation patterns may effect glycosidic geometry through residue interaction of iduronic acid (IdoA)-glucosamine (GlcN) or GlcN-IdoA45, 46. Intriguingly, previous studies showed that removal the 6,O sulfate group of the glucosamine influences the IdoA-GlcN glycosidic linkage but not that of GlcN-IdoA45. Removal of sulfation on each position of iduronic acid or glucosamine may result in various inter-residue glycosidic bond changes. Additionally, these different conformations may explain why several intermolecular hydrogen-bond interactions between IdoA and GlcN result in various idoA structure formations. 46, 47. Minimal differences in CCS were detected for extended conformers of de-6, O (409.9 Å2) and de-N- sulfated (413.2 Å2) species retaining one sulfation at the L-iduronic acid and glucosamine at the 4− charge state. At the 5− charge state, the CCS for de-2,O sulfated octasaccharides without sulfation at the L-iduronic acid showed minimal differences (9 Å2 CCS difference), and there were no differences in CCS between de-6,O and de-N-sulfated. The isomers in the 5− charge state may exhibit bulky conformers due to charge effects, thus providing little effect on isomer separations.
Table 1.
Collision cross sections of heparin octasaccharides in the 4− and 5− charge states. The data are from three individual measurements.
| Collision Cross Sections (Å2) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Charge state | De-N-sulfated | De-6,O-sulfated | De-2,O-sulfated | N-Acetylated | dodecasulfated | |||
| 4− | 338.8±1.4 | 372.4±1.4 | 371.5±1.4 | 383.0±1.4 | 379.3±1.4 | 400.0±1.4 | 413.0±1.4 | 418.7±1.4 |
| 5− | 413.2±1.7 | 409.9±1.7 | 419.9±1.7 | 435.2±1.7 | 441.9±1.7 | |||
In addition to charge state effects on ion mobility measurements, substituent groups also had an effect as seen when acetylation replaced sulfation at the N-position of glucosamine. Although the glucosamine species showed only one conformation, two conformations were observed for the N-acetylated compound. The CCSs for this latter compound were 413.0 Å2 and 400.0 Å2. It is possible that substitution by the acetyl group caused rotation about the glycosidic bond causing more or less coordination with the iduronic acid on the reducing end,48, 49 hence the appearance of more than one conformation.
MS/MS analysis of heparin octasaccharides
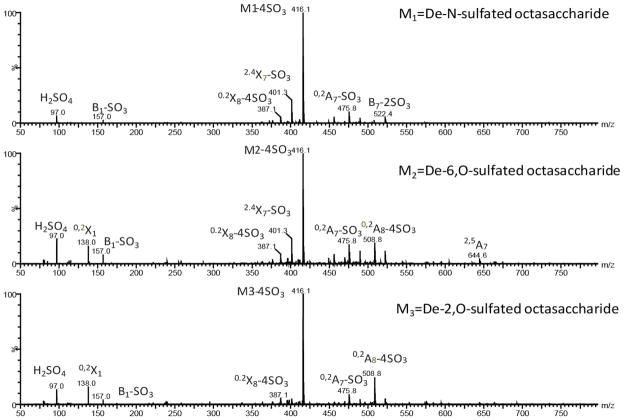
In order to validate the structure and to verify the sulfate position on the three isomeric species, collision induced dissociation was performed at 27V for the m/z 496.0 ion at the 4− charge state for de-6,O-, de-2,O-, and de-N-sulfated heparin octasaccharides(Figure 3). Resulting product ions from both glycosidic and cross-ring cleavage were identified (Figure 3 and Table 2) 50. The most intense fragment ion observed was at m/z 416.1 for all three species and correlates to a neutral loss of 4 sulfates resulting in the 8S-4SO3 ion. Observation of this neutral loss is consistent with our previous studies using tandem MS analysis coupled with isotope labeling experiments 51. At 27V collision energy, complete eradication of the precursor ion was observed. At collision energies less than 27V, glycosidic and cross-ring cleavages were absent and losses of less than 4 sulfates were observed. Since the 5− charge state produced no differences in ATDs, no further experiments were pursued as there was no analytical benefit.
Figure 3.
MS/MS spectra of (a) de-N-sulfated, (b) de-6,O-sulfated, and (C) de-2,O-sulfated heparin octasaccharides. The products ions are labeled using the Domon and Costello nomenclature50. The precursor ion at m/z=496.0 is subject to 27V collision energy in the transfer cell.
Table 2.
Identification of ions from MS/MS spectra of eight sulfated heparin octasaccharide isomers.
| Mass to Charge ratio | Identification of Ions | ||
|---|---|---|---|
| De-N-sulfated octasaccharide | De-6,O-sulfated octasaccharide | De-2,O-sulfated octasaccharide | |
| 97.0 | H2SO4 | H2SO4 | H2SO4 |
| 138.0 | 0,2X0 | 0,2X0 | |
| 157.0 | B1-SO3 | B1-SO3 | B1 |
| 387.1 | 0,2X7-4SO3 | 0,2X7-4SO3 | 0,2X7-4SO3 |
| 401.3 | 2,4X6-SO3 | 2,4X6-SO3 | 2,4X6-2SO3 |
| 416.1 | M-4SO3 | M-4SO3 | M-4SO3 |
| 449.1 | Y7-5SO3 | Y7-5SO3 | Y7-6SO3 |
| 456.1 | 2,4X6-4SO3 | 2,4X6-4SO3 | 2,4X6-5SO3 |
| 470.1 | Z7-4SO3 | Z7-4SO3 | Z7-5SO3 |
| 475.8 | Y7-4SO3 | Y7-4SO3 | Y7-5SO3 |
| 489.8 | B6 | B6 | B6 |
| 507.8 | 0,2A7-SO3 | ||
| 508.8 | 0,2A8-4SO3 | 0,2A8-4SO3 | |
| 522.4 | B7-2SO3 | B7-2SO3 | B7-SO3 |
| 575.6 | B5-SO3 | B5-SO3 | B5 |
| 644.7 | 2,4A6 | 2,4A6 | |
In order to discriminate between the de-2,O-, de-6,O-, and de-N sulfated octasaccharides, we sought diagnostic fragment ions from the collision induced dissociation of the m/z 496.0 ion. Fragment ions at m/z 138.0 represent the 0,2X0 ions of de-2,O-sulfated and de-6,O-sulfated species with one remaining sulfate at the N-position of glucosamine. However, for the de-N sulfated octasaccharide, subsequent tandem MS of the m/z 496.0 ion yielded an absence of the m/z 138.0 ion. Likewise, the complementary ion to that of the m/z 138.0 ion was also observed as a diagnostic marker. The 0,2A8-4SO3 ion at m/z 508.8 was present in both the de-2,O and de-6,O sulfated, but not in that of the de-Nsulfated octasaccharide. These two fragment ions, along with the 0,2A7-SO3 ion at m/z 507.8 differentiated the de-N-sulfated from the de-2,O- or de-6,O-sulfated species(Table 2).
In order to classify the de-2,O- from de-6,O sulfated species, the lone diagnostic ion resulted from the 2,4A6 ion at m/z 644.7. This ion was generated from a cross-ring cleavage of glucosamine and is specific to the de-6,O-sulfated species and does not occur with the de-2,O sulfated octasaccharide which actually retains two sulfated groups on the glucosamine. Additionally, sulfation loss appeared to be more prevalent in both de-6,O- and de-N- sulfated species than in the de-2,O. Ions at m/z = 157.0, 522.4, and 575.6 were present in both de-6,O- and de-N- sulfated species and correspond to the B1-SO3, B7-2SO3, and B5-SO3, respectively. However, for the de-2,O-sulfated octasaccharide, those same ions correspond to B1, B7-SO3, and B5, thus only one sulfate loss was observed for B7 and no other ions.
Conclusions
Analytical methods aimed at the study of heparin structure can be very challenging due to isomeric heterogeneity. In this study, we prepared specific biologically relevant heparin octasaccharides with different sulfation or acetylation patterns. We showed that sulfation or acetylation substitution induces a conformational change in heparin octasaccharide structure. Data indicate that IMMS is capable of identifying and separating the heparin octasaccharide isomers based on their structure formed from removing sulfate groups at specific residue sites. In order to distinguish the isomers, ATD and CID were used and allowed for the verification of each isomer. Additionally, we showed that metal ions coordination to the octasaccharide induces a conformational change. Our findings may provide a platform for future research to identify the essential structures necessary for heparin interactions, binding, and its role in signaling pathways.
Supplementary Material
Acknowledgments
We also acknowledge NIH #GM47356 for supporting this research.
References
- 1.Varki A. Essentials of glycobiology. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2009. p. xxix.p. 784. [PubMed] [Google Scholar]
- 2.Powell AK, Yates EA, Fernig DG, Turnbull JE. Glycobiology. 2004;14(4):17r–30r. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 3.Rabenstein DL. Nat Prod Rep. 2002;19(3):312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 4.Bishop JR, Schuksz M, Esko JD. Nature. 2007;446(7139):1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 5.Capila I, Linhardt RJ. Angew Chem Int Edit. 2002;41(3):391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Gray E, Mulloy B, Barrowcliffel TW. Thromb Haemostasis. 2008;99(5):807–818. doi: 10.1160/TH08-01-0032. [DOI] [PubMed] [Google Scholar]
- 7.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Science. 1996;271(5252):1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 8.Sisu E, Tripathy S, Mallet JM, Driguez PA, Herault JP, Sizun P, Herbert JM, Petitou M, Sinay P. Biochimie. 2003;85(1–2):91–99. doi: 10.1016/s0300-9084(03)00054-3. [DOI] [PubMed] [Google Scholar]
- 9.Yates EA, Santini F, De Cristofano B, Payre N, Cosentino C, Guerrini M, Naggi A, Torri G, Hricovini M. Carbohyd Res. 2000;329(1):239–247. doi: 10.1016/s0008-6215(00)00144-0. [DOI] [PubMed] [Google Scholar]
- 10.Yu Y, Sweeney MD, Saad OM, Crown SE, Hsu AR, Handel TM, Leary JA. J BIOL CHEM. 2005;280(37):32200–32208. doi: 10.1074/jbc.M505738200. [DOI] [PubMed] [Google Scholar]
- 11.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Proc Natl Acad Sci U S A. 2003;100(4):1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai UR, Wang HM, Linhardt RJ. Biochemistry. 1993;32(32):8140–8145. doi: 10.1021/bi00083a012. [DOI] [PubMed] [Google Scholar]
- 13.Shively JE, Conrad HE. Biochemistry. 1976;15(18):3932–3942. doi: 10.1021/bi00663a005. [DOI] [PubMed] [Google Scholar]
- 14.Rudd TR, Skidmore MA, Guimond SE, Guerrini M, Cosentino C, Edge R, Brown A, Clarke DT, Torri G, Turnbull JE, Nichols RJ, Fernig DG, Yates EA. Carbohydrate Research. 2008;343(12):2184–2193. doi: 10.1016/j.carres.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Khan S, Rodriguez E, Patel R, Gor J, Mulloy B, Perkins SJ. Journal of Biological Chemistry. 2011;286(28):24842–24854. doi: 10.1074/jbc.M111.226027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Jones CJ, Beni S, Limtiaco JFK, Langeslay DJ, Larive CK. Annu Rev Anal Chem. 2011;4:439–465. doi: 10.1146/annurev-anchem-061010-113911. [DOI] [PubMed] [Google Scholar]
- 17.Saad OM, Leary JA. Anal Chem. 2003;75(13):2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- 18.von Helden G, Wyttenbach T, Bowers MT. Science. 1995;267(5203):1483–1485. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- 19.Schenauer MR, Leary JA. International Journal of Mass Spectrometry. 2009;287(1–3):70–76. doi: 10.1016/j.ijms.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenn LS, McLean JA. Physical Chemistry Chemical Physics. 2011;13(6):2196–2205. doi: 10.1039/c0cp01414a. [DOI] [PubMed] [Google Scholar]
- 21.Liu YS, Clemmer DE. Anal Chem. 1997;69(13):2504–2509. doi: 10.1021/ac9701344. [DOI] [PubMed] [Google Scholar]
- 22.Koomen JM, Ruotolo BT, Gillig KJ, McLean JA, Russell DH, Kang MJ, Dunbar KR, Fuhrer K, Gonin M, Schultz JA. Anal Bioanal Chem. 2002;373(7):612–617. doi: 10.1007/s00216-002-1363-2. [DOI] [PubMed] [Google Scholar]
- 23.Plasencia MD, Isailovic D, Merenbloom SI, Mechref Y, Clemmer DE. J Am Soc Mass Spectr. 2008;19(11):1706–1715. doi: 10.1016/j.jasms.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun WJ, May JC, Gillig KJ, Russell DH. International Journal of Mass Spectrometry. 2009;287(1–3):39–45. [Google Scholar]
- 25.Tao L, McLean JR, McLean JA, Russell DH. Journal of the American Society for Mass Spectrometry. 2007;18(7):1232–1238. doi: 10.1016/j.jasms.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. International Journal of Mass Spectrometry. 2007;261(1):1–12. [Google Scholar]
- 27.Schenauer MR, Meissen JK, Seo Y, Ames JB, Leary JA. Anal Chem. 2009;81(24):10179–10185. doi: 10.1021/ac902186h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ototani N, Yosizawa Z. J Biochem. 1981;90(5):1553–1556. doi: 10.1093/oxfordjournals.jbchem.a133625. [DOI] [PubMed] [Google Scholar]
- 29.Hasan J, Shnyder SD, Clamp AR, McGown AT, Bicknell R, Presta M, Bibby M, Double J, Craig S, Leeming D, Stevenson K, Gallagher JT, Jayson GC. Clin Cancer Res. 2005;11(22):8172–8179. doi: 10.1158/1078-0432.CCR-05-0452. [DOI] [PubMed] [Google Scholar]
- 30.Baumann H, Scheen M, Huppertz B, Keller R. Carbohydrate Research. 1998;308(3–4):381–388. doi: 10.1016/s0008-6215(98)00097-4. [DOI] [PubMed] [Google Scholar]
- 31.Inoue Y, Nagasawa K. Carbohydrate Research. 1976;46(1):87–95. doi: 10.1016/s0008-6215(00)83533-8. [DOI] [PubMed] [Google Scholar]
- 32.Seo Y, Schenauer MR, Leary JA. International Journal of Mass Spectrometry. 2011;303(2–3):191–198. doi: 10.1016/j.ijms.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saad OM, Leary JA. Anal Chem. 2005;77(18):5902–5911. doi: 10.1021/ac050793d. [DOI] [PubMed] [Google Scholar]
- 34.Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA. Glycobiology. 2005;15(8):818–826. doi: 10.1093/glycob/cwi064. [DOI] [PubMed] [Google Scholar]
- 35.Williams JP, Scrivens JH. Rapid Commun Mass Spectrom. 2008;22(2):187–196. doi: 10.1002/rcm.3346. [DOI] [PubMed] [Google Scholar]
- 36.Weiss J. Ion chromatography. 2. VCH; Weinheim; New York: 1995. p. xi.p. 465. [Google Scholar]
- 37.Chan AKC, Stevic I, Parmar N, Paredes N, Berry LR. Cell Biochem Biophys. 2011;59(3):171–178. doi: 10.1007/s12013-010-9129-5. [DOI] [PubMed] [Google Scholar]
- 38.Manning GS. Accounts Chem Res. 1979;12(12):443–449. [Google Scholar]
- 39.Vanboeckel CAA, Vanaelst SF, Wagenaars GN, Mellema JR, Paulsen H, Peters T, Pollex A, Sinnwell V. Recl Trav Chim Pay B. 1987;106(1):19–29. [Google Scholar]
- 40.Heftmann E. Chromatography: fundamentals and applications of chromatography and related differential migration methods. 6. Elsevier; Amsterdam; Boston: 2004. [Google Scholar]
- 41.Ferro DR, Provasoli A, Ragazzi M, Torri G, Casu B, Gatti G, Jacquinet JC, Sinay P, Petitou M, Choay J. J Am Chem Soc. 1986;108(21):6773–6778. doi: 10.1038/322215b0. [DOI] [PubMed] [Google Scholar]
- 42.Guerrini M, Guglieri S, Beccati D, Torri G, Viskov C, Mourier P. Biochem J. 2006;399:191–198. doi: 10.1042/BJ20060656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulloy B, Forster MJ, Jones C, Drake AF, Johnson EA, Davies DB. Carbohydrate Research. 1994;255:1–26. doi: 10.1016/s0008-6215(00)90968-6. [DOI] [PubMed] [Google Scholar]
- 44.Mulloy B, Forster MJ. Glycobiology. 2000;10(11):1147–1156. doi: 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- 45.Rudd TR, Skidmore MA, Guimond SE, Cosentino C, Torri G, Fernig DG, Lauder RM, Guerrini M, Yates EA. Glycobiology. 2009;19(1):52–67. doi: 10.1093/glycob/cwn103. [DOI] [PubMed] [Google Scholar]
- 46.Pol-Fachin L, Verli H. Carbohydrate Research. 2008;343(9):1435–1445. doi: 10.1016/j.carres.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 47.Murphy KJ, Mclay N, Pye DA. J Am Chem Soc. 2008;130(37):12435–12444. doi: 10.1021/ja802863p. [DOI] [PubMed] [Google Scholar]
- 48.Rudd TR, Guimond SE, Skidmore MA, Duchesne L, Guerrini M, Torri G, Cosentino C, Brown A, Clarke DT, Turnbull JE, Fernig DG, Yates EA. Glycobiology. 2007;17(9):983–993. doi: 10.1093/glycob/cwm062. [DOI] [PubMed] [Google Scholar]
- 49.Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. Carbohydrate Research. 1996;294:15–27. doi: 10.1016/s0008-6215(96)90611-4. [DOI] [PubMed] [Google Scholar]
- 50.Domon B, Costello CE. Biochemistry. 1988;27(5):1534–1543. doi: 10.1021/bi00405a021. [DOI] [PubMed] [Google Scholar]
- 51.Saad OM, Leary JA. J Am Soc Mass Spectr. 2004;15(9):1274–1286. doi: 10.1016/j.jasms.2004.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.