Abstract
Understanding the interactions of nanomaterials with the immune system is essential for the engineering of new macromolecular systems for in vivo applications. Systematic study of immune activation is challenging due to the complex structure of most macromolecular probes. We present here the use of engineered gold nanoparticles to determine the sole effect of hydrophobicity on the immune response of splenocytes. The gene expression profile of a range of cytokines (immunological reporters) was analyzed against the calculated LogP of the nanoparticle headgroups, with an essentially linear increase in immune activity with the increase in hydrophobicity observed in vitro. Consistent behavior was observed with in vivo mouse models, demonstrating the importance of hydrophobicity in immune system activation.
Navigating the response of the immune system is a major issue in the design of nanomaterials for in vivo applications. For example, avoiding immune system detection is an important consideration in gene and drug delivery,1 whereas in the case of adjuvants for vaccine therapies, immune activation is desired.2 Therefore, deeper understanding of how nanomaterials elicit immune responses is essential for the optimization of these systems for biomedical applications.3
A key issue in understanding immune system activation by macromolecular probes is determining interactions of these materials with the innate immune system, the first line of defense of the body and the gatekeeper to full immunoresponse.4 Innate immune activation is associated with the recognition of conserved molecular motifs related with pathogens (pathogen-associated molecular patterns, PAMPs)5 as well as non-specific danger-associated molecular patterns (DAMPs).6 Hydrophobicity per se is considered to be a DAMP.7 Under healthy conditions, hydrophobic cellular materials (“hyppos”) are hidden from the external environment. During necrotic cell disruption or protein denaturation, however, these hyppos become exposed, and by interaction with membranes and specific surface receptors, an innate immune response is generated. This response has been hypothesized to be the origin of the need for oil-based adjuvants in vaccine treatments8.
Quantifying the relationship between hydrophobicity and immune response is experimentally challenging. In aqueous environments, structural changes and aggregation accompany variations in the hydrophobic content of synthetic9 and biomolecular agents (e.g. proteins and lipids).10 As a result, immune response to the hydrophobicity of these materials is also influenced by structural differences in the probe, complicating the structure-activity correlation of these systems.11
In recent studies, nanoparticles with well-defined surfaces have been used to probe the interactions of nanomaterials with biological systems.12 We have developed a family of gold nanoparticles (AuNPs, Figure 1) designed to explore structure-activity relationships (SAR) at biological interfaces.13 A key point in the design of these AuNPs is the use of a passivating non-interacting tetra(ethylene glycol) spacer to remove background effects arising from the core and hydrophobic ligand interior, while also preventing aggregation even in complex biofluids such as serum.14 Using these particles, different functional groups can then be displayed at the AuNP surface and their effects isolated using this inherently multivalent platform. This design hence provides structural uniformity, thus offering means to utilize specific surface attributes for SAR purposes.15 We report here the use of this AuNP model to quantify the interplay between hydrophobicity and immune activation of splenocytes.
Figure 1.
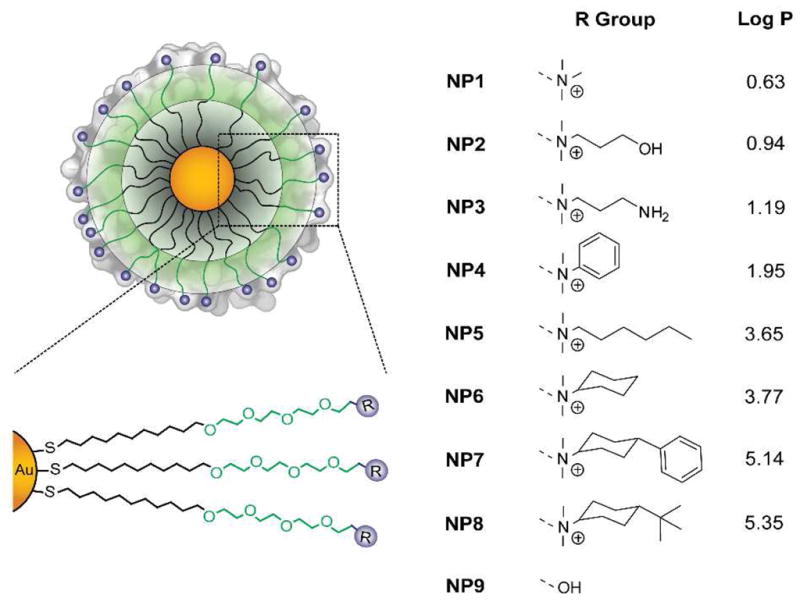
Chemical structure of the monolayer protected 2nm core diameter gold nanoparticles. The passivating tetra(ethylene glycol) spacer (green area) removes possible background effects from the nanoparticle hydrophobic interior (gray zone). To generate the profiles for the SAR studies, functionalities (blue) are tuned at the ligand termini to control the surface hydrophobicity. LogP represents the calculated hydrophobic values of the headgroups.
SAR studies at the nanomaterial level provide an efficient tool in the analysis of nanoparticles properties.16 When other structural parameters are controlled, nanoparticle properties can be described and established based on descriptors of their surfaces.17 Given that NP1-8 differ only in their surface functionality (Figure 1) and their physicochemical properties are similar (both at room temperature and at 37°C, Fig. S3, S4, S5 and S6), we used the computationally predicted n-octanol/water partition coefficient18 of the ligand headgroup (R groups, Figure 1) as the quantitative descriptor of relative nanoparticle surface hydrophobicity. LogP values were estimated using MacroModel (Maestro 8.0).19 The calculations were performed at 298K using the Merck Molecular Force Field (MMFF94).
In our initial studies we explored the immune response of NP1-8 with splenocytes, profiling cytokine mRNA levels to provide a direct assessment of immune response.20 This measurement can be done since protein expression follows gene expression for the cytokines under study.21 Splenocytes were selected as the experimental model of study as they are the reservoir of immune cells packed in the largest lymphoid organ in the body. These cells are comprised of mostly B-lymphocytes, but also include T cells and monocytes.22 Taken together, these jointly representing both the innate and the adoptive arms of the immune system.23 Splenocytes harvested from mice were exposed to each nanoparticle (10μM) under in vitro conditions. After 2 hours, the cells were washed and lysed. Quantitative RT-PCR was employed to quantify the mRNA expression level associated with each one of the cytokines; primers for IL-2, IL-6, IL-10, TNFα, IFNγ, and the interferon responsive genes OAS1, STAT1 and IFNβ were used to preferably amplify them from the cDNA library, and normalized against housekeeping genes HPRT1 and GAPDH.20
As shown in Figure 2A, the plot of cytokine expression against LogP reveals an essentially linear correlation between hydrophobicity and immune response, with the exception of NP1. This trend was observed for each of the cytokines under study, with variations only observed in the relative level of expression (Figure S1), indicating a selective type of immune response.24 The distinct behavior of NP1 can be explained by its highly exposed charge, capable of inducing alternate responses through electrostatic interactions15 or by contact with specific amino acids.25
Figure 2.
Cytokine gene expression as a function of nanoparticle headgroup LogP. (A) TNFα (a representative pro-inflammatory cytokine) in vitro gene expression and (B) IL-10 (a representative anti-inflammatory cytokine) in vivo gene expression as a function of the calculated AuNP headgroup LogP. The gene expression values are normalized by dividing the observed response against the values of a positive control (LPS) under the same experimental set. A minimally interacting neutral particle (NP9) was used as a negative control. NPs were used at a concentration of 10μM for in vitro and 5mg/kg for in vivo studies. Data taken after 2h (in vitro) and 1.5h (in vivo) after the exposure to AuNPs. In vivo correlation is lost at 6h (Figure S2).
In vivo response to nanomaterials is much more complex than in vitro systems.26 We probed immune response to NP1-8 using mouse models. For that purpose, mice (12 week old) were injected intravenously (100μl) via the tail vein. Each group of mice (n = 6 mice per group) received a single dose of a specific nanoparticle at 5mg/kg. At 1.5 and 6 hours post intravenous administration, the mice were sacrificed and splenocytes harvested and treated as before to assess cytokine mRNA expression levels. Figure 2B presents the tendency of cytokine expression against LogP in vivo. At lower LogP values, increasing hydrophobicity elicits increased immune response. However, with high degrees of hydrophobicity the dependence is less evident, and a maximum in immune response is observed. This leveling off can be explained by the expected changes in biodistribution for hydrophobic nanoparticles, in particular the poor biodistribution expected for highly hydrophobic particles.27 Nonetheless, it is clear that upon availability of hydrophobic portions in the system, immune response is generated (Figure S2, correlation is lost at 6 hours time).24
In summary, we have demonstrated a direct, quantitative correlation between hydrophobicity and immune system activation, an important determinant for nanomedical and nanoimmunological applications. This correlation provides a promising starting point for determining the specific molecular mechanisms of immune cell activation,28 an issue of importance for understanding the evolution of the innate immune system.29 Moreover, these probes present both a tool for harnessing the immune system and a probe for quantifying the role of hydrophobicity in immune response.30
Supplementary Material
Acknowledgments
This work was supported by the grants from the ISF (#181/10), Lewis foundation for blood cancer and the Kenneth Rainin Foundation to DP. VMR acknowledges support from the NIH (GM07717) and the Center for Hierarchical Manufacturing (CMMI-1025020).
Footnotes
ASSOCIATED CONTENT
Supporting Information. In vitro and in vivo cytokines expression plots, and nanoparticles characterization data (DLS, TEM, ZP). These materials are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) LaVan DA, McGuire T, Langer R. Biotech. 2003;21:1184–1191. doi: 10.1038/nbt876. [DOI] [PubMed] [Google Scholar]; (b) Kumar MNVR. J Pharm Pharmaceut Sci. 2000;3:234–258. [Google Scholar]
- 2.(a) Akagi T, Wang X, Uto T, Baba M, Akashi M. Biomaterials. 2007;28:3427–3436. doi: 10.1016/j.biomaterials.2007.04.023. [DOI] [PubMed] [Google Scholar]; (b) Maltzahn GV, Park JH, Lin KY, Singh N, Schwoppe C, Mesters R, Berdel WE, Ruoslahti E, Sailor MJ, Bhatia SN. Nat Mat. 2011;10:545–552. doi: 10.1038/nmat3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Mizrahy S, Raz SB, Hasgaard M, Liu H, Soffer-Tsur N, Cohen K, Dvash R, Landsman-Milo D, Bremer MGEG, Moghimi SM, Peer D. J Control Release. 2011;156:231–238. doi: 10.1016/j.jconrel.2011.06.031. [DOI] [PubMed] [Google Scholar]; (b) Massich MD, Giljohann DA, Seferos DS, Ludlow LE, Horvath CM, Mirkin CA. Mol Pharm. 2009;6:1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Janeway CA, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]; (b) Gallucci S, Lolkema M, Matzinger P. Nat Med. 1999;5:1249–1255. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- 5.Janeway CA., Jr Cold Spring Harb Symp Quant Biol. 1989;54:1–13. [PubMed] [Google Scholar]
- 6.Matzinger P. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 7.Seong SY, Matzinger P. Nat Rev Immunol. 2004;4:469–478. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Jiang J, Li Z, Zhang J, Wang H, Qin Z. J Immunother. 2010;33:167–177. doi: 10.1097/CJI.0b013e3181bed2ba. [DOI] [PubMed] [Google Scholar]
- 9.Pike JK, Ho T, Wynne KJ. Chem Mater. 1996;8:856–860. [Google Scholar]
- 10.(a) Rose GD, Wolfenden R. Annu Rev Bioph Biom. 1993;22:381–385. doi: 10.1146/annurev.bb.22.060193.002121. [DOI] [PubMed] [Google Scholar]; (b) Chen Z, Ward R, Tian Y, Baldelli S, Opdahl A, Shen YR, Somorjai GA. J Am Chem Soc. 2000;122:10615–10620. [Google Scholar]
- 11.Lonez C, Vandenbranden M, Ruysschaert JM. Prog Lip Res. 2008;47:340–347. doi: 10.1016/j.plipres.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 12.(a) Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]; (b) Choi CHJ, Alabi CA, Webster P, Davis ME. P Natl Acad Sci USA. 2010;107:1235–1240. doi: 10.1073/pnas.0914140107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, Frangioni JV. Nat Nanotech. 2010;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Moyano DF, Rotello VM. Langmuir. 2011;27:10376–10385. doi: 10.1021/la2004535. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Miranda OR, Chen HT, You CC, Mortenson DE, Yang XC, Bunz UHF, Rotello VM. J Am Chem Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman AS. J Control Release. 2008;132:153–163. doi: 10.1016/j.jconrel.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Moyano DF, Rana S, Bunz UHF, Rotello VM. Faraday Discuss. 2011;152:33–42. doi: 10.1039/c1fd00024a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Puzyn T, Rasulev B, Gajewicz A, Hu X, Dasari TP, Michalkova A, Hwang HM, Toropov A, Leszczynska D, Leszczynski J. Nat Nanotechnol. 2011;6:175–180. doi: 10.1038/nnano.2011.10. [DOI] [PubMed] [Google Scholar]; (b) Xia XR, Monteiro-Riviere NA, Riviere JE. Nat Nanotechnol. 2011;5:671–675. doi: 10.1038/nnano.2010.164. [DOI] [PubMed] [Google Scholar]
- 17.(a) Fourches D, Pu D, Tassa C, Weissleder R, Shaw SY, Mumper RJ, Tropsha A. ACS Nano. 2010;4:5703–5712. doi: 10.1021/nn1013484. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Puzyn T, Leszczynska D, Leszczynski J. Small. 2009;5:2494–2509. doi: 10.1002/smll.200900179. [DOI] [PubMed] [Google Scholar]
- 18.Toropov AA, Leszczynska D, Leszczynski J. Comput Biol Chem. 2007;31:127–128. doi: 10.1016/j.compbiolchem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Maestro, version 8.0. Schrödinger, LLC; New York, NY: 2007. [Google Scholar]
- 20.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shebl FM, Pinto LA, García-Piñeres A, Lempicki R, Williams M, Harro C, Hildesheim A. Cancer Epidem Biomar. 2010;19:978–981. doi: 10.1158/1055-9965.EPI-10-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong AM, Premont RT, Richardson RM, Yu YRA, Lefkowitz RJ, Patel DD. P Natl Acad Sci USA. 2002;99:7478–7483. doi: 10.1073/pnas.112198299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwasaki A, Medzhitov R. Science. 2010;327:292–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matzinger P, Kamala T. Nat Rev Immunol. 2011;11:221–230. doi: 10.1038/nri2940. [DOI] [PubMed] [Google Scholar]
- 25.(a) Ma Z, Li J, He F, Wilson A, Pitt B, Li S. Biochem Bioph Res Co. 2005;330:755–759. doi: 10.1016/j.bbrc.2005.03.041. [DOI] [PubMed] [Google Scholar]; (b) Kedmi R, Ben-Arie N, Peer D. Biomaterials. 2010;31:6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 26.(a) Cho EC, Xie J, Wurm PA, Xia Y. Nano Lett. 2009;9:1080–1084. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]; (b) Arvizo RR, Miranda OR, Moyano DF, Walden CA, Giri K, Bhattacharya R, Robertson JD, Rotello VM, Reid JM, Mukherjee P. PLoS One. 2011;6:e24374. doi: 10.1371/journal.pone.0024374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(a) Zhu Z-J, Carboni R, Quercio MJ, Jr, Yan B, Miranda OR, Anderton DL, Arcaro KF, Rotello VM, Vachet RW. Small. 2010;6:2261–2265. doi: 10.1002/smll.201000989. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Leeson PD, Springthorpe B. Nat Rev Drug Discov. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- 28.Akira S, Takeda K. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 29.Kimbrell DA, Beutler B. Nat Rev Genet. 2001;2:256–267. doi: 10.1038/35066006. [DOI] [PubMed] [Google Scholar]
- 30.Kono H, Rock KL. Nat Rev Immunol. 2004;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



