Abstract
Sox11 is a high mobility group (HMG) containing transcription factor that is significantly elevated in peripheral neurons in response to nerve injury. In vitro and in vivo studies support a central role for Sox11 in adult neuron growth and survival following injury. Brain-derived neurotrophic factor (BDNF) is a pleiotropic growth factor that has effects on neuronal survival, differentiation, synaptic plasticity and regeneration. BDNF transcription is elevated in the DRG following nerve injury in parallel with Sox11 allowing for the possible regulation by Sox11. To begin to assess the possible influence of Sox11 we used reverse transcriptase PCR assays to determine the relative expression of the nine (I-IXa) noncoding exons and one coding exon (exon IX) of the BDNF gene after sciatic nerve axotomy in the mouse. Exons with upstream promoter regions containing the Sox binding motif 5′-AACAAAG-3′ (I, IV, VII and VIII) were increased at 1d or 3d following axotomy. Exons 1 and IV showed the greatest increase and only exon 1 remained elevated at 3d. Luciferase assays showed that Sox11 could activate the most highly regulated exons, I and IV, and that this activation was reduced by mutation of putative Sox binding sites. Exon expression in injured DRG neurons had some overlap with Neuro2a cells that overexpress Sox11, showing elevation in exon IV and VII transcripts. These findings indicate cell type and contextual specificity of Sox11 in modulation of BDNF transcription.
Keywords: sensory neuron, nerve regeneration, BDNF, neurotrophin, sry
INTRODUCTION
The SRY (Sex Determining Region-Y)-related HMG box transcription factor family is comprised of twenty proteins that have diverse roles in development, cell-type specification and tissue remodeling (Penzo-Mendez 2010). Sox11 is a member of the group C Sox factors, which also includes Sox4 and Sox12, and functions as a transcriptional activator (Dy et al. 2008; Penzo-Mendez 2010). In development Sox11 promotes sensory neuroblast proliferation, neuronal survival and axon outgrowth (Bergsland et al. 2006; Lin et al. 2011). Expression in adult dorsal root ganglia (DRG) sensory neurons is normally low but increases within 24h in response to peripheral nerve injury (Jankowski et al. 2006; Tanabe et al. 2003). The injury-evoked increase in Sox11 coupled with its developmental role suggests it is an important modulator of genes involved in neuronal survival and growth. Such a role is supported by studies using RNAi-mediated knockdown of Sox11 that showed increased cell death and diminished neurite growth in treated neurons (Jankowski et al. 2006; Jankowski et al. 2009).
As an injury-induced transcription factor a possible target of Sox11 is the gene encoding brain-derived neurotrophic factor (BDNF). BDNF, a member of the neurotrophin family of growth factors, is essential for the survival, differentiation, and growth of specific populations of developing sensory neurons (Ernfors et al. 1995; Ernsberger 2009; LeMaster et al. 1999; Patel and Krimm 2010; Valdes-Sanchez et al. 2010). BDNF in adult systems has both autocrine and paracrine effects. It provides homeostatic trophic support to peripheral and central neurons, modulates synaptic plasticity related to processes such as memory, learning and pain signaling (Cowansage et al. 2010; Obata and Noguchi 2006) and has a role in regeneration following nerve injury (Cho et al. 1998; Ha et al. 2001; Karchewski et al. 2002). Nerve injury evokes a rapid elevation in BDNF gene transcription (within 24h) that correlates with nerve remyelination, neurite growth and expression of regeneration-associated genes (Geremia et al. 2010; Zhang et al. 2000).
Given the intrinsic role of BDNF in neuron development, plasticity and regeneration, the regulation of BDNF gene transcription has been of significant interest (Pruunsild et al. 2011). In both human and rodent the structure of the BDNF gene is complex with expression regulated by multiple promoters in a tissue-, context- and activity-dependent manner (Kim et al. 2001; Liu et al. 2006). For example, in the rat, which has eight non-coding exons and one protein coding exon (Aid et al. 2007), the promoter linked to exon I was most highly expressed after spinal nerve ligation (Kim et al. 2001; Kobayashi et al. 2008). To examine if a similar pattern of activity occurs in the mouse we examined expression of the nine recently identified BDNF exons in lumbar DRG at early times (1d and 3d) after sciatic nerve axotomy. Given the potential transcriptional modulation of BDNF by the injury-evoked factor Sox11, we also used bioinformatics, luciferase reporter and mutagenesis assays to assess the role of Sox11 as a transcriptional modulator of the subset of exons activated in response to peripheral nerve injury.
MATERIALS AND METHODS
Animal models
Experiments were conducted using male C57BL/6J (Jackson Laboratory, Bar Harbor, ME) or Swiss Webster (Hilltop Lab Animals, Scottdale, PA) mice between 6-10 wks of age. Both strains produced similar outcomes. Animals were housed in group-cages and maintained in a temperature-controlled environment on a 12h light/dark cycle with food and water provided ad libitum. Sciatic nerve cuts were performed on the left flank of mice anesthetized and maintained under isoflurane anesthesia (Abbott Laboratories, Abbott Park, IL). Subcutaneous injections of buprenorphine were provided postoperatively. All studies were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at the University of Pittsburgh and the Guide for the Care and Use of Laboratory Animals.
RNA isolation and reverse transcriptase-PCR
DRGs at lumber spinal levels L3-L5 were rapidly removed from euthanized mice at 0d, 1d or 3d following sciatic nerve axotomy (n=4 per group). Naïve mice were used for controls. Tissue was collected in 2ml microfuge tubes on dry ice and immediately homogenized in Qiagen Buffer RLT (Qiagen, Valenia, CA) using a Polytron tissue grinder (Kinematica, Bohemia, NY). RNA was purified on RNeasy columns, reverse-transcribed using the Invitrogen cDNA kit following the manufacturer’s protocols (Invitrogen, Carlsbad, CA) and an aliquot used as template in a 25 uL SYBR green labeled PCR reaction in an ABI 7000 thermocycler (Applied Biosystems, Life Technologies, Carlsbad, CA). Ct values were normalized to GAPDH (for in vivo studies) or actin (for Neuro2a studies) mRNA levels and fold change calculated using the ddCt method. PCR primer sequences were obtained from the literature or designed using MacVector 9.0.2 software (MacVector, Inc, Cary, NC) and are listed in Table 1.
Table 1.
PCR primers used in this study.
| Primer name | Sequence 5′ → 3′ | Reference |
|---|---|---|
| BDNFpro1-408F | TTC AGA AAG AGG TTA GAG CCT G | * |
| BDNFpro1-3435R | GCA GTA AAT CCA GTG TTG CG | * |
| BDNFpro4-12192F | CAT CCT TGT CAC TCT GCT CAT CG | * |
| BDNFpro4-15036R | CCA GTC CTA GCA AAT TCA CGC AC | * |
| BDNFexon1F | CCT GCA TCT GTT GGG GAG AC | Zajac et al. |
| BDNFexon1R | GCC TTG TCC GTG GAC GTT TA | Zajac et al. |
| BDNFexon2F | CTA GCC ACC GGG GTG GTG TAA | Zajac et al. |
| BDNFexon2R | AGG ATG GTC ATC ACT CTT CTC | Zajac et al. |
| BDNFexon3F | CTT CCT TGA GCC CAG TTC C | Zajac et al. |
| BDNFexon3R | CCG TGG ACG TTT ACT TCT TTC | Zajac et al. |
| BDNFexon4F | CAG AGC AGC TGC CTT GAT GTT | Zajac et al. |
| BDNFexon4R | GCC TTG TCC GTG GAC GTT TA | Zajac et al. |
| BDNFexon5F | CCA TAA CCC CGC ACA CTC TG | * |
| BDNFexon5R | TGG TCA TCA CTC TTC TCA CCT GG | * |
| BDNFexon6F | CTG GGA GGC TTT GAT GAG AC | Zajac et al. |
| BDNFexon6R | GCC TTC ATG CAA CCG AAG TA | Zajac et al. |
| BDNFexon7F | CTT ACT TAC AGG TCC AAG GTC AAC G | * |
| BDNFexon7R | CAG AGG GTC GGA TAC AGG CTG | * |
| BDNFexon8F | TCC CAT CTA CCC ACA CAC TTT TAT G | * |
| BDNFexon8R | TGT TCG GCT CCA CTG AGG CG | * |
| BDNFexon9AF | GCC ACA TGC TGT CCC CGA G | * |
| BDNFexon9AR | GCC AAG TTG CCT TGT CCG TG | * |
| SOX11F | ATC AAG CGG CCC ATG AAC | Jankowski et al. |
| SOX11R | TGC CCA GCC TCT TGG AGA T | Jankowski et al. |
| GAPDH 756F | ATG TGT CCG TCG TGG ATC TGA | Jankowski et al. |
| GAPDH 904R | GCT GTT GAA GTC GCA GGA GAC A | Jankowski et al. |
Primers designed by authors.
Neuro2a cell culture
The mouse neuroblastoma cell line Neuro2a (ATCC clone number CCL-131, Manassas, VA) (Olmsted et al. 1970) was maintained in Eagle’s minimal essential medium (MEM) containing 10% fetal bovine serum (MEMS) and 1% penicillin/streptomycin in an incubator set at 37°C and 5% CO2.
Plasmid and HSV viral vectors
pCMV-Sox11 plasmid was made by subcloning mouse Sox11 (from IMAGE clone #63299841) into pCMV-IRES-EGFP (Jankowski et al. 2006). The IRES-EGFP sequence was removed by restriction digest to create pCMV-Sox11, which contains 166 bp of 5′UTR and 1,093 bp of 3′UTR plus the SV40 polyA cassette from the vector. pCMV-IRES-EGFP plasmid lacking the IRES-EGFP sequence was used as a control in luciferase reporter assays. BDNF exon reporter plasmids were made by inserting fragments containing either 3,027 bp upstream of BDNF exon I (pGL2BDNFpro1) or 2,844 bp upstream of exon IV (pGL2BDNFpro1) into pGL2-Basic luciferase reporter vector (Promega, Madison, WI). Sequences were first PCR cloned from C57BLK/6J mouse liver genomic DNA into the TOPO-TA cloning vector (Invitrogen). Plasmids were sequenced to confirm identity and orientation and subcloned into the pGL2-Basic vector. The Renilla luciferase expression vector pRL-TK (Promega) was used for normalizing expression.
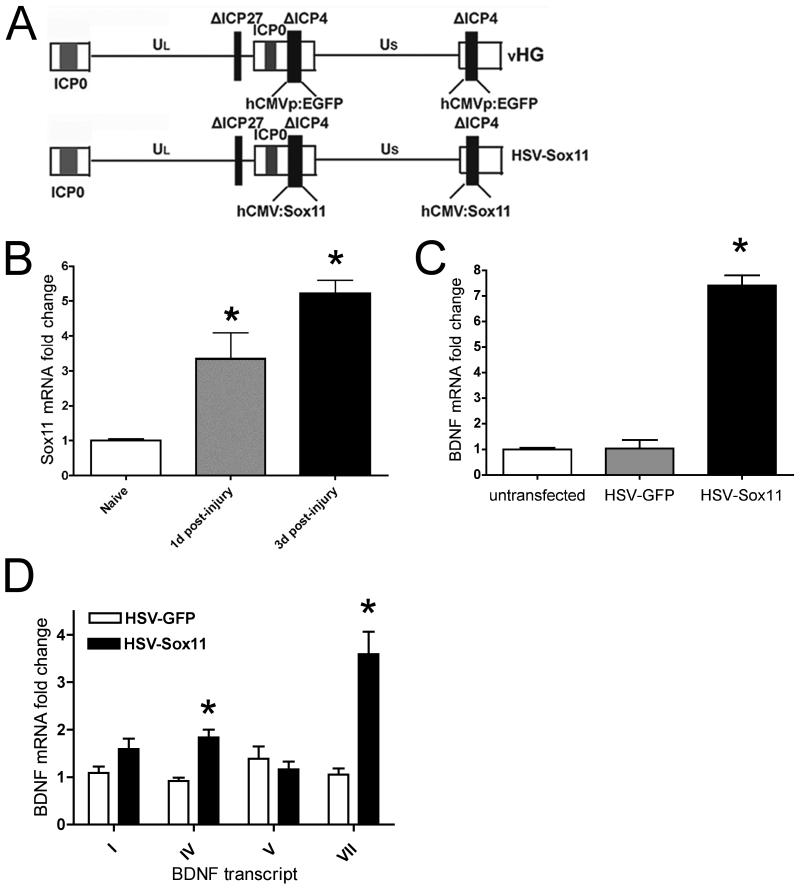
HSV constructs were generated following procedures described in (Goins et al. 2008). The control vector (HSV-GFP) contains two copies of a hCMVp:EGFP cassette targeted to the deleted (Δ) infected cell protein 4 (ICP4) loci of the backbone vector (vHG) (see Fig. 3A). An insert containing the human CMV promoter driving Sox11 (hCMV:Sox11) was also targeted to the ICP4 loci. Propagation and purification of high titer stocks of the HSV-Sox11 vector were produced as previously described (Goins et al. 2008; Goins et al. 2002) and stored at −80°C until used. Neuro2a cells were incubated for 2h with HSV vectors at a multiplicity of infection (MOI) of 2.5 after which fresh medium was added.
Figure 3.
A. Components of HSV vector used for GFP and Sox11 gene transfer. The control vHG vector contains deletions (Δ) of ICP4 and ICP27 and a hCMVp:eGFP cassette. The hCMV:Sox11 cassette was also targeted to the two ICP4 loci. B. Real time PCR analysis shows time dependent increase in Sox11 mRNA resulting from sciatic nerve axotomy. C. HSV-Sox11 infection of Neuro2a cells increases BDNF mRNA by 7-fold relative to control cultures or cells transfected with HSV-GFP. Experiments in B and C used n=4 animals. D. Fold-change in BDNF exon expression in HSV-BDNF infected Neuro2a cells. Values are relative to untransfected Neuro2a cells and normalized to actin mRNA. All data were analyzed using a two-way ANOVA with a Bonferroni post-hoc test with p < 0.05.
Luciferase assays
On the day prior to transfection Neuro2a cells were plated in 6-well plates at 2 × 105 cells per well. The culture medium was changed the next day and cells transfected using Mirus TransIT-Neural transfection reagent (Mirus Biosciences, Madison, WI). Plasmid concentrations used per well were: 1450 ng pGL-BDNF promoter vectors; 50 ng pRL-TK, 150 ng pCMV-Sox11 or empty pCMV vector. Cells were harvested 24h after transfection by lysing in Passive Lysis Buffer (Promega). Lysates were centrifuged, the supernatant removed to a clean tube and measures of fluorescence performed using a Turner 20/20n automatic luminometer with dual injectors. Normalized values (RLUpGL2/RLUpRL-TK) were used for statistical analysis using GraphPad Prism 4 software (La Jolla, CA). Each experiment was performed in triplicate and repeated at least 3 times.
Site-directed mutagenesis assays
pGL-BDNF plasmids with mutations in putative Sox11 binding sites in BDNF exons 1 or 4 were generated using the Quik-Change II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA), according to the manufacturer’s protocol. Primers used to generate mutations were: BDNFpro1mut-2927-pGL2 F: 5′- CCA TGT GAA CAC AAA CCC CGC AAA AAT CCT TCC TC -3′; R: 5′- GAG GAA GGA TTT TTG CGG GGT TTG TGT TCA CAT GG -3′. BDNFpro4mut-479-pGL2 F: 5′- GGC AAC GTT TAA CCC CGC AGC AAT CCA GG -3′; R: 5′- CCT GGA TTG CTG CGG GGT TAA ACG TTG CC -3′. Clones with mutated residues were verified by DNA sequencing by the University of Pittsburgh Genomics and Proteomics Core Laboratory. Transfection, luciferase assays and data analysis was done as described above.
RESULTS
BDNF exon expression is differentially regulated following nerve injury
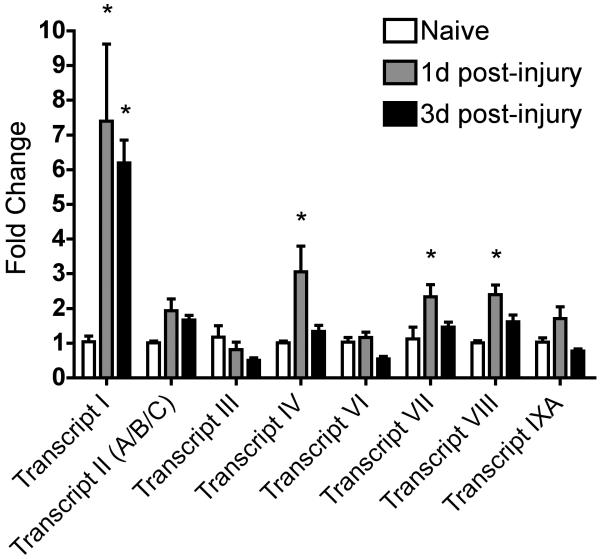
In previous studies using rat DRG neurons an increase in BDNF mRNA was measured by 1d after peripheral nerve axotomy (Karchewski et al. 2002; Kim et al. 2001). To examine this early, injury-induced rise in BDNF transcription in the mouse we used real time PCR (RT PCR) assays to determine the relative expression of BDNF exons in lumbar ganglia at 1d and 3d post sciatic nerve axotomy. The mouse BDNF gene has at least eight noncoding exons and one coding exon (exon IX; Fig. 1) (Aid et al. 2007). BDNF transcripts typically have 5′ exons spliced to the coding exon IX (although exon IX can also be independently transcribed). PCR primers described in (Zajac et al. 2010) or designed by our laboratory were used to amplify regions specific to each of these recently identified exons (Table 1). RT-PCR analysis detected transcripts for each BDNF exon, with the exception of exon V, in DRG of naïve and axotomized mice (Fig. 2). At 1d post-injury transcripts I, IV, VII, and VIII were significantly increased compared with naïve controls (Table 2). At 3d post-injury, transcripts IV, VII, and VIII returned to control levels and BDNF transcript I remained significantly elevated. BDNF transcripts II, III, VI, and IXA did not show significant change at 1d or 3d post-sciatic nerve injury.
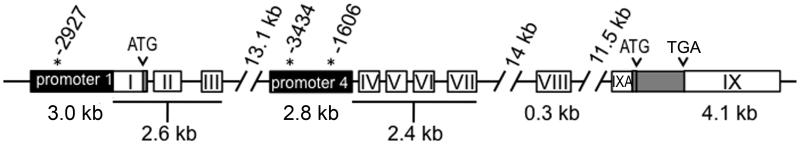
Figure 1.
Diagram of the mouse BDNF gene using nomenclature from Aid et al. (2007). Numbered exons are shown as white boxes; lines demarcate introns. Grey box indicates the coding exon with attached 3′ UTR (exon IX). Numbers indicate distance from coding region ATG start site. Numbers with asterisks indicate putative AACAAAG motif Sox binding sites within promoter regions (black boxes) of exon 1 and exon 4 used in luciferase expression assays (see Fig. 4). Figure is not drawn to scale.
Figure 2.
Plot shows relative fold-change in mRNA level of BDNF exons in lumbar DRG at 1d or 3d after sciatic nerve axotomy. n = 4 animals per time point. Asterisks indicate significance with p < 0.05 using two-way ANOVA with Bonferroni post-hoc test.
Table 2.
Fold change in transcript-specific BDNF expression following sciatic nerve injury.
| Exon | Change 1d post-injury | Change 3d post-injury | ||||
|---|---|---|---|---|---|---|
| dCt | Fold change | p-value | dCt | Fold change | p-value | |
| I | −2.50 | 5.67 | <0.001 | −2.60 | 6.09 | <0.001 |
| II | −0.87 | 1.82 | ns | −0.73 | 1.66 | ns |
| III | 0.51 | 0.70 | ns | 0.99 | 0.50 | ns |
| IV | −1.44 | 2.70 | <0.01 | −0.38 | 1.30 | ns |
| VI | −0.18 | 1.13 | ns | 0.91 | 0.53 | ns |
| VII | −1.17 | 2.25 | <0.05 | −0.53 | 1.45 | ns |
| VIII | −1.23 | 2.35 | <0.05 | −0.66 | 1.58 | ns |
| IXA | −0.70 | 1.62 | ns | 0.39 | 0.77 | ns |
ns, no significant change.
Sox11 modulates BDNF expression in an exon specific manner
As an initial assessment to determine if injury-induced expression of Sox11 (Fig. 3B) could function as a driver of BDNF transcription, a bioinformatic analysis of Sox binding sites within 5 Kbp of the transcriptional start of each BDNF exon was done using the general Sox binding motif 5′-(A/T)(A/T)CAA(A/T)G-3′ (Badis et al. 2009; Harley et al. 1994). Table 3 lists the possible Sox motifs associated with each exon. The 5′ flanking regions of all BDNF exons were found to have at least one potential Sox binding site in either forward or reverse orientations. Interestingly, only exons that have at least one 5′-AACAAAG-3′ motif (exon I, IV, VII and VIII) displayed upregulation at 1d or 3d after nerve injury.
Table 3.
Bioinformatic analysis of ~5Kbp sequence proximal to BDNF exons shows potential Sox binding sites on (+) and (−) DNA strands. Only exons with an upstream AACAAAG sequence (in bold) were expressed following nerve injury.
| Exon # | # Sox Sites (+ strand) |
Location/sequence | # Sox Sites (− strand) |
Location/sequence |
|---|---|---|---|---|
| Exon I | 3 | −2927 (AACAAAG) | 5 | −1632 (TTCAAAG) |
| −4422 (AACAATG) | −1685 (TACAAAG) | |||
| −4973 (TTCAAAG) | −2822 (TTCAAAG) | |||
| −2953 (ATCAATG) | ||||
| −4138 (ATCAAAG) | ||||
|
| ||||
| Exon II | 1 | −326 (TTCAAAG) | 0 | |
|
| ||||
| Exon III | 0 | - | 1 | −253 (TACAAAG) |
|
| ||||
| Exon IV | 5 | −172 (TACAAAG) | 4 | −1051 (AACAAAG) |
| −479 (AACAAAG) | −1397 (AACAAAG) | |||
| −1606 (AACAAAG) | −2238 (ATCAATG) | |||
| −3434 (AACAAAG) | −3514 (AACAATG) | |||
| −3915 (TTCAAAG) | ||||
|
| ||||
| Exon V | All Sox sites on 5kb promoter also upstream of Exon IV | |||
|
| ||||
| Exon VI | All Sox sites on 5kb promoter also upstream of Exon IV | |||
|
| ||||
| Exon VII | 4 | −550 (AACAATG) | 4 | −649 (ATCAAAG) |
| −2145 (TACAAAG) | −3024 (AACAAAG) | |||
| −2452 (AACAAAG) | −3190 (AACAAAG) | |||
| −4280 (AACAAAG) | −4157 (ATCAATG) | |||
|
| ||||
| Exon VIII |
5 | −862 (AACAAAG) | 2 | −2468 (TTCAAAG) |
| −1872 (TACAAAG) | −3333 (TTCAATG) | |||
| −3071 (TTCAATG) | ||||
| −3951 (TACAAAG) | ||||
| −4539 (TTCAAAG) | ||||
|
| ||||
| Exon IXA |
4 | −416 (ATCAAAG) | 4 | −303 (AACAATG) |
| −3020 (TTCAAAG) | −371 (ATCAATG) | |||
| −4626 (TACAATG) | −560 (TACAAAG) | |||
| −4760 (ATCAAAG) | −2006 (TTCAAAG) | |||
Overexpression of Sox11 increases expression of BDNF exon IV and VII in Neuro2a cells
Regulation of BDNF exon expression by Sox11 was then examined by assaying transcript expression in Neuro2a cells that overexpressed Sox11 protein. Cells were treated with nonreplicating herpes simplex virus (HSV) vectors that express Sox11 (HSV-Sox11) or green fluorescent protein (HSV-GFP) (Fig. 3A). Exon activation was assessed using RT-PCR measures of total BDNF (exon IX; Fig. 3C) and individual BDNF transcript levels. At 24h post-infection a 7-fold increase in total BDNF mRNA level was measured. At the exon level an increase in transcripts containing exons IV and VII was measured relative to cells infected with the control HSV-GFP vector (Fig. 3D). Transcripts containing exons II, III, VI, VIII and IXA were expressed at very low levels and not reliably detected.
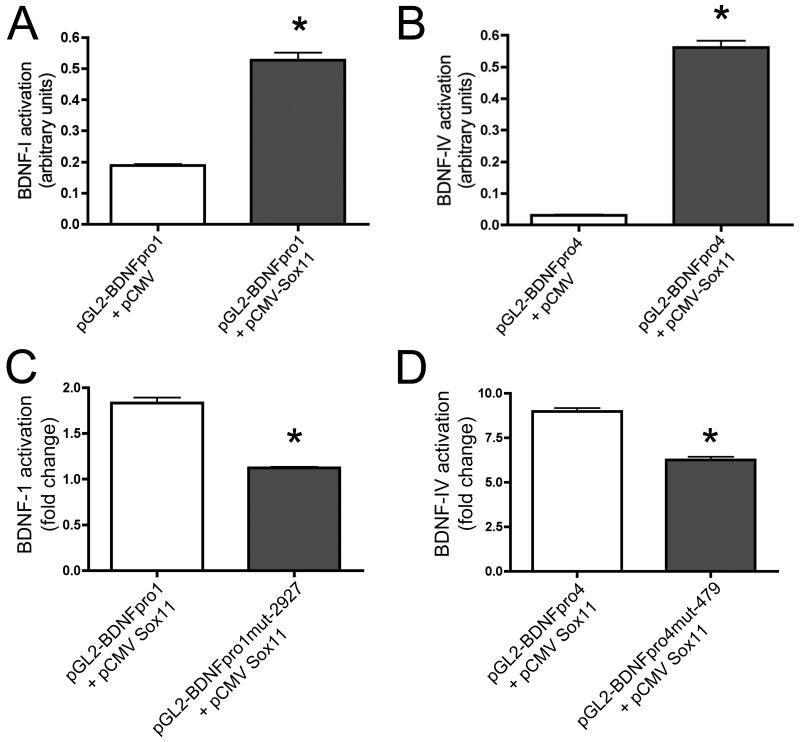
To determine if Sox11 could drive individual exon expression, luciferase reporter assays in Neuro2a cells were carried out. Promoter regions upstream of exons I and IV were tested since these exons showed the greatest increase in response to axotomy (Fig. 2). The pGL2BDNFpro1 reporter plasmid, containing a 3,027 bp insert corresponding to sequence immediately upstream of mouse BDNF exon I, and pGL2BDNFpro4, with 2,844 bp of sequence immediately upstream of mouse BDNF exon IV, were tested (Fig. 1). Cotransfection of pGL2-BDNFproI with pCMV control vector or pCMV-Sox11 showed exon 1 can weakly drive luciferase expression alone (Fig. 4A) and that pCMV-Sox11 increased activation 2.5-fold over baseline. Interestingly, a significantly greater 23-fold increase in promoter activation was determined in cells cotransfected with pCMV-Sox11 and pGL2-BDNFpro4 plasmids (Fig. 4B), suggesting a much stronger interaction of Sox11 with the exon 4 promoter region. To further investigate these interactions we repeated the luciferase assays using exon promoter-luciferase plasmids that contained mutagenized Sox sites (pGL2-BDNFpro1mut-2927 and pGL-BDNFpro4mut-479). Site-directed mutagenesis of core residues of the single AACAAAG site in exon 1 (nt-2927; see Table 3) significantly reduced luciferase activity in Neuro2a cells relative to cells transfected with the wildtype pGL2-BDNFpro1 plasmid (Fig. 4C). Mutation of one of the two AACAAAG sites in the exon 4 promoter construct (nt-479) also produced a significant reduction in reporter activity (Fig. 4D). These findings suggest Sox11 directly activates BDNF transcription and that disruption of even a single Sox site can impair promoter activation.
Figure 4.
Plot shows increase in luciferase activity in Neuro2a cultures cotransfected with pCMV-Sox11 and luciferase reporter plasmids pGL2-BDNFpro1 (A) containing 3.8 Kbp of sequence upstream of exon 1 or pGL2-BDNFpro4, (B) containing 2.9 Kbp upstream of exon 4. For both experiments n=3 and p<0.0001 using Student’s t-test. Mutation of one of the putative AACAAAG Sox sites in the promoter of exon 1 (C) or exon 4 (D) reduced Sox11 stimulated reporter gene expression. Data were analyzed using a 2-way ANOVA with a Bonferroni post hoc test; asterisks indicate p<0.005 and n=3.
DISCUSSION
This study determined the pattern of BDNF transcript expression in the mouse dorsal root ganglia at 1d and 3d following sciatic nerve axotomy. Several reports have shown that elevation of BDNF expression (up to 83% of the injured neuron population) occurs within one day after peripheral nerve injury (Cho et al. 1998; Ernfors et al. 1993; Karchewski et al. 2002; Michael et al. 1999; Sebert and Shooter 1993; Tonra et al. 1998). This increase appears across all neuron subtypes, i.e., sciatic nerve axotomy induces an expansion in BDNF mRNA expression across small, medium and large diameter neurons that are trkA-, trkB- and trkC-positive (Karchewski et al. 2002; Michael et al. 1999). The rapid expression of BDNF and its persistent elevation for up to 4 weeks post axotomy (Michael et al. 1999) indicates it has a key role in regulating both early and late injury responses.
In the mouse we found that an increase in noncoding exon expression on day 1 post injury occurred in a differential manner with increases only in exons I, IV, VII and VIII. Exons I and IV showed the greatest change consistent with studies in rat that used multiple nerve injury and inflammation models (Kobayashi et al. 2008). These studies in rat also assayed expression of newly identified BDNF transcripts (Aid et al. 2007), and showed that at two weeks after L5-selective spinal nerve ligation (a model of chronic nerve injury), total BDNF mRNA and transcripts associated with exons I, II, III, IV, VI, and IXA increase in the L4 DRG. However, in the L5 DRG total BDNF mRNA level was unchanged, BDNF exon I was increased and exon IV was decreased (Kobayashi et al. 2008). It was also found that at 2d after complete Freund’s adjuvant (CFA)-induced footpad inflammation, exons I, II, III, IV, V, VIII and IXA increase in the L5 DRG. In an in vitro analysis (Matsuoka et al. 2007), cultures of rat DRG neurons grown with NGF (which increases following nerve injury), caused an increase in exons I, II, III, IV, VI, and IX, supporting the role of NGF as a regulator of BDNF transcription (Apfel et al. 1996). Furthermore, exon VI (in the current terminology) was dramatically upregulated in the distal sciatic nerve following axotomy (Funakoshi et al. 1993; Timmusk et al. 1995). Collectively these studies indicate that injury or inflammation induce BDNF gene transcription via activity of multiple exons in a time and spatially dependent manner. In the present study we measured increased transcription for exons I, IV, VII and VIII at 1d following axotomy and found that this activation was transient, declining by day 3 after injury. Consistent with previous studies that found exon I persistently upregulated following injury, this exon showed the most robust increase on day 1 with sustained expression on day 3. Therefore, activation of exon I most likely accounts for the observed rise in BDNF in the DRG at later days following injury (Geremia et al. 2007; Karchewski et al. 2002). Interestingly, recent studies in rat hippocampal neurons show that transcripts containing exon I (or exon IV) are restricted to cell somata and proximal dendrites, whereas all other transcripts are dendritically-targeted (Chiaruttini et al. 2009). Whether a similar distribution occurs in sensory neurons is unclear.
Another goal of this study was to examine a possible role for Sox11 in regulating injury-induced BDNF gene expression. Temporally, the rise in BDNF transcription by 1d following injury overlaps with the increase in Sox11 in injured DRG neurons (this study and (Jankowski et al. 2006)). In addition, overexpression of Sox11 in Neuro2a cells increased mRNAs associated with promoter regions linked to exons IV and VII, a pattern that partially overlaps with exons activated in the DRG in response to injury (exons I, IV, VII and VIII). Luciferase reporter assays showed Sox11 increased exon I promoter activity 2.5-fold and exon IV promoter activity 23-fold. Mutagenesis of the single AACAAAG Sox site in the exon 1 reporter blocked activation whereas mutation of one of the two AACAAAG sites in the exon IV reporter caused only a moderate reduction in activity. Collectively, these data suggest that Sox11 may be necessary but not sufficient for expression of exon 1, whereas it may be sufficient but not necessary for expression of exon IV.
Several questions remain concerning the mechanism of Sox11 regulation of BDNF. For example, luciferase assays indicate a Sox11-dependent activation of exon I, a promoter strongly upregulated by injury but unchanged by Sox11 transfection. This suggests exon I transcription is regulated by additional elements not included in our reporter system. Another possibility is that Sox11 may only upregulate the exon I promoter in the context of the injured cell. Injury signaling and neuronal regeneration involves epigenetic modification and transcriptional regulation of a wide array of genes including transcription factors (Jacob et al. 2011; Michaelevski et al. 2010; Qureshi and Mehler 2011; Raivich and Makwana 2007; Stam et al. 2007; Uchida et al. 2010). Therefore, it is possible that injury signaling may be required for the expression or recruitment of binding partners of Sox11, or for the specific activation of BDNF exon I which is frequently epigenetically silenced (Aid et al. 2007; Hara et al. 2009; Tian et al. 2009). Although more detailed analysis of Sox11 regulation is certainly required, these data provide a foundation for further investigation of Sox11 regulation of BDNF transcription.
Other transcription factors have been shown to regulate BDNF expression in an exon and stimulus specific manner. These include the cAMP-responsive element binding protein (CREB; (Greenberg et al. 2009; Pruunsild et al. 2011; Shieh et al. 1998; Tabuchi et al. 2002)), calcium-responsive transcription factor (CaRF; (Tao et al. 2002)), aryl hydrocarbon receptor nuclear translocator 2 (ARNT2)(Pruunsild et al. 2011) as well as transcriptional repressor proteins, e.g., REST/NRSF (Zuccato et al. 2003) and methyl-CpG binding protein-2 (MeCP2), which promote transcription following release from the BDNF sequence (Chen et al. 2003; Martinowich et al. 2003). Whether Sox11 interacts with these other regulators is unknown. Sox factors are known to work in multicomponent regulatory complexes that include POU and Oct proteins (Lefebvre et al. 2007; Tanaka et al. 2004; Wiebe et al. 2003). Binding of the Sox11 HMG domain to DNA is also predicted to induce conformational changes whereby the major groove is compressed, the minor groove widens and the DNA helix is bent. These architectural changes could increase accessibility of the DNA to other DNA-binding proteins. Thus, it is possible that Sox11-mediated activation of promoters, including the BDNF promoters, may be important to the initiation of long-term epigenetic regulatory mechanisms that coordinate the transcriptional response to injury in neurons. Further analysis of Sox11 interactions on the BDNF gene is required to assess this possibility as well as identify possible coactivators.
The ability of BDNF to enhance neuronal regeneration is likely due to several downstream effects, including stimulation of intrinsic growth pathways (Geremia et al. 2010), increased mRNA trafficking to neurites (Chiaruttini et al. 2009; Willis et al. 2005) and promotion of Schwann cell myelination (Ng et al. 2007). BDNF upregulation also mediates the pro-regenerative effects of some experimental treatments, such as low-intensity electrical stimulation after injury (Geremia et al. 2007). However, BDNF is also anterogradely transported to the dorsal horn of the spinal cord following injury (Michael et al. 1997; Tonra et al. 1998), where it promotes synaptic plasticity and contributes to chronic pain signaling (Obata and Noguchi 2006). Thus, elevation in Sox11 in response to persistent nerve injury could lead to long-term activation of BDNF transcription and contribute to chronic pain. The relationship between Sox11, BDNF expression and pain hypersensitivity are issues that remain to be investigated.
ACKNOWLEDGEMENTS
We thank Dr. Ting Wang and Christopher Sullivan for technical advice and assistance and professor John C. Salerno (Kennesaw State University, Kennesaw, GA) for providing expertise and assistance in site-directed mutagenesis studies.
This work was supported by NINDS grant #NS059003 to KMA.
REFERENCES
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85(3):525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfel SC, Wright DE, Wiideman AM, Dormia C, Snider WD, Kessler JA. Nerve growth factor regulates the expression of brain-derived neurotrophic factor mRNA in the peripheral nervous system. Mol Cell Neurosci. 1996;7(2):134–142. doi: 10.1006/mcne.1996.0010. [DOI] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CF, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324(5935):1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20(24):3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci U S A. 2009;106(38):16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Kim JK, Park HC, Kim DS, Ha SO, Hong HS. Changes in brain-derived neurotrophic factor immunoreactivity in rat dorsal root ganglia, spinal cord, and gracile nuclei following cut or crush injuries. Exp Neurol. 1998;154(1):224–230. doi: 10.1006/exnr.1998.6936. [DOI] [PubMed] [Google Scholar]
- Cowansage KK, LeDoux JE, Monfils MH. Brain-derived neurotrophic factor: a dynamic gatekeeper of neural plasticity. Curr Mol Pharmacol. 2010;3(1):12–29. doi: 10.2174/1874467211003010012. [DOI] [PubMed] [Google Scholar]
- Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36(9):3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P, Kucera J, Lee KF, Loring J, Jaenisch R. Studies on the physiological role of brain-derived neurotrophic factor and neurotrophin-3 in knockout mice. Int J Dev Biol. 1995;39(5):799–807. [PubMed] [Google Scholar]
- Ernfors P, Rosario CM, Merlio JP, Grant G, Aldskogius H, Persson H. Expression of mRNAs for neurotrophin receptors in the dorsal root ganglion and spinal cord during development and following peripheral or central axotomy. Brain Res Mol Brain Res. 1993;17(3-4):217–226. doi: 10.1016/0169-328x(93)90005-a. [DOI] [PubMed] [Google Scholar]
- Ernsberger U. Role of neurotrophin signalling in the differentiation of neurons from dorsal root ganglia and sympathetic ganglia. Cell Tissue Res. 2009;336(3):349–384. doi: 10.1007/s00441-009-0784-z. [DOI] [PubMed] [Google Scholar]
- Funakoshi H, Frisen J, Barbany G, Timmusk T, Zachrisson O, Verge VM, Persson H. Differential expression of mRNAs for neurotrophins and their receptors after axotomy of the sciatic nerve. J Cell Biol. 1993;123(2):455–465. doi: 10.1083/jcb.123.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia NM, Gordon T, Brushart TM, Al-Majed AA, Verge VM. Electrical stimulation promotes sensory neuron regeneration and growth-associated gene expression. Exp Neurol. 2007;205(2):347–359. doi: 10.1016/j.expneurol.2007.01.040. [DOI] [PubMed] [Google Scholar]
- Geremia NM, Pettersson LM, Hasmatali JC, Hryciw T, Danielsen N, Schreyer DJ, Verge VM. Endogenous BDNF regulates induction of intrinsic neuronal growth programs in injured sensory neurons. Exp Neurol. 2010;223(1):128–142. doi: 10.1016/j.expneurol.2009.07.022. [DOI] [PubMed] [Google Scholar]
- Goins WF, Krisky DM, Wechuck JB, Huang S, Glorioso JC. Construction and production of recombinant herpes simplex virus vectors. Methods Mol Biol. 2008;433:97–113. doi: 10.1007/978-1-59745-237-3_6. [DOI] [PubMed] [Google Scholar]
- Goins WF, Krisky DM, Wolfe DP, Fink DJ, Glorioso JC. Development of replication-defective herpes simplex virus vectors. Methods Mol Med. 2002;69:481–507. doi: 10.1385/1-59259-141-8:481. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Xu B, Lu B, Hempstead BL. New insights in the biology of BDNF synthesis and release: implications in CNS function. J Neurosci. 2009;29(41):12764–12767. doi: 10.1523/JNEUROSCI.3566-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SO, Kim JK, Hong HS, Kim DS, Cho HJ. Expression of brain-derived neurotrophic factor in rat dorsal root ganglia, spinal cord and gracile nuclei in experimental models of neuropathic pain. Neuroscience. 2001;107(2):301–309. doi: 10.1016/s0306-4522(01)00353-0. [DOI] [PubMed] [Google Scholar]
- Hara D, Miyashita T, Fukuchi M, Suzuki H, Azuma Y, Tabuchi A, Tsuda M. Persistent BDNF exon I-IX mRNA expression following the withdrawal of neuronal activity in neurons. Biochem Biophys Res Commun. 2009;390(3):648–653. doi: 10.1016/j.bbrc.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22(8):1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Lebrun-Julien F, Suter U. How Histone Deacetylases Control Myelination. Mol Neurobiol. 2011 doi: 10.1007/s12035-011-8198-9. [DOI] [PubMed] [Google Scholar]
- Jankowski MP, Cornuet PK, McIlwrath S, Koerber HR, Albers KM. SRY-box containing gene 11 (Sox11) transcription factor is required for neuron survival and neurite growth. Neuroscience. 2006;143(2):501–514. doi: 10.1016/j.neuroscience.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski MP, McIlwrath SL, Jing X, Cornuet PK, Salerno KM, Koerber HR, Albers KM. Sox11 transcription factor modulates peripheral nerve regeneration in adult mice. Brain Res. 2009;1256:43–54. doi: 10.1016/j.brainres.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchewski LA, Gratto KA, Wetmore C, Verge VM. Dynamic patterns of BDNF expression in injured sensory neurons: differential modulation by NGF and NT-3. Eur J Neurosci. 2002;16(8):1449–1462. doi: 10.1046/j.1460-9568.2002.02205.x. [DOI] [PubMed] [Google Scholar]
- Kim DS, Lee SJ, Cho HJ. Differential usage of multiple brain-derived neurotrophic factor promoter in rat dorsal root ganglia following peripheral nerve injuries and inflammation. Brain Res Mol Brain Res. 2001;92(1-2):167–171. doi: 10.1016/s0169-328x(01)00154-1. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Yokoyama M, Matsuoka Y, Omori M, Itano Y, Kaku R, Morita K, Ichikawa H. Expression changes of multiple brain-derived neurotrophic factor transcripts in selective spinal nerve ligation model and complete Freund’s adjuvant model. Brain Res. 2008;1206:13–19. doi: 10.1016/j.brainres.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39(12):2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaster AM, Krimm RF, Davis BM, Noel T, Forbes ME, Johnson JE, Albers KM. Overexpression of brain-derived neurotrophic factor enhances sensory innervation and selectively increases neuron number. J Neurosci. 1999;19(14):5919–5931. doi: 10.1523/JNEUROSCI.19-14-05919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Lee VM, Wang Y, Lin JS, Sock E, Wegner M, Lei L. Sox11 regulates survival and axonal growth of embryonic sensory neurons. Dev Dyn. 2011 doi: 10.1002/dvdy.22489. [DOI] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067(1):1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Yokoyama M, Kobayashi H, Omori M, Itano Y, Morita K, Mori H, Nakanishi T. Expression profiles of BDNF splice variants in cultured DRG neurons stimulated with NGF. Biochem Biophys Res Commun. 2007;362(3):682–688. doi: 10.1016/j.bbrc.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Michael GJ, Averill S, Nitkunan A, Rattray M, Bennett DL, Yan Q, Priestley JV. Nerve growth factor treatment increases brain-derived neurotrophic factor selectively in TrkA-expressing dorsal root ganglion cells and in their central terminations within the spinal cord. J Neurosci. 1997;17(21):8476–8490. doi: 10.1523/JNEUROSCI.17-21-08476.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael GJ, Averill S, Shortland PJ, Yan Q, Priestley JV. Axotomy results in major changes in BDNF expression by dorsal root ganglion cells: BDNF expression in large trkB and trkC cells, in pericellular baskets, and in projections to deep dorsal horn and dorsal column nuclei. Eur J Neurosci. 1999;11(10):3539–3551. doi: 10.1046/j.1460-9568.1999.00767.x. [DOI] [PubMed] [Google Scholar]
- Michaelevski I, Segal-Ruder Y, Rozenbaum M, Medzihradszky KF, Shalem O, Coppola G, Horn-Saban S, Ben-Yaakov K, Dagan SY, Rishal I, Geschwind DH, Pilpel Y, Burlingame AL, Fainzilber M. Signaling to transcription networks in the neuronal retrograde injury response. Sci Signal. 2010;3(130):ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng BK, Chen L, Mandemakers W, Cosgaya JM, Chan JR. Anterograde transport and secretion of brain-derived neurotrophic factor along sensory axons promote Schwann cell myelination. J Neurosci. 2007;27(28):7597–7603. doi: 10.1523/JNEUROSCI.0563-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata K, Noguchi K. BDNF in sensory neurons and chronic pain. Neurosci Res. 2006;55(1):1–10. doi: 10.1016/j.neures.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Olmsted JB, Carlson K, Klebe R, Ruddle F, Rosenbaum J. Isolation of microtubule protein from cultured mouse neuroblastoma cells. Proc Natl Acad Sci U S A. 1970;65(1):129–136. doi: 10.1073/pnas.65.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AV, Krimm RF. BDNF is required for the survival of differentiated geniculate ganglion neurons. Dev Biol. 2010;340(2):419–429. doi: 10.1016/j.ydbio.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42(3):425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T. Identification of cis-Elements and Transcription Factors Regulating Neuronal Activity-Dependent Transcription of Human BDNF Gene. J Neurosci. 2011;31(9):3295–3308. doi: 10.1523/JNEUROSCI.4540-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. The emerging role of epigenetics in stroke: III. Neural stem cell biology and regenerative medicine. Arch Neurol. 2011;68(3):294–302. doi: 10.1001/archneurol.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G, Makwana M. The making of successful axonal regeneration: genes, molecules and signal transduction pathways. Brain Res Rev. 2007;53(2):287–311. doi: 10.1016/j.brainresrev.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Sebert ME, Shooter EM. Expression of mRNA for neurotrophic factors and their receptors in the rat dorsal root ganglion and sciatic nerve following nerve injury. J Neurosci Res. 1993;36(4):357–367. doi: 10.1002/jnr.490360402. [DOI] [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20(4):727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Stam FJ, MacGillavry HD, Armstrong NJ, de Gunst MC, Zhang Y, van Kesteren RE, Smit AB, Verhaagen J. Identification of candidate transcriptional modulators involved in successful regeneration after nerve injury. Eur J Neurosci. 2007;25(12):3629–3637. doi: 10.1111/j.1460-9568.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Sakaya H, Kisukeda T, Fushiki H, Tsuda M. Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I. The Journal of biological chemistry. 2002;277(39):35920–35931. doi: 10.1074/jbc.M204784200. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Bonilla I, Winkles JA, Strittmatter SM. Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J Neurosci. 2003;23(29):9675–9686. doi: 10.1523/JNEUROSCI.23-29-09675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Molecular and cellular biology. 2004;24(20):8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, West AE, Chen WG, Corfas G, Greenberg ME. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33(3):383–395. doi: 10.1016/s0896-6273(01)00561-x. [DOI] [PubMed] [Google Scholar]
- Tian F, Hu XZ, Wu X, Jiang H, Pan H, Marini AM, Lipsky RH. Dynamic chromatin remodeling events in hippocampal neurons are associated with NMDA receptor-mediated activation of Bdnf gene promoter 1. J Neurochem. 2009;109(5):1375–1388. doi: 10.1111/j.1471-4159.2009.06058.x. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Lendahl U, Funakoshi H, Arenas E, Persson H, Metsis M. Identification of brain-derived neurotrophic factor promoter regions mediating tissue-specific, axotomy-, and neuronal activity-induced expression in transgenic mice. J Cell Biol. 1995;128(1-2):185–199. doi: 10.1083/jcb.128.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonra JR, Curtis R, Wong V, Cliffer KD, Park JS, Timmes A, Nguyen T, Lindsay RM, Acheson A, DiStefano PS. Axotomy upregulates the anterograde transport and expression of brain-derived neurotrophic factor by sensory neurons. J Neurosci. 1998;18(11):4374–4383. doi: 10.1523/JNEUROSCI.18-11-04374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida H, Sasaki K, Ma L, Ueda H. Neuron-restrictive silencer factor causes epigenetic silencing of Kv4.3 gene after peripheral nerve injury. Neuroscience. 2010;166(1):1–4. doi: 10.1016/j.neuroscience.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Valdes-Sanchez T, Kirstein M, Perez-Villalba A, Vega JA, Farinas I. BDNF is essentially required for the early postnatal survival of nociceptors. Dev Biol. 2010;339(2):465–476. doi: 10.1016/j.ydbio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Wiebe MS, Nowling TK, Rizzino A. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. The Journal of biological chemistry. 2003;278(20):17901–17911. doi: 10.1074/jbc.M212211200. [DOI] [PubMed] [Google Scholar]
- Willis D, Li KW, Zheng JQ, Chang JH, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25(4):778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac MS, Pang TY, Wong N, Weinrich B, Leang LS, Craig JM, Saffery R, Hannan AJ. Wheel running and environmental enrichment differentially modify exon-specific BDNF expression in the hippocampus of wild-type and pre-motor symptomatic male and female Huntington’s disease mice. Hippocampus. 2010;20(5):621–636. doi: 10.1002/hipo.20658. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Luo XG, Xian CJ, Liu ZH, Zhou XF. Endogenous BDNF is required for myelination and regeneration of injured sciatic nerve in rodents. Eur J Neurosci. 2000;12(12):4171–4180. [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35(1):76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]