Abstract
In recent years, glycated hemoglobin (HbA1c) has been increasingly accepted as a functional metric of mean blood glucose in the treatment of diabetic patients. Importantly, HbA1c provides an alternate measure of total glycemic exposure due to the representation of blood glucose throughout the day, including post-prandially. In this article, we propose and demonstrate the potential of Raman spectroscopy as a novel analytical method for quantitative detection of HbA1c, without using external dyes or reagents. Using the drop coating deposition Raman (DCDR) technique, we observe that the non-enzymatic glycosylation (glycation) of the hemoglobin molecule results in subtle but discernible and highly reproducible changes in the acquired spectra, which enable the accurate determination of glycated and non-glycated hemoglobin using standard chemometric methods. The acquired Raman spectra display excellent reproducibility of spectral characteristics at different locations in the drop and show a linear dependence of the spectral intensity on the analyte concentration. Furthermore, in hemolysate models, the developed multivariate calibration models for HbA1c show a high degree of prediction accuracy and precision - with a limit of detection that is nearly a factor of 15 smaller than the lowest physiological concentrations encountered in clinical practice. The excellent accuracy and reproducibility achieved in this proof-of-concept study opens substantive avenues for characterization and quantification of the glycosylation status of (therapeutic) proteins, which are widely used for biopharmaceutical development. We also envision that the proposed approach can provide a powerful tool for high-throughput HbA1c sensing in multi-component mixtures and potentially in hemolysate and whole blood lysate samples.
INTRODUCTION
Diabetes mellitus, characterized by the defective regulation of blood glucose, is the most common disorder of the endocrine system affecting approximately 25.8 million people in the US alone (2010).1,2 Given the lack of suitable therapeutic options, effective glycemic control is imperative in avoiding acute and chronic complications such as diabetic coma, and microvascular and macrovascular complications.3 To this end, several research groups, including our own laboratory, have pursued the development of a non-invasive blood glucose sensor using a wide variety of optical and spectroscopic modalities.4–12 Nevertheless, these attempts to monitor glucose levels directly through Raman, infrared or other optical modalities have faced significant challenges and none have been translated to clinical practice. Given this scenario, glycated hemoglobin (HbA1c) presents a new promising target for performing long-term diabetes monitoring. While monitoring blood glucose remains the gold standard for continuous monitoring and evaluation of treatment options, HbA1c has gained approval in the medical community in assessing the long-term history of glycemic control.13, 14 HbA1c is formed by the non-enzymatic glycosylation (glycation) of hemoglobin exposed to blood glucose15 and therefore has a strong correlation with the average glucose concentrations in the bloodstream in the preceding three month period (life span of the erythrocytes).16 Due to this strong correlation, HbA1c levels have been regularly used for monitoring long-term glucose control in established diabetics and has been recently approved for screening for diabetes (HbA1c≥6.5%) and pre-diabetes (5.7%≤HbA1c≤6.4%) in the US.17
Presently, HbA1c is distinguished from non-glycated hemoglobin using assay techniques such as high-performance liquid chromatography (HPLC), isoelectric focusing and immunoassay. However, the presence of hemoglobin variants and other clinical factors such as uremia may interfere with HbA1c determinations.18 As an alternate method for HbA1c detection, Ishikawa and co-workers have recently reported the application of surface enhanced resonance Raman spectroscopy (SERRS).19 Though promising in approach, precise quantification of the analyte of interest (HbA1c) using SERSS is difficult (as also noted by the authors) due to poor spectral reproducibility and generation of spurious background signals.20
Here, we propose and demonstrate, for the first time to the best of our knowledge, the application of drop coating deposition Raman (DCDR) spectroscopy for the sensitive and selective detection of HbA1c, without addition of any exogenous dyes or reagents. Previously, pioneering studies by Ben-Amotz and co-workers have shown that DCDR provides significant signal amplification by pre-concentration of the analytes of interest.21, 22 In DCDR, spectra are acquired from the coffee-ring pattern of analytes deposited from a drying drop, which is a formed as a result of the interplay of contact line pinning, solvent evaporation and capillary flow.23 Evidently, the coffee ring pattern results in significant constituent pre-concentration and has been employed by several recent investigators, notably Stone and Morris research groups, to provide strong and reproducible Raman signals for different bio-analytes without considerable loss of their solution conformation.24–27 It is worth noting that analogous thin film studies for blood analytes using infrared absorption spectroscopy have also been performed (for example, see the works of Petrich, Shaw and co-workers28, 29).
In this article, we show that glycation of hemoglobin results in subtle changes in the acquired Raman spectra that facilitate the accurate classification of glycated and non-glycated (pure) hemoglobin using conventional multivariate techniques. On close examination, the acquired Raman spectra reveal excellent reproducibility of spectral characteristics at different locations in the coffee-ring pattern and show a linear response (between the spectral intensity and the analyte concentration). Furthermore, in hemolysate models comprising of both non-glycated and glycated hemoglobin components, the developed multivariate calibration models for HbA1c show a high degree of prediction accuracy and precision - with a limit of detection that is nearly a factor of 15 smaller than the lowest physiological concentrations encountered in clinical practice. Given the promising sensitivity, linearity of response and rapid measurement possibility, this method should provide an ideal complement for the gold standard analytical/diagnostic techniques used for detection of glycated proteins. The proposed approach can be readily extended to quantification of glycation status of proteins in mixtures, which is of great significance because such modification (glycosylation) can alter the stability, pharmacokinetics and immunogenicity of glycoprotein-based biopharmaceuticals. Independently, it will also provide a means of high-throughput HbA1c sensing in multi-component mixtures and, in the future, in hemolysates and whole blood lysates.
MATERIALS AND METHODS
Our present goal is to establish the foundation of Raman spectroscopy for quantitative HbA1c measurements as a route to expanding the range of Raman-based analytical methods. To this end, experimental studies were undertaken to accomplish the following specific objectives: (1) evaluate the ability to distinguish between Hb and HbA1c samples (i.e. the specificity of our approach); (2) quantify the prediction accuracy and precision of these analytes in two-component mixture samples; and (3) investigate the reproducibility of the measurements in mixture samples from the resultant dried coffee-ring patterns. To fulfill objective (1), we performed spectroscopic measurements on single protein drop-coated samples, namely those obtained from the aqueous solutions of Hb and HbA1c. This study was employed to investigate the spectral differences, if any, and the possibility of exploiting these differences for selective fingerprinting of the glycemic marker. In order to accomplish objective (2), two-component mixtures (i.e. mixtures containing both Hb and HbA1c in different proportions) were studied. This system represents a hemolysate model, the product resulting from the lysis of erythrocytes, and adds an important layer of complexity to studies in single protein or protein/buffer solutions. Moreover, it provides an ideal platform to study some of the intricacies of spectroscopic measurements prior to analyzing more complex, multi-component mixtures. Finally, to study objective (3), 2D spatial Raman mapping was performed on the drop-coated two-component mixture samples.
Experimental
The Raman system used for these experiments was originally described in one of our laboratory’s previous publications.30 Briefly, a tunable CW Ti:Sapphire laser (3900S, Spectra-Physics) set at 785 nm excitation wavelength was used for the experiment. The Ti:Sapphire laser was pumped using a frequency-doubled Nd:YAG laser (Millennia 5sJ, Spectra-Physics). The Raman back-scattered light from the sample was transmitted by a multimode fiber to a spectrograph (Kaiser Holospec f/1.8i) and a liquid nitrogen-cooled CCD (LN/CCD 1340/400-EB, Roper Scientific). The laser power was measured at the sample to be ca. 3 mW. A water immersion objective lens (Olympus UPLSAPO60XWIR 60X/1.20) was used to both focus the laser to a spot size of approximately 1 μm at the drop-coated substrate surface and to collect the back-scattered Raman light. It is worth noting that although the pumped Ti:Sapphire laser was used here as a part of our regular Raman microscope setup, a simpler laser such as a stabilized diode system can also adequately function as an excitation source for the purpose of these investigations.
Hemoglobin powder (in lyophilized form) was obtained from Sigma-Aldrich (St. Louis, MO, USA) and frozen HbA1c liquid from Lee Biosolutions, Inc. (St. Louis, MO, USA). For the single protein samples, aqueous solutions of hemoglobin were prepared in the range of 10–155 μM (the typical physiological range is between 1.55–2.7 mM or, ca. 10–17.5 g/dL. Similarly, HbA1c samples were formulated with concentrations in the range of 4–34 μM (typical physiological values are between 4 and 25% of the aforementioned hemoglobin values with the critically important range being 5–10%). For all sample preparations, PESTANAL water (34478, Fluka) was used to ensure higher reproducibility of the measurements.22, 24 For the two-component mixture study, 16 total samples were formulated by pipetting different volumes of Hb and HbA1c from their respective stock solutions. The final concentration ranges for Hb and HbA1c in the two-component mixture models were ca. 2–100 μM and 5–25 μM, respectively (with the minimum glycated hemoglobin percentage in these hemolysate models being ca. 5%).
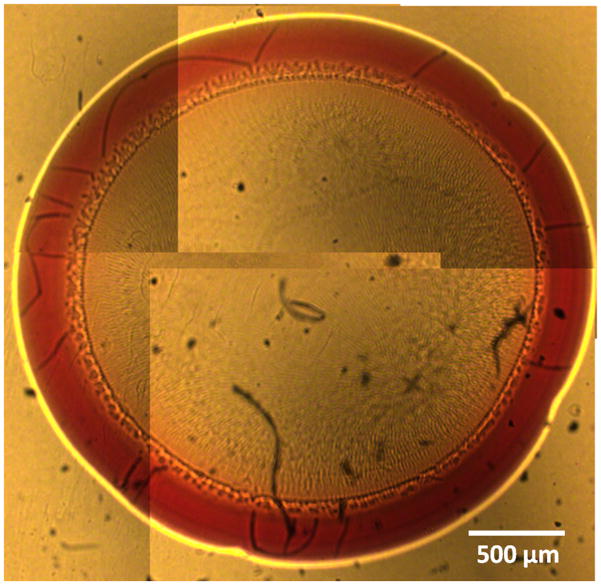
Prior to Raman spectroscopic study, aliquots (4 μL) of these solutions (i.e. the single proteins and the two-component mixtures) was pipetted on to the quartz coverslips and air-dried for approximately 20 minutes under temperature and humidity-controlled laboratory conditions. Figure 1 shows the ring pattern obtained by air-drying a representative two-component mixture sample. The width of the protein rings produced after solvent evaporation scaled roughly linearly with protein concentration in the range of 50–350 μm.
Figure 1.
Composite photograph showing the drop coating ring pattern produced by air-drying Hb and HbA1c mixture solution on a quartz substrate. The proteins are observed to be concentrated on the ring with little or no material left in the interior region. Here, the ring width is ca. 300 μm and the ring diameter is 3.5 mm.
All spectra presented in this paper were acquired with a 20-s acquisition time from the approximate geometric centre of the deposited rings. For the quantification study, multiple spectra were collected from each point (5 spectra per point) and multiple such points (3 points per sample at uniform separation in the direction of the arc of the ring) were probed to ascertain the measurement precision. The coverslips used in this study were all made of quartz in order to avoid the strong fluorescence of glass. For the 2D spatial Raman mapping, a total of 121 spectra were collected over a 44×44 μm field of view with inter-point distance of 4 μm.
All acquired Raman spectra were subject to vertical binning and cosmic ray removal prior to further data analysis (no background fluorescence removal was undertaken). It is worth noting that the spectra of the bare substrate and that obtained from the center of the drop-coated deposit (not to be confused with the center of the ring) were essentially identical and did not exhibit characteristic Raman features of any of the sample components. Consequently, further references to the (drop-coated) ring should be taken to mean the annular region where the analyte(s) are concentrated, unless otherwise noted. Also, laser power-dependent studies display no evidence for optical and/or thermal damage of either the analytes or substrates used in this study. Indeed, the power at the sample plane was intentionally kept at low levels to ensure no thermal/photochemical damage to the drop-coated protein rings.
Data Analysis
To evaluate the ability of the proposed method in regard to specific discrimination between non-glycated and glycated hemoglobin samples, principal component analysis (PCA) was employed on the entire spectral dataset acquired from single protein samples using the Statistics Toolbox of MATLAB R2010b (Math Works, Natick, MA). Specifically, the entire dataset was constituted by 75 and 60 spectra acquired from 5 and 4 separate Hb and HbA1c samples at different concentrations, respectively. As noted above, 15 spectra were acquired per sample with 5 measurements each at different points in the protein ring to test its homogeneity (or lack thereof). PCA is a powerful tool for exploratory data analysis and the linear multivariate PCA models are developed using orthogonal basis vectors (principal components, PC) thereby reducing the high-dimensional spectral data onto a lower dimensional space.31,32 In this article, we have employed logistic regression33,34 on the relevant principal components to obtain a separation plane between the samples and to quantify the specificity of our proposed approach.
Moreover, in order to characterize the capability of DCDR to provide quantitative measurements of these bio-analytes in hemolysate model systems, we have employed partial least squares (PLS) regression.35 For the hemolysate model systems, 240 spectra acquired from a total of 16 samples were used for data analysis. Due to the relatively limited number of distinct samples, a leave-one-sample-out cross-validation routine was performed to test the predictive power of the data. In the leave-one-sample-out cross-validation employed here, one sample (i.e. 15 spectra) is left out at a time from the calibration data set and the developed model is used to compute the concentration associated with the spectra of the left out sample. This calibration procedure is repeated until all samples have been left out in turn. In particular, for each prediction step, the calibration step for the hemolysate models was based on 225 spectra.
The root-mean-square error of cross-validation (RMSECV) (i.e. square root of the average of the squares of the differences between predicted and reference concentrations) was computed to assess the prediction accuracy of the models. Moreover, we have also evaluated the relative predictive determinant (RPD) metric to classify and appropriately compare the overall prediction quality of the individual calibration models for the two components, which have very different concentration ranges in this study. RPD is defined as the ratio of the standard deviation of the reference concentration in the sample population (σR) to the standard error of prediction (the standard deviation of the differences between predicted and reference values) (σR-P):
| (1) |
In addition to the accuracy, the precision of the developed calibration model for HbA1c was estimated by calculating the average relative standard deviation of predicted concentrations, RSD:
| (2) |
where Nconc is the number of distinct concentrations in the dataset, p is the number of spectra per concentration and σck is the standard deviation obtained at concentration ck.
Finally, we also evaluated the limit of detection (LOD) of our system for HbA1c measurements. The LOD, as per the IUPAC definition36, is calculated from the best-fit line attained between the predicted concentrations and reference concentrations37:
| (3) |
where sy/x is the standard deviation of the residuals and is a measure of the average deviation of the predicted values from the regression line.
RESULTS AND DISCUSSION
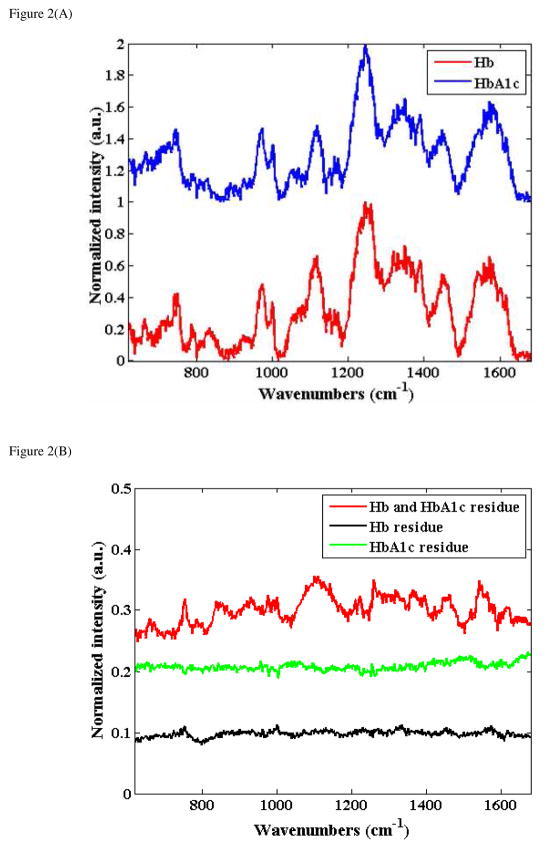
Figure 2(A) shows representative spectra acquired from the drop-coated single protein Hb and HbA1c samples derived from ca. 39 μM and 34 μM solutions, respectively (the spectra are normalized and offset for the sake of clarity). The spectra shown in this figure were obtained by averaging 5 spectra obtained from each of these samples. Not unexpectedly, no detectable signal could be recorded from the corresponding aqueous Hb and HbA1c solutions. Nevertheless, the acquired DCDR spectra were more representative of the protein in solution form (when concentrated to several mM range) than that obtained from the pure protein powder (i.e. sample bought from the manufacturer). While the two spectra grossly appear to have similar profiles, careful visual inspections exhibit subtle but discernible and highly reproducible spectral shape differences.
Figure 2.
Figure 2(A) Representative Raman spectra acquired from the drop-coated single protein Hb and HbA1c samples derived from ca. 39 μM and 34 μM solutions, respectively (the spectra are normalized and offset for the sake of clarity).
Figure 2(B): Spectra computed from the difference between: normalized Hb (39 μM) and HbA1c (34 μM) spectra shown in Fig. 2(A) (red); normalized Hb spectra from drop-coated rings derived from 39 μM and 19.5 μM (green); normalized HbA1c spectra from drop-coated rings derived from 34 μM and 17 μM (black).
To understand these differences that are embedded in the spectra in a more explicit manner, the difference spectra were plotted in Fig. 2(B). Here, we show the spectra obtained from computing the difference between the normalized Hb and HbA1c spectra (Fig. 2(A)) (red) as well as those obtained from the difference between normalized Hb spectra at 39 μM and 19.5 μM (green) and normalized HbA1c spectra at 34 μM and 17 μM (black). The lack of spectral features in the difference spectra from the latter two (green and black plots) reveal the highly reproducible nature of the measurements (and the deposits) obtained from the same analyte. Importantly, the presence of subtle but distinct features in the difference spectra between Hb and HbA1c deposits highlight the sensitivity of the proposed approach to very small structural variations in the protein molecules. (Identical features were obtained in the spectra calculated from the differences between sample spectra recorded from other concentrations of Hb and HbA1c deposits.) It is worth mentioning that these spectral features do not show a direct correspondence with those acquired from glucose deposits (which expectedly resemble the features obtained from a saturated glucose solution)38 nor from any one of these individual analytes. The variations in the two protein spectra can then be attributed to structural changes in hemoglobin molecule related to the binding of a glucose moiety. Such glycation-induced changes to the hemoglobin molecule have been well-characterized in the literature39 and are reported to decrease the α-helix content40 and weaken the heme-globin interaction.41
Our observations, here, are consistent with previous reports of the preservation of protein structure between the solution form and the drop-coated deposits.22 Indeed, the maintenance of the secondary structure of such deposits have been verified using circular dichroism (CD) spectroscopy42 and by FTIR investigations.43 Notably, our use of NIR excitation and quartz substrates coupled with measurements from the DCDR deposits (in contrast to pure protein powder measurements) substantially reduces the presence of any fluorescence background. This enables the application of multivariate chemometric methods without necessitating the incorporation of background removal procedures and without having to address the enhancement of the noise floor from the presence of a large fluorescence background.
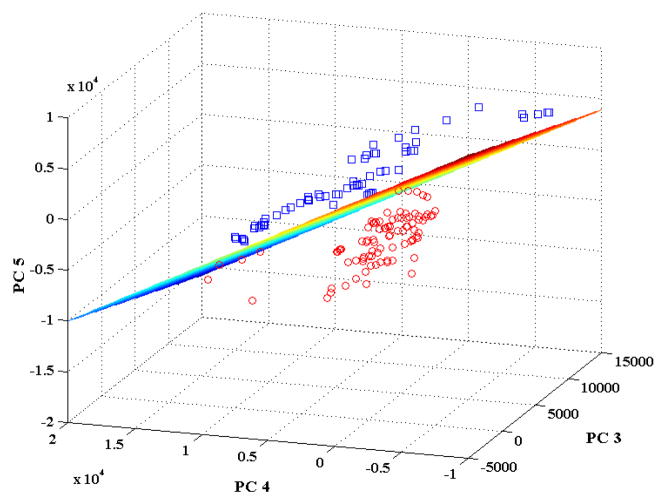
To further establish the ability of the proposed approach in selectively detecting the glycemic marker (HbA1c), we have performed multivariate classification on DCDR spectra acquired from the Hb and HbA1c deposits. Figure 3 shows the scores plot for the DCDR spectra dataset corresponding to principal components 3, 4 and 5. As PCA seeks to explain the net variance in the dataset, the first two PCs in this case (which are largely representative of the background and certain common features of the two analytes) do not provide the maximum discriminatory power between the 2 classes. However, as evident from Fig. 3, PCs 3–5 provide the capability of distinguishing spectra derived from the above analytes. As explained in the Data Analysis section, logistic regression was subsequently used on the scores of PC 3, 4 and 5 to quantify the discrimination capability. Based on these three scores, we obtained the following optimal separation plane:
| (4) |
Figure 3.
Scores plot corresponding to principal components 3, 4 and 5 for the spectral dataset acquired from the single protein Hb and HbA1c drop-coated rings. The Hb and HbA1c samples are indicated by red circles and blue squares, respectively.
From Fig. 3, one can observe that this separation plane is able to distinguish between non-glycated and glycated samples with 100% accuracy. Furthermore, using logistic regression in a leave-one-out cross-validation protocol on the set of 135 spectra, we were also able to obtain 100% classification accuracy between the glycated and non-glycated hemoglobin samples. To validate this result, a control study was done on the same 135 spectra (i.e. from 75 Hb and 60 HbA1c samples), where the sample classes “non-glycated hemoglobin” and “glycated hemoglobin” were randomly assigned - irrespective of their true class labels. Multiple such iterations were performed and the classification accuracy of these newly developed logistic regression models were observed to consistently lie in the range of 25–55%, showing the lack of predictive ability of such models. In essence, this confirms that our actual logistic regression model (Eq. (4)) is based on reproducible variations in the Raman fingerprint characteristics of non-glycated and glycated hemoglobin samples. Clearly, the model is not confounded even at substantially different concentration levels and by potential sample-to-sample variations (stemming from the imprecision in preparation and measurements). Thus, Fig. 2 in combination with Fig. 3 establish the excellent specificity of our approach in detecting the glycated variant from its non-glycated counterpart (the first specific study aim as outlined in the Materials and Methods section).
Given the excellent performance of PCA in discriminating the analytes from the spectra, partial least squares (PLS) regression was employed to demonstrate the predictive power of the proposed approach. Specifically, this was performed on the hemolysate model systems to assess the prediction accuracy and precision of the proposed approach. For the PLS regression analysis, the number of loading vectors which give the minimum RMSECV was determined to be in the range of 3–15. Subsequently, to minimize any possibility of overfitting (i.e. inclusion of spurious correlations and noise components), the calibration model corresponding to the minimum number of loading vectors, which provides a less than 5% deviation from the minimum RMSECV, was selected.
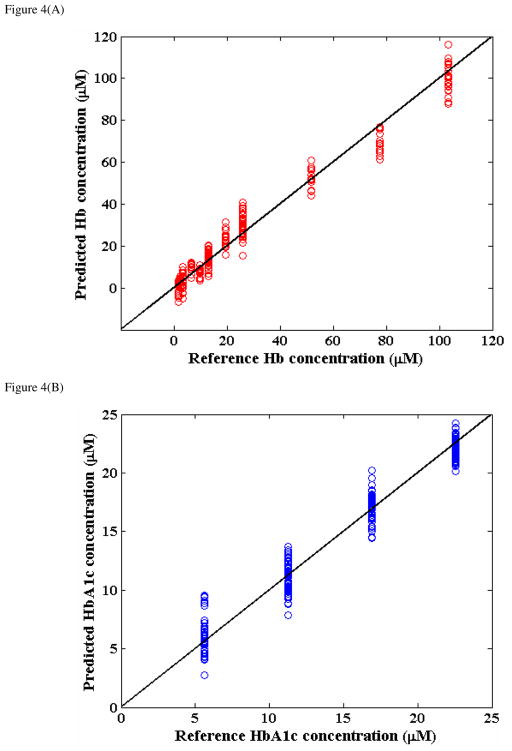
Figure 4(A) and (B) provide the results of leave-one-sample-out cross-validation analysis for the Hb and HbA1c constituents in the hemolysate models, respectively. This figure plots the predicted analyte concentration on the y-axis and the reference analyte concentration on the x-axis. The solid line indicates y=x and is provided to visualize the closeness of the measurements to an ideal linear response. As seen from the plot, the 16 hemolysate model samples had 11 distinct Hb and 4 distinct HbA1c concentration values with the range of glycated contributions in these samples covering and exceeding the normal physiological range of 5–10%. From Fig. 4, the RMSECV values for Hb and HbA1c were computed to be 5.44 μM and 1.27 μM. Evidently, our measurements show near-ideal linearity (between the predicted and reference concentration values) with the correlation coefficients of 0.99 and 0.98 for Hb and HbA1c, respectively.
Figure 4.
Prediction results obtained from partial least squares (PLS) regression on the hemolysate model samples: (A) Hb predictions; (B) HbA1c predictions.
In order to put the quantification capability of our models in perspective (and also to provide a common standard for comparison of the Hb and HbA1c models), we computed the RPD values for the two datasets. In general, for spectroscopic applications in the industry, a RPD value of 5 is typically considered to be good for quality control while a value larger than 6.5 can be used for process monitoring44. Here, the RPD values for the Hb and HbA1c predictions were calculated to be ca. 6.3 and 4.8. This result indicates that the predictive power of the Hb model is superior to that of the HbA1c model, which is to be expected because of the higher spectral SNR resulting from the larger values of the Hb concentrations. Further, we can infer that both the Hb and HbA1c PLS models provide adequate predictive power, even for predictions at these concentration levels, which are significantly lower than their corresponding physiological ranges.
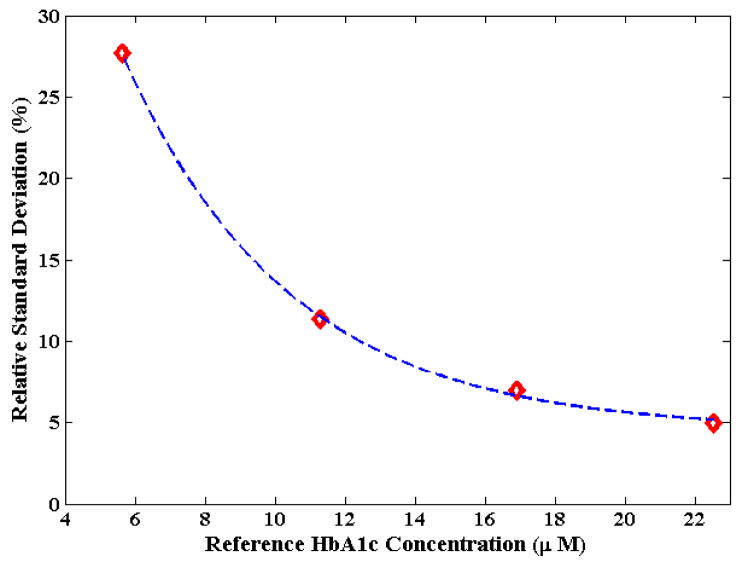
Another notable feature in Fig. 4 is the reasonably tight distribution of the predicted values for each distinct concentration point in the dataset. This indicates good reproducibility of the PLS regression models for both sets of DCDR analyte measurements. Quantitatively, the precision (relative standard deviation, RSD) over the entire 5–25 μM concentration range for HbA1c was calculated to be 12.7% (which is in the range of clinical acceptability). The RSD values as a function of the reference HbA1c concentrations in the hemolysate models are shown in the precision profile of Fig. 5. Expectedly, the RSD value deteriorates with decreasing concentration in an exponential manner.
Figure 5.
Plot of precision as a function of reference HbA1c concentration. The red diamonds gives the values computed from the experimental measurements and the dotted blue curve represents the best-fit exponential curve.
Additionally, since the RSD is expected to be 33% at the limit of detection (as per the IUPAC definition), one can extrapolate this precision profile to compute the LOD. Here, using the exponential best-fit line in Fig. 5 as our guide, the LOD for HbA1c measurements in our system was estimated to be 4.6 μM. The LOD, as mentioned above, can also be calculated from the regression sum of squares using Eq. (3). Using this method, we computed a LOD of 3.8 μM for HbA1c measurements, which is close to the aforementioned graphically estimated value. The small disparity in these two values can be ascribed to the non-ideal exponential fit for the precision profile. Importantly, though, these numbers establish that the LOD of our system is nearly a factor of 15 smaller than the lowest HbA1c absolute concentration values encountered in clinical practice (ca. 60 μM). It should be noted that these proof-of-concept results should not be interpreted as representing the best possible signal-to-noise ratio or lowest analyte detection limits, which are likely to be improved on further system and modeling optimization. For example, application of feature selection coupled with incorporation of advanced chemometric methods is expected to boost the prediction performance of the calibration model.
Finally, we performed a more detailed investigation of the topography of the analyte deposits and their impact, if any, on the reproducibility of our measurements. Based on the above precision computations, one would expect that there would be substantial overlap observed between the replicate measurements from a single drop-coated deposit. However, the critical question that remains unanswered at this juncture is what are the distributions of the two analytes across the ring width and, consequently, what is the ideal spot for DCDR measurements.
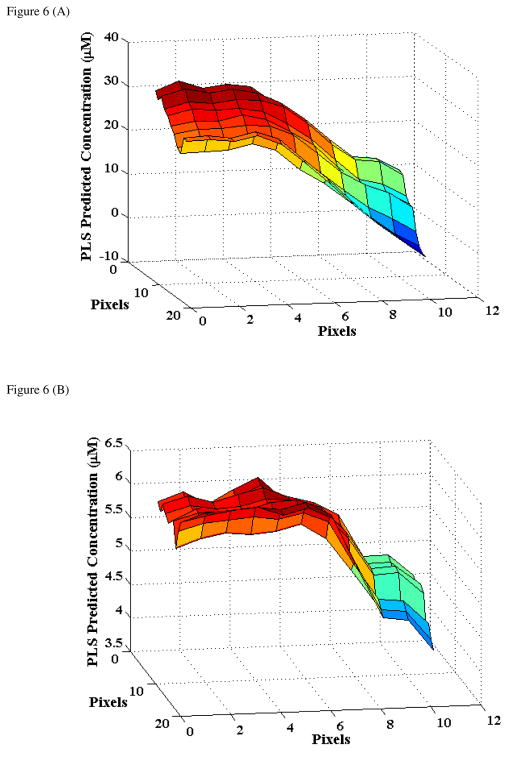
To characterize this in greater details, we performed a 2D spatial Raman mapping on a representative hemolysate model deposit (where Hb concentration was 25.9 μM and the corresponding HbA1c concentration was 5.6 μM). Based on the physics of the ring formation and prior observations by other investigators (for example, Kopecky et al.45), we mapped from the inner part of the coating ring to its central portion in order to avoid the lack of reproducibility usually associated with the outer perimeter of the ring (arising primarily from the desiccation of the analytes in this region). Using the previously developed calibration models, the spectral dataset was converted to a 2D matrix of concentration predictions for both Hb and HbA1c to allow the visualization of potential differences in distribution (organization) of the molecules in the edge region.
Figure 6 shows the results of our 2D mapping study over a 44×44 μm field of view (pixel-to-pixel distance of 4 μm). From Fig. 6, we observe the high degree of consistency between point-to-point measurements, especially at a constant radial distance from the centre of the deposit. Specifically, we computed that the point-to-point deviation at any radial distance is less than 5% of the mean value (3×3 pixel average) for both analytes. Significantly, we also find that the reference values of the analyte concentrations are reproduced with high fidelity at the centre of the ring, i.e. the centre of the ring displays a consistent value of ca. 25.5 μM and 5.5 μM for Hb and HbA1c, respectively.
Figure 6.
2D spatial Raman mapping based concentration prediction results for a representative hemolysate model for: (A) Hb and (B) HbA1c. The reference Hb and HbA1c concentrations are 25.9 μM and 5.6 μM, respectively, for this sample. The field of view is 44×44 μm with a pixel-to-pixel distance of 4 μm.
Interestingly, we find that the distribution profiles for both Hb and HbA1c are remarkably similar in the ring deposit. We suspect that the similarity in profiles stems from their comparable molecular masses, which reduces any separation in the protein distribution that may otherwise have been caused by the flow in the drying droplet. This result is closely related to our previous precision results in the hemolysate models. It is evident that inhomogeneous distributions for the two analytes in the ring deposit would likely have adversely impacted the acquired Raman spectra, which in turn would have introduced significant uncertainty in the PLS predictions beyond the currently obtained levels. Nevertheless, in measurements of small analyte concentrations (where chances of inhomogeneity increase) it is advisable to average spectra acquired from multiple points in the ring center to increase signal-to-noise ratio as well as to compensate for potential variations in protein distribution. It is worth noting that the field of view in Fig. 6 forms about half the total ring width of this deposit. Our mapping results from the center-to-outside region of the ring displayed a similar - but not identical - profile as that of Fig. 6, albeit with slightly higher point-to-point variations.
In summary, we have proposed and evaluated a novel analytical method for quantitative detection of HbA1c in single protein solutions as well as in hemolysate models. Given its linear response, high prediction accuracy and precision, we envision that the proposed approach can serve as a complementary tool to other more established analytical techniques, such as HPLC and immunoassay measurements, for detection of glycemic markers. Based on the promising results obtained here, this method should provide an ideal tool for characterization and quantification of the glycosylation status of other proteins. This is particularly significant given the substantive implications of protein glycosylation for the stability, pharmacokinetics and immunogenicity of glycoprotein-based biopharmaceuticals.46, 47
Further, while the studies reported in this article provide proof-of-concept demonstrations, the milestones along the path from laboratory research to clinical diagnostic application for HbA1c detection are clear. The next step is to perform these measurements in hemolysates obtained from whole blood samples in normal human subjects as well as diabetic patients. The whole blood samples (drawn by standard venipuncture into a EDTA tube) will be subject to centrifugation to separate out the cellular components from the plasma and subsequently lysed (using a suitable lysing reagent or changing tonicity of cell suspension) before performing DCDR. Since red blood cells greatly outnumber white blood cells and platelets, and hemoglobin forms ca. 95% of the dry mass of the RBC, one anticipates little or no spectral interference from any other Raman-active constituent. Once validated in the hemolysate samples, HbA1c determination in blood lysates can be performed by applying the lysing reagent directly to the whole blood sample thereby eliminating the centrifugation step. The spectral interference in this case is likely to be higher due to the presence of other serum analytes such as albumin, globulin (e.g. IgG) and transferrin (almost all other blood analytes have significantly less concentration48). Nevertheless, given the strong characteristic Raman signature of hemoglobin (both glycated and non-glycated forms) and the substantially lower concentration of other analytes in comparison to the physiological values of hemoglobin concentration, one can reasonably infer that the prediction accuracy and precision will not undergo much deterioration. In particular, the Raman spectra of albumin, IgG and transferrin are well-characterized49–51 and do not exhibit much interference with the Raman signature of hemoglobin. Additionally, to address any potential spectral interference from other analytes, appropriate feature selection mechanisms in combination with advanced classification tools52 will be pursued in our future work. For example, while PCA is adequate in determination of the analytes in the samples studied here, a more sophisticated protocol such as those based on the Minimum Noise Fraction Transform may be required to handle the potentially lower signal-to-noise ratios in the spectroscopic data acquired from clinical samples.53 Using such an approach (where data are forward transformed, components that correspond mostly to signal are selected and used in inverse transformation), investigators have previously shown that it is possible to achieve a high classification accuracy even with a 10-fold reduction in SNR.54 This and related noise-reduction techniques may play a crucial role in our future work, as the fluorescence from other analytes in hemolysate samples can introduce significant noise in comparison to the Raman signal from the analyte of interest.
CONCLUDING REMARKS
In the present study, we report drop coating deposition Raman-based selective detection and quantification of HbA1c, an important glycemic marker, at significantly lower concentration levels in comparison with typically observed physiological values. Our investigations have also systematically considered the detection of HbA1c in hemolysate model systems. The spectra obtained from micro-liter aliquots of the samples were highly reproducible (as revealed from both point measurements and spectral mapping) and did not suffer from significant fluorescence background commonly associated with conventional Raman scattering. The proposed method is quantitative, rapid and does not necessitate the addition of external reagents, dyes or labels. We anticipate that our ability to detect and quantify this important glycemic marker at remarkably low concentrations will provide molecular-level insight into protein glycation mechanisms, in combination with information revealed by other vibrational spectroscopy approaches (e.g. 2D-IR absorption spectroscopy).
Additionally, in a concurrent study, we are seeking to establish the effectiveness of DCDR for identifying and quantifying glycated albumin, which provides clinicians with an intermediate term marker for glycemic control (~14–17 days). Glycated albumin has also been reported to have a stronger correlation with the presence and severity of coronary artery disease (resulting from diabetic vascular complications) in comparison to HbA1c.55 Moreover, determination of both glycemic markers (namely HbA1c and glycated albumin), simultaneously, would provide very useful data, because of the different ‘lifetimes’ of these molecules and because of the different interferences. If the two values are not concordant, it would necessitate further investigations before interpreting either value as reflecting the true long-term glucose profile. Our eventual goal is to establish the clinical feasibility of performing Raman measurements for the detection of alternate glycemic markers. Once clinical feasibility is established, we envision developing a miniaturized Raman spectrometer fitted with an appropriate stage where the drop-coated plates (prepared following the centrifugation and lysis of whole blood samples) may be placed for routine diagnostic measurements. Since HbA1c provides a long-term history of glycemic control, it is sufficient for such a device to be placed in the clinic as opposed to a continuous glucose monitor, which needs to be tailored for home users, thereby significantly relaxing the device design requirements and constraints.
Acknowledgments
The authors wish to thank the NIH National Center for Research Resources for their grant P41-RR02594, at the MIT Laser Biomedical Research Center.
References
- 1.Centers for Disease Control and Prevention. National Diabetes Fact Sheet: national estimates and general information on diabetes and prediabetes in the United States. 2011. [Google Scholar]
- 2.Brownlee M. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 3.Saudek CD, Herman WH, Sacks DB, Bergenstal RM, Edelman D, Davidson MB. J Clin Endocrinol Metab. 2008;93:2447–2453. doi: 10.1210/jc.2007-2174. [DOI] [PubMed] [Google Scholar]
- 4.Cameron BD, Gorde HW, Satheesan B, Cote GL. Diab Tech Thera. 1999;1:135–143. doi: 10.1089/152091599317341. [DOI] [PubMed] [Google Scholar]
- 5.Shafer-Peltier KE, Haynes CL, Glucksberg MR, Van Duyne RP. J Am Chem Soc. 2003;125:588–593. doi: 10.1021/ja028255v. [DOI] [PubMed] [Google Scholar]
- 6.Chaiken J, Finney W, Knudson PE, Weinstock RS, Khan M, Bussjager RJ, Hagrman D, Hagrman P, Zhao YW, Peterson CM, Peterson K. J Biomed Opt. 2005;10:031111. doi: 10.1117/1.1922147. [DOI] [PubMed] [Google Scholar]
- 7.Arnold MA, Burmeister JJ, Small GW. Anal Chem. 1998;70:1773–1781. doi: 10.1021/ac9710801. [DOI] [PubMed] [Google Scholar]
- 8.Heise HM, Marbach R, Koschinsky TH, Gries FA. Artif Organs. 1994;18:439–447. doi: 10.1111/j.1525-1594.1994.tb02230.x. [DOI] [PubMed] [Google Scholar]
- 9.Barman I, Kong CR, Dingari NC, Dasari RR, Feld MS. Anal Chem. 2010;82:9719–9726. doi: 10.1021/ac101754n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barman I, Kong CR, Singh GP, Dasari RR, Feld MS. Anal Chem. 2010;82:6104–6114. doi: 10.1021/ac100810e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingari NC, Barman I, Singh GP, Kang JW, Dasari RR, Feld MS. Anal Bioanal Chem. 2011;400:2871–2880. doi: 10.1007/s00216-011-5004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingari NC, Barman I, Kang JW, Kong CR, Dasari RR, Feld MS. J Biomed Opt. 2011;16:087009. doi: 10.1117/1.3611006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koenig R, Peterson C, Jones R, Saudek C, Lehrman M, Cerami A. N Engl J Med. 1976;295:417–420. doi: 10.1056/NEJM197608192950804. [DOI] [PubMed] [Google Scholar]
- 14.Cagliero E, Levina EV, Nathan DM. Diabetes Care. 1999;22:1785–1789. doi: 10.2337/diacare.22.11.1785. [DOI] [PubMed] [Google Scholar]
- 15.Rahbar S. Clinica Chimica Acta. 1968;22:296–298. doi: 10.1016/0009-8981(68)90372-0. [DOI] [PubMed] [Google Scholar]
- 16.Mortensen HB, Christophersen C. Clinica Chimica Acta. 1983;134:317–326. doi: 10.1016/0009-8981(83)90370-4. [DOI] [PubMed] [Google Scholar]
- 17.Diabetes Care. 2010;33:S4–S10. [Google Scholar]
- 18.Little RR, Goldstein DE. Anal Chem. 1995;67:393–397. doi: 10.1021/ac00108a020. [DOI] [PubMed] [Google Scholar]
- 19.Syamala Kiran M, Itoh T, Yoshida K, Kawashima N, Biju V, Ishikawa M. Anal Chem. 2010;82:1342–1348. doi: 10.1021/ac902364h. [DOI] [PubMed] [Google Scholar]
- 20.Sackmann M, Bom S, Balster T, Materny A. Journal of Raman Spectroscopy. 2007;38:277–282. [Google Scholar]
- 21.Zhang D, Mrozek MF, Xie Y, Ben-Amotz D. Appl Spectrosc. 2004;58:929–933. doi: 10.1366/0003702041655430. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz C, Zhang D, Xie Y, Ribbe AE, Ben-Amotz D. Anal Biochem. 2006;353:157–166. doi: 10.1016/j.ab.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 23.Deegan RD, Bakajin O, Dupont TF, Huber G, Nagel SR, Witten TA. Nature. 1997;389:827. doi: 10.1103/physreve.62.756. [DOI] [PubMed] [Google Scholar]
- 24.Filik J, Stone N. Analyst. 2007;132:544–550. doi: 10.1039/b701541k. [DOI] [PubMed] [Google Scholar]
- 25.Esmonde-White KA, Le Clair SV, Roessler BJ, Morris MD. Appl Spectrosc. 2008;62:503–511. doi: 10.1366/000370208784344370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halvorson RA, Vikesland PJ. Environ Sci Technol. 2011;45:5644–5651. doi: 10.1021/es200255y. [DOI] [PubMed] [Google Scholar]
- 27.Kocisova E, Prochazka M. J Raman Spectrosc. 2011;42:1606–1610. [Google Scholar]
- 28.Rohleder DR, Kocherscheidt G, Gerber K, Kiefer W, Kohler W, Mocks J, Petrich WH. J Biomed Opt. 2005;10:031108. doi: 10.1117/1.1911847. [DOI] [PubMed] [Google Scholar]
- 29.Shaw RA, Rigatto C, Reslerova M, Ying SL, Man A, Schattka B, Battrell CF, Matthewson J, Mansfield C. Analyst. 2009;134:1224–1231. doi: 10.1039/b821442e. [DOI] [PubMed] [Google Scholar]
- 30.Kang JW, Lue N, Kong CR, Barman I, Dingari NC, Goldfless SJ, Niles JC, Dasari RR, Feld MS. Biomed Opt Exp. 2011;2:2484–2492. doi: 10.1364/BOE.2.002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brereton RG. Chemometrics: Data Analysis for the Laboratory and Chemical Plant. John Wiley and Sons; Chichester, West Sussex, UK: 2003. [Google Scholar]
- 32.Myakalwar AK, Sreedhar S, Barman I, Dingari NC, Rao SV, Kiran PP, Tewari SP, Kumar GM. Talanta. 2011;87:53–59. doi: 10.1016/j.talanta.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haka AS, Shafer-Peltier KE, Fitzmaurice M, Crowe J, Dasari RR, Feld MS. Proc Natl Acad Sci. 2005;102:12371–12376. doi: 10.1073/pnas.0501390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha A, Barman I, Dingari NC, McGee S, Volynskaya Z, Galindo LH, Liu W, Plecha D, Klein N, Dasari RR, Fitzmaurice M. Biomed Opt Exp. 2011;2:2792–2803. doi: 10.1364/BOE.2.002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wold S, Martin H, Wold H. Lecture Notes in Mathematics. Springer-Verlag; Heidelberg: 1983. [Google Scholar]
- 36.Currie LA. Anal Chim Acta. 1999;391:103–134. [Google Scholar]
- 37.Anderson DJ. Clin Chem. 1989;35:2152–2153. [PubMed] [Google Scholar]
- 38.Zhang D, Xie Y, Mrozek MF, Ortiz C, Jo Davisson V, Ben-Amotz D. Anal Chem. 2003;75:5703–5709. doi: 10.1021/ac0345087. [DOI] [PubMed] [Google Scholar]
- 39.Sen S, Kar M, Roy A, Chakraborti AS. Biophys Chem. 2005;113:289–298. doi: 10.1016/j.bpc.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Cussimanio BL, Booth AA, Todd P, Hudson BG, Khalifah RG. Biophys Chem. 2003;105:743–755. doi: 10.1016/s0301-4622(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 41.GhoshMoulick R, Bhattacharya J, Roy S, Basak S, Dasgupta AK. Biochim Biophys Acta. 2007;1774:233–242. doi: 10.1016/j.bbapap.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 42.Safar J, Roller PP, Ruben GC, Gajdusek DC, Gibbs CJ., Jr Biopolymers. 1993;33:1461–76. doi: 10.1002/bip.360330915. [DOI] [PubMed] [Google Scholar]
- 43.Oberg KA, Fink AL. Anal Biochem. 1998;256:92–106. doi: 10.1006/abio.1997.2486. [DOI] [PubMed] [Google Scholar]
- 44.Williams P. In: Near-infrared technology in the agricultural and food industries. 2. Williams P, Norris K, editors. AACC Inc; St. Paul, MN, USA: 2001. [Google Scholar]
- 45.Kopecky V, Jr, Baumruk V. Vibrat Spectrosc. 2006;42:184–187. [Google Scholar]
- 46.Brewster VL, Ashton L, Goodacre R. Anal Chem. 2011;83:6074–6081. doi: 10.1021/ac2012009. [DOI] [PubMed] [Google Scholar]
- 47.Greer F. Eur Biopharm Rev. 2007;6:106–111. [Google Scholar]
- 48.Dati F, Schumann G, Thomas L, et al. Eur J Clin Chem Clin Biochem. 1996;34:517–520. [PubMed] [Google Scholar]
- 49.Bellocq AM, Lord R, Mendelsohn R. Biochim Biophys Acta. 1972;257:280. doi: 10.1016/0005-2795(72)90280-2. [DOI] [PubMed] [Google Scholar]
- 50.Pezolet M, Pigeon-Gosselin M, Coulombe L. Biochim Biophys Acta. 1976;453:502–512. doi: 10.1016/0005-2795(76)90146-x. [DOI] [PubMed] [Google Scholar]
- 51.Gaber BP, Miskowski V, Spiro TG. J Am Chem Soc. 1974;96:6868–6873. doi: 10.1021/ja00829a010. [DOI] [PubMed] [Google Scholar]
- 52.de Paula AR, Silveira L, Pacheco MTT. Analyst. 2009;134:1203–1207. doi: 10.1039/b821248a. [DOI] [PubMed] [Google Scholar]
- 53.Bhargava R, Wang SQ, Koenig JL. Appl Spec. 2000;54:1690–1706. [Google Scholar]
- 54.Reddy RK, Bhargava R. Analyst. 2010;135:2818–2825. doi: 10.1039/c0an00350f. [DOI] [PubMed] [Google Scholar]
- 55.Pu LJ, Lu L, Shen WF, Zhang Q, Zhang RY, Zhang JS, Hu J, Yang ZK, Ding FH, Chen QJ, Shen J, Fang DH, Lou S. Circ J. 2007;71:1067–73. doi: 10.1253/circj.71.1067. [DOI] [PubMed] [Google Scholar]