Abstract
There is growing evidence that the metabolic network is an integral regulator of cellular physiology. Dynamic changes in metabolite concentrations, metabolic flux, or network topology act as reporters of biological or environmental signals, and are required for the cell to trigger an appropriate biological reaction. Changes in the metabolic network are recognized by specific sensory macromolecules and translated into a transcriptional or translational response. The protein family of sirtuins, discovered more than 30 years ago as regulators of silent chromatin, seems to fulfill the role of a metabolic sensor during aging and conditions of caloric restriction. The archetypal sirtuin, yeast silent information regulator2 (SIR2), is an NAD+ dependent protein deacetylase that interacts with metabolic enzymes glyceraldehyde-3-phosphate dehydrogenase and alcohol dehydrogenase, as well as enzymes involved in NAD(H) synthesis, that provide or deprive NAD+ in its close proximity. This influences sirtuin activity, and facilitates a dynamic response of the metabolic network to changes in metabolism with effects on physiology and aging. The molecular network downstream Sir2, however, is complex. In just two orders, Sir2’s metabolism related interactions span half of the yeast proteome, and are connected with virtually every physiological process. Thus, although it is fundamental to analyze single molecular mechanisms, it is at the same time crucial to consider this genome-scale complexity when correlating single molecular events with complex phenotypes such as aging, cell growth, or stress resistance.
Keywords: sirtuins, metabolic network, glycolysis, caloric restriction, pentose phosphate pathway, aging, redox state, nicotinamide
Robustness and Sensing of the Metabolic Network
Metabolite concentrations are driving forces for enzymatic reactions (Bruice, 2002), which implies that there is evolutionary pressure to keep them in a narrow, controlled range. Indeed, intracellular metabolite concentrations are extraordinarily robust against external as well as genetic perturbations (Blank et al., 2005; Ishii et al., 2007). Important for this robustness is a modular structure of the metabolic network. Modularity of substructures grants network stability, as perturbations may affect the module but not necessarily the flux of the entire system (Parter et al., 2007; Kreimer et al., 2008). Indeed, system-wide consequences on metabolite concentration levels are virtually limited to perturbations that affect common, network-interconnecting metabolic co-factors (i.e., ATP, SAM, or NADH; Blank et al., 2005; Kuepfer et al., 2005).
Maintaining metabolite concentrations requires molecular sensing and monitoring of the network, which occurs both at the level of (a) concentration and (b) metabolic flux (Grüning et al., 2010; Heinemann and Sauer, 2010). An illustrative example for network regulation at the concentration level is purine biosynthesis. Two intermediates, 5′-phosphoribosyl-5-amino-4-imidazole carboxamide (AICAR) and succinyl-AICAR (S-AICAR), bind to the transcription factors Pho2, Pho4, and Bas1. An increase in their concentration induces dimerization of these transcription factors and triggers expression of genes involved in their own biosynthesis (Pinson et al., 2009). This feed-forward mechanism may be required to react appropriately to energy shortage; it may allow the cell to decipher if rapid proliferation or starvation is the reason for the energy shortage.
AICAR and S-AICAR represent so called reporter metabolites, metabolites whose concentration controls a transcriptional response, or in the original definition, metabolic intermediates that are surrounded by transcriptional changes (Patil and Nielsen, 2005; Cakir et al., 2006) The TOR pathway also contains an example of the second considerably less understood type of metabolite control, that of flux-based monitoring of the metabolic network. In mammalian cells the mTOR interactor Rheb is bound to the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Under low glucose conditions, GAPDH prevents Rheb from binding to mTOR and thereby inhibits mTORC1 signaling. High glycolytic flux suppresses the interaction between GAPDH and Rheb and thus allows Rheb to activate TOR signaling (Lee et al., 2009).
The Metabolic Network and the Aging Process
These systems of metabolome regulation play important roles during the aging process. Metabolism is regarded as a cause of aging, as it is responsible for the increase in molecular and cellular damage observed in senescent cells. Although it is clear that the damage on macromolecules contributes to cell death and apoptosis and is age dependent, it is subject to ongoing debate if this damage is indeed the basis, or rather a consequence of the aging process, or both (Muller et al., 2007; Blagosklonny, 2008; Gruber et al., 2008; Blagosklonny and Hall, 2009).
Yeast aging research distinguishes between replicative and chronological aging. The first measure, also called mother cell-specific aging, describes how many cell cycles a yeast cell completes before senescence, or in other words, how many daughters bud from each mother (Mortimer and Johnston, 1959). The second, termed “chronological aging,” defines how long a yeast culture endures at 30°C in a non-dividing state (Fabrizio and Longo, 2003), or in its special case of “hibernating lifespan,” at 4°C (Postma et al., 2009). Although it is in general assumed that replicative aging shares more features with the aging process in humans (Laun et al., 2006), both measures depend on the metabolic network. The accumulation of oxidatively damaged proteins during both chronological and replicative aging is undisputed, however, at the same time there exist several yeast mutants which are strongly oxidant resistant, but show massively shortened lifespans [such as peroxiredoxin tsa1-B7; Timmermann et al., 2010, or triosephosphate isomerase (tpi1) mutants; Ralser et al., 2007). This indicated that the intense relationship between oxidative damage and aging is rather complex, as it is the function of sirtuins, as discussed below.
Among oxidizing molecules that originate in biosynthetic mechanisms, research predominantly focused on reactive oxygen species (ROS). A major source of ROS in the cell is superoxide , which leaks from complex III and complex I (in yeast only complex III, as it lacks complex I) of the mitochondrial respiratory chain (Cadenas and Davies, 2000; Breitenbach et al., 2012). In fact, superoxide production is quantitatively quite significant, as 1–2% of consumed oxygen is interconverted into this agent (Cadenas and Davies, 2000). However, there are other important sources of ROS, such as the mitochondrial external NADH dehydrogenases Nde1, Nde2 and the mitochondrial internal NADH dehydrogenase Ndi1, which feed the respiratory chain without proton pumping (Luttik et al., 1998; Li et al., 2006), or the degradation of fatty acids (Martin et al., 2007). Moreover, other oxidizing agents, such as reactive nitrogen species (RNS) also contribute significantly to macromolecular damage (Novo and Parola, 2008). Although RNS are less well studied in yeast than in plants or mammals, they seem to play a role in the yeast aging process, at least it was reported that they accumulate in senescent RAS mutants (Wilhelm et al., 2006).
Production of these oxidizing agents is a natural, inevitable consequence of metabolic activity. In young and healthy cells, natural ROS production is unproblematic as all living cells are evolutionarily adapted and are able to clear this ROS through a series of enzymatic and non-enzymatic antioxidant systems (Sies, 1997; Cadenas and Davies, 2000). These stabilize ROS levels; for instance, respiring and non-respiring yeast cells possess the same levels of superoxide and hydrogen peroxide (Gruning et al., 2011). The in quantitative terms most important (but not the only) redox buffer is the glutathione system (Meister and Anderson, 1983; Grant et al., 1996). Glutathione, a cysteine containing tri-peptide of non-ribosomal origin, clears ROS by being oxidized. The oxidized form is then recycled by glutathione reductase, an enzyme which restores GSH under NADPH consumption (Massey and Williams, 1965). Regulation of the NADPH supply is critical for this process. This metabolite is consumed in large amounts under oxidative stress conditions, but cannot be provided in large excess, as this would shift the redox balance toward reduction (“reductive stress”), which is another pathogenic situation (Rajasekaran et al., 2007; Brandes et al., 2009).
The dynamic increase in NADP+ reduction during oxidative stress has largely been attributed to increased activity of the pentose phosphate pathway (PPP), a catabolic carbohydrate pathway alternative to glycolysis (Wamelink et al., 2008). Although other cellular reactions are capable of maintaining a normal NADPH/NADP+ ratio under anaerobic growth conditions, they fail to do so upon contact with an oxidant or when they respire at high rates. A H2O2 treatment of zwf1Δ yeast – lacking the first NADP+ reducing enzyme of the PPP (glucose 6 phosphate dehydrogenase Zwf1) – causes an immediate collapse of the NADPH/NADP+ ratio resulting in massive cellular sensitivity to oxidants (Castegna et al., 2011).
Regulation of Dynamic PPP Activation
The dynamic activation of the PPP is a vivid example for the adaptation of the metabolic network to changing environmental conditions. Upon the addition of an oxidant, PPP activity is increased in temporally discrete steps. This regulation involves both the transcriptome and metabolome level, beginning with the latter (Larochelle et al., 2006; Chechik et al., 2008; Ralser et al., 2009a). In addition, glycolysis and the PPP are self-adapting to prevent oxidative stress when cells switch from anaerobic to oxidative metabolism. Low activity of yeast pyruvate kinase [PYK; an enzyme which catalyzes the last glycolytic step converting phosphoenol-pyruvate (PEP) to pyruvate] in respiring cells causes accumulation of its substrate PEP. PEP in turn acts as feedback inhibitor of triosephosphate isomerase (Tpi1). The resultant block in glycolysis drives flux into the PPP (Gruning et al., 2011). This mechanism helps to balance the increased ROS production during oxidative metabolism, and keeps the ROS level similar to that of non-respiring cells. However, interruption of this feedback circuit makes cells vulnerable to oxidative stress: when respiration is induced in zwf1Δ yeast, ROS accumulate, and damage macromolecules and organelles by oxidation (Gruning et al., 2011). Recently, it has become clear that this mechanism is conserved in mammalian cells, and might explain central features of the Warburg effect, which is a decrease in oxidative respiration during cancer formation (Warburg, 1956). Also cancer cells suffer from oxidative damage, and redirect the metabolic flux via PYK for inducing ROS clearance (Anastasiou et al., 2011; Grüning and Ralser, 2011).
The discovery of yeast sirtuins
The metabolic shift from glycolysis to the PPP is one of the best studied examples, where a metabolic re-configuration induces adaptation of the transcriptome, and the glycolysis/PPP transition functions as a “metabolic signaling cascade” induced under oxidative stress (Kruger et al., 2011). The response in the transcriptome points to the existence of regulating macromolecules that detect these changes in metabolic activity and translate this information into a transcriptional response. Sirtuins are such proteins, they react to changes in metabolic activity and are closely connected with metabolites and enzymes of glycolysis.
The papers dealing with the sirtuin protein family recently exceeded 2200. The authors who first discovered the Saccharomyces cerevisiae SIR2 gene, from which the name, sirtuin is derived, certainly could not have dreamed that their discovery would within just a few decades lead to such an enormous “gene rush” in the scientific community around the world. For the most part, this development is caused by the association of sirtuins in the aging process. However, the exact nature of this connection is still unclear, and the debate about whether or not the sirtuins are part of a public mechanism of aging (term meaning cross-species aging mechanism; Martin et al., 1996) is still ongoing. Prominently, the involvement of sirtuins in caloric restriction (also called “calorie restriction” or “dietary restriction”) intervention in the aging process is central to this debate, as it is the mechanism of action of resveratrol, a plant stilbene which was believed to extend S. cerevisiae replicative lifespan though activating Sir2 (Lin et al., 2000; not further discussed in this review).
The SIR (silent information regulator) mutants and genes were discovered by Rine et al. (1979). Four different complementation groups of unlinked recessive yeast mutations were found that all led to the same phenotype: the mutants were extragenic suppressors of the matα1-5 mutation. The suppressor mutation caused cells to regain functional MATα genetic information and to mate again normally. This effect was completely dependent on the presence of MATα genetic information at the distant HML locus near the left telomere of chromosome III. Viewed from 2012, it is not surprising that the sir mutations were shown to influence not only mating, but also sporulation efficiency. Herskowitz and many others have established that mating of alpha strains needs the functional gene product Matα1 and sporulation requires the functional gene products Mata1 and Matα2. In most laboratory strains, the HML locus near the left telomere of chromosome III contains functional but silent copies of the alpha1 and alpha2 genes, while the HMR locus located near the right telomer of chromosome III, contains functional but silent copies of the a1 and a2 genes. The first identified mutant sir allele (sir1-1) suppressed all known mating deficient and sporulation-deficient mutations located in the mating type locus. All four SIR genes are needed independently for transcriptional repression of HML and HMR. Importantly, it was later shown (Moazed et al., 1997; Liou et al., 2005) that three of the four Sir proteins (Sir2/Sir3/Sir4) form a tight heteromeric complex in vitro; Sir1 is not part of this complex. (Please see Huang, 2002; for a Review of proteins that are needed for silencing and for the pleiotropic functions of Sir2).
We know that the sir mutations relieve the transcriptional repression of genetic information at HML and HMR, and from work in the ensuing decades that the Sir gene products bind to controlling segments in the DNA of HML and HMR, and are part of the chromatin at those loci. In addition, the Sir proteins are found at telomeres of all chromosomes and at the rDNA locus on chromosome XII (Strahl-Bolsinger et al., 1997; Mekhail and Moazed, 2010). The mutations were found at the time to have no other phenotypes apart form silent mating type information expression. Herskowitz and Oshima (1981) wrote that “it is clear that sir mutants are not grossly pleiotropic.” This statement is no longer true, as will be discussed below.
It was shown that Sir proteins interact genetically with histone H4, indicating that transcriptional silencing includes H4 modification and that co-operation of H4 with Sir2 is needed for silencing (Imai et al., 2000). The first hint of a special role for Sir2 that differs from the other three Sir proteins came from the work of Easton-Esposito and her colleagues (Gottlieb and Esposito, 1989; Fritze et al., 1997). They showed that Sir2 (but not Sir 1, Sir3, or Sir4) suppresses non-allelic recombination between the tandem copies of rDNA on chromosome XII. The authors concluded that Sir2 is a limiting component required for chromatin modeling at the rDNA locus. This was a very important step toward our present understanding of Sir2. Guarente (1999) reviewed the role of the Sir proteins in silencing, chromatin organization at telomers, silent mating type loci, the NOR (nucleolar organizer), in repair of double strand breaks, recombination, cell cycle regulation, and mother cell-specific aging.
The discovery of Sir2’s role in aging and the metabolic network
Among other groups, the lab of Guarente (1999) started work on yeast mother cell-specific aging in the 1990s. While it is to the present day, for technical reasons, not possible to directly select for replicatively long-lived yeast mutants, several indirect protocols for mutation isolation have been used. After conventional chemical mutagenesis of haploid yeast cells and applying starvation stress, replicatively long-lived mutants were isolated among the survivors of extended periods of starvation (Kennedy et al., 1995). One of them was located within SIR4 and was shown to be a “semi-dominant” missense mutation. This led to the possibility that the Sir complex could have a special role in replicative aging. Consequently, all components of this complex were tested for aging phenotypes (Kaeberlein et al., 1999). It was shown that yeast replicative lifespan depended on the presence of SIR2, SIR3, and SIR4, but not on SIR1. Again, SIR2 was shown to have a special role that is slightly different from that of SIR3 and SIR4. Comparing the deletion mutants of the three genes (sir2Δ, sir3Δ, and sir4Δ) it appears that sir2Δ had the largest effect (−50%). Conversely, overexpression constructs increased lifespan, again with Sir2 overexpression having the largest effect. It appeared that the effects of sir3Δ and sir4Δ could be fully compensated by deleting HMLα in the used haploid MATa strain, while the same effect was not seen in sir2Δ yeast (Kaeberlein et al., 1999). The authors explained these results with Sir2-triggered repression of minicircles (ERCs), formed by non-allelic recombination of the tandem repeated copies of rDNA in the NOR on chromosome XII. These minicircles behave like non-centromeric plasmids, therefore accumulate in mother cells but are very inefficiently transmitted to daughter cells. In the mother cells, they can become so abundant with replicative age that either the sheer mass of the circular DNA may have an adverse effect on the cells (Sinclair and Guarente, 1997) or they may titrate away the Sir2 which is then limiting for the other important functions like silencing of HML and HMR. This most likely contributes to the observed mating defect of old haploid mother cells. Other functions compromised in these cells are binding and silencing to telomeres and adjacent chromosomal regions, affecting among other things unwanted non-homologous end-joining (NHEJ), perhaps leading to replication problems and genome instability in the old mother cells (Gottschling, 2000; McMurray and Gottschling, 2004). FOrk Blocking less 1 (Fob1) is a non-essential recombination factor needed for keeping the sequences of the rDNA repeat units constant. Preventing the formation of minicircles by deleting fob1, increases the replicative lifespan of yeast by about 6 generations (Defossez et al., 1999). However, the SIR mutations (in particular sir2Δ) have effects on lifespan even in the absence of minicircles, and the gain in lifespan in fob1Δ strains is relatively small (Defossez et al., 1998; Borghouts et al., 2004; Heeren et al., 2009). This underlines the fact that replicative aging of yeast and aging in general is a very multicausal process and depends not only on the genetic makeup of the cell but also on many external conditions. Thus, there is no single and “most important” mechanism of aging.
Minicircles have been described only in the yeast S. cerevisiae and its close relatives, but not in the other eukaryotic model systems of aging, Drosophila melanogaster, Caenorhabditis elegans, and Mus musculus. We therefore call minicircles a “private mechanism of aging” (the term meaning species-specific going back to Martin, 1997). However, in the non-yeast model systems of aging, the SIR2 gene and its protein family does exist and is even split into a number of paralogs (i.e., seven in humans) some of which are essential for life in mammals (McBurney et al., 2003; Chen et al., 2005).
Despite the absence of minicircles, lifespan extending properties for Sir2 homologs have been reported for C. elegans and D. melanogaster. However, recently, these studies received a serious setback as these lifespan phenotypes were lost by out-crossing the Sir2 overexpression alleles (Burnett et al., 2011), or at least, were overestimated in one original report (Tissenbaum and Guarente, 2001) due to co-segregation of an unlinked second mutation which alters lifespan as well (Viswanathan and Guarente, 2011). We think that the question of sirtuins involvement in lifespan extension in non-yeast species is undecided at present. The careful genetic crosses of Burnett et al. above everything show one fact: due to the extreme (and not fully analyzed) pleiotropy and the large number of target genes of the deacetylase Sir2, it is extremely important to investigate co-segregation of the aging phenotype with the overexpression mutation to determine hidden factors in the genetic background.
The situation is also different for the involvement of Sir2 in the second measure of yeast aging, chronological lifespan. This measure is quite different to replicative aging as it describes the survival of a stationary yeast population rather then the survival of an individual cell. Indeed, gene sets which extend replicative, chronological, and also hibernating lifespan are only marginally overlapping (Laun et al., 2006; Postma et al., 2009). Nonetheless, systematic analysis of chronological aging led to the discovery of aging factors conserved from yeast to mammals, including the TOR pathway (Fabrizio and Longo, 2003; Powers et al., 2006). Sir2 has however shortening effects on this measure: Lack of Sir2 along with calorie restriction and/or mutations in the yeast AKT homolog, Sch9, or Ras pathways caused a dramatic chronological lifespan extension (Fabrizio et al., 2005).
Irrespective of this debate, studies on Sir2’s involvement in the aging process stimulated a massive amount of research on Sirtuins and improved our understanding of their regulatory potential in humans. In animal species SIRT1 shows the highest degree of sequence similarity with yeast Sir2, and displayed aging-related physiological effects in spite of the absence of ERCs (Tissenbaum and Guarente, 2001; Rogina and Helfand, 2004; Chen et al., 2005; Herranz and Serrano, 2010). These effects were explained by the high number and great variety of protein targets of Sir2 deacetylase activity (Westphal et al., 2007). In mammals, considering the targets that have so far been discovered, they connect sirtuins activity to metabolic regulation, stress resistance and diseases of old age like diabetes and cancer.
Asymmetric segregation, caloric restriction and Sir2
The literature on Sir2 homologs and aging is closely interrelated with (i) asymmetric segregation, and (ii) caloric restriction. These seem to be unrelated processes from the mechanistic point of view, but both protect cells from oxidative damage.
Asymmetric segregation in yeast S. cerevisiae was found in aged mother cells that retain oxidatively damaged proteins; daughter cells are formed from a fresh set of proteins. This is Sir2 dependent; sir2Δ mothers are unable to protect their daughters from the transmission of damaged proteins during cytokinesis (Aguilaniu et al., 2003). Thus, sir2Δ daughters suffer much earlier from molecular damage as do wild-type cells.
It is not yet clear if Sir2 homologs play a similar role in other species. Thus, it cannot be excluded that Sir2’s modulation of asymmetric segregation is again a yeast’s private mechanism of aging. In contrast, however, the second treatment associated with Sir2’s anti aging effects, caloric restriction, has been found to increase lifespan and improve health in all tested species from yeast to humans (Sohal and Weindruch, 1996). Deleting or down-regulating the respective Sir2 homologous gene in the aforementioned species abrogates effects of caloric restriction on lifespan and on the preventive effect of caloric restriction on diseases of aging, like diabetes (Chen et al., 2005; Lamming et al., 2005; Qiu et al., 2010a). However, the question of whether the yeast SIR2 gene is needed for the increase the replicative lifespan by a reduction of glucose is an open one till the present day. One laboratory maintains that the effect of this mild CR is effective independently of Sir2, and is in the absence of Sir2 dependent on Hst2, a Sir2 paralog (Lamming et al., 2004, 2005; Smith et al., 2007). The HST2 gene is a member of a small family of four yeast genes which is highly homologous and functionally related to Sir2 (Derbyshire et al., 1996). However, another laboratory insists that life span extension by CR is strictly dependent on the presence of the SIR2 gene (Lin et al., 2000).
Metabolic Effects of Caloric Restriction in Yeast
This lifespan extension in regard to Sir2 has been studied by reducing glucose concentration from the usual 2% to the lower 0.5% in S. cerevisiae media (Lin et al., 2000). Although this treatment is referred as caloric restriction, it is – if any – a mild form of it, and it is worth speculating that 0.5% glucose may closer resemble the nutrient availability in the yeast’s natural environment. Interestingly, it was described that an induction of respiration is required for the lifespan extending effects of a growth on 0.5% glucose, and that Sir2 is involved in this process (Lin et al., 2002). Importantly, S. cerevisiae is a “Crabtree” positive yeast: high glucose concentrations (but not other carbon sources such as galactose, raffinose or glycerol) repress S. cerevisiae respiration. Analyzing chronological lifespan, it was shown that in contrast to glucose, inducing CR by reducing the content of galactose, raffinose, or glycerol plus ethanol did not enhance lifespan. Moreover, the lifespan extending effects of diminished glucose concentration were not observed in the Crabtree negative yeast Kluyveromyces lactis (Oliveira et al., 2008). In this context, it is worth to mention that yeast strains with extended hibernating lifespan (mutants that survived 5 years in the cold room) contain a high number of strains which cannot survive without respiratory activity (Postma et al., 2009). Moreover, when respiration was induced by PYK or by shifting to galactose, yeast cells developed a strong increase in resistance to different oxidants (Gruning et al., 2011), but in the Crabtree negative K. lactis, limitation of glucose supply did not protect against ROS as it does in S. cerevisiae (Oliveira et al., 2008). Recently, also nitric oxide (NO) production was associated with these lifespan extending effects of low glucose levels. A reduction of glucose concentration induces S. cerevisiae NO production; and NO donors such as GSNO (S-nitrosoglutathione) were sufficient to extend lifespan (Li et al., 2011). Thus, it is conceivable that the lifespan extending effects of reducing glucose to 0.5% result from circumventing glucose repression causing an activation of mitochondrial metabolism, respiration, and the cellular antioxidant machinery.
Additionally, the nutrient-responsive signaling kinase TOR (target of rapamycin), and the concentrations of NAD+ play a role in this process. Although it was reported that tor1 mutants have an increased lifespan independently from Sir2 activity (Kaeberlein et al., 2005), other results supported the notion that the TOR mediated lifespan extension is a direct result of enhanced NAD+ salvage pathway activity and Sir2 targeting (Silva et al., 2009). It was shown that rapamycin and nitrogen starvation leads to enhanced Sir2 association with rDNA via the inhibition of TOR complex 1 (TORC1; Ha and Huh, 2011). To undergo these reactions, Sir2 required the salvage pathway Nicotinamidase Pnc1 and Net1, the core subunit of the RENT complex, indicating that TORC1 activity results in increased silencing of RNA polymerase genes at rDNA loci, enhanced histone deacetylation and reduced homologous recombination at rDNA repeats. This allowed to conclude on a lowered number of ERCs as basis for this lifespan extension (Ha and Huh, 2011). Indeed, NAD+ and nicotinamide are directly involved in the functional activity of Sir2 in relation to the metabolic network.
The Enzymatic Function of Sir2 and Links to the Metabolic Network
Sir2 has deacetylase and ADP-ribosyl-transferase activity, both of which are required for deacetylating proteins. Sir2 catalyzes a NAD+-dependent ε-N-acetyl-lysine deacetylation from proteins (including histones, class III HDACs), resulting in deacetylated protein, nicotinamide, and the molecule 2′O-acetyl-ADP-ribose (OAADPR; Moazed, 2001; Jackson and Denu, 2002). It is an open field of research if Sir2 can for this reason sense the NAD+/NADH ratio (Yu and Auwerx, 2009; Yu et al., 2009). First Sir2 activity may be directly regulated by changes in the cellular redox potential. This notion is perhaps naïve, as the main determinant of the redox potential of the cell is the glutathione redox couple according to the Nernst equation. The measured changes in NAD+/NADH ratio during CR have only minor influence on this measure (Schafer and Buettner, 2001). In this line, Anderson et al. (2003) showed that reducing the glucose intake decreases nuclear NAD+ levels in vivo, but that yeast Sir2 as well as its mammalian homolog SirT1 are not affected by this alteration in NAD+/NADH ratios.
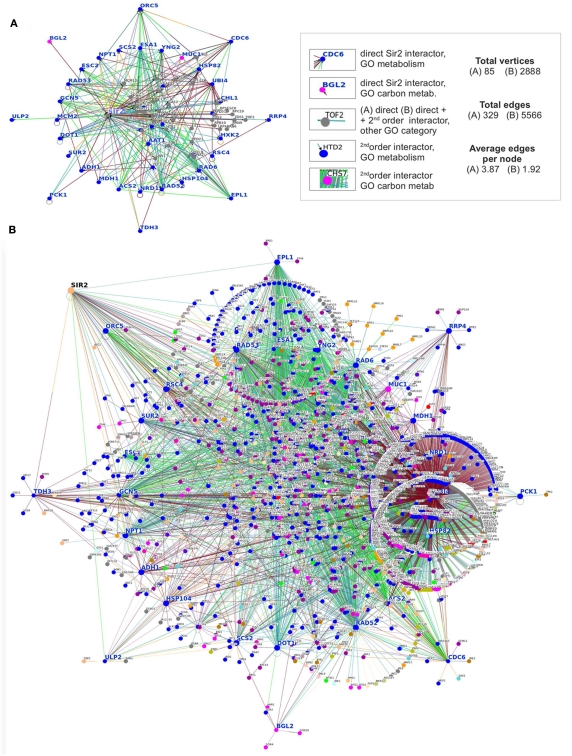
These assumptions however require a more differentiated elaboration as the overall redox state measure does not necessarily reflect the cofactor presence in the protein’s microenvironment. A significant part of NAD+ is protein bound, as it is presumably Sir2, and therefore protein complexes may feed Sir2 directly with its cofactor. Recently, Sir2 was the basis for several genome-scale screens to identify genetic and physical interactors. These include a genome-wide screen to identify anti-silencer factors (Raisner and Madhani, 2008), and proteomic analyses that use methods of label-free protein quantification (Rye et al., 2011). Together with other genes identified in earlier targeted and untargeted screens, they form a large network which is indicative of the global function of Sir2. Based on the information stored in the BioGrid database as of December 2011 (Stark et al., 2006), we generated a metabolism-centric protein interaction network for Sir2 (Figure 1). This network contains all direct genetic and physical interaction partners of the enzyme, and interactors that are associated with the gene ontology (GO) term “metabolism” or “carbohydrate metabolism” are highlighted. These two GO terms have been assigned to 40 genes out of the total 84 annotated Sir2 interactors (Figure 1A). To illustrate the large interconnectivity that comes with these 40 genes, the network was expanded with their direct interactors, which yielded a “metabolism-centric” Sir2 interactions network (Figure 1B). The inclusion of the second order Sir2 interactors considerably increased the network and its interconnectivity; the network is formed of 2888 vertices interconnected by 5566 edges. Thus, in just two steps, the metabolic interaction network of Sir2 interconnects half of the yeast proteome.
Figure 1.
A metabolism-centric interaction network for Sir2. (A) Network of 84 direct interactors of Sir2; the 40 genes associated with the GO term “metabolism” and/or “carbohydrate metabolism” are highlighted in blue or purple, respectively. (See Table A1 in Appendix for GO terms associated with these Sir2 interactors). (B) The network (A) expanded with the direct interactors of the 40 genes with annotated metabolic function. This network containing first and second order metabolism related Sir2 interactors interconnects with 2888 vertices half of the yeast proteome. We apologize that due to this complexity, not all genes names may be readable. (A,B) Physical- and genetic-interactors for Sir2 were obtained from the BioGrid database as of December 2011 and illustrated with OSPrey (Stark et al., 2006).
Sir2 Interacting Enzymes Which Generate NAD+ in Close Proximity Modulate Its Biological Function
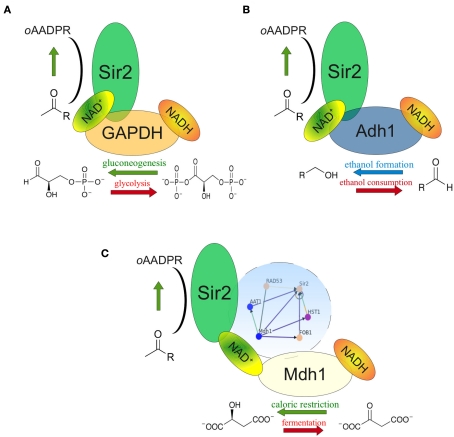
In Table A1 in Appendix we list the genes of the direct (first order) interactors of Sir2, and their associated GO terms. Interestingly, this list contains three proteins that are among the major NADH/NAD+ consumers of the cell. Two of them – alcohol dehydrogenase Adh1 and the predominant yeast GAPDH Tdh3 – are found in physical complex with Sir2 (Gavin et al., 2002). In both cases, low-scale genetic interaction studies confirmed that they influence the biological activity of Sir2 (Figure 2). The first protein, the predominant yeast GAPDH Tdh3, is a central enzyme of glycolysis and reduces NAD+ to NADH in its catabolic role, but oxidizes NADH in its gluconeogenetic function (Tristan et al., 2011). A genetic screen conducted in the lab of Scott Holmes revealed that overexpression of Tdh3 rescues lethality caused by GAL1-promoter driven overexpression of Sir2, indicating a modulation of Sir2 activity by yeast GAPDH (Matecic et al., 2002). Later, our investigations revealed that overexpression of Tdh3 and E. coli GAPDH increases the rate of mitotic recombination in a Sir2 dependent manner (Ralser et al., 2009b). This phenotype was dependent on the catalytic activity of Sir2, and, interestingly, a function of metabolic activity: The recombination frequency in yeast overexpressing Tdh3 increased from 3 times over wild-type to more than 10 times in exponential batch cultures, the condition that exhibits the highest glycolytic flux (Ralser et al., 2009b).
Figure 2.
NAD+ feeding or depletion in the Sir2 microenvironment. (A) The NAD(H) metabolizing glycolytic enzyme GAPDH (Tdh3) is found in the same protein complex as Sir2. At high glycolytic flux, GAPDH deprives NAD+ in close proximity to Sir2; during gluconeogenesis it generates NAD+. Genetic interplay of Tdh3 and Sir2 influences the rate of mitotic recombination. (B) The predominant cytoplasmic alcohol dehydrogenase (Adh11) also complexes with Sir2. Adh1 overexpression increased the NAD+/NADH ratio and Sir2 activity as well as prolonged yeast replicative lifespan. (C) Mitochondrial malate dehydrogenase Mdh1 is up-regulated during caloric restriction. Its overexpression increases NADH oxidation and prolongs replicative lifespan in a Sir2 dependent manner. Complex formation between Sir2 and Mdh1 has not been reported, but both proteins are interconnected through a dense genetic interaction network involving Rad53, Aat1, Fob1, and Hst1 (blue circle).
Similar observations were made with alcohol dehydrogenase ADH1, another component of the Sir2 protein complex (Gavin et al., 2002; Figure 2B). Adh1 catalyzes the conversion of acetaldehyde to ethanol, regenerating NAD+. Overexpression of the Adh1 enzyme increased both the NAD+/NADH ratio and the activity of Sir2. Moreover, this treatment caused a 30% extension in yeast replicative life span (Reverter-Branchat et al., 2007). Thus, it is conceivable that in the close proximity within a protein complex, metabolic enzymes GAPDH (Tdh3) and alcohol dehydrogenase (Adh1) provide or deprive NAD+ that is used by Sir2. As the catalytic reactions catalyzed by Tdh3 and Adh1 are reversible, this catabolic function does not only rely on the overall metabolic flux, but also on the reaction equilibrium in the microenvironment: these enzyme may provide or metabolize NAD+ dependent on their localization.
The situation of the third metabolic enzyme, malate dehydrogenase Mdh1 is different, as it localizes to mitochondria, were, to our knowledge, yeast Sir2 is not localized (Blander and Guarente, 2004) Mdh1p, is involved in the citrate cycle and component of the malate-aspartate NADH shuttle. However, it has been reported that Mdh1 is over-expressed under conditions of calorie restriction (Lee and Lee, 2008), and that ectopic overexpression of Mdh1 causes extension of replicative lifespan. Remarkably, this lifespan extension was Sir2 dependent (Easlon et al., 2008). Although these proteins do not co-localize, they are part of a heavily interconnected interaction sub-network involving proteins Sir2, Mdh1, Rad53, Aat1, Fob1, and Hst1 (Figure 2C). Thus, although physical association of the two proteins may be unlikely as they predominantly localize to different compartments, they are involved in a network of close genetic interactions, and their interplay is involved in yeast replicative lifespan. Nonetheless, it remains possible that either Sir2 or Mdh1 do co-localize under conditions which have not yet been defined.
Metabolic regulation by Sir2 has been associated with NAD(H) biosynthesis and salvage pathways. These differ in some, but important, enzymatic steps among the eukaryotic lineage. While mammals prevalently use nicotinamide as the main NAD+ source, single-cellular eukaryotes like S. cerevisiae have focused on utilizing tryptophan and nicotinic acid as precursors for NAD(H) biosynthesis (Rongvaux et al., 2003). Due to the lack of a homolog to the mammalian Nicotinamide phosphoribosyltransferase (NAMPT) enzyme, the budding yeast is unable to synthesize NAD(H) directly from the precursor nicotinamide. Hence, nicotinamide gets converted to nicotinic acid, a substrate used by Npt1 (nicotinic acid phosphoribosyltransferase), to produce nicotinic acid mononucleotide (NaMN). This metabolite is further converted to NAD+ by two additional enzymatic steps involving Nma1/Nma2 that generate deamido-NAD, and Qns1, which generates NAD+ from deamido-NAD (Rongvaux et al., 2003). Although S. cerevisiae does not possess NAMPT or nicotinamide mononucleotide (NMN), it was shown that components of the NAD+ biosynthesis and salvage pathways interact with Sir2 (Gallo et al., 2004).
Interestingly, also the quite recently discovered nicotinamide riboside salvage pathway is involved in the regulation of Sir2, and affects replicative lifespan. The eukaryotic nicotinamide riboside kinase (Nrk) converts nicotinamide riboside to NAD+ by phosphorylation and adenylylation (Tempel et al., 2007). This pathway promoted Sir2 dependent repression of recombination, improved gene silencing, and extended lifespan without calorie restriction, but was dependent on the rate of NAD+ synthesis (Belenky et al., 2007).
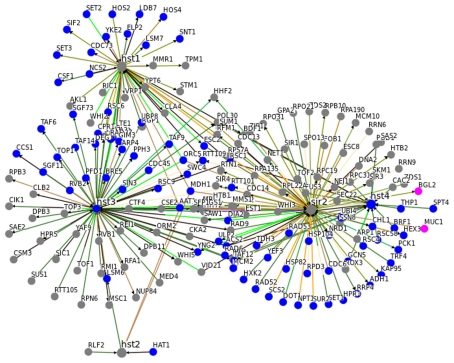
In general, this close association with central metabolic pathways seem to be specific for Sir2. In Figure 3, we illustrate the interaction network of SIR2 and its homologous genes HST1-4. Although they are closely interconnected, the common interactors with a GO annotation metabolism have been associated with assembly or disassembly of the chromatin and, rather then with energy or intermediate-yielding metabolic pathways.
Figure 3.
An interaction network for SIR2 and HST1-4 indicates metabolism-specificity for SIR2. Genetic and physical interaction data for SIR2 and HST1, HST2, HST3, and HST4 was obtained from yeast BioGrid database (01/2012; Stark et al., 2006). Genes associated with GO terms metabolism are highlighted in blue, those for carbon metabolism in pink. Sir2 has the highest number of metabolism related interactors. Moreover, metabolism related HST1-4 interactors predominantly contain proteins involved in assembly, disassembly and repair of chromatin and DNA, rather then enzymes of primary and intermediary metabolism. (See Table A1 in Appendix for GO terms associated with Sir2 interactors).
Sir2 Catalyzed Cleavage of NAD+ as Regulatory Metabolic Reaction
Another interesting, but still in its entire magnitude elusive mechanism in metabolic regulation concerns the putative signaling function of the cleavage products of NAD+ (Lu and Lin, 2010; Qiu et al., 2010b). These are formed when Sir2 is active as a protein deacetylase. At low efficiency, the ADP ribose moiety can be transferred to the protein substrates like histone H4, but the majority yields nicotinamide and a unique metabolite, 2′O-acetyl-ADP-ribose (OAADPR; Imai et al., 2000; Tanner et al., 2000; Moazed, 2001; Tanny and Moazed, 2001; Bitterman et al., 2002; Jackson and Denu, 2002). There is experimental evidence that OAADPR represents a reporter metabolite with signaling function (Borra et al., 2002). In yeast, where NAD+ is concentrated is the millimolar range, OAADPR levels of 0.5 μM have been detected (Lee et al., 2008). Furthermore, Lee et al. (2008) provided strong evidence that SIR2 and its paralogous genes are the only source of this molecule in yeast, and that the Nudix hydrolase Ysa1 is involved in its degradation in vivo.
OAADPR has biological activity, although its exact molecular function remains elusive. Microinjection of OAADPR blocked oocyte maturation and cell division in Asterina miniata (Borra et al., 2002). Additional in vitro studies demonstrated that OAADPR binds to the cation channel TRPM2 (Grubisha et al., 2006) and the histone variant histone macroH2A1.1 (Kustatscher et al., 2005; Tong and Denu, 2010). Moreover, OAADPR promotes the association of multiple copies of Sir3 with Sir2/Sir4 and induces a structural rearrangement in the SIR complex (Liou et al., 2005). These studies suggest OAADPR functions as a signaling molecule and second messenger; however, there is a paucity of information that directly links cellular OAADPR with sirtuin functions.
More details are known about the second molecule, nicotinamide, which is a precursor of nicotinic acid (niacin/vitamin B3). Nicotinamide acts as inhibitor of Sir2. First, it had been reported that yeast silencing, rDNA recombination, and replicative lifespan in sir2Δ yeast depend on nicotinamide concentration (Bitterman et al., 2002). Then, it was demonstrated that the endogenous level of nicotinamide limits Sir2 activity in wild-type yeast cells, and that this property can be antagonized by isonicotinamide, which caused an increase in Sir2 deacetylation activity (Sauve et al., 2005). This property may be explained by a Sir2 catalyzed transglycosidation reaction, were nicotinamide intercepts an ADP-ribosyl-enzyme-acetyl peptide intermediate regenerating NAD+ (Jackson et al., 2003). Thus, high deacetylase activity in the Sir2 microenvironment is likely prevented by efficient product inhibition of Sir2.
Taken together, the Sir2’s catalytic activity interferes in multiple ways with cellular metabolism, and the other way around, changes in metabolism modulate Sir2 activity. The action of Sir2 through its deacetylase activity is better understood, but also the indirect action of Sir2 through its catabolic products OAADPR and nicotinamide seems of high biological relevance. The analysis of the latter under in vivo conditions is very challenging, as it induces a very complex biological response, which demands for further intensive research both at the molecular/biochemical as well as at the systems biology level.
Summary and Conclusion
Stability and dynamics of metabolic networks require the interaction of the small molecule world with the genome and proteome. Sirtuins, at the forefront its founding member Sir2, belong to proteins which fulfill this function in a broad context. They interact with metabolites, proteins and nucleic acids, and spread metabolic signals through these molecular universes. However the complexity of these networks and their interconnectivity makes it difficult to associate unique molecular mechanisms with complex phenotypes such as an extended lifespan. Therefore, there remain several questions to be answered how Sir2 in yeast and the sirtuins in mammals regulate metabolism and thereby increase lifespan and cardiovascular health. Mechanistically, the function of Sir2 in increasing yeast replicative (but not chronological) lifespan is largely overlapping with caloric restriction, and is related to the activity and the stability of the metabolic network. Moreover, Sir2 is present in protein complexes containing major NAD+ producing enzymes in the cell, indicating that this enzyme is supplied or deprived from NAD+ in its microenvironment, dependent on the metabolic activity and flux. While these considerations are intuitively convincing, what is still needed and in most cases still missing, is a detailed chain of events based on biochemistry, which would explain the function of Sir2 and sirtuins in metabolic regulation and consequently in regulation of redox state, the aging process, and the diseases of aging like cancer, cardiovascular disease, and neurodegenerative disease.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kate Campbell (University Cambridge) for critical reading of the manuscript, Aleksey G. Kazantsev (Massachusetts General Hospital) and Hans Lehrach (Max Planck Institute for Molecular Genetics) for support and stimulating discussions. We acknowledge funding from the Max Planck Society, the European Research council (Starting Grant StG-260809 Metabolic Regulators to Markus Ralser). Markus Ralser is a Wellcome Trust Research career development and Wellcome-Beit prize fellow.
Appendix
Table A1.
Data analyzed by GO slim mapper, http://www.yeastgenome.org/, 02/02/2012.
| GOID | GO term | Frequency | Genome frequency | Gene(s) |
|---|---|---|---|---|
| RESULTS FOR THE MAPPING OF 39 GENES TO THE YEAST GO-SLIM FUNCTION | ||||
| 3677 | DNA binding | 5 out of 39 genes, 12.8% | 369 of 6311 genes, 5.8% | MCM2, CDC6, RAD52, ORC5, RAD53 |
| 42393 | Histone binding | 4 out of 39 genes, 10.3% | 39 of 6311 genes, 0.6% | DOT1, GCN5, YNG2, RSC4 |
| 16491 | Oxidoreductase activity | 4 out of 39 genes, 10.3% | 276 of 6311 genes, 4.4% | SUR2, TDH3, MDH1, ADH1 |
| 16746 | Transferase activity, transferring acyl groups | 4 out of 39 genes, 10.3% | 116 of 6311 genes, 1.8% | EPL1, GCN5, YNG2, ESA1 |
| 3723 | RNA binding | 4 out of 39 genes, 10.3% | 753 of 6311 genes, 11.9% | SET1, MDH1, YEF3, NRD1 |
| 16887 | ATPase activity | 4 out of 39 genes, 10.3% | 230 of 6311 genes, 3.6% | CDC6, RSC4, YEF3, HSP82 |
| 16874 | Ligase activity | 3 out of 39 genes, 7.7% | 181 of 6311 genes, 2.9% | SLX5, RAD6, ACS2 |
| 3674 | Molecular function unknown | 3 out of 39 genes, 7.7% | 1990 of 6311 genes, 31.5% | ESC2, RRP4, MUC1 |
| 8168 | Methyltransferase activity | 2 out of 39 genes, 5.1% | 90 of 6311 genes, 1.4% | DOT1, SET1 |
| 16829 | Lyase activity | 2 out of 39 genes, 5.1% | 84 of 6311 genes, 1.3% | PCK1, PAP2 |
| 16301 | Kinase activity | 2 out of 39 genes, 5.1% | 199 of 6311 genes, 3.2% | HXK2, RAD53 |
| 988 | Protein binding transcription factor activity | 2 out of 39 genes, 5.1% | 127 of 6311 genes, 2.0% | GCN5, RPD3 |
| 8289 | Lipid binding | 1 out of 39 genes, 2.6% | 92 of 6311 genes, 1.5% | SCS2 |
| 16810 | Hydrolase activity, acting on carbon–nitrogen (but not peptide) bonds | 1 out of 39 genes, 2.6% | 61 of 6311 genes, 1% | RPD3 |
| 8135 | Translation factor activity, nucleic acid binding | 1 out of 39 genes, 2.6% | 44 of 6311 genes, 0.7% | YEF3 |
| 3729 | mRNA binding | 1 out of 39 genes, 2.6% | 68 of 6311 genes, 1.1% | MDH1 |
| 16757 | Transferase activity, transferring glycosyl groups | 1 out of 39 genes, 2.6% | 100 of 6311 genes, 1.6% | NPT1 |
| 16779 | Nucleotidyltransferase activity | 1 out of 39 genes, 2.6% | 113 of 6311 genes, 1.8% | PAP2 |
| 5198 | Structural molecule activity | 1 out of 39 genes, 2.6% | 356 of 6311 genes, 5.6% | HPR1 |
| 16798 | Hydrolase activity, acting on glycosyl bonds | 1 out of 39 genes, 2.6% | 47 of 6311 genes, 0.7% | BGL2 |
| 8233 | Peptidase activity | 1 out of 39 genes, 2.6% | 137 of 6311 genes, 2.2% | ULP2 |
| 51082 | Unfolded protein binding | 1 out of 39 genes, 2.6% | 66 of 6311 genes, 1.0% | HSP82 |
| 4386 | Helicase activity | 1 out of 39 genes, 2.6% | 80 of 6311 genes, 1.3% | CHL1 |
| 3924 | GTPase activity | 1 out of 39 genes, 2.6% | 58 of 6311 genes, 0.9% | CDC6 |
| 30234 | Enzyme regulator activity | 1 out of 39 genes, 2.6% | 219 of 6311 genes, 3.5% | KAP95 |
| 8565 | Protein transporter activity | 1 out of 39 genes, 2.6% | 52 of 6311 genes, 0.8% | KAP95 |
| 32182 | Small conjugating protein binding | 1 out of 39 genes, 2.6% | 43 of 6311 genes, 0.7% | SLX5 |
| Other | Other | 3 out of 39 genes, 7.7% | MCD1, AAT1, UBI4 | |
| RESULTS FOR THE MAPPING OF 39 GENES TO THE YEAST GO-SLIM PROCESS | ||||
| 6974 | Response to DNA damage stimulus | 13 out of 39 genes, 33.3% | 281 of 6311 genes, 4.5% | MCM2, MCD1, SLX5, HPR1, ESC2, DOT1, EPL1, RAD6, YNG2, RAD52, PAP2, ESA1, RAD53 |
| 6281 | DNA repair | 12 out of 39 genes, 30.8% | 232 of 6311 genes, 3.7% | MCM2, MCD1, HPR1, ESC2, DOT1, EPL1, RAD6, YNG2, RAD52, PAP2, ESA1, RAD53 |
| 6325 | Chromatin organization | 10 out of 39 genes, 25.6% | 223 of 6311 genes, 3.5% | DOT1, EPL1, RAD6, GCN5, YNG2, SET1, RSC4, ACS2, RPD3, ESA1 |
| 6366 | Transcription from RNA polymerase II promoter | 9 out of 39 genes, 23.1% | 438 of 6311 genes, 6.9% | HPR1, EPL1, RAD6, GCN5, SET1, RSC4, NRD1, RPD3, ESA1 |
| 278 | Mitotic cell cycle | 9 out of 39 genes, 23.1% | 288 of 6311 genes, 4.6% | MCM2, MCD1, ESC2, RAD6, ULP2, CDC6, RSC4, RPD3, CHL1 |
| 16570 | Histone modification | 9 out of 39 genes, 23.1% | 98 of 6311 genes, 1.6% | DOT1, EPL1, RAD6, GCN5, YNG2, SET1, ACS2, RPD3, ESA1 |
| 6310 | DNA recombination | 8 out of 39 genes, 20.5% | 149 of 6311 genes, 2.4% | MCM2, HPR1, ESC2, DOT1, RAD6, RAD52, RPD3, CHL1 |
| 51726 | Regulation of cell cycle | 7 out of 39 genes, 17.9% | 183 of 6311 genes, 2.9% | ESC2, DOT1, RAD6, ULP2, RPD3, ESA1, RAD53 |
| 6260 | DNA replication | 7 out of 39 genes, 17.9% | 140 of 6311 genes, 2.2% | MCM2, CDC6, ORC5, RPD3, CHL1, RAD53, HSP82 |
| 43543 | Protein acylation | 6 out of 39 genes, 15.4% | 64 of 6311 genes, 1.0% | EPL1, GCN5, YNG2, SET1, ACS2, ESA1 |
| 18193 | Peptidyl-amino acid modification | 6 out of 39 genes, 15.4% | 83 of 6311 genes, 1.3% | EPL1, GCN5, YNG2, SET1, ACS2, ESA1 |
| 6354 | Transcription elongation, DNA-dependent | 5 out of 39 genes, 12.8% | 72 of 6311 genes, 1.1% | HPR1, GCN5, RSC4, RPD3, ESA1 |
| 70647 | Protein modification by small protein conjugation or removal | 5 out of 39 genes, 12.8% | 149 of 6311 genes, 2.4% | SLX5, RAD6, ULP2, UBI4, KAP95 |
| 5975 | Carbohydrate metabolic process | 4 out of 39 genes, 10.3% | 275 of 6311 genes, 4.4% | HXK2, TDH3, PCK1, ADH1 |
| 6091 | Generation of precursor metabolites and energy | 4 out of 39 genes, 10.3% | 163 of 6311 genes, 2.6% | HXK2, TDH3, MDH1, ADH1 |
| 51052 | Regulation of DNA metabolic process | 4 out of 39 genes, 10.3% | 71 of 6311 genes, 1.1% | CDC6, RPD3, CHL1, HSP82 |
| 51186 | Cofactor metabolic process | 4 out of 39 genes, 10.3% | 164 of 6311 genes, 2.6% | MDH1, ACS2, ADH1, NPT1 |
| 48285 | Organelle fission | 4 out of 39 genes, 10.3% | 124 of 6311 genes, 2% | MCD1, ESC2, ULP2, CHL1 |
| 51321 | Meiotic cell cycle | 4 out of 39 genes, 10.3% | 162 of 6311 genes, 2.6% | DOT1, RAD6, RAD52, RPD3 |
| 32200 | Telomere organization | 4 out of 39 genes, 10.3% | 67 of 6311 genes, 1.1% | SLX5, SET1, RAD52, HSP82 |
| 7059 | Chromosome segregation | 3 out of 39 genes, 7.7% | 131 of 6311 genes, 2.1% | MCD1, ESC2, CHL1 |
| 33043 | Regulation of organelle organization | 3 out of 39 genes, 7.7% | 143 of 6311 genes, 2.3% | SET1, ULP2, HSP82 |
| 6605 | Protein targeting | 3 out of 39 genes, 7.7% | 266 of 6311 genes, 4.2% | SCS2, KAP95, HSP82 |
| 6629 | Lipid metabolic process | 3 out of 39 genes, 7.7% | 262 of 6311 genes, 4.2% | SUR2, SCS2, KAP95 |
| 55086 | Nucleobase-containing small molecule metabolic process | 3 out of 39 genes, 7.7% | 183 of 6311 genes, 2.9% | ADH1, NPT1, RAD53 |
| 51169 | Nuclear transport | 3 out of 39 genes, 7.7% | 163 of 6311 genes, 2.6% | HPR1, SCS2, KAP95 |
| 8213 | Protein alkylation | 2 out of 39 genes, 5.1% | 41 of 6311 genes, 0.6% | DOT1, SET1 |
| 43934 | Sporulation | 2 out of 39 genes, 5.1% | 126 of 6311 genes, 2% | SET1, UBI4 |
| 31399 | Regulation of protein modification process | 2 out of 39 genes, 5.1% | 73 of 6311 genes, 1.2% | SET1, KAP95 |
| 51049 | Regulation of transport | 2 out of 39 genes, 5.1% | 58 of 6311 genes, 0.9% | SCS2, RAD6 |
| 6520 | Cellular amino acid metabolic process | 2 out of 39 genes, 5.1% | 240 of 6311 genes, 3.8% | AAT1, ADH1 |
| 6401 | RNA catabolic process | 2 out of 39 genes, 5.1% | 106 of 6311 genes, 1.7% | RRP4, PAP2 |
| 70271 | Protein complex biogenesis | 2 out of 39 genes, 5.1% | 222 of 6311 genes, 3.5% | KAP95, HSP82 |
| 43144 | snoRNA processing | 2 out of 39 genes, 5.1% | 37 of 6311 genes, 0.6% | RRP4, PAP2 |
| 71554 | Cell wall organization or biogenesis | 1 out of 39 genes, 2.6% | 211 of 6311 genes, 3.3% | BGL2 |
| 6397 | mRNA processing | 1 out of 39 genes, 2.6% | 185 of 6311 genes, 2.9% | HPR1 |
| 6360 | Transcription from RNA polymerase I promoter | 1 out of 39 genes, 2.6% | 63 of 6311 genes, 1% | RPD3 |
| 45333 | Cellular respiration | 1 out of 39 genes, 2.6% | 90 of 6311 genes, 1.4% | MDH1 |
| 6457 | Protein folding | 1 out of 39 genes, 2.6% | 88 of 6311 genes, 1.4% | HSP82 |
| 42221 | Response to chemical stimulus | 1 out of 39 genes, 2.6% | 351 of 6311 genes, 5.6% | HXK2 |
| 6414 | Translational elongation | 1 out of 39 genes, 2.6% | 332 of 6311 genes, 5.3% | YEF3 |
| 7005 | Mitochondrion organization | 1 out of 39 genes, 2.6% | 333 of 6311 genes, 5.3% | HSP82 |
| 6353 | Transcription termination, DNA-dependent | 1 out of 39 genes, 2.6% | 34 of 6311 genes, 0.5% | NRD1 |
| 15931 | Nucleobase-containing compound transport | 1 out of 39 genes, 2.6% | 116 of 6311 genes, 1.8% | HPR1 |
| 6869 | Lipid transport | 1 out of 39 genes, 2.6% | 49 of 6311 genes, 0.8% | SCS2 |
| 55085 | Transmembrane transport | 1 out of 39 genes, 2.6% | 186 of 6311 genes, 2.9% | SCS2 |
| 51603 | Proteolysis involved in cellular protein catabolic process | 1 out of 39 genes, 2.6% | 192 of 6311 genes, 3.0% | RAD6 |
| 6997 | Nucleus organization | 1 out of 39 genes, 2.6% | 54 of 6311 genes, 0.9% | KAP95 |
| 7124 | Pseudohyphal growth | 1 out of 39 genes, 2.6% | 65 of 6311 genes, 1.0% | MUC1 |
| 8643 | Carbohydrate transport | 1 out of 39 genes, 2.6% | 38 of 6311 genes, 0.6% | HXK2 |
| 6364 | rRNA processing | 1 out of 39 genes, 2.6% | 294 of 6311 genes, 4.7% | RRP4 |
| 1403 | Invasive growth in response to glucose limitation | 1 out of 39 genes, 2.6% | 52 of 6311 genes, 0.8% | MUC1 |
| 9408 | Response to heat | 1 out of 39 genes, 2.6% | 47 of 6311 genes, 0.7% | RPD3 |
| 48308 | Organelle inheritance | 1 out of 39 genes, 2.6% | 51 of 6311 genes, 0.8% | SCS2 |
| 6970 | Response to osmotic stress | 1 out of 39 genes, 2.6% | 75 of 6311 genes, 1.2% | HSP82 |
References
- Aguilaniu H., Gustafsson L., Rigoulet M., Nystrom T. (2003). Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299, 1751–1753 10.1126/science.1080418 [DOI] [PubMed] [Google Scholar]
- Anastasiou D., Poulogiannis G., Asara J. M., Boxer M. B., Jiang J. K., Shen M., Bellinger G., Sasaki A. T., Locasale J. W., Auld D. S., Thomas C. J., Vander M. G., Heiden, Cantley L. C. (2011). Inhibition of pyruvate kinase m2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283 10.1126/science.1211485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. M., Latorre-Esteves M., Neves A. R., Lavu S., Medvedik O., Taylor C., Howitz K. T., Santos H., Sinclair D. A. (2003). Yeast life-span extension by calorie restriction is independent of NAD fluctuation. Science 302, 2124–2126 10.1126/science.1088697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky P., Racette F. G., Bogan K. L., McClure J. M., Smith J. S., Brenner C. (2007). Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129, 473–484 10.1016/j.cell.2007.03.024 [DOI] [PubMed] [Google Scholar]
- Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A. (2002). Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277, 45099–45107 10.1074/jbc.M205670200 [DOI] [PubMed] [Google Scholar]
- Blagosklonny M., Hall M. (2009). Growth and aging: a common molecular mechanism. Aging 1, 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny M. V. (2008). Aging: ROS or TOR. Cell Cycle 7, 3344–3354 10.4161/cc.7.17.6626 [DOI] [PubMed] [Google Scholar]
- Blander G., Guarente L. (2004). The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435 10.1146/annurev.biochem.73.011303.073651 [DOI] [PubMed] [Google Scholar]
- Blank L. M., Kuepfer L., Sauer U. (2005). Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 6, R49. 10.1186/gb-2005-6-6-r49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C., Benguria A., Wawryn J., Jazwinski S. M. (2004). Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics 166, 765–777 10.1534/genetics.166.2.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra M. T., O’Neill F. J., Jackson M. D., Marshall B., Verdin E., Foltz K. R., Denu J. M. (2002). Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem. 277, 12632–12641 10.1074/jbc.M111830200 [DOI] [PubMed] [Google Scholar]
- Brandes N., Schmitt S., Jakob U. (2009). Thiol-based redox switches in eukaryotic proteins. Antioxid. Redox Signal 11, 997–1014 10.1089/ars.2008.2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenbach M., Laun P., Dickinson J. R., Klocker A., Rinnerthaler M., Dawes I. W., Aung-Htut M. T., Breitenbach-Koller L., Caballero A., Nystrom T., Buttner S., Eisenberg T., Madeo F., Ralser M. (2012). The role of mitochondria in the aging processes of yeast. Subcell. Biochem. 57, 55–78 10.1007/978-94-007-2561-4_3 [DOI] [PubMed] [Google Scholar]
- Bruice T. C. (2002). A view at the millennium: the efficiency of enzymatic catalysis. Acc. Chem. Res. 35, 139–148 10.1021/ar0001665 [DOI] [PubMed] [Google Scholar]
- Burnett C., Valentini S., Cabreiro F., Goss M., Somogyvari M., Piper M. D., Hoddinott M., Sutphin G. L., Leko V., McElwee J. J., Vazquez-Manrique R. P., Orfila A. M., Ackerman D., Au C., Vinti G., Riesen M., Howard K., Neri C., Bedalov A., Kaeberlein M., Soti C., Partridge L., Gems D. (2011). Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477, 482–485 10.1038/nature10296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas E., Davies K. J. (2000). Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 29, 222–230 10.1016/S0891-5849(00)00317-8 [DOI] [PubMed] [Google Scholar]
- Cakir T., Patil K. R., Onsan Z., Ulgen K. O., Kirdar B., Nielsen J. (2006). Integration of metabolome data with metabolic networks reveals reporter reactions. Mol. Syst. Biol. 2, 50. 10.1038/msb4100085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castegna A., Scarcia P., Agrimi G., Palmieri L., Rottensteiner H., Spera I., Germinario L., Palmieri F. (2011). Identification and functional characterization of a novel mitochondrial carrier for citrate and oxoglutarate in Saccharomyces cerevisiae. J. Biol. Chem. 285, 17359–17370 10.1074/jbc.M109.097188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechik G., Oh E., Rando O., Weissman J., Regev A., Koller D. (2008). Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nat. Biotechnol. 26, 1251–1259 10.1038/nbt.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Steele A. D., Lindquist S., Guarente L. (2005). Increase in activity during calorie restriction requires Sirt1. Science 310, 1641. 10.1126/science.1120385 [DOI] [PubMed] [Google Scholar]
- Defossez P. A., Park P. U., Guarente L. (1998). Vicious circles: a mechanism for yeast aging. Curr. Opin. Microbiol. 1, 707–711 10.1016/S1369-5274(98)80119-7 [DOI] [PubMed] [Google Scholar]
- Defossez P. A., Prusty R., Kaeberlein M., Lin S. J., Ferrigno P., Silver P. A., Keil R. L., Guarente L. (1999). Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3, 447–455 10.1016/S1097-2765(00)80472-4 [DOI] [PubMed] [Google Scholar]
- Derbyshire M. K., Weinstock K. G., Strathern J. N. (1996). HST1, a new member of the SIR2 family of genes. Yeast 12, 631–640 [DOI] [PubMed] [Google Scholar]
- Easlon E., Tsang F., Skinner C., Wang C., Lin S. J. (2008). The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 22, 931–944 10.1101/gad.1648308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Gattazzo C., Battistella L., Wei M., Cheng C., McGrew K., Longo V. D. (2005). Sir2 blocks extreme life-span extension. Cell 123, 655–667 10.1016/j.cell.2005.08.042 [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Longo V. D. (2003). The chronological life span of Saccharomyces cerevisiae. Aging Cell 2, 73–81 10.1046/j.1474-9728.2003.00033.x [DOI] [PubMed] [Google Scholar]
- Fritze C. E., Verschueren K., Strich R., Easton Esposito R. (1997). Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16, 6495–6509 10.1093/emboj/16.21.6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo C. M., Smith D. L., Jr., Smith J. S. (2004). Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 24, 1301–1312 10.1128/MCB.24.3.1301-1312.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A. C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Hofert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 10.1038/415141a [DOI] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R. E. (1989). A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56, 771–776 10.1016/0092-8674(89)90681-8 [DOI] [PubMed] [Google Scholar]
- Gottschling D. E. (2000). Gene silencing: two faces of SIR2. Curr. Biol. 10, R708–R711 10.1016/S0960-9822(00)00714-4 [DOI] [PubMed] [Google Scholar]
- Grant C. M., MacIver F. H., Dawes I. W. (1996). Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29, 511–515 10.1007/BF02426954 [DOI] [PubMed] [Google Scholar]
- Gruber J., Schaffer S., Halliwell B. (2008). The mitochondrial free radical theory of ageing – where do we stand? Front. Biosci. 13, 6554–6579 10.2741/3174 [DOI] [PubMed] [Google Scholar]
- Grubisha O., Rafty L. A., Takanishi C. L., Xu X., Tong L., Perraud A. L., Scharenberg A. M., Denu J. M. (2006). Metabolite of SIR2 reaction modulates TRPM2 ion channel. J. Biol. Chem. 281, 14057–14065 10.1074/jbc.M513741200 [DOI] [PubMed] [Google Scholar]
- Grüning N. M., Lehrach H., Ralser M. (2010). Regulatory crosstalk of the metabolic network. Trends Biochem. Sci. 35, 220–227 10.1016/j.tibs.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Grüning N.-M., Ralser M. (2011). Cancer: sacrifice for survival. Nature 480, 190–191 10.1038/480190a [DOI] [PubMed] [Google Scholar]
- Gruning N. M., Rinnerthaler M., Bluemlein K., Mulleder M., Wamelink M. M., Lehrach H., Jakobs C., Breitenbach M., Ralser M. (2011). Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 14, 415–427 10.1016/j.cmet.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (1999). Diverse and dynamic functions of the Sir silencing complex. Nat. Genet. 23, 281–285 10.1038/15458 [DOI] [PubMed] [Google Scholar]
- Ha C. W., Huh W. K. (2011). Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 39, 1336–1350 10.1093/nar/gkq895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeren G., Rinnerthaler M., Laun P., von Seyerl P., Kössler S., Klinger H., Hager M., Bogengruber E., Jarolim S., Simon-Nobbe B., Schüller C., Carmona-Gutierrez D., Breitenbach-Koller L., Mück C., Jansen-Dürr P., Criollo A., Kroemer G., Madeo F., Breitenbach M. (2009). The mitochondrial ribosomal protein of the large subunit, Afo1p, determines cellular longevity through mitochondrial back-signaling via TOR1. Aging 1, 622–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann M., Sauer U. (2010). Systems biology of microbial metabolism. Curr. Opin. Microbiol. 13, 337–343 10.1016/j.mib.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Herranz D., Serrano M. (2010). SIRT1: recent lessons from mouse models. Nat. Rev. Cancer 10, 819–823 10.1038/nrc2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I., Oshima Y. (1981). “Control of cell type in yeast,” in The Molecular Biology of the Yeast Saccharomyces. Life Cycle and Inheritance, eds Strathern J. N., Jones E. W., Broach J. R. (Cold Spring Harbor: Cold Spring Harbor Laboratory Press; ), 181–209 [Google Scholar]
- Huang Y. (2002). Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 30, 1465–1482 10.1093/nar/30.3.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 10.1038/35001622 [DOI] [PubMed] [Google Scholar]
- Ishii N., Nakahigashi K., Baba T., Robert M., Soga T., Kanai A., Hirasawa T., Naba M., Hirai K., Hoque A., Ho P. Y., Kakazu Y., Sugawara K., Igarashi S., Harada S., Masuda T., Sugiyama N., Togashi T., Hasegawa M., Takai Y., Yugi K., Arakawa K., Iwata N., Toya Y., Nakayama Y., Nishioka T., Shimizu K., Mori H., Tomita M. (2007). Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316, 593–597 10.1126/science.1132067 [DOI] [PubMed] [Google Scholar]
- Jackson M. D., Denu J. M. (2002). Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta-NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 277, 18535–18544 10.1074/jbc.M203523200 [DOI] [PubMed] [Google Scholar]
- Jackson M. D., Schmidt M. T., Oppenheimer N. J., Denu J. M. (2003). Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J. Biol. Chem. 278, 50985–50998 10.1074/jbc.M306552200 [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L. (1999). The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 10.1101/gad.13.19.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R. W., III, Steffen K. K., Westman E. A., Hu D., Dang N., Kerr E. O., Kirkland K. T., Fields S., Kennedy B. K. (2005). Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 10.1126/science.1115535 [DOI] [PubMed] [Google Scholar]
- Kennedy B. K., Austriaco N. R., Jr., Zhang J., Guarente L. (1995). Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80, 485–496 [DOI] [PubMed] [Google Scholar]
- Kreimer A., Borenstein E., Gophna U., Ruppin E. (2008). The evolution of modularity in bacterial metabolic networks. Proc. Natl. Acad. Sci. U.S.A. 105, 6976–6981 10.1073/pnas.0712149105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger A., Gruning N. M., Wamelink M. M., Kerick M., Kirpy A., Parkhomchuk D., Bluemlein K., Schweiger M. R., Soldatov A., Lehrach H., Jakobs C., Ralser M. (2011). The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid. Redox Signal. 15, 311–324 10.1089/ars.2010.3797 [DOI] [PubMed] [Google Scholar]
- Kuepfer L., Sauer U., Blank L. M. (2005). Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res. 15, 1421–1430 10.1101/gr.3992505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A. G. (2005). Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 12, 624–625 10.1038/nsmb956 [DOI] [PubMed] [Google Scholar]
- Lamming D. W., Latorre-Esteves M., Medvedik O., Wong S. N., Tsang F. A., Wang C., Lin S. J., Sinclair D. A. (2005). HST2 mediates SIR2-independent life-span extension by calorie restriction. Science 309, 1861–1864 10.1126/science.1113611 [DOI] [PubMed] [Google Scholar]
- Lamming D. W., Wood J. G., Sinclair D. A. (2004). Small molecules that regulate lifespan: evidence for xenohormesis. Mol. Microbiol. 53, 1003–1009 10.1111/j.1365-2958.2004.04209.x [DOI] [PubMed] [Google Scholar]
- Larochelle M., Drouin S., Robert F., Turcotte B. (2006). Oxidative stress-activated zinc cluster protein Stb5 has dual activator/repressor functions required for pentose phosphate pathway regulation and NADPH production. Mol. Cell. Biol. 26, 6690–6701 10.1128/MCB.02450-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laun P., Rinnerthaler M., Bogengruber E., Heeren G., Breitenbach M. (2006). Yeast as a model for chronological and reproductive aging – a comparison. Exp. Gerontol. 41, 1208–1212 10.1016/j.exger.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Lee M. N., Ha S. H., Kim J., Koh A., Lee C. S., Kim J. H., Jeon H., Kim D. H., Suh P. G., Ryu S. H. (2009). Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol. Cell. Biol. 29, 3991–4001 10.1128/MCB.00165-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Tong L., Denu J. M. (2008). Quantification of endogenous sirtuin metabolite O-acetyl-ADP-ribose. Anal. Biochem. 383, 174–179 10.1016/j.ab.2008.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. L., Lee C. K. (2008). Transcriptional response according to strength of calorie restriction in Saccharomyces cerevisiae. Mol. Cells 26, 299–307 [PubMed] [Google Scholar]
- Li B., Skinner C., Castello P. R., Kato M., Easlon E., Xie L., Li T., Lu S. P., Wang C., Tsang F., Poyton R. O., Lin S. J. (2011). Identification of potential calorie restriction-mimicking yeast mutants with increased mitochondrial respiratory chain and nitric oxide levels. J. Aging Res. 2011, 673185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sun L., Liang Q., Wang J., Mo W., Zhou B. (2006). Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Mol. Biol. Cell 17, 1802–1811 10.1091/mbc.E05-04-0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. J., Defossez P. A., Guarente L. (2000). Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 10.1126/science.289.5487.2126 [DOI] [PubMed] [Google Scholar]
- Lin S. J., Kaeberlein M., Andalis A. A., Sturtz L. A., Defossez P. A., Culotta V. C., Fink G. R., Guarente L. (2002). Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418, 344–348 10.1038/nature00904 [DOI] [PubMed] [Google Scholar]
- Liou G. G., Tanny J. C., Kruger R. G., Walz T., Moazed D. (2005). Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121, 515–527 10.1016/j.cell.2005.03.035 [DOI] [PubMed] [Google Scholar]
- Lu S.-P., Lin S.-J. (2010). Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim. Biophys. Acta 1804, 1567–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttik M. A., Overkamp K. M., Kotter P., de Vries S., van Dijken J. P., Pronk J. T. (1998). The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem. 273, 24529–24534 10.1074/jbc.273.38.24529 [DOI] [PubMed] [Google Scholar]
- Martin C. E., Oh C. S., Jiang Y. (2007). Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 1771, 271–285 [DOI] [PubMed] [Google Scholar]
- Martin G. M. (1997). The Werner mutation: does it lead to a “public” or “private” mechanism of aging? Mol. Med. 3, 356–358 [PMC free article] [PubMed] [Google Scholar]
- Martin G. M., Austad S. N., Johnson T. E. (1996). Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat. Genet. 13, 25–34 10.1038/ng0596-25 [DOI] [PubMed] [Google Scholar]
- Massey V., Williams C. H., Jr. (1965). On the reaction mechanism of yeast glutathione reductase. J. Biol. Chem. 240, 4470–4480 [PubMed] [Google Scholar]
- Matecic M., Stuart S., Holmes S. G. (2002). SIR2-induced inviability is suppressed by histone H4 overexpression. Genetics 162, 973–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney M. W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J. R., Lansdorp P. M., Lemieux M. (2003). The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23, 38–54 10.1128/MCB.23.1.38-54.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray M. A., Gottschling D. E. (2004). Aging and genetic instability in yeast. Curr. Opin. Microbiol. 7, 673–679 10.1016/j.mib.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Meister A., Anderson M. E. (1983). Glutathione. Annu. Rev. Biochem. 52, 711–760 10.1146/annurev.bi.52.070183.003431 [DOI] [PubMed] [Google Scholar]
- Mekhail K., Moazed D. (2010). The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 11, 317–328 10.1038/nrg2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. (2001). Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13, 232–238 10.1016/S0955-0674(00)00202-7 [DOI] [PubMed] [Google Scholar]
- Moazed D., Kistler A., Axelrod A., Rine J., Johnson A. D. (1997). Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. U.S.A. 94, 2186–2191 10.1073/pnas.94.6.2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Johnston J. R. (1959). Life span of individual yeast cells. Nature 183, 1751–1752 10.1038/1831751a0 [DOI] [PubMed] [Google Scholar]
- Muller F. L., Lustgarten M. S., Jang Y., Richardson A., Van Remmen H. (2007). Trends in oxidative aging theories. Free Radic. Biol. Med. 43, 477–503 10.1016/j.freeradbiomed.2007.03.034 [DOI] [PubMed] [Google Scholar]
- Novo E., Parola M. (2008). Redox mechanisms in hepatic chronic wound healing and fibrogenesis. Fibrogenesis Tissue Repair 1, 5. 10.1186/1755-1536-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira G. A., Tahara E. B., Gombert A. K., Barros M. H., Kowaltowski A. J. (2008). Increased aerobic metabolism is essential for the beneficial effects of caloric restriction on yeast life span. J. Bioenerg. Biomembr. 40, 381–388 10.1007/s10863-008-9159-5 [DOI] [PubMed] [Google Scholar]
- Parter M., Kashtan N., Alon U. (2007). Environmental variability and modularity of bacterial metabolic networks. BMC Evol. Biol. 7, 169. 10.1186/1471-2148-7-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil K. R., Nielsen J. (2005). Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. U.S.A. 102, 2685–2689 10.1073/pnas.0406811102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson B., Vaur S., Sagot I., Coulpier F., Lemoine S., Daignan-Fornier B. (2009). Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev. 23, 1399–1407 10.1101/gad.521809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma L., Lehrach H., Ralser M. (2009). Surviving in the cold: yeast mutants with extended hibernating lifespan are oxidant sensitive. Aging 1, 957–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers R. W., III, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S. (2006). Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20, 174–184 10.1101/gad.1381406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Brown K., Hirschey M. D., Verdin E., Chen D. (2010a). Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 12, 662–667 10.1016/j.cmet.2010.11.015 [DOI] [PubMed] [Google Scholar]
- Qiu X., Brown K. V., Moran Y., Chen D. (2010b). Sirtuin regulation in calorie restriction. Biochim. Biophys. Acta 1804, 1576–1583 10.1016/j.bbapap.2009.09.015 [DOI] [PubMed] [Google Scholar]
- Raisner R. M., Madhani H. D. (2008). Genomewide screen for negative regulators of sirtuin activity in Saccharomyces cerevisiae reveals 40 loci and links to metabolism. Genetics 179, 1933–1944 10.1534/genetics.108.088443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran N. S., Connell P., Christians E. S., Yan L. J., Taylor R. P., Orosz A., Zhang X. Q., Stevenson T. J., Peshock R. M., Leopold J. A., Barry W. H., Loscalzo J., Odelberg S. J., Benjamin I. J. (2007). Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell 130, 427–439 10.1016/j.cell.2007.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser M., Wamelink M. M., Kowald A., Gerisch B., Heeren G., Struys E. A., Klipp E., Jakobs C., Breitenbach M., Lehrach H., Krobitsch S. (2007). Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 6, 10. 10.1186/jbiol61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser M., Wamelink M. M., Latkolik S., Jansen E. E., Lehrach H., Jakobs C. (2009a). Metabolic reconfiguration precedes transcriptional regulation in the antioxidant response. Nat. Biotechnol. 27, 604–605 10.1038/nbt0709-604 [DOI] [PubMed] [Google Scholar]
- Ralser M., Zeidler U., Lehrach H. (2009b). Interfering with glycolysis causes Sir2-dependent hyper-recombination of Saccharomyces cerevisiae plasmids. PLoS ONE 4, e5376. 10.1371/journal.pone.0005376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter-Branchat G., Cabiscol E., Tamarit J., Sorolla M. A., Angeles de la Torre M., Ros J. (2007). Chronological and replicative life-span extension in Saccharomyces cerevisiae by increased dosage of alcohol dehydrogenase 1. Microbiology 153(Pt 11), 3667–3676 10.1099/mic.0.2007/009340-0 [DOI] [PubMed] [Google Scholar]
- Rine J., Strathern J. N., Hicks J. B., Herskowitz I. (1979). A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics 93, 877–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B., Helfand S. L. (2004). Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. U.S.A. 101, 15998–6003 10.1073/pnas.0404184101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A., Andris F., Van Gool F., Leo O. (2003). Reconstructing eukaryotic NAD metabolism. Bioessays 25, 683–690 10.1002/bies.10297 [DOI] [PubMed] [Google Scholar]
- Rye P. T., Frick L. E., Ozbal C. C., Lamarr W. A. (2011). Advances in label-free screening approaches for studying sirtuin-mediated deacetylation. J. Biomol. Screen. 16, 1217–1226 10.1177/1087057111418653 [DOI] [PubMed] [Google Scholar]
- Sauve A. A., Moir R. D., Schramm V. L., Willis I. M. (2005). Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol. Cell 17, 595–601 10.1016/j.molcel.2004.12.032 [DOI] [PubMed] [Google Scholar]
- Schafer F. Q., Buettner G. R. (2001). Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30, 1191–1212 10.1016/S0891-5849(01)00480-4 [DOI] [PubMed] [Google Scholar]
- Sies H. (1997). Oxidative stress: oxidants and antioxidants. Exp. Physiol. 82, 291–295 [DOI] [PubMed] [Google Scholar]
- Silva R. M., Duarte I. C. N., Paredes J. A., Lima-Costa T., Perrot M., Boucherie H., Goodfellow B. J., Gomes A. C., Mateus D. D., Moura G. R., Santos M. A. S. (2009). The yeast PNC1 longevity gene is up-regulated by mRNA mistranslation. PLoS ONE 4, e5212. 10.1371/journal.pone.0005212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D. A., Guarente L. (1997). Extrachromosomal rDNA circles – a cause of aging in yeast. Cell 91, 1033–1042 10.1016/S0092-8674(00)80493-6 [DOI] [PubMed] [Google Scholar]
- Smith D. L., Jr., McClure J. M., Matecic M., Smith J. S. (2007). Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell 6, 649–662 10.1111/j.1474-9726.2007.00326.x [DOI] [PubMed] [Google Scholar]
- Sohal R. S., Weindruch R. (1996). Oxidative stress, caloric restriction, and aging. Science 273, 59–63 10.1126/science.273.5271.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B. J., Reguly T., Boucher L., Breitkreutz A., Tyers M. (2006). BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M. (1997). SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11, 83–93 10.1101/gad.11.1.83 [DOI] [PubMed] [Google Scholar]
- Tanner K. G., Landry J., Sternglanz R., Denu J. M. (2000). Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 97, 14178–14182 10.1073/pnas.250422697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J. C., Moazed D. (2001). Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. U.S.A. 98, 415–420 10.1073/pnas.031563798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel W., Rabeh W. M., Bogan K. L., Belenky P., Wojcik M., Seidle H. F., Nedyalkova L., Yang T., Sauve A. A., Park H. W., Brenner C. (2007). Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol. 5, e263. 10.1371/journal.pbio.0050263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermann B., Jarolim S., Russmayer H., Kerick M., Michel S., Kruger A., Bluemlein K., Laun P., Grillari J., Lehrach H., Breitenbach M., Ralser M. (2010). A new dominant peroxiredoxin allele identified by whole-genome re-sequencing of random mutagenized yeast causes oxidant-resistance and premature aging. Aging (Albany N. Y.) 2, 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H. A., Guarente L. (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230 10.1038/35065638 [DOI] [PubMed] [Google Scholar]
- Tong L., Denu J. M. (2010). Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochim. Biophys. Acta 1804, 1617–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristan C., Shahani N., Sedlak T. W., Sawa A. (2011). The diverse functions of GAPDH: views from different subcellular compartments. Cell. Signal. 23, 317–323 10.1016/j.cellsig.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M., Guarente L. (2011). Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature 477, E1–E2 10.1038/nj7364-1 [DOI] [PubMed] [Google Scholar]
- Wamelink M. M., Struys E. A., Jakobs C. (2008). The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: a review. J. Inherit. Metab. Dis. 31, 703–717 10.1007/s10545-008-1015-6 [DOI] [PubMed] [Google Scholar]
- Warburg O. (1956). Origin of cancer cells. Science 123, 309–314 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- Westphal C. H., Dipp M. A., Guarente L. (2007). A therapeutic role for sirtuins in diseases of aging? Trends Biochem. Sci. 32, 555–560 10.1016/j.tibs.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Wilhelm J., Fuksova H., Schwippelova Z., Vytasek R., Pichova A. (2006). The effects of reactive oxygen and nitrogen species during yeast replicative ageing. Biofactors 27, 185–193 10.1002/biof.5520270116 [DOI] [PubMed] [Google Scholar]
- Yu J., Auwerx J. (2009). The role of sirtuins in the control of metabolic homeostasis. Ann. N. Y. Acad. Sci. 1173(Suppl. 1), E10–E19 10.1111/j.1749-6632.2009.04952.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Qin W., Zhuang G., Zhang X., Chen G., Liu W. (2009). Monitoring oxidative stress and DNA damage induced by heavy metals in yeast expressing a redox-sensitive green fluorescent protein. Curr. Microbiol. 58, 504–510 10.1007/s00284-008-9331-5 [DOI] [PubMed] [Google Scholar]