SUMMARY
Streptococcus mutans is generally recognized as a causative agent of human dental caries. The production of mutacins (bacteriocins) by S. mutans is considered to be an important factor in the colonization and establishment of S. mutans in the dental biofilm. Two types of mutacins have been characterized: the lantibiotics and the non-lantibiotics. The lantibiotics generally have a wider spectrum of activity than the non-lantibiotics, which make them attractive targets for development into new antimicrobial modalities. The non-lantibiotics are much more prevalent among strains of S. mutans and play a significant role in both community and population level interactions in the dental biofilm. These interactions are directly mediated through the ComCDE two-component system and the newly characterized LytTR Regulation Systems HdrRM and BrsRM. These systems coordinate natural competence development and mutacin production as a means to acquire transforming DNA either by killing closely related streptococcal species in the vicinity of S. mutans, or through an altruistic suicide mechanism among a subpopulation of competent cells within the S. mutans community. As more S. mutans strains are sequenced, it is anticipated that additional mutacins with novel functions will be discovered, which may yield further insights into the ecological role of mutacins within the oral biofilm.
Keywords: mutacin, Streptococcus mutans, bacteriocin
INTRODUCTION
Streptococcus mutans is a gram-positive, facultative anaerobic bacterium that inhabits the human oral cavity and is commonly recognized as a primary pathogen in dental caries. One of its principal virulence factors is the production of bacteriocins (peptide antibiotics) referred to as mutacins (Hamada & Ooshima, 1975a). In fact, S. mutans is one of the most prolific producers of bacteriocins within the entire Streptococcus genus. For example, the sequenced reference strain UA159 was originally considered to be a mutacin-negative wild type. However, since the time of its sequencing, this strain has been shown to actively produce three separate bacteriocins and it likely encodes at least several more! Hence, it is fair to say that it is probably difficult, if not impossible, to identify a true mutacin-negative wild type strain of S. mutans. This suggests that mutacin production likely plays a fundamental role in the survival of the species. Mutacins may help S. mutans to compete with other streptococci in early dental biofilms and allow it to establish or sustain colonization of the tooth surface (Kuramitsu et al., 2007). On the other hand, because the oral cavity is one of the primary portals of entry for human pathogens, it is conceivable that the production of mutacins could even play an important role in protecting the host from many harmful infections, similar to observations made with other oral streptococci [for reviews, see (Burton et al., 2011, Tagg, 2004, Tagg & Dierksen, 2003)]. In the following review, we examine the current state of mutacin research and give particular attention to mutacin gene regulation and the potential roles of mutacins in S. mutans ecology, as these details have yet to be compiled in a review format.
History of mutacins
The term “mutacin” was first coined by Hamada and Ooshima (Hamada & Ooshima, 1975b) and it refers specifically to the bacteriocins produced by S. mutans. Multiple examples of mutacins from both of the major classes of bacteriocins (the lantibiotics and non-lantibiotics) have been characterized to date (Table 1). The lantibiotic mutacins tend to have a wide spectrum of activity against gram-positive bacteria, including non-producing strains of S. mutans, whereas the non-lantibiotic mutacins have thus far proven to be primarily active against closely related species. The mutacin repertoire of individual strains of S. mutans also tends to be highly strain-specific. For this reason, mutacin types had been previously used as an epidemiological tool to track the transmission of S. mutans before the dawn of the molecular age (Baca et al., 1990, Davey & Rogers, 1984, Rogers, 1980).
Table 1.
Characterized mutacins
| Name | Mature peptide Sequence | Producer strains | Regulator |
|---|---|---|---|
| Lantibiotics* | |||
| Mutacin I | FSSLSLCSLGCTGVKNPSFNSYCC | UA140, CH43 | MutR |
| Mutacin II | NRWWQGVVPTVSYECRMNSWQHVFTCC | T8 | MutR |
| Mutacin III/1140 | FKSWSLCTPGCARTGSFNSYCC | UA787, JH1140 | MutR |
| Mutacin B-NY266 | FKSWSFCTPGCAKTGSFNSYCC | NY266 | |
| Mutacin Smb | STPACAIGVVGITVAVTGISTACTSRCINK (SmbA) GTTVVNSTFSIVLGNKGYICTVTVECMRNCSK (SmbB) |
GS5 | ComE, HdrR?, BrsR? |
| Mutacin K8 | MGKGAVGTISHECRYNSWAFLATCCS | K8 | MukR? |
| Non-lantibiotics | |||
| Mutacin IV | KVSGGEAVAAIGICATASAAIGGLAGATLVTPYCVGTWGLIRSH (NlmA) DKQAADTFLSACGGAASGFTYCASNGVWHPYILAGCAGVGAVGSVVFPH (NlmB) |
UA140, UA159, 25175, L13 | ComE, HdrR, BrsR |
| Mutacin V | GRGWNCAAGIALGAGQGYMATAGGTAF LGPYAIGTGAFGAIAGGIGGALNSCG |
UA159, UA140 | ComE, HdrR, BrsR |
| Mutacin VI | GMIRCALGTAGSAGLGFVGGMGAGTVT LPVVGTVSGAALGGWSGAAVGAATFC |
UA159 | ComE, HdrR, BrsR |
| Mutacin N | SRQAADTFLSGAYGAAKGVTARASTGVYVVPATLVALGVYGAGLNIAFP | N | ComE? |
Bold letters denote residues involved in thioether linkages in the mature peptide.
Lantibiotic mutacins
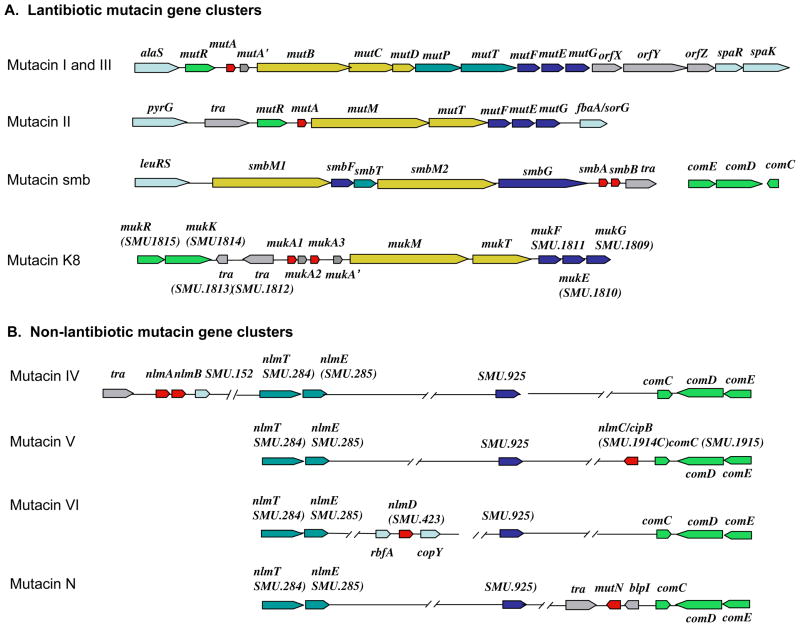
The currently characterized lantibiotic mutacins include mutacin I (Qi et al., 2000), mutacin II (Novak et al., 1994), mutacin III/mutacin 1140 (these two mutacins have identical sequences. See Hillman et al., 1998, Qi et al., 1999b), mutacin B-NY266 (Mota-Meira et al., 1997), mutacin K8 (Robson et al., 2007), and mutacin Smb (Yonezawa & Kuramitsu, 2005). The term “lantibiotic” refers to the characteristic dehydrated amino acids and intramolecular thioether bonds that create lanthionine and methyllanthionine residues within the bacteriocin molecule (Jung, 1991). These unusual structures result from extensive post-translational modifications of the prepeptide and are crucial for the inhibitory activity of the bacteriocin. They also confer tremendous stability to the peptide making them highly resistant to inactivation under a wide range of environmental extremes. Lantibiotics (also referred to as Class I bacteriocins) may consist of a single peptide, such as mutacins I, II, III, 1140, B-NY266, and K8, or two peptides such as mutacin Smb. Based upon their tertiary structures, lantibiotics are classified either as Type A (linear) or Type B (globular) (Jung, 1991). All characterized lantibiotic mutacins belong to the Type A class, which kills susceptible cells primarily through membrane pore formation. Recent studies have demonstrated that some Type A lantibiotics, such as nisin as well as mutacin 1140, use lipid II as docking molecules for pore formation (Hasper et al., 2006). Consequently, this also results in the simultaneous inhibition of cell wall synthesis, likely compounding the bacteriocidal activity. This mode of action may explain why lantibiotics have such a wide spectrum of activity against gram-positive bacteria. Genes for the lantibiotic precursor, the modification of the prepeptide, transport/processing of the propeptide, and immunity against self-toxicity are encoded within the same gene cluster (Fig. 1) and transcribed in a coordinated fashion. The production of lantibiotics requires multiple dedicated enzymes and occurs through conserved mechanisms that have been reviewed previously. Interested readers are directed to these excellent reviews for additional information (Bierbaum & Sahl, 2009, Guder et al., 2000, Kupke & Gotz, 1996, Sahl et al., 1995, Willey & van der Donk, 2007).
Fig. 1.
Gene clusters of characterized lantibiotic mutacins. Green: mutacin transcription regulators; red: structural genes for the prepro-mutacin peptides; yellow: modification enzymes for the prepeptide; dark green: transportation and processing enzymes for the propeptide; dark blue: immunity proteins; gray: transposases or other genes not required for mutacin production; light blue: flanking genes outside the locus.
Non-lantibiotic mutacins
In contrast to the lantibiotic mutacins, which are produced by only a small minority of isolates (Bekal-Si Ali et al., 2002, Kamiya et al., 2005), the non-lantibiotic mutacin genes are present in every S. mutans strain analyzed thus far. Typically, individual strains encode multiple non-lantibiotic mutacin genes. Unlike the lantibiotics, the non-lantibiotic mutacins are not post-translationally modified and their antimicrobial spectra are much narrower than that of the lantibiotic mutacins. For example, mutacin IV is active against numerous non-mutans streptococcal species, but is inactive against the mutans streptococci, staphylococci, or enterococci (Qi et al., 2001). Currently, four non-lantibiotic mutacins have been characterized (mutacins IV, V, VI, and mutacin N) (Balakrishnan et al., 2000, Hale et al., 2005b, Qi et al., 2001, Xie et al., 2010), but S. mutans likely encodes many additional uncharacterized non-lantibiotic mutacin-like peptides. One of the characteristic features that are often used to identify potential mutacins from genome data is the presence of a conserved peptide leader region that terminates in a double-glycine (GG) motif. The leader peptide targets the prepeptide to a membrane bound ABC transporter apparatus, which is responsible for both secreting the peptide and cleaving the leader sequence at the GG processing site (Nes et al., 1996). Similar machinery is also used for the secretion of a variety of peptide pheromones, such as the S. mutans competence-inducing peptide ComC (Petersen & Scheie, 2000). Unlike the lantibiotic mutacins, genes for the production of the non-lantibiotic mutacins (the prepeptide, ABC transporter, and immunity) may not be localized within the same gene cluster. Also, one ABC transporter may be responsible for the production of multiple non-lantibiotic mutacins (Hale et al., 2005a) (Fig. 3). Intoxication by non-lantibiotic bacteriocins is generally considered to result due to the dissipation of the proton motif force via pore formation (Hechard & Sahl, 2002). For Class IIa bacteriocins, the mannose specific PTS has been shown to function as a receptor required for bacteriocin susceptibility (Nes et al., 2007). The relative susceptibility can even be further subdivided based upon the particular phylogeny of mannose PTS (Kjos et al., 2009). Assuming other non-lantibiotics also utilize particular protein receptors, this may account for their relatively narrow spectrum of activity.
Fig. 3.
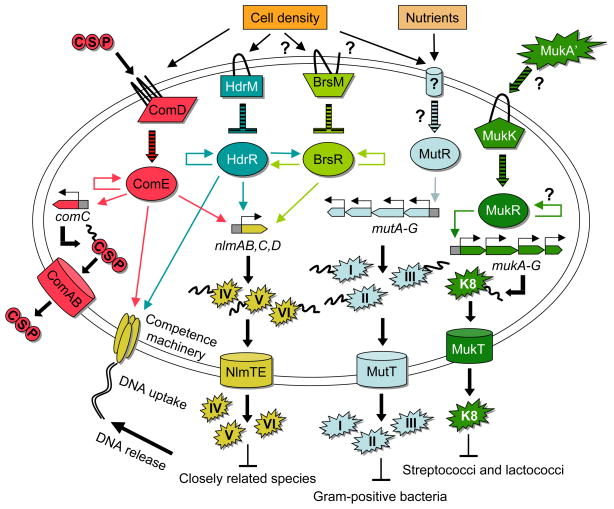
Summary of the regulation and ecology of mutacins. Components belonging to individual regulatory or genetic pathways are grouped by color. Colored line arrows indicate transcription regulation, whereas striped block arrows indicate post-transcriptional regulation. Of the extrinsic factors regulating mutacins, cell density is the predominant stimulus. It directly or indirectly affects the expression of the ComCDE system, the HdrRM and possibly BrsRM LytTR Regulatory Systems, as well as uncharacterized factor(s) controlling the expression of the lantibiotic mutacins I, II, and III. CSP is the signal directly sensed by the ComCDE system and MukA’ has been suggested to be the signal sensed by the MukRK sensory system. Nutrient composition also impacts the regulation of the lantibiotic mutacins through an uncharacterized mechanism. The ComCDE, HdrRM, and BrsRM regulatory systems are all autoinducing and activate the expression of the competence coordinated bacteriocins mutacins IV, V, and VI. These mutacins kill related oral species containing homologous chromosomes. The DNA released from intoxicated cells can be imported into the mutacin producer strain through the competence system. The competence system is itself induced by the ComCDE and HdrRM systems. Hence, these two systems can coordinate the production of particular mutacins with the development of natural genetic competence. The lantibiotic bacteriocins mutacins I, II, and III are regulated primarily by cell density and nutrients through an unknown mechanism. The mutacin operon transcription activator MutR is responsible for ensuring the transcription of the mutacin structural gene as well as all of the machinery required to posttranslationally modify and excrete the peptide and to provide immunity against self-intoxication. These lantibiotics kill most Gram-positive organisms due to interactions with the conserved lipid II molecule. The lantibiotic mutacin K8 locus also encodes similar machinery as mutacins I, II, and III. However, the K8 locus also encodes the MukRK sensory system that is presumably essential for inducing the expression of the muk genes. The K8 inhibitory spectrum does not appear to be as broad as mutacins I, II, and III and it is unknown which, if any, molecules are used as mutacin K8 receptors.
REGULATORY SYSTEMS CONTROLLING MUTACIN GENE EXPRESSION
Mutacins are regulated by multiple genetic and environmental factors that frequently overlap. For this reason, we discuss the regulation of mutacin gene expression based upon the different categories of regulation. A summary of the mutacin regulatory interaction network is provided in Fig. 3.
Regulation by Rgg-like regulators (MutR)
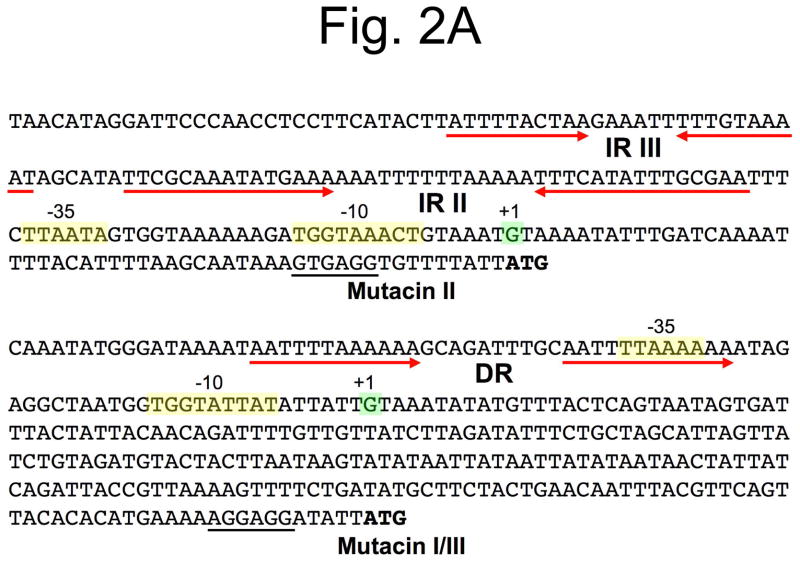
Of the mutacin gene clusters characterized to date, the mutacin I, II, and III loci have each been shown to encode homologous Rgg-family transcription regulators referred to as mutR (Fig. 1) (Chen et al., 1999, Qi et al., 1999b, Qi et al., 2001). Though the entire mutacin 1140 locus has yet to be sequenced, a partial open reading frame annotated as ORF X was identified in the beginning of the published sequence (Hillman et al., 1998). A BLASTN search of this sequence yields a match with 100% identity to the latter portion of the mutR open reading frame of mutacin III (unpublished results), which indicates that mutacin 1140 is highly likely to encode mutR as well. In addition, for each of these mutacin loci, the location of mutR is conserved as the first gene in the cluster (Fig. 1). From studies of mutacins I and II, it is known that MutR is an essential activator of mutacin operon gene expression. In a mutR background, mutacin activity is undetectable by the deferred antagonism assay, while transcription of the downstream mutacin operon is severely reduced (Kreth et al., 2004, Qi et al., 1999a). Studies of the mutacin II operon promoter region have identified three inverted repeat sequences (IR I-III) and three direct repeats (DR I-III) that may play a role in regulating the expression of the operon via MutR (Qi et al., 1999a). Of particular note, IR II and III are located upstream of the −35 site (Fig. 2) and bear some similarities to regulatory elements found in the promoter region of the staphylococcal lantibiotic epidermin (Qi et al., 1999a). In S. epidermidis, these repeats are bound by the epidermin operon activator protein EpiQ (Peschel et al., 1993). In the mutacin I and III operons, direct repeat sequences can also be found in similar locations as mutacin II (Fig. 2). Interestingly, for mutacins I and III, the putative -35 sequences overlap with the direct repeat sequences, which is inconsistent with the role of MutR as a transcription activator. Thus, additional biochemical studies will be required to fully address the role of MutR for these mutacins. Furthermore, studies of mutR transcription in mutacin II (Qi et al., 1999a) and mutacin I (Merritt and Qi unpublished) have determined that mutR expression is relatively invariant, whereas the mutacin operon is subject to extreme changes in gene expression (Kreth et al., 2005b, Merritt et al., 2005). This suggests that mutacin operon gene expression is highly unlikely to be controlled at the level of mutR transcription and additional uncharacterized regulatory elements are likely to play a major role in mutacin operon regulation.
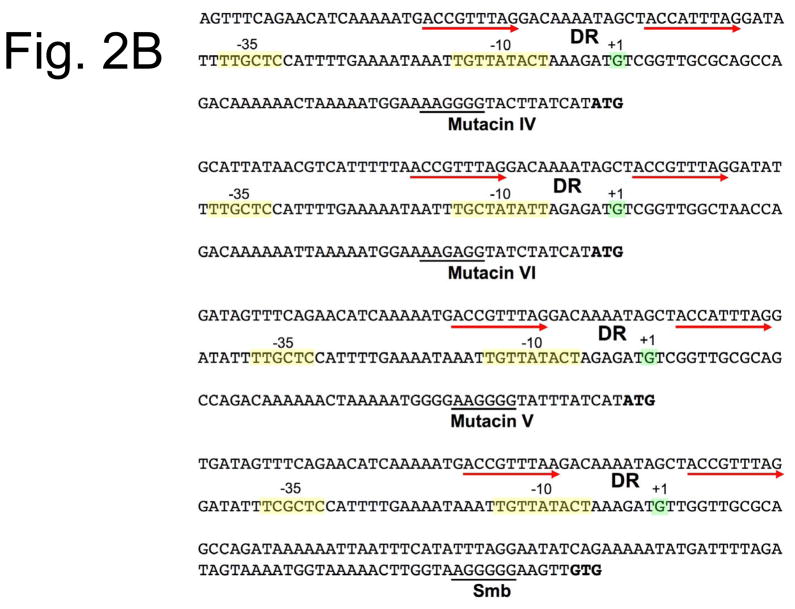
Fig. 2.
Comparison of the promoter regions for mutacin genes. A) The promoter regions upstream of the mutacin structural genes are shown for mutacins I, II, and III. The initiation codons are listed in bold and the ribosome binding sites are underlined. The transcription start sites are highlighted in green, while the −10 and −35 sequences are highlighted in yellow. Both inverted repeat (IR) and direct repeat sequences (DR) are marked with red arrows. B) Sequences of the upstream intergenic regions of mutacins IV, VI, V, and the Smb operon. The direct repeat sequences for mutacins IV and VI were confirmed as BrsR binding sites in strain UA140, while the mutacin V direct repeats were shown to be bound by ComE in strain UA159. The transcription start sites for mutacins IV and VI were identified by rapid amplification of cDNA ends (RACE) PCR, while those of mutacin V and the Smb operon are inferred based upon their sequence similarity to mutacins IV and VI.
Regulation by two-component signal transduction systems
Two component signal transduction systems are commonly utilized by bacteria to regulate both non-lantibiotic and lantibiotic bacteriocin gene expression (Eijsink et al., 2002, Kleerebezem, 2004). In S. mutans, the ComCDE two-component signal transduction system plays a direct role in the regulation of a variety of mostly non-lantibiotic bacteriocins, while the ScnRK-like sensory system (SMU.1814 – SMU.1815, also referred to as MukRK) probably serves an analogous function for the lantibiotic mutacin K8 (Fig. 1).
The S. mutans ComCDE two-component regulatory system responds to cell density and certain environmental stresses by excreting a peptide signal molecule called CSP (encoded by the comC gene) (Li et al., 2001, Perry et al., 2009). When the optimal CSP concentration has accumulated in the environment, it will induce the autophosphorylation of the ComD sensor kinase and the subsequent phosphotransfer to the ComE response regulator (Cheng et al., 1997, Havarstein et al., 1996, Pestova et al., 1996). Phosphorylated ComE will then activate gene expression from its target bacteriocin promoters, which results in greatly increased bacteriocin production (Kreth et al., 2005a, Kreth et al., 2006, Perry et al., 2009, van der Ploeg, 2005, Yonezawa & Kuramitsu, 2005). Both in silico analyses and in vitro DNA binding studies have identified conserved direct repeat regulatory elements in the promoters of the bacteriocin genes controlled by the ComCDE system (Fig. 2). These repeats match to the proposed consensus pattern recognized by members of the LytTR family of transcription regulators and serve as the binding site for ComE (Hung et al., 2011, Kreth et al., 2007, Nikolskaya & Galperin, 2002, van der Ploeg, 2005). Accordingly, ComE is also a member of the LytTR family. For each of the bacteriocins regulated by ComE, the LytTR consensus direct repeats were shown to occur just upstream of weak −35 sequences, which indicates a role for ComE in the transcription activation of these genes (Kreth et al., 2007, van der Ploeg, 2005, Xie et al., 2010). Likewise, most members of the LytTR family are also known to function as transcription activators (Galperin, 2008).
The entire lantibiotic mutacin K8 locus is highly homologous to that of the lantibiotic SA-FF22 of Streptococcus pyogenes. In the SA-FF22 locus, the ScnR response regulator is essential for the production of SA-FF22 (Hynes & Ferretti, 1995) and the mutacin K8 locus encodes a similar ScnRK-like two-component system referred to as MukRK (Fig. 1) (Robson et al., 2007). However, it has not been tested experimentally whether this two-component system is also essential for mutacin K8 production. Interestingly, the genes encoding the MukRK sensory system and the MukFEG immunity proteins are clustered in many S. mutans strains that do not encode the structural genes for mutacin K8, such as the sequenced reference strain UA159. Given that S. pyogenes strains containing the SA-FF22 locus exhibit immunity to mutacin K8 (Robson et al., 2007), it would be interesting to determine whether mutacin K8 non-producer strains such as UA159 have retained the mukRK and mukFEG genes in order to detect and prevent the toxicity associated with mutacin K8 production from other strains. This may also give further insight into the regulation of the mutacin K8 locus.
Regulation by LytTR Regulatory Systems
Recently, a newly described class of gene regulatory system in S. mutans was shown to play a direct role in the regulation of the same bacteriocin genes controlled by the ComCDE system (Table 1) (Okinaga et al., 2010a, Xie et al., 2010). Interestingly, these systems (referred to as LytTR Regulatory Systems) do not require ComCDE to exert their effect (Okinaga et al., 2010a, Okinaga et al., 2010b). Therefore, they appear to form fully independent, parallel regulatory pathways for mutacin gene expression (Fig. 3). So far, the HdrRM and BrsRM LytTR Regulatory Systems of S. mutans are the only characterized examples of LytTR Regulatory Systems, but numerous uncharacterized homologous systems can be found widely distributed throughout the phylum Firmicutes (unpublished). A LytTR Regulatory System consists of a membrane bound inhibitor protein (M) that antagonizes the activity of an associated LytTR family transcription regulator (R). The genes encoding both proteins are cotranscribed (Merritt et al., 2007). Currently, it is unknown which cellular or environmental signals activate LytTR Regulatory Systems, but they can be constitutively activated by simply mutating the membrane bound inhibitor protein. Consequently, a mutation of hdrM or brsM results in a constitutively active HdrR or BrsR, respectively. Activated HdrR and BrsR transcription regulator proteins will then cause greatly increased mutacin gene expression and mutacin production (Okinaga et al., 2010a, Xie et al., 2010). Given that both HdrR and BrsR are members of the LytTR family, it is also not surprising that they too interact with the same direct repeat sequences bound by ComE (Fig. 2). Mutagenesis of these repeats negates the increased bacteriocin expression phenotypes of an hdrM or brsM mutation, while BrsR DNA binding studies have confirmed that the LytTR consensus repeats are essential for a stable protein-DNA interaction (Okinaga et al., 2010a, Xie et al., 2010). Multiple lines of evidence indicate that both HdrR and BrsR function as transcription activators utilized to compensate for the weak −35 sequences found within the mutacin promoters.
ENVIRONMENTAL INFLUENCE UPON MUTACIN PRODUCTION
Mutacin production is highly influenced by environmental conditions. It has yet to be demonstrated how these environmental factors are able to influence mutacin gene expression, but it is assumed that they must control the activity of the aforementioned mutacin regulatory systems. Interestingly, mutacins share a large degree of overlap in the environmental cues that positively or negatively influence their gene expression, even though they are directly controlled by a wide variety of separate regulatory systems. Overall, the two most influential environmental factors are cell density and nutrition. In the following sections, we describe the current knowledge regarding the role of these two environmental factors on mutacin production. These factors are also incorporated into the mutacin regulatory network illustrated in Fig. 3.
The role of cell density
A variety of environmental factors have been shown to influence the production of mutacins, primarily through altered transcription of mutacin structural genes or operons. By far, the most potent inducer of mutacin transcription seems to be related to cell density, particularly conditions of extreme high cell density. Studies of the non-lantibiotic mutacin IV and the lantibiotic mutacin I have demonstrated that the relatively weak transcription of both of these mutacins in liquid culture can be pushed considerably higher if the cells are centrifuged and incubated as a cell pellet (Kreth et al., 2005b, Kreth et al., 2005a, Merritt et al., 2005). A similar result can also be obtained by spotting planktonic cells onto agar plates and incubating for 1 – 3 hrs (unpublished), while mutacin transcription can be increased even further still if the cells are allowed to develop into colonies on a plate (Kreth et al., 2005b). Thus, there is a direct correlation between the level of mutacin transcription and increasing cell density. Though, mutacin transcription at any growth stage in batch culture is generally low relative to the level of mutacin gene expression on a plate. Ecologically, this type of extreme high cell density regulation is well suited for mutacins, since bacteriocins are both energetically expensive to produce and are only effective when there is a high density of producer cells. For example, most bacteriocins function in the nanomolar range (Sang & Blecha, 2008). In batch culture or during growth in saliva, excreted mutacins would be quickly diluted, resulting in the loss of their inhibitory activity. In contrast, plate colonies or oral biofilm communities routinely yield cell densities up to 1000-fold higher than batch cultures (Evaldson et al., 1982). Under these conditions, considerably smaller quantities of metabolic resources are required to maintain an inhibitory local concentration of mutacin. In addition, the physiochemical properties of an agar plate or a biofilm likely reduce the diffusion rate of bacteriocins away from the producer cells (Chan et al., 2004). This should further concentrate the mutacins as they are excreted and improve the range at which they are effective. Consequently, it appears that the regulatory machinery controlling mutacin transcription has evolved a mechanism to promote mutacin production in the environment that provides the greatest benefit to the producer. How mutacin transcription is coordinated with conditions of extreme high cell density remains a significant open question.
Nutrient availability and mutacin production
Despite the strong influence of cell density upon mutacin transcription, other factors such as nutrient composition can play a major role in determining whether particular mutacins are produced. For mutacin I, the addition of ~5 mM phosphate to the growth medium significantly delayed the temporal pattern of mutacin production, whereas production was permanently inhibited at concentrations >10 mM (Nguyen et al., 2009). This effect was demonstrated to occur as a result of significantly decreased mutacin operon transcription. However, it has yet to be determined whether phosphate has any influence upon the production or activity of the mutacin I transcription activator MutR. In general, easily fermentable carbohydrate sources and yeast extract stimulate the production of numerous mutacins (Delisle, 1975, Nicolas et al., 2004, Nicolas et al., 2006, Qi et al., 1999a). In some cases, this effect is maximized only when these nutrients are supplied in limiting quantities (Qi & Kreth, 2010). Studies of mutacin I have demonstrated that diluted growth medium results in much greater mutacin operon transcription and mutacin production than a more concentrated growth medium supplemented with additional carbohydrate (Kreth et al., 2005b). These studies have led to the suggestion that mutacin production is optimal when the cells are subjected to mild stress.
ECOLOGICAL ROLE OF MUTACIN
Mutacins play an interesting role in the ecology of dental plaque. The frequent co-occurrence of mutacin loci and transposase genes (Fig. 1) implicates horizontal gene transfer as the primary driving force dictating the highly strain-specific repertoire of mutacin genes in S. mutans isolates. It also attests to the selective advantage mutacins provide to the producer. As studies probe deeper into mutacin regulation and function, mounting evidence suggests that they serve multiple functions both on the population and the community levels. The simplest and probably most obvious of these is their role in community level interactions. Many early colonizing species vie with S. mutans for a limited supply of available ecological niches. This naturally creates a scenario that is ripe for competitive behaviors among the organisms. Compared with S. mutans, many competitor species such as S. sanguinis colonize available tooth surfaces earlier, grow faster, and are more resistance to oxygen and oxidative stress (Kreth et al., 2009). Without some counterbalancing competitive advantage, S. mutans would have little chance of survival. Despite the greater aciduricity and acidogenicity of S. mutans, in vitro competition studies with S. sanguinis clearly demonstrate that bacteriocin activity from mutacins I and IV is required for S. mutans to maintain its dominance within dual species biofilms (Kreth et al., 2005b). Similarly, in vivo studies of humans infected with a non-acidogenic/non-cariogenic mutacin 1140 producer strain indicate that its increased mutacin production is sufficient to support reliable colonization and persistence within new hosts (Hillman, 2002). Presumably, the broad-spectrum inhibitory activity of mutacin 1140 and related lantibiotics plays a significant role in increasing the colonization potential of the producers. Though, surprisingly, studies indicate that only a minority of S. mutans strains actually harbor these lantibiotic clusters (Kamiya et al., 2005, Bekal-Si Ali et al., 2002), whereas most strains encode multiple non-lantibiotic mutacins that exhibit very narrow activity spectra (Kamiya et al., 2008).
Thus, the ecological function of these narrow-spectrum mutacins is perhaps more complex. For example, a large number of these mutacins are coordinately regulated with the development of genetic competence (Kreth et al., 2006, van der Ploeg, 2005), while no such coordination exists for the wide-spectrum mutacins I, II, III, and 1140. A comparison of the oral bacterial species susceptible to the competence-coordinated mutacins IV, V, VI, and Smb reveals a strong bias for organisms closely related to S. mutans, such as the “mitis” streptococci (Qi et al., 2001, Hale et al., 2005b, Xie et al., 2010, Paul & Slade, 1975). In a highly diverse biofilm, why might such a bias exist? Indeed, some of these organisms, such as S. sanguinis, are numerically more abundant and colonize the tooth surface before S. mutans, but the same could be said for various Actinomyces species. Yet, actinomycetes are infrequent targets of mutacin activity. This discrepancy may be partially explained by the fact that actinomycete genomes exhibit weak homology with S. mutans. In vitro co-culture studies of S. mutans and S. gordonii have demonstrated that the induction of the competence system can result in the interspecies transfer of transforming DNA from S. gordonii to S. mutans (Kreth et al., 2005a). Furthermore, this phenomenon was stimulated by the ability to produce the competence-coordinated bacteriocin mutacin IV. In contrast, the competence-independent lantibtiotic mutacin I played little or no role in this process. This has led to the hypothesis that competence-coordinated bacteriocins serve a dual purpose. The first function is to attack competitor species to either invade occupied niches or to protect niches already occupied by S. mutans. The second function is to provide transforming DNA from closely related organisms that could be used for genome repair and/or the acquisition of novel fitness-enhancing traits. Liberated DNA from distantly related species would be highly unlikely to recombine with the S. mutans chromosome. Consequently, there is probably little selective advantage to acquire such DNA. By selecting which organisms are targeted for attack, S. mutans can maximize the potential utility of the released DNA. Further support for this notion may be found among the competence-coordinated bacteriocin gene sequences. As previously discussed, the promoters of these bacteriocins all share highly conserved regulatory elements (Fig. 2) that are essential for induction by the ComCDE system or LytTR Regulatory Systems. In addition to mutacin regulation, the ComCDE and LytTR Regulatory Systems also have the ability to simultaneously induce competence development through the competence-specific sigma factor ComX (Okinaga et al., 2010a, Aspiras et al., 2004). In this way, these systems serve as the principal intermediaries to coordinate mutacin expression and competence development (Fig. 3). Despite the extensive homology found within the promoter regions of the competence-coordinated mutacins (Fig. 2), their structural genes exhibit considerable diversity, even encoding bacteriocins of multiple classes. This suggests that the selective pressure to maintain a regulatory connection with the competence system is quite strict, whereas the mutacin open reading frames seem to have much greater flexibility to evolve. One can speculate that this flexibility within the coding portion of the genes serves to prevent target organisms from developing resistance to these bacteriocins (Kjos et al., 2011).
In addition to community level interactions, recent studies are beginning to offer insight into a previously unexplored aspect of mutacin production: the role of competence-coordinated mutacins in population level interactions. Studies of the ComCDE two-component system and LytTR Regulatory Systems have demonstrated that the competence-coordinated bacteriocins mutacin IV and V each have the potential to kill the producer strains in a process that resembles a suicide-like pathway (Perry et al., 2009, Xie et al., 2010). In the case of mutacin V, this process appears to occur intracellularly (Perry et al., 2009), which implicates a unique killing mechanism. Presumably, such activity serves an altruistic function that benefits the S. mutans population during conditions of severe stress. This ability would not be surprising given that analogous altruistic behaviors are a common feature of microbial communities (Thomas & Hancock, 2009, Rice & Bayles, 2003). However, the use of bacteriocins for this purpose appears to be a novel strategy that could potentially form the basis for a new chapter in mutacin research.
FUTURE PERSPECTIVES
With the advent of more affordable next-generation sequencing technologies, it seems inevitable that the pan-genome of S. mutans will be assembled. At this stage, the true breadth of the mutacin repertoire will be revealed. This should offer many interesting opportunities for future mutacin research. Current studies have largely focused on two mutacin categories: the wide-spectrum lantibiotics and the competence-coordinated mutacins. However, even with the limited genome sequence information that is currently available, one can identify additional uncharacterized mutacin-like genes that do no fit into either of these categories. Perhaps these genes share some features that could place them into a third category of mutacin. For the mutacins already characterized, many significant questions remain regarding their role in oral biofilm ecology. If the competence-independent lantibiotic mutacins, such as mutacins I, II, and III are such efficient weapons, why are they only encoded by a small subset of S. mutans isolates? What is the selective advantage for strains such as UA159 to encode the immunity genes for particular mutacins, but not the structural genes? There is even some indication that mutacins may have additional functions in S. mutans biology. In addition to their potential role in cellular suicide, mutacins also play a role in the proper development of genetic competence. Noticeably lower transformation frequencies are observed when mutations are introduced into the genes encoding mutacin V and mutacin VI (Dufour et al., 2011, Perry et al., 2009). Clearly, the connections between competence development and mutacin production are intimately interwoven in S. mutans. Lastly, there is a longstanding interest to commercialize the wide-spectrum lantibiotic mutacins. They hold great promise as therapeutics for Gram-positive infections, particularly with the increasing threat of vancomycin resistant enterococci (VRE), methicillin resistant Staphylococcus aureus (MRSA), or perhaps even the newly emerging strains of vancomycin-resistant S. aureus (VRSA). Unfortunately, the complex chemistry that gives these mutacins their potent activity, also makes them difficult (i.e. expensive) to synthesize chemically. Therefore, if the commercial potential of these bacteriocins is ever going to be realized, either new breakthroughs will be required for chemical synthesis or novel strategies will need to be devised to greatly increase the yield of mutacins produced in batch cultures. As previously described, mutacin transcription is heavily dependent upon extreme high cell density growth conditions, but such an environment is currently impractical to replicate using industrial-scale fermentors. In addition to the wide-spectrum mutacins, the numerous narrow-spectrum mutacins may hold commercial potential as well. For example, we have found penicillin resistant S. pneumoniae to be highly sensitive to mutacin VI (unpublished results). Therefore, it may be feasible to use this mutacin to treat pneumococcal otitis media cases in order to reserve the use of beta-lactams or other wide-spectrum antibiotics for more serious infections. Targeted mutacin activity would also have the advantage of being minimally disruptive to the human flora, unlike the vast majority of commonly used antibiotics. With the continued discovery and characterization of new mutacins, it is becoming increasingly evident that there is a potential wealth of useful antimicrobial compounds located right beneath our noses.
Acknowledgments
This work was supported by an NIDCR DE018893 grant to JM.
References
- Aspiras MB, Ellen RP, Cvitkovitch DG. ComX activity of Streptococcus mutans growing in biofilms. FEMS microbiology letters. 2004;238:167–174. doi: 10.1016/j.femsle.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Baca P, Liebana J, Piedrola G. Epidemiological application of a new bacteriocin typing scheme for Streptococcus mutans. Community dentistry and oral epidemiology. 1990;18:194–196. doi: 10.1111/j.1600-0528.1990.tb00055.x. [DOI] [PubMed] [Google Scholar]
- Balakrishnan M, Simmonds RS, Carne A, Tagg JR. Streptococcus mutans strain N produces a novel low molecular mass non-lantibiotic bacteriocin. FEMS microbiology letters. 2000;183:165–169. doi: 10.1111/j.1574-6968.2000.tb08952.x. [DOI] [PubMed] [Google Scholar]
- Bekal-Si Ali S, Hurtubise Y, Lavoie MC, LaPointe G. Diversity of Streptococcus mutans bacteriocins as confirmed by DNA analysis using specific molecular probes. Gene. 2002;283:125–131. doi: 10.1016/s0378-1119(01)00875-7. [DOI] [PubMed] [Google Scholar]
- Bierbaum G, Sahl HG. Lantibiotics: mode of action, biosynthesis and bioengineering. Current pharmaceutical biotechnology. 2009;10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- Burton JP, Wescombe PA, Cadieux PA, Tagg JR. Beneficial microbes for the oral cavity: time to harness the oral streptococci? Beneficial microbes. 2011;2:93–101. doi: 10.3920/BM2011.0002. [DOI] [PubMed] [Google Scholar]
- Chan C, Burrows LL, Deber CM. Helix induction in antimicrobial peptides by alginate in biofilms. The Journal of biological chemistry. 2004;279:38749–38754. doi: 10.1074/jbc.M406044200. [DOI] [PubMed] [Google Scholar]
- Chen P, Qi F, Novak J, Caufield PW. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Applied and environmental microbiology. 1999;65:1356–1360. doi: 10.1128/aem.65.3.1356-1360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Campbell EA, Naughton AM, Johnson S, Masure HR. The com locus controls genetic transformation in Streptococcus pneumoniae. Molecular microbiology. 1997;23:683–692. doi: 10.1046/j.1365-2958.1997.2481617.x. [DOI] [PubMed] [Google Scholar]
- Davey AL, Rogers AH. Multiple types of the bacterium Streptococcus mutans in the human mouth and their intra-family transmission. Archives of oral biology. 1984;29:453–460. doi: 10.1016/0003-9969(84)90026-8. [DOI] [PubMed] [Google Scholar]
- Delisle AL. Production of bacteriocins in a liquid medium by Streptococcus mutans. Antimicrobial agents and chemotherapy. 1975;8:707–712. doi: 10.1128/aac.8.6.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D, Cordova M, Cvitkovitch DG, Levesque CM. Identification of a Novel Role Associated with a Streptococcal Bacteriocin: Regulation of the Competence Pathway. Journal of bacteriology. 2011 doi: 10.1128/JB.05968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijsink VG, Axelsson L, Diep DB, Havarstein LS, Holo H, Nes IF. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie van Leeuwenhoek. 2002;81:639–654. doi: 10.1023/a:1020582211262. [DOI] [PubMed] [Google Scholar]
- Evaldson G, Heimdahl A, Kager L, Nord CE. The normal human anaerobic microflora. Scandinavian journal of infectious diseases. 1982;35:9–15. [PubMed] [Google Scholar]
- Galperin MY. Telling bacteria: do not LytTR. Structure. 2008;16:657–659. doi: 10.1016/j.str.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guder A, Wiedemann I, Sahl HG. Posttranslationally modified bacteriocins--the lantibiotics. Biopolymers. 2000;55:62–73. doi: 10.1002/1097-0282(2000)55:1<62::AID-BIP60>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Hale JD, Heng NC, Jack RW, Tagg JR. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. Journal of bacteriology. 2005a;187:5036–5039. doi: 10.1128/JB.187.14.5036-5039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JD, Ting YT, Jack RW, Tagg JR, Heng NC. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Applied and environmental microbiology. 2005b;71:7613–7617. doi: 10.1128/AEM.71.11.7613-7617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Ooshima T. Inhibitory spectrum of a bacteriocinlike substance (mutacin) produced by some strains of Streptococcus mutans. Journal of dental research. 1975a;54:140–145. doi: 10.1177/00220345750540010801. [DOI] [PubMed] [Google Scholar]
- Hamada S, Ooshima T. Production and properties of bacteriocins (mutacins) from Streptococcus mutans. Archives of oral biology. 1975b;20:641–648. doi: 10.1016/0003-9969(75)90131-4. [DOI] [PubMed] [Google Scholar]
- Hasper HE, Kramer NE, Smith JL, Hillman JD, Zachariah C, Kuipers OP, de Kruijff B, Breukink E. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science. 2006;313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- Havarstein LS, Gaustad P, Nes IF, Morrison DA. Identification of the streptococcal competence-pheromone receptor. Molecular microbiology. 1996;21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- Hechard Y, Sahl HG. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie. 2002;84:545–557. doi: 10.1016/s0300-9084(02)01417-7. [DOI] [PubMed] [Google Scholar]
- Hillman JD. Genetically modified Streptococcus mutans for the prevention of dental caries. Antonie van Leeuwenhoek. 2002;82:361–366. [PubMed] [Google Scholar]
- Hillman JD, Novak J, Sagura E, Gutierrez JA, Brooks TA, Crowley PJ, Hess M, Azizi A, Leung K, Cvitkovitch D, Bleiweis AS. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infection and immunity. 1998;66:2743–2749. doi: 10.1128/iai.66.6.2743-2749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung DC, Downey JS, Ayala EA, Kreth J, Mair R, Senadheera DB, Qi F, Cvitkovitch DG, Shi W, Goodman SD. Characterization of DNA Binding Sites of the ComE Response Regulator from Streptococcus mutans. Journal of bacteriology. 2011;193:3642–3652. doi: 10.1128/JB.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes WL, Ferretti JJ. Developments in biological standardization. Vol. 85. 1995. A response regulator gene controls production of the lantibiotic streptococcin A-FF22; pp. 635–637. [PubMed] [Google Scholar]
- Jung G. Lantibiotics: a survey. In: Jung G, Sahl HG, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: ESCOM Science Publishers; 1991. pp. 1–35. [Google Scholar]
- Kamiya RU, Hofling JF, Goncalves RB. Frequency and expression of mutacin biosynthesis genes in isolates of Streptococcus mutans with different mutacin-producing phenotypes. Journal of medical microbiology. 2008;57:626–635. doi: 10.1099/jmm.0.47749-0. [DOI] [PubMed] [Google Scholar]
- Kamiya RU, Napimoga MH, Hofling JF, Goncalves RB. Frequency of four different mutacin genes in Streptococcus mutans genotypes isolated from caries-free and caries-active individuals. Journal of medical microbiology. 2005;54:599–604. doi: 10.1099/jmm.0.45870-0. [DOI] [PubMed] [Google Scholar]
- Kjos M, I, Nes F, Diep DB. Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology (Reading, England) 2009;155:2949–2961. doi: 10.1099/mic.0.030015-0. [DOI] [PubMed] [Google Scholar]
- Kjos M, I, Nes F, Diep DB. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Applied and environmental microbiology. 2011;77:3335–3342. doi: 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides. 2004;25:1405–1414. doi: 10.1016/j.peptides.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Kreth J, Hung DC, Merritt J, Perry J, Zhu L, Goodman SD, Cvitkovitch DG, Shi W, Qi F. The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology (Reading, England) 2007;153:1799–1807. doi: 10.1099/mic.0.2007/005975-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Bordador C, Shi W, Qi F. Transcriptional analysis of mutacin I (mutA) gene expression in planktonic and biofilm cells of Streptococcus mutans using fluorescent protein and glucuronidase reporters. Oral microbiology and immunology. 2004;19:252–256. doi: 10.1111/j.1399-302X.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA and cell biology. 2009;28:397–403. doi: 10.1089/dna.2009.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Molecular microbiology. 2005a;57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. Journal of bacteriology. 2005b;187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Zhu L, Shi W, Qi F. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS microbiology letters. 2006;265:11–17. doi: 10.1111/j.1574-6968.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- Kupke T, Gotz F. Post-translational modifications of lantibiotics. Antonie van Leeuwenhoek. 1996;69:139–150. doi: 10.1007/BF00399419. [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. Journal of bacteriology. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt J, Kreth J, Shi W, Qi F. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Molecular microbiology. 2005;57:960–969. doi: 10.1111/j.1365-2958.2005.04733.x. [DOI] [PubMed] [Google Scholar]
- Merritt J, Zheng L, Shi W, Qi F. Genetic characterization of the hdrRM operon: a novel high-cell-density-responsive regulator in Streptococcus mutans. Microbiology (Reading, England) 2007;153:2765–2773. doi: 10.1099/mic.0.2007/007468-0. [DOI] [PubMed] [Google Scholar]
- Mota-Meira M, Lacroix C, LaPointe G, Lavoie MC. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS letters. 1997;410:275–279. doi: 10.1016/s0014-5793(97)00425-0. [DOI] [PubMed] [Google Scholar]
- Nes IF, Diep DB, Havarstein LS, Brurberg MB, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus. Journal of bacteriology. 2007;189:1189–1198. doi: 10.1128/JB.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Zhang Z, Huang IH, Wu C, Merritt J, Shi W, Qi F. Genes involved in the repression of mutacin I production in Streptococcus mutans. Microbiology (Reading, England) 2009;155:551–556. doi: 10.1099/mic.0.021303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G, Auger I, Beaudoin M, Halle F, Morency H, LaPointe G, Lavoie MC. Improved methods for mutacin detection and production. Journal of microbiological methods. 2004;59:351–361. doi: 10.1016/j.mimet.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Nicolas G, Morency H, LaPointe G, Lavoie MC. Mutacin H-29B is identical to mutacin II (J–T8) BMC microbiology. 2006;6:36. doi: 10.1186/1471-2180-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya AN, Galperin MY. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic acids research. 2002;30:2453–2459. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak J, Caufield PW, Miller EJ. Isolation and biochemical characterization of a novel lantibiotic mutacin from Streptococcus mutans. Journal of bacteriology. 1994;176:4316–4320. doi: 10.1128/jb.176.14.4316-4320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okinaga T, Niu G, Xie Z, Qi F, Merritt J. The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. Journal of bacteriology. 2010a;192:1844–1852. doi: 10.1128/JB.01667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okinaga T, Xie Z, Niu G, Qi F, Merritt J. Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Molecular oral microbiology. 2010b;25:165–177. doi: 10.1111/j.2041-1014.2010.00574.x. [DOI] [PubMed] [Google Scholar]
- Paul D, Slade HD. Production and properties of an extracellular bacteriocin from Streptococcus mutans bacteriocidal for group A and other streptococci. Infection and immunity. 1975;12:1375–1385. doi: 10.1128/iai.12.6.1375-1385.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Molecular microbiology. 2009;72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A, Augustin J, Kupke T, Stevanovic S, Gotz F. Regulation of epidermin biosynthetic genes by EpiQ. Molecular microbiology. 1993;9:31–39. doi: 10.1111/j.1365-2958.1993.tb01666.x. [DOI] [PubMed] [Google Scholar]
- Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Molecular microbiology. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- Petersen FC, Scheie AA. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral microbiology and immunology. 2000;15:329–334. doi: 10.1034/j.1399-302x.2000.150511.x. [DOI] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Applied and environmental microbiology. 1999a;65:652–658. doi: 10.1128/aem.65.2.652-658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Applied and environmental microbiology. 1999b;65:3880–3887. doi: 10.1128/aem.65.9.3880-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Applied and environmental microbiology. 2000;66:3221–3229. doi: 10.1128/aem.66.8.3221-3229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Applied and environmental microbiology. 2001;67:15–21. doi: 10.1128/AEM.67.1.15-21.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Kreth J. Characterization of anti-competitor activities produced by oral bacteria. Methods in molecular biology (Clifton, N J) 2010;666:151–166. doi: 10.1007/978-1-60761-820-1_11. [DOI] [PubMed] [Google Scholar]
- Rice KC, Bayles KW. Death’s toolbox: examining the molecular components of bacterial programmed cell death. Molecular microbiology. 2003;50:729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x. [DOI] [PubMed] [Google Scholar]
- Robson CL, Wescombe PA, Klesse NA, Tagg JR. Isolation and partial characterization of the Streptococcus mutans type AII lantibiotic mutacin K8. Microbiology (Reading, England) 2007;153:1631–1641. doi: 10.1099/mic.0.2006/003756-0. [DOI] [PubMed] [Google Scholar]
- Rogers AH. Bacteriocin typing of Streptococcus mutans strains isolated from family groups. Australian dental journal. 1980;25:279–283. doi: 10.1111/j.1834-7819.1980.tb05201.x. [DOI] [PubMed] [Google Scholar]
- Sahl HG, Jack RW, Bierbaum G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. European journal of biochemistry/FEBS. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- Sang Y, Blecha F. Antimicrobial peptides and bacteriocins: alternatives to traditional antibiotics. Animal health research reviews/Conference of Research Workers in Animal Diseases. 2008;9:227–235. doi: 10.1017/S1466252308001497. [DOI] [PubMed] [Google Scholar]
- Tagg JR. Prevention of streptococcal pharyngitis by anti-Streptococcus pyogenes bacteriocin-like inhibitory substances (BLIS) produced by Streptococcus salivarius. The Indian journal of medical research. 2004;119(Suppl):13–16. [PubMed] [Google Scholar]
- Tagg JR, Dierksen KP. Bacterial replacement therapy: adapting ‘germ warfare’ to infection prevention. Trends in biotechnology. 2003;21:217–223. doi: 10.1016/S0167-7799(03)00085-4. [DOI] [PubMed] [Google Scholar]
- Thomas VC, Hancock LE. Suicide and fratricide in bacterial biofilms. The International journal of artificial organs. 2009;32:537–544. doi: 10.1177/039139880903200902. [DOI] [PubMed] [Google Scholar]
- van der Ploeg JR. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. Journal of bacteriology. 2005;187:3980–3989. doi: 10.1128/JB.187.12.3980-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey JM, van der Donk WA. Lantibiotics: peptides of diverse structure and function. Annual review of microbiology. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Xie Z, Okinaga T, Niu G, Qi F, Merritt J. Identification of a novel bacteriocin regulatory system in Streptococcus mutans. Molecular microbiology. 2010;78:1431–1447. doi: 10.1111/j.1365-2958.2010.07417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Kuramitsu HK. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrobial agents and chemotherapy. 2005;49:541–548. doi: 10.1128/AAC.49.2.541-548.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]