Summary
Although eukaryotic nuclei contain distinct architectural structures associated with noncoding RNAs (ncRNAs), their potential relationship to regulated transcriptional programs remains poorly understood. Here, we report that methylation/demethylation of Polycomb 2 protein (Pc2) controls relocation of growth control genes between Polycomb bodies (PcGs) and interchromatin granules (ICGs) in response to growth signals. This movement is the consequence of binding of methylated and unmethylated Pc2 to the ncRNAs, TUG1 and MALAT1/NEAT2, located in PcGs and ICGs, respectively. These ncRNAs mediate assembly of multiple co-repressors/co-activators, and can alter the histone marks read by Pc2 in vitro. Additionally, binding of NEAT2 to unmethylated Pc2 promotes E2F1 SUMOylation, leading to activation of the growth control gene program. These observations delineate a molecular pathway linking the actions of subnuclear structure-specific ncRNAs and non-histone protein methylation to relocation of transcription units in the three-dimensional space of the nucleus, thus achieving coordinated gene expression programs.
Keywords: Noncoding RNA, Nuclear Architecture, Subnuclear Structures, Suv39h1, Pc2, Methylation, KDM4C, Polycomb Body, Interchromatin Granules, E2F1, SUMOylation, Ubiquitination
Introduction
Regulated programs of gene expression are defined by the concerted interplay of transcriptional factors as well as other modulators such as epigenomic and signaling mediators. Recent studies have begun to elucidate the mechanistic control of gene expression by modulation of nuclear architecture and noncoding RNA (ncRNA). (Mercer et al., 2009; Misteli, 2010; Wilusz et al., 2009), suggesting the crosstalk between subnuclear architectural features and ncRNA as a possible mechanism for controlling gene expression. Thus, a growing number and type of molecular strategies combinatorially contribute to the precise control of gene expression involved in proliferation, differentiation, and maturation of cells.
In regulation of cell proliferation, the E2F family of transcription factors plays a critical role in organizing early cell cycle progression by coordinating early cell cycle events with the transcription of genes required for entry into S phase (Nevins, 1992). The transcriptional activity of E2F1 is regulated principally by association with different factors including the Rb–E2F complexes, which predominate in quiescent cells, acting as repressors of transcription (Bagchi et al., 1991). Other levels of control that influence E2F1 activity include its post-translational modifications, protein stabilization (Campanero and Flemington, 1997) and regulation of its intracellular location (de la Luna et al., 1996).
At the level of “epigenomic control”, a large and growing class of proteins, known as chromatin modifiers, constrains expression by adapting regions of the genome to maintain either gene silencing or gene activation (Kouzarides, 2007). Histone methylation, together with other site-specific modifications, plays an important role in the generation of specific molecular marks in chromatin, which are recognized by regulatory proteins, thus determining dynamic transitions between transcriptionally active and inactive states (Jenuwein and Allis, 2001). Recent data also show that the regulatory role of lysine methylation on proteins is not restricted to the “histone code” with this modification modulating activation, stabilization, and degradation of non-histone proteins, thus influencing numerous cell processes (Huang and Berger, 2008). A number of histone methyltransferases (HMTases) have been identified and characterized, which can methylate specific lysine residues of histones and certain non-histone substrates (Chuikov et al., 2004; Sampath et al., 2007; Subramanian et al., 2008). Suv39h1 is a H3 lysine 9 methyltransferase containing the conserved SET domain and post-SET domains (Aagaard et al., 1999). It is documented that Suv39h1-mediated histone methylation functions in transcription repression and gene silencing by recruitment of Polycomb proteins and forming and spreading heterochromatin (Lachner et al., 2001).
Genetic study of Suv39h1 transgenic mice has demonstrated that overexpression of Suv39h1 accelerates homeostatic cell proliferation (Czvitkovich et al., 2001), while Suv39h1/h2-double-null mice have reduced viability during embryonic development and reduced growth as adult animals (Peters et al., 2001). Furthermore, Suv39h1 forms a complex with pRb/E2F1 and plays a role in the dynamic regulation of specific cell-cycle genes needed for entry into the S phase (Ait-Si-Ali et al., 2004; Nielsen et al., 2001). These observations suggest that Suv39h1 may function as a transcriptional co-repressor of the E2F1 transcription factor. Indeed, Suv39H1 selectively interacts with vertebrate chromobox family (Cbx) proteins, including heterochromatin protein 1 (HP1) and Cbx4 (Pc2) (Sewalt et al., 2002). Although Cbx proteins are thought to function as repressor in heterochromatin formation and gene silencing, recent evidence has demonstrated that Cbx proteins also play a critical role in cell cycle progression (Satijn et al., 1997). However, the role of Pc2 in cell cycle regulation remains largely unknown.
The notion that ncRNAs can have numerous molecular functions is rapidly evolving (Eddy, 2001; Ponting et al., 2009). Even though long ncRNAs represent a large class of transcriptional units and appear to be evolutionarily conserved, their specific roles in critically modulating regulated gene expression remains poorly understood.
Here, we report a critical role of Suv39h-mediated Pc2 methylation in regulating the cellular proliferation program, causing a relocation of growth control gene loci from Polycomb bodies (PcGs) to interchromatin granules upon serum stimulation. We show that noncoding RNAs TUG1 and NEAT2 cause the growth control genes to relocate from the repressive environment of PcG bodies, where they interact with co-repressor complexes to the gene activation milieu of the interchromatin granules, by selectively interacting with methylated and unmethylated Pc2 present on growth control gene promoters, respectively. Interaction with NEAT2 regulates Pc2-dependent E2F1 SUMOylation, which licenses the recruitment of an H2B monoubiquitinase, CDCA7L, and biochemically switches the Pc2 chromodomain from preferring repressive histone marks to interacting with activation-associated histone marks. Broadly, our data highlight combinational actions of transcription factor/co-regulators, non-histone protein methylation, and noncoding RNAs resident in distinct subnuclear architectural structures as a central strategy to achieve coordinated programs of regulated gene expression, with implications for our understanding of the epigenomic programming in homeostasis and disease.
Results
Pc2 is a Substrate for Suv39h1, a Lysine-specific Methyltransferase
We searched for non-histone candidate substrates for Suv39h1 and found that Suv39h1 methylated Pc2 in vitro (Figures 1A and S1A–S1C). Under our assay conditions, Suv39h1-mediated methylation of Pc2 appeared to be quite specific. Other proteins such as SUMO1, SUMO activating enzyme E1, E2, and E2F1 did not serve as Suv39h1 substrates (Figure 1A). Moreover, a Suv39h1 hyperactive mutant (H320R) (Rea et al., 2000) further enhanced the methylation of Pc2, while an enzymatically-inactive Suv39h1 mutant (H324L) (Rea et al., 2000) failed to use Pc2 as substrate (Figures 1B and S1A). Trypsin digestion of immunoprecipitated FLAG-Pc2 from 293T cells co-expressed with wild-type (wt) Suv39h1 (Figure 1C, arrow), followed by mass spectrometry (MS), revealed that lysine 191 of Pc2 was di-methylated, which was not observed in cells overexpressed with enzymatically-inactive Suv39h1 mutant (H324L) (Figure 1C and data not shown). A single K191R point mutation completely abolished Pc2 methylation, suggesting that K191 serves as a sole methylation site targeted by Suv39h1 (Figures 1D and S1D).
Figure 1. Suv39h1 Methylates Pc2 at K191.
(A) Pc2 is a non-histone substrate for Suv39h1. In vitro methylation assay was performed by incubating GST-Suv39h1 with recombinant His-Pc2, His-E2F1, histone H3, E1, E2, and SUMO1 in the presence of [3H]-SAM. (B) The enzymatic activity of Suv39h1 is required for Pc2 methylation. In vitro methylation assay was performed by incubating GST-Suv39h1 (wt, H320R or H324L mutant) with His-Pc2 in the presence of [3H]-SAM. (C) Suv39h1 methylates Pc2 at lysine 191 in vivo. FLAG-Pc2 immunoprecipitated from 293T cells (indicated by red arrow in left panel) was subjected to MOLDI-TOF analysis (right panel) and modified peptides are indicated by the red box. (D) Suv39h1 methylates Pc2 at K191 in vitro. In vitro methylation assay was performed by incubating GST-Suv39h1 with wt Pc2 or K191R mutant in the presence of [3H]-SAM. (E) Validation of Pc2K191me2 and Pc2K191 antibodies. Methylated and unmethylated His-Pc2 was subjected to autoradiography (top panel) or immunoblotting with antibodies indicated (middle panels). (F and G) Suv39h1 methylates Pc2 at K191 in vivo. Cell lysates from HeLa cells transfected with siRNAs (F) or expression vectors (G) as indicated were immunoblotted with antibodies indicated. For A, B and D, the reaction products were separated by SDS-PAGE followed by Coomassie blue staining (CBB) (left panel) or autoradiography (right panel).
See also Figure S1.
To investigate the role of Pc2 methylation in vivo, we generated rabbit polyclonal antibodies that specifically recognized di-methylated Pc2K191 and unmethylated Pc2 (Figure S1E). The anti-Pc2K191-di-methyl antibody (referred to as Pc2K191me2 antibody) specifically detected bacterially-purified Pc2 protein methylated by Suv39h1 in vitro, but was totally unreactive against unmethylated Pc2 protein while anti-Pc2K191 antibody (referred to as Pc2K191 antibody) specifically recognized unmethylated Pc2 protein, with no cross-reaction to methylated Pc2 protein when equivalent amounts of methylated and unmethylated Pc2 were used (Figure 1E). We concluded that the Pc2K191me2 and Pc2K191 antibodies specifically recognized K191-methylated and unmethylated Pc2, respectively. To address the question of whether Pc2 is methylated by Suv39h1 in vivo, we knocked down Suv39h1 and other histone H3K9 methyltransferases including Suv39h2, G9a, EuHMT, and ESET by specific siRNAs in HeLa cells and observed that the Pc2K191me2 level was specifically reduced only by Suv39h1 siRNA compared to control siRNA, with a concurrent increase of unmethylated Pc2 level (Figures 1F and S1F). To rule out any potential off-target effects of Suv39h1 siRNA, wt Suv39h1 or its enzymatically-inactive mutant form (H324L) was transiently expressed in HeLa cells. Immunoblotting analysis using Pc2K191me2 antibody detected increased level of Pc2 methylation in extracts derived from cells overexpressing the wt Suv39h1, while cells equivalently transfected with the enzymatically-inactive H324L mutant displayed minimal, if any, Pc2 methylation (Figure 1G). The expression levels of Suv39h1 proteins and the amount of total proteins in extracts were similar (Figure 1G). We therefore conclude that Suv39h1 methylates Pc2 at K191 in vivo.
Growth Control Gene Activation Requires KDM4C-Mediated Pc2K191me2 Demethylation
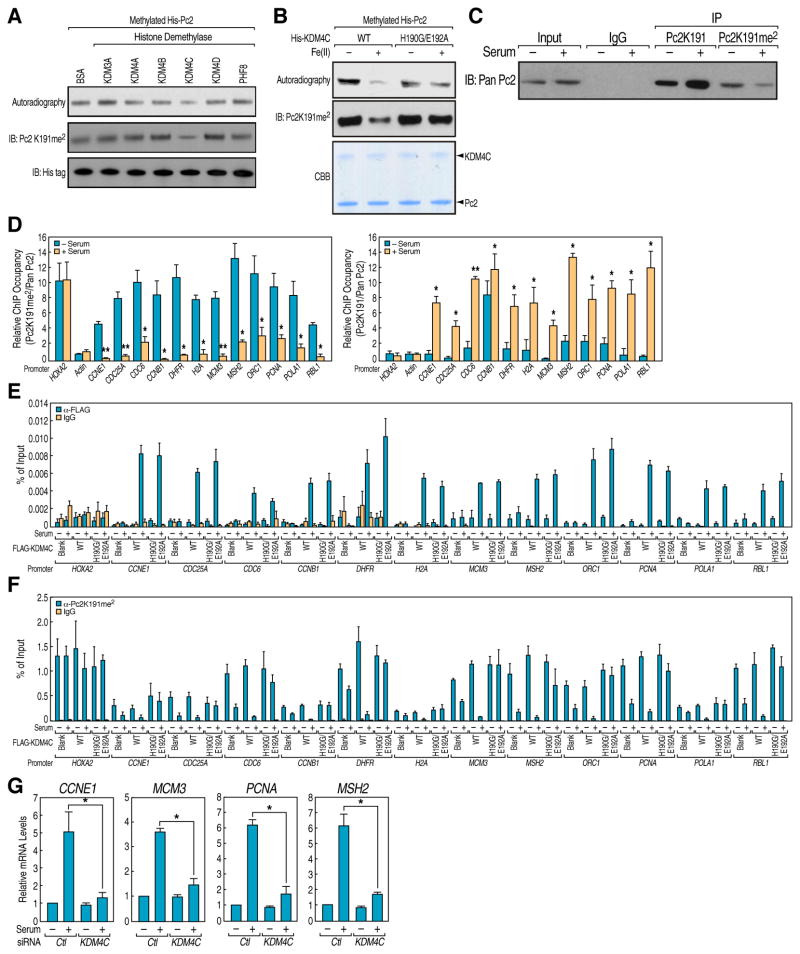
Interestingly, Pc2 K191 exists in a sequence context resembling that of histone H3K9, which led us to search for candidate H3K9 demethylases that may demethylate Pc2K191me2. By performing in vitro demethylation assay using active FLAG-H3K9 demethylases purified from 293T cells (Figures S2A and S2B), we found that, in addition to histone H3K9me2, KDM4C demethylated the dimethyl Pc2K191 (Figure 2A). Under our assay conditions, KDM4C-mediated demethylation of Pc2K191me2 appeared to be quite specific, as other demethylases, including KDM3A, KDM4A, KDM4B, KDM4C, KDM4D and PHF8, did not serve as Pc2K191me2 demethylases although each of the purified demethylases exhibited robust enzymatic activity on an H3K9me2 substrate (Figure S2B). Moreover, the enzymatically-inactive KDM4C-H190G/E192A mutant (Cloos et al., 2006) failed to demethylate Pc2K191me2 (Figures 2B and S2C).
Figure 2. Growth Control Gene Activation Requires KDM4C-Mediated Pc2K191me2 Demethylation.
(A) KDM4C demethylates Pc2 in vitro. Histone Demethylase assay was performed by incubating purified FLAG-histone demethylases with in vitro methylated His-Pc2. Changes in substrate methylation levels were analyzed by autoradiography (upper panel) and Pc2K191me2 antibodies (middle panel). Immunoblotting with antibodies against His tag was used to demonstrate equal loading (bottom panel). (B) Enzymatic activity of KDM4C is required for Pc2K191me2 demethylation. His-KDM4C wt or H190G/E192A mutant was incubated with in vitro methylated His-Pc2 in the presence or absence of Fe (II). The reactions were separated by SDS-PAGE followed by autoradiography (upper panel), immunoblotting with Pc2K191me2 antibodies (middle panel) or Coomassie blue staining (lower panel). (C) Serum-induced demethylation of Pc2K191me2 in vivo. HeLa cells were serum-starved followed by restimulation and the cell lysates were immunoprecipitated with Pc2K191 or Pc2K191me2 antibodies followed by immunoblotting with pan Pc2 antibodies. (D) Serum-induced demethylation of Pc2 on growth control gene promoters. HeLa cells were serum-starved followed by restimulation and ChIP analyses were performed using Pc2K191, Pc2K191me2 and pan Pc2 antibodies on indicated regions. For each primer pair, Pc2K191me2 (left panel) and Pc2K191 ChIP values (right panel) were normalized to the corresponding total pan Pc2 ChIP values. (E and F) Serum-induced KDM4C recruitment is responsible for Pc2K191me2 demethylation on growth control gene promoters. HeLa cells transfected with blank vector, FLAG-KDM4C wt or H190G/E192A mutant were serum-starved followed by restimulation and ChIP analyses using FLAG antibody (E) or Pc2K191me2 antibody (F) were performed on indicated regions. (G) KDM4C is required for serum-induced growth control gene expression. HeLa cells transfected with control or validated KDM4C siRNAs were serum-starved followed by restimulation and relative mRNA levels of indicated genes were determined by qRT-PCR. Mean±SEM, *p<0.05.
See also Figure S2.
To investigate the potential role of Pc2 in cell cycle and growth control, we next analyzed whether KDM4C-mediated demethylation of Pc2 regulates the transcriptional activation of E2F1 target genes. First, we examined whether Pc2K191me2 level is physiologically regulated under the conditions of E2F1 activation. Immunoprecipitation experiments with Pc2K191me2 and Pc2K191 antibodies revealed that Pc2K191me2 levels exhibited a strong decrease in response to serum stimulation, concurrent with increased levels of unmethylated Pc2 (Figure 2C). We further examined the methylation status of Pc2 on E2F1 target gene promoters previously identified by E2F1 ChIP-Seq (Liu et al., 2010), finding that Pc2K191me2 specifically enriched on E2F1 target gene promoters, in contrast to no significant occupancy on the Actin promoter, which is not regulated by Polycomb proteins (Figure 2D, left panel). Importantly, Pc2K191me2 levels dramatically decreased upon serum stimulation, which correlates with the increased levels of unmethylated Pc2 (Figure 2D, right panel). Under the same conditions, the HoxA2 promoter, on which Pc2 was consistently enriched, did not exhibit Pc2K191me2 demethylation (Figure 2D).
ChIP assay in HeLa cells transfected with KDM4C wt or H190G/E192A mutant revealed that serum stimulation resulted in increased recruitment of wt KDM4C on E2F1 target gene promoters (Figure 2E), which correlated with a decreased Pc2K191me2 level (Figure 2F). The KDM4C-H190G/E192A mutant largely inhibited Pc2K191me2 demethylation, even though being equally recruited on the same set of promoters, (Figures 2E and 2F). Indeed, siRNA-mediated reduction of the levels of KDM4C also impaired the expression of E2F1 target genes upon serum stimulation in HeLa cells (Figures 2G and S2D). To further evaluate the alternative possibility that the actions of KDM4C that critically dictated growth control gene expression might reside in its histone H3K9 demethylase activity, rather than actions on Pc2K191me2, we took advantage of the previous observations that different H3K9 demethylases were utilized in a promoter-specific fashion to provide H3K9 demethylase activity (Fodor et al., 2006; Qi et al., 2010; Yamane et al., 2006). We found that growth control gene promoters did not exhibit a KDM4C-dependence on H3K9me2 demethylation (Figure S2E, bottom panel), yet demethylase activity of KDM4C was required for the demethylation of Pc2K191me2 and for activation for these growth control genes (Figures S2E, top panel and 2G). Taken together, these data argue strongly that it is the specific demethylation of the repressive dimethyl Pc2K191 mark, caused by recruitment of KDM4C, rather than H3K9 demethylation, that is required for the transcriptional activation of E2F1 target genes.
Pc2 Methylation/Demethylation Controls the Cell Growth Program
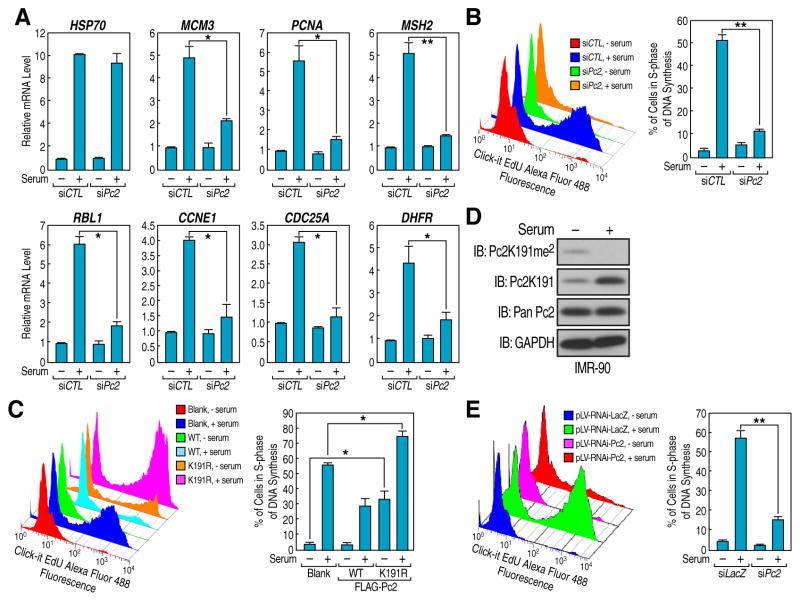
Considering the involvement of KDM4C-mediated Pc2K191me2 demethylation on E2F1 target gene promoters in response to serum, we tested the possibility that Pc2 methylation/demethylation is required for controlling the growth control gene expression and cell proliferation. Knockdown of Pc2 in HeLa cells dramatically decreased serum-induced growth control gene expression and cell proliferation (Figures 3A, 3B and S3A). In accord with this finding, exogenous expression of Pc2 K191R mutant noticeably increased cell proliferation even at the basal level, further indicating that Pc2 is a potent regulator of the cell growth program (Figure 3C).
Figure 3. Pc2 Methylation/Demethylation Controls Mitogenic Signal-Induced Cell Proliferation.
(A and B) Pc2 is essential for serum-induced growth control gene expression and cell proliferation. HeLa cells transfected with Pc2 siRNAs were serum-starved followed by restimulation and mRNA level of indicated genes (A) and cell proliferation (B) were analyzed, respectively. (C) Pc2K191 methylation is a key regulator for cell proliferation. HeLa cells transfected with Pc2 plasmids (wt or K191R mutant) were serum-starved followed by restimulation and cell proliferation were analyzed. (D and E) IMR-90 cells were serum-starved followed by restimulation and Pc2K191 methylation level (D) and cell proliferation (E) was analyzed by immunoblotting and flow cytometry, respectively. For B, C and E, cell proliferation was analyzed by monitoring the EDU incorporation into DNA synthesis in S-phase cells. Mean±SEM, *p<0.05 and **p<0.01.
See also Figure S3.
The potential requirement of Pc2 in mitogenic signal-induced proliferation was next investigated in primary human fibroblasts. IMR-90 primary human lung fibroblasts became growth arrested with serum starvation and proliferated once released into serum-containing medium (Nahle et al., 2002). Immunoblotting showed that, although global Pc2 protein levels were not affected, serum stimulation of IMR-90 cells was largely accompanied by Pc2K191me2 demethylation (Figure 3D). Pc2 shRNAs (shPc2-1 and -2) dramatically reduced Pc2 protein levels when stably transduced into IMR-90 cells and impaired serum-induced cell proliferation (Figures 3E and S3B). Collectively, our data suggest that methylated Pc2 may represent an important anti-mitogenic signal and, under certain stimuli, a stress-induced modification mark required for growth control gene repression and cell cycle arrest, while unmethylated Pc2 is essential for physiological growth control gene expression and cell proliferation.
Serum Induction Causes Relocation of E2F1-Regulated Genes between PcG Bodies and ICGs
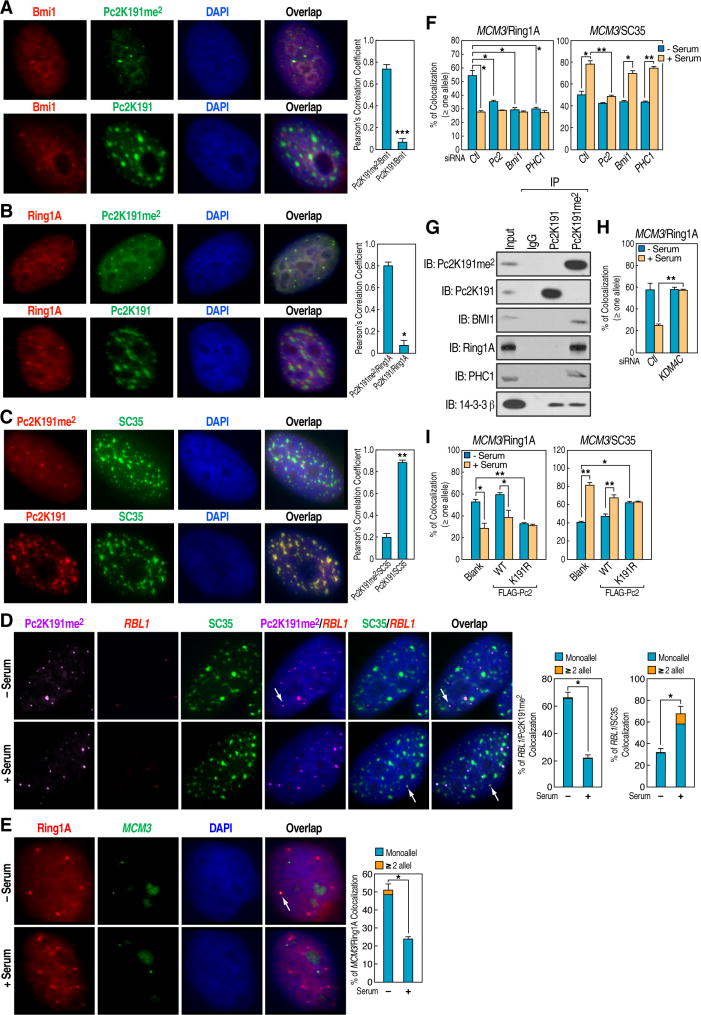
We speculated that, given that the involvement of Pc2 in stable gene repression (Lund and van Lohuizen, 2004), E2F1-regulated growth control genes might relocate within the cell nuclei from transcriptional repressive PcG bodies to a transcriptionally permissive environment upon serum stimulation. We first examined the subnuclear localization of methylated and unmethylated Pc2. We observed that methylated Pc2 was concentrated in several defined domains that were faithfully co-stained with Bmi1 and Ring1A (Figures 4A and 4B), two well-established PcG body markers (Hernandez-Munoz et al., 2005). In contrast, unmethylated Pc2 showed strong colocalization only with SC35, a marker for the interchromatin granules/nuclear speckles (Fu and Maniatis, 1990) (Figure 4C). Under our immunostaining conditions colocalization of unmethylated Pc2 with interchromatin granules appeared to be quite specific. Other nuclear bodies, including nuclear stress bodies, paraspeckles, Cajal bodies, and PML bodies, failed to exhibit co-staining with unmethylated Pc2 (data not shown). We then tested whether Pc2-associated E2F1 target transcription units relocated from PcG bodies to interchromatin granules upon serum stimulation by performing Immuno-FISH. Interestingly, gene loci encoding growth control genes (RBL1, MCM3, PCNA and MSH2) were found to predominantly colocalize with PcG bodies, as marked by Pc2K191me2 staining (or Ring1A, See below) in serum-starved HeLa cells, and these loci largely colocalized with SC35-staining interchromatin granules in the presence of serum (Figures 4D and S4A–S4C). Under the same conditions, colocalization of E2F1 target gene loci with PcG bodies was reproducibly observed in Immuno-FISH experiments using Ring1A, instead of Pc2K191me2, staining as a marker for PcG bodies (Figure 4E). In contrast, unmethylated Pc2 specifically colocalized with activated E2F1 target gene loci upon serum stimulation (Figures S4D–S4G). It was previously shown that interchromatin granules occupy the large space in nuclei, which results in a 14–26% “unspecific” colocalization with genes under basal conditions (Szczerbal and Bridger, 2010). It is to be expected that any given gene loci may also exhibit a similar ~15–25% “unspecific” colocalization with interchromatin granules. To further support this assumption, we performed Immuno-FISH on a cell cycle unrelated gene locus BCR, finding an average of ~10% and 30% colocalization of this locus with PcG bodies and interchromatin granules in serum-starved cells, respectively, and that these colocalization percentages did not change upon serum stimulation (Figure S4H). Thus, given the assay background, the actual percent of relocation of growth control genes from PcG bodies to interchromatin granules would be considerably greater if this background were to be subtracted.
Figure 4. Transition of E2F1-regulated Genes from PcG Bodies to Interchromatin Granules following Serum Induction.
(A–C) Methylated and unmethylated Pc2 exhibit distinct immunostaining patterns with respect to Bmi1 (A) or Ring1A (B) as a marker of PcG bodies, and SC35 (C) of interchromatin granules. (D and E) Serum-induced relocation of growth control gene locus between PcG bodies and interchromatin granules. HeLa cells were serum-starved followed by restimulation and Immuno-FISH analyses were performed using antibodies against Pc2K191me2 (D) or Ring1A (E) and FISH probes targeting RBL1 (D) or MCM3 (E). (F) Effect of knockdown of Pc2, Bmi1 and PHC1 on relocation of MCM3 locus between PcG bodies and interchromatin granules. HeLa cells microinjected with indicated siRNAs were serum-starved followed by restimulation and Immuno-FISH analyses were performed using antibodies and FISH probes as indicated. (G) Interaction between endogenous Pc2 and other core components of PRC1 complex demonstrated by co-immunoprecipitation of Pc2K191me2 with Bmi1, Ring1A and PHC1 in HeLa cell extracts. The input represents 10% of the protein amount used for immunoprecipitation. (H) KDM4C is required for serum-induced dissociation of MCM3 locus from PcG bodies. HeLa cells microinjected with control siRNA or validated siRNA against KDM4C were serum-starved followed by restimulation and Immuno-FISH analyses were performed using Ring1A antibodies and probes targeting MCM3 locus. (I) Demethylated Pc2 is required for relocation of MCM3 locus. HeLa cells microinjected with blank vector, Pc2 wt or K191R mutant were serum-starved followed by restimulation and Immuno-FISH analyses were performed using Ring1A, SC35 antibodies and probes targeting MCM3 locus. Statistical analyses on colocalization of MCM3 locus with Ring1A staining (left panel) and SC35 staining (right panel) were shown. For A, B, C, D and E, left panel shows representative images and right panel shows statistical analyses. Mean±SEM, *p<0.05 and **p<0.01.
See also Figure S4.
As expected, knockdown of Pc2 impaired the colocalization of MCM3 with both PcG bodies and interchromatin granules (Figure 4F). Interestingly, other core components of PRC1 complex including Bmi1 and PHC1 (Simon and Kingston, 2009) specifically interacted with methylated Pc2 (Figure 4G) and siRNA knockdown of these components disrupted the colocalization of MCM3 with PcG bodies, but not interchromatin granules, while having no obvious effects on maintenance of PcG bodies (Figures 4F, S4I and S4J), suggesting that other components of PRC1 complex may facilitate the repression of growth control genes mediated by methylated Pc2 in PcG bodies.
We next tested whether demethylation of Pc2K191me2 is required for the relocation process in HeLa cells microinjected with specific siRNAs against KDM4C, and we found that depletion of KDM4C largely abrogated dissociation of MCM3 gene locus from PcG bodies in the presence of serum stimulation (Figure 4H). In contrast, microinjection of plasmid encoding the Pc2K191R mutant in serum-starved cells resulted in dissociation of MCM3 gene locus from the Ring1A-stained PcG bodies, while enhancing their colocalization with SC35-positive interchromatin granules at basal level (Figure 4I). Together, these findings reveal an unexpected role of Pc2 in transcriptional regulation of growth control genes by promoting the relocation of these gene loci from the transcriptional repressive nuclear environment of PcG bodies to the transcriptionally-permissive environment of interchromatin granules. Given the observation that siRNA knockdown of KDM4C exerted no effect on serum-induced demethylation of H3K9me2 on growth control gene promoters (Figure S2E), we concluded that the KDM4C-dependent demethylation of Pc2K191me2, rather than histones, appear to serve as the key determinant of the relocation of growth control genes in response to growth signals.
Methylation Modulates Pc2 Association with Noncoding RNAs
It has been reported that noncoding RNA may serve as a component to form subnuclear bodies and maintain their integrity (Bond and Fox, 2009). Given that methylated and unmethylated Pc2 are localized to PcGs and interchromatin granules, respectively and previous observations that the chromodomain is capable of binding RNA (Akhtar et al., 2000), we therefore tested the possibility that Pc2 might bind specific ncRNAs in these subnuclear structures. By performing in vivo Cross-Linking and Immuno-Precipitation procedures (CLIP) using either anti- Pc2K191me2 or Pc2K191 antibodies, we observed that specific RNA–methylated Pc2 and RNA-unmethylated Pc2 complexes were formed after the UV cross-linking, which was significantly disrupted upon adding the blocking peptides for Pc2K191me2 and Pc2K191 antibodies in cell lysates (Figure 5A). Sequence analysis of 94 different clones identified TUG1, a noncoding RNA interacting with EZH2 identified by RIP assay (Khalil et al., 2009) and repressing cell cycle genes, as the most enriched RNA that interacted with methylated Pc2 (Figure S5A). Surprisingly, in the absence of K191 methylation, the predominant RNA associated with Pc2 was NEAT2/MALAT1 (Figure S5B), an exclusive signature noncoding RNA for interchromatin granules (Hutchinson et al., 2007). Likewise, purified biotinylated TUG1 RNA (nt 117-3390) harboring the binding sequence identified in the CLIP assay specifically selected methylated Pc2 from HeLa cell nuclear extract, while biotinylated NEAT2 RNA (nt 2281-5611) specifically bound unmethylated Pc2 (Figures 5B, S5C and S5D). Electrophoretic mobility-shift assays (EMSA) were next used to analyze whether a direct switch of Pc2 RNA binding preference is observed in response to K191 methylation. Incubation of the TUG1 RNA probe with methylated Pc2 resulted in specific gel retardation (Figure 5C, first panel, lane 3), while incubation of only a 49-nt NEAT2 RNA probe with unmethylated Pc2 specifically resulted in gel shift (Figure 5C, third panel, lane 4). Under these conditions, no shift was observed when the corresponding TUG1 and NEAT2 DNA probes or cold probes were used (Figures 5C and S5E). We, therefore, conclude that the interactions of methylated or non-methylated Pc2 with noncoding RNAs are controlled by a specific covalent modification of Pc2.
Figure 5. NcRNAs-Pc2 Interactions Underlie Relocation and Transcriptional Activation of Growth Control Genes.
(A) Methylated and unmethylated Pc2 bind to RNAs. CLIP assay using Pc2K191me2 (left panel) or Pc2K191 (right panel) antibodies were performed and specific protein-RNA complexes (indicated by red box) were visualized by autoradiography. (B) Validation of interactions of TUG1 and NEAT2 with Pc2. In vitro transcribed biotinylated TUG1 RNA (left panel) or NEAT2 RNA (right panel) were incubated with HeLa nuclear extract. The elution of Monoavadin beads was subjected to immunoblotting with indicated antibodies. (C) The selective binding of Pc2 to TUG1 or NEAT2 depends on K191 methylation state. RNA gel shift assay was performed using synthesized TUG1, NEAT2 RNA and their corresponding DNA oligos and recombinant His-Pc2 with or without in vitro methylation. (D–F) TUG1 and NEAT2 function as modulators of Pc2 chromodomain for “reading” the histone code. MODified Histone Peptide ArrayR was incubated with Recombinant His-Pc2 chromodomain in the presence of yeast tRNA (D), in vitro transcribed TUG1 (117-3390) (E), or NEAT2 (2281-5611) (F) RNA fragments and subjected to immunoblotting with indicated antibodies. The Myc tag antibody was added to detect Myc tagged peptides on position P21 as positive control. The binding specificity calculated by Array Analyses Software based on two arrays (images shown in Figures S5F–S5H) was shown. (G) TUG1 is localized in PcG bodies. HeLa cells microinjected with molecular beacon probes targeting TUG1 were subjected to immunostaining with Pc2K191me2 antibodies. (H) TUG1 and NEAT2 are required for serum-induced relocation of the MCM3 locus. HeLa cells microinjected with validated siRNA targeting TUG1 or NEAT2 were serum-starved followed by restimulation and Immuno-FISH analyses were performed using Ring1A, SC35 antibodies and probes targeting MCM3 locus. Statistical analyses on colocalization of MCM3 loci with Ring1A staining (left panel) and SC35 staining (right panel) are shown. (I and J) Effect of TUG1 or NEAT2 knockdown on serum-induced growth control gene expression and cell proliferation. HeLa cells transfected with TUG1 or NEAT2 siRNA were serum-starved followed by restimulation and relative mRNA levels of indicated genes and cell proliferation were determined by qRT-PCR (I) and flow cytometry (J). (K) A list of TUG1 and NEAT2 ncRNA- associated proteins identified from two independent MS experiments. Mean±SEM, *p<0.05, **p<0.01 and ***p<0.001.
See also Figure S5 and Table S1–S3.
ncRNA-Pc2 Interactions Underlie Relocation and Transcriptional Regulation of Growth-Control Genes
Pc2 has been characterized as a “reader” for the histone code through its chromodomain (Bernstein et al., 2006). To gain insight into whether binding of ncRNAs affect the histone code reading ability of Pc2, we carried out a modified histone peptide array analysis in an open-ended manner. Consistent with a previous publication (Bernstein et al., 2006), bacterially purified chromodomain of Pc2 specifically recognized histone H3K9me3 (Figures 5D and S5F). Strikingly, in the presence of TUG1 ncRNA, the Pc2 chromodomain now instead read both H4R3me2s and H3K27me2 (Figures 5E and S5G), which are associated with global gene repression (Xu et al., 2010). In contrast, by binding to NEAT2 RNA, Pc2 switched from an H3K9me3 reader to preferentially binding H2AK5ac and H2AK13ac, marks of gene activation (Figures 5F and S5H). As controls, TUG1 (117-3390) or NEAT2 (2281-5611) alone failed to bind any histone marks (Figures S5I and S5J). Collectively, our results now suggest a mechanism by which ncRNAs can regulate “reading” of the histone code by chromatin-binding domains of various regulatory cofactors. In this case, ncRNAs interacting with a chromodomain switches preferential binding from repressive to activation histone marks.
Interchromatin granule-specific localization of NEAT2 (Hutchinson et al., 2007) led us to examine the cellular localization of TUG1 by utilizing a molecular beacon targeting TUG1. Single-cell nuclear microinjection of the TUG1 molecular beacon followed by immunostaining suggested that part of TUG1 localization overlapped with methylated Pc2 staining, suggesting the localization of TUG1 within the PcG body (Figure 5G). Thus, the subnuclear structure-specific localization of TUG1 and NEAT2 may indicate that they serve as the actual docking sites responsible for relocation of Pc2-bound growth control gene promoters. Indeed, knockdown of TUG1 caused a dramatic redistribution of growth control gene promoters out of PcG bodies, (Figures 5H and S5K), which correlated with increased gene expression (Figure 5I) and cell proliferation, even in the absence of serum (Figure 5J). Conversely, removal of NEAT2 disrupted the colocalization of MCM3 gene locus with interchromatin granules (Figures 5H and S5K), caused loss of serum-induced gene expression (Figure 5I) and of cell proliferation (Figure 5J), although structures of the PcG bodies and nuclear speckles were not discernibly affected by knocking down TUG1 (Figure S5L), NEAT2 (Clemson et al., 2009) and serum treatment (Figure S5M).
We next identified the proteins associated with TUG1 and NEAT2 by RNA pull-down experiments. The experiment performed here calls for washes with buffer containing 1000 mM salt, which definitively result in relatively low backgrounds in pull-downs and do not disrupt association of TUG1 and NEAT2 with proteins. Mass spectrometry analysis on proteins pulled down by biotinylated TUG1 and NEAT2 revealed that TUG1 RNA specifically bound to a number of proteins involved in transcriptional repression, including histone methyltransferases/demethylases and chromatin modifiers (Figure 5K, Tables S1 and S2), while NEAT2 RNA bound to functionally distinctive cofactors associated with gene activation including transcriptional co-activators, active histone marks associated histone methyltransferases/demethylases, and pre-mRNA splicing factors (Figure 5K, Tables S1 and S3), suggesting that ncRNAs may impose a chromatin remodeling environment by selectively interacting with chromatin modifier proteins. Intriguingly, TUG1 interacted with Coronin2A, which is a co-repressor based on actin oligomerization (Huang et al., 2011), while LSD1 interacted with NEAT2, consistent with previous observation that LSD1 colocalizes with transcription units in interchromatin granules (Hu et al., 2008).
Unmethylated Pc2 Mediates Serum-Induced E2F1 SUMOylation
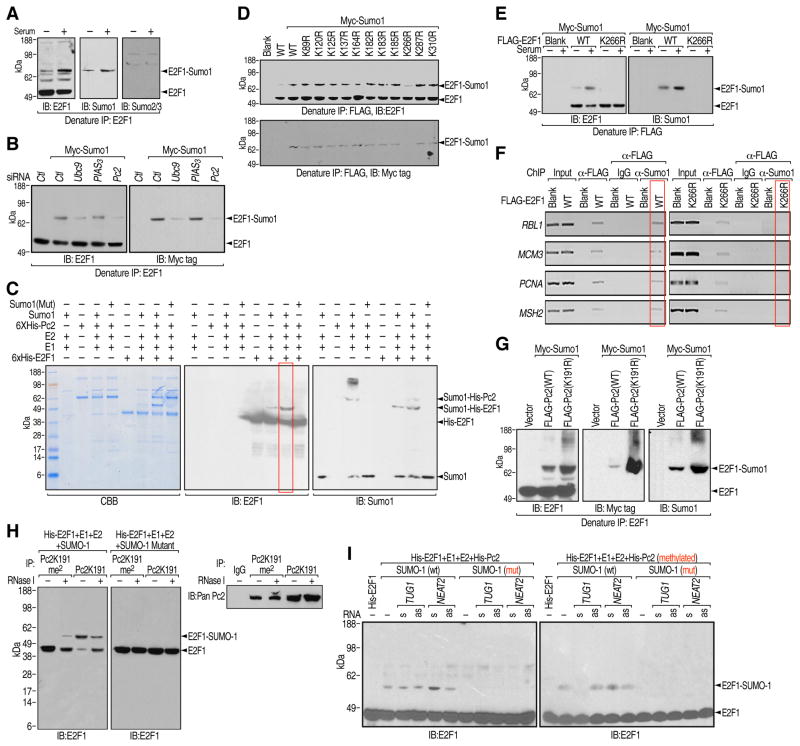
We next examined the functional roles of unmethylated Pc2 in growth control gene activation. Immunoblotting of E2F1 immunoprecipitates from denatured cell lysates unexpectedly revealed an upshifted E2F1 band induced by serum stimulation, which was not due to an E2F1-associated protein, but rather a post-translational modification (Figures 6A and S6A). Immunoblotting using antibody specific against SUMO1 and SUMO2/3 confirmed that E2F1 and E2F6, but not E2F4, was SUMO1 modified (Figures 6A, S6B and S6C). In searching for cellular components responsible for E2F1 SUMOylation, we observed a decreased E2F1 SUMOylation with depletion of Ubc9 and Pc2, but not other candidate E3 ligases tested (Figures 6B and S6D). When Pc2 was co-transfected with Myc-SUMO1, we observed a significant increase in E2F1 SUMOylation in vivo (Figure S6E). Pc2 directly mediated enhanced E2F1 SUMOylation was further confirmed in vitro (Figure 6C). To map the SUMOylation site, charge-conserving E2F1 lysine to arginine point mutants were tested for SUMOylation, revealing that the E2F1 K266R mutant showed deficient SUMOylation both in vivo and in vitro (Figures 6D, 6E and S6F). Although E2F1 and E2F1-K266R localized equally on the promoters of E2F1 target genes, the E2F1-K266R mutant largely inhibited serum-induced gene expression, which was not due to any effects of K266R mutation on E2F1 stability or its interaction with Rb (Figures 6F, S6G and S6H). Together, these results have revealed a requirement for Pc2-mediated E2F1 K266 SUMOylation or stabilization of this SUMOylation for the transcriptional activation of growth control genes.
Figure 6. Pc2-mediated E2F1 SUMOylation Is Required for Growth Control Gene Activation.
(A) Serum-induced E2F1 SUMOylation. HeLa cells were serum-starved followed by restimulation and the cell lysates were immunoprecipitated with E2F1 antibodies under denaturing conditions followed by immunoblotting with antibodies indicated. (B) Pc2 promotes E2F1 SUMOylation in vivo. HeLa cells were transfected with indicated siRNAs/plasmids and the cell lysates were immunoprecipitated with E2F1 antibodies under denaturing conditions followed by immunoblotting with E2F1 (left panel) or Myc tag (right panel) antibodies. (C) Pc2 promotes E2F1 SUMOylation in vitro. Bacterially-expressed His-Pc2, His-E2F1 were incubated with recombinant E1, E2 and SUMO1 or SUMO1 mutant. The reactions were separated by SDS-PAGE followed by Coomassie blue staining (left panel), immunoblotting with E2F1 (middle panel) or SUMO1 (right panel) antibodies. (D and E) E2F1 is SUMOylated at lysine 266. HeLa cells were transfected with wt or single K mutated E2F1 (D) or K266R mutant (E) as indicated and the cell lysates were immunoprecipitated with FLAG antibodies under denaturing conditions followed by immunoblotting with E2F1 (upper panel) or Myc tag (bottom panel) antibodies (D) and E2F1 (left panel) or SUMO1 (right panel) antibodies (E). (F) E2F1 SUMOylation on growth control gene promoters. HeLa cells were transfected with indicated plasmids followed by two-step ChIP analyses using FLAG and SUMO1 antibodies on indicated regions. (G) K191 methylation inhibits Pc2-mediated E2F1 SUMOylation in vivo. HeLa cells were transfected with indicated plasmids and the cell lysates were immunoprecipitated with E2F1 antibodies under denaturing conditions followed by immunoblotting with E2F1 (left panel), Myc tag (middle panel) or SUMO1 (right panel) antibodies. (H) Pc2-mediated E2F1 SUMOylation is RNA dependent. Immunoprecipitates of Pc2K191me2 or Pc2K191 from HeLa cells (right panel) were incubated with His-tagged E2F1, recombinant E1, E2 and SUMO1 or SUMO1 mutant with or without RNase I treatment. The reaction products were subjected to immunoblotting using antibodies targeting E2F1 (left panel). (I) TUG1 and NEAT2 regulate Pc2-mediated E2F1 SUMOylation. Bacterially-expressed His-E2F1 were incubated with recombinant E1, E2, SUMO1 (wt or mutant), and His- Pc2 (unmethylated or methylated) in the presence of synthesized sense or antisense TUG1 or NEAT2 RNA oligos. The reactions were subjected to immunoblotting using antibodies against E2F1.
See also Figure S6.
The observation that expression of the Pc2K191R mutant, which exerted no effects on endogenous levels of Ubc9, noticeably enhanced E2F1 SUMOylation (Figures 6G and S6I) led us to explore whether ncRNAs might modulate E2F1 SUMOylation mediated by Pc2. We first tested whether Pc2-regulated E2F1 SUMOylaton was RNA dependent. Treatment of Pc2K191me2 immunoprecipitates with RNase I slightly enhanced E2F1 SUMOylation, whereas the same treatment impaired E2F1 SUMOylation mediated by Pc2K191 immunoprecipitates (Figure 6H). Consistently, the inhibitory activity of TUG1 on E2F1 SUMOylation was observed in performing an in vitro SUMOylation assay, with the addition of TUG1 RNA abolishing the enhanced E2F1 SUMOylation mediated by methylated Pc2 (Figure 6I, right panel, lane 3). Taken together, our findings suggest that Pc2-mediated E2F1 SUMOylation at K266, which was modulated by interactions with ncRNAs, is required for the activation of cell growth control genes in response to serum.
CDCA7L Links E2F1 SUMOylation and H2B Ubiquitination in Growth Control Gene Activation
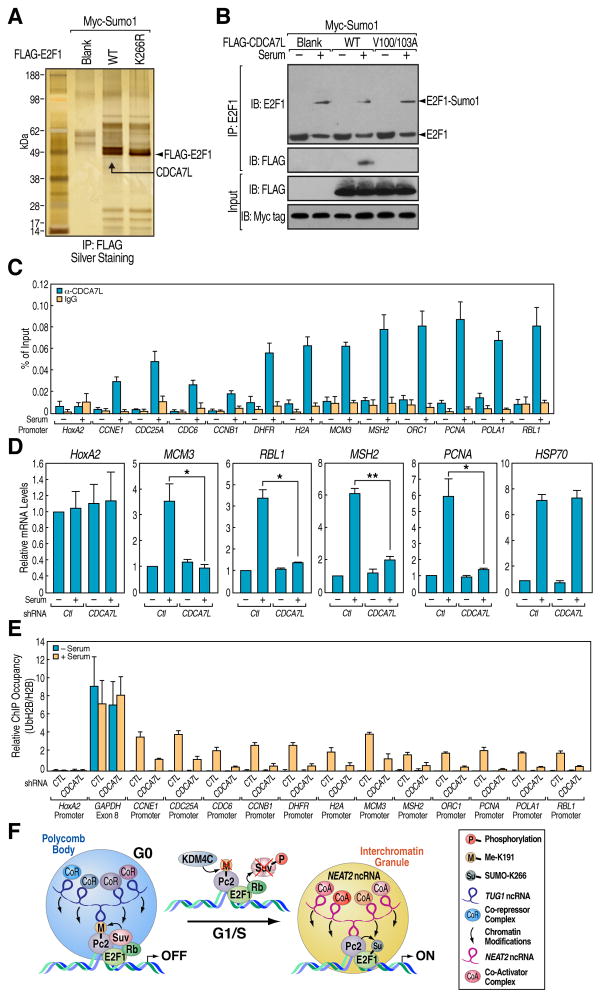
The functional binding of SUMOylated proteins to the SUMO-interacting Motif (SIM)-containing proteins plays critical roles in many biological processes including transcriptional activation, cell growth/differentiation and DNA damage response (Kerscher, 2007). Thus, we searched for the potential SIM-containing proteins that can recognize the E2F1 K266 SUMOylation and its functional role in regulating growth control gene activation. Analysis of a protein exclusively co-purified with wt E2F1, but not the K266R mutant, by MS identified it as CDCA7L/JPO2/RAM2 (Figures 7A, S7A and S7B), a cell cycle regulator with a conserved RING1 domain (Huang et al., 2005). The specific interaction between SUMOylated E2F1 and CDCA7L was mediated by a SIM motif located at N-terminal of CDCA7L, and mutations in this motif (V100/103A) abolished the interaction with SUMOylated E2F1 (Figure 7B). Consistent with the specific recruitment of CDCA7L on the E2F1 target gene promoters upon serum stimulation (Figure 7C), knockdown of CDCA7L by shRNAs blocked serum-induced expression of E2F1 target genes (Figures 7D and S7C). CDCA7L possesses a conserved RING finger domain, a characteristic of RING-class E3 ubiquitin protein ligases (Deshaies and Joazeiro, 2009). We therefore screened potential substrates of CDCA7L by both in vivo and in vitro ubiquitination assay, finding that CDCA7L strongly stimulated H2B monoubiquitination at K120 (Figures S7D–S7F). These results indicate that the RING finger of CDCA7L is required for the monoubiquitination of H2B, and perhaps additional substrates, critical for transcriptional activation of growth control genes.
Figure 7. CDCA7L Interacts with SUMOylated E2F1 and Functions as a H2B Monoubiquitinase to Promote Growth Control Gene Activation.
(A) CDCA7L interacts with K266 SUMOylated E2F1. FLAG-E2F1 immunoprecipitates from HeLa cells were separated by SDS-PAGE followed by silver staining. The band indicated was cut and subjected to MS analysis. (B) CDCA7L interacts with SUMOylated E2F1 via its SUMO-interacting motif. HeLa cells transfected with indicated plasmids were serum-starved followed by restimulation and the cells lysates were immunoprecipitated with E2F1 antibodies followed by immunoblotting with antibodies indicated. (C) Serum-induced recruitment of CDCA7L on growth control gene promoters. ChIP analyses were performed in serum-starved and restimulated HeLa cells using CDCA7L antibodies on indicated regions. (D) CDCA7L is required for growth control gene activation. HeLa cells transfected with shRNA against CDCA7L were serum-starved followed by restimulation and relative mRNA levels of indicated genes were determined by qRT-PCR. Mean±SEM, *p<0.05 and **p<0.01. (E) CDCA7L is required for serum-induced histone H2B ubiquitination on growth control gene promoters. HeLa cells transfected with shRNA against CDCA7L were serum-starved followed by restimulation and ChIP analyses were performed using antibodies targeting UbH2B and H2B on indicated regions. For each primer pair, UbH2B values were normalized to the corresponding total H2B ChIP. (F) Model: Interplay between ncRNAs and methylated vs. unmethylated Pc2 determines intranuclear localization of growth control genes in PcG bodies or interchromatin granules, which dictates gene repression vs. activation events.
See also Figure S7.
Analysis of UbH2B and H2B enrichment by ChIP assay revealed that ubiquitination of H2B was enhanced at the E2F1 binding sites upon serum stimulation, which was diminished with knockdown of CDCA7L (Figure 7E). This finding is consistent with the previous notion that UbH2B correlates with gene activation and transcriptional elongation (Minsky et al., 2008). Together, these data revealed that a series of covalent modifications initiated by Pc2-mediated SUMOylation of E2F1 leads to the subsequent H2B ubiquitination and the activation of a cohort of growth control genes.
Discussion
Orchestrating precise spatial and temporal patterns of gene expression is crucial for the normal development of all organisms and maintenance of homeostasis. DNA-binding transcription factors, chromatin modifiers and dynamic positioning of particular loci are all essential to establish precise transcriptional regulation and achieve the correct patterns of gene expression. Here, we have elucidated how a non-histone methylation event and the resultant specific ncRNA associations spatially modulate signal-induced nuclear architecture alteration, which underlies regulation of transcription units controlling cell growth programs both in normal and transformed cells.
Our studies have revealed that the methylation/demethylation of a single protein associated with E2F1-bound growth control genes, Pc2, is responsible for the physical relocation of these transcription units from PcG bodies, in which their expression is repressed, to interchromatin granules, in which their expression is activated (Figure 7F). Unexpectedly, this reflects the differential interactions of methylated vs. unmethylated Pc2 with two distinct ncRNAs that are located in distinct subnuclear architectural compartments (Figure 7F). In the case of Pc2, methylation controls the protein’s interaction with two distinct ncRNAs, TUG1 and NEAT2, which results in the exclusive subnuclear localization of methylated and unmethylated Pc2 in PcG bodies and interchromatin granules, respectively. Methylated Pc2 may represent an important antimitogenic signal and, under certain stimuli, a stress-induced modification mark required for growth-control gene repression and senescence, whereas unmethylated Pc2 is essential for physiological growth-control gene expression and cell proliferation.
TUG1, previously reported to interact with the PRC2 complex (Khalil et al., 2009), is localized in PcG bodies to serve as a scaffold by interacting with Suv39h1-methylated Pc2, both relocating growth control transcription units and bringing them in proximity with specific co-repressor machinery. In contrast, NEAT2 interacts with several proteins associated with gene activation. These ncRNAs promote the relocation of growth control genes in response to mitogenic signals and are likely to facilitate the repression/activation of the encoded transcription units based both on their association with multiply co-regulating complexes and actions to modulate histone marks preferred by “readers” of the histone code. Indeed, the loss of TUG1 causes signal-independent activation of growth by failure to recruit growth control gene loci to PcG bodies, while, conversely, the loss of NEAT2 inhibits growth, even in the presence of growth signals, with a failure to relocate the growth control genes to interchromatin granules.
This dynamic process is apparently dependent on a histone demethylases KDM4C that facilitates signal-dependent relocation of growth control genes between distinct subnuclear architectural structures, with two ncRNAs serving as subnuclear molecular scaffolds. Our data have thus identified the Pc2/ncRNAs interactions as a key mechanism that determines the relative positioning of a growth control gene regulatory program with respect to subnuclear architectural structures; however, we cannot absolutely exclude the possibility of a rapid assembly of subnuclear architectural structures in situ on the regulated promoters.
Consistent with our ncRNA pull-down results, the promoters interacting with TUG1 are postulated to present multiple epigenomic regulators that might also modulate histone marks, including the H3K9 methyltransferases RIZ1, the H3K27 methyltransferase EZH2, and the H3K4 demethylase JARID1A. In contrast, the promoters associated with NEAT2 in interchromatin granules exhibit a low levels of H3K9me2/3, but high levels of H3K36me3 and H3K4me3, perhaps regulated by interaction of NEAT2 with LSD1, SET2 and MLL. These data suggest the potential importance of these two ncRNAs as platforms for organizing a coordinated repression or activation program respectively.
Taken together, our findings highlight the large network of long ncRNAs, together with their associated protein complexes, as key components of the subnuclear architectural structures, each acting as a “sensor” for specific cohorts of regulated gene programs in response to signal-induced modifications that license gene regulatory regions-ncRNA interactions. While Pc2 methylation appears to be the key determinant for relocation of growth control genes, based on a methylation/demethylation dependent switch in preference for TUG1 or NEAT2 interactions, respectively, we are tempted to speculate that other ncRNAs recognize covalent modifications of other gene-associated proteins, exerting their ncRNA scaffold functions in distinct subnuclear compartments to act as sensors for many regulated signaling pathways, and as modulators of “reading” marks on histone tails for a variety of chromodomain-containing proteins and other histone code “readers.”
Future exploration of interactions between subnuclear structure-specific ncRNAs and functionally-distinctive protein cohorts is likely to uncover fresh insights into central strategies in development, homeostasis and disease.
Experimental Procedures
Cell Culture and Synchronization
Human cervical cancer cell line HeLa, embryonic kidney cell line HEK293T, and human lung normal fibroblast IMR-90 were obtained from American Type Culture Collection (ATCC) and were cultured under standard conditions. Transfection of HeLa cells with siRNA and plasmid DNA were performed using Lipofectamine2000™ (Invitrogen). To induce growth control gene expression and cell proliferation, HeLa cells transfected with plasmids or siRNAs as indicated were starved in serum-free media for 48 hours followed by stimulation with complete serum media for additional 16 hours. IMR-90 fibroblasts were incubated in DMEM (Gibco) medium supplemented with a suboptimal concentration of FBS (0.1%) for 4 days. Normal serum level (10%) was restored and cells were harvested at 24 hours after release from low serum. A detailed description of methods is included in the Extended Experimental Procedures.
Antibody Generation
The rabbit anti-dimethyl-Pc2-specific antibody (Pc2K191me2) was raised against a synthetic peptides CPDLGA (di-methyl-K) SHPPDKWAQ based on the human Pc2 sequence flanking lysine 191 (Abgent). The rabbit anti-unmethylated Pc2 antibody (Pc2K191) was obtained at the same time against peptide CPDLGAKSHPPDKWAQ (Abgent). The Pc2K191me2 and Pc2K191 antibodies were used for immunoprecipitation, immunoblotting, ChIP, CLIP and immunofluorescence.
Cell Proliferation Assay
Cell proliferation was measured by using Click-iT™ EdU Cell Proliferation Assay Kit (Invitrogen) following the manufacturer’s protocol. Data were recorded on BD™ LSR II Flow Cytometer and analyzed by FlowJo flow cytometry analysis software (Tree Star, Inc.).
Immunostaining and Colocalization Analyses
HeLa cells cultured on sterile glass coverslips were synchronized with serum free media for 24 hours before they were stimulated with serum-containing media for 16 hours. The cells were rinsed with PBS twice and fixed with 4% paraformaldehyde at room temperature for 10 min. Then the cells were washed three times with PBS/0.01% Triton X-100 and permeabilized with PBS/0.5%Triton X-100 for 10 min. After brief rinse in PBS, the cells were blocked by 1% BSA in PBS for 30 min followed by incubation with primary antibodies as indicated for 1 hour. After washing three times with PBS, the cells were incubated with Alexa Fluor 488 donkey anti-mouse IgG or Alexa Fluor 594 donkey anti-Rabbit IgG for additional 1 hour. The cells were then washed three times with PBS, and mounted in Prolong Gold antifade reagent with DAPI. Immunofluorescence images were acquired with a Zeiss Axioplan 2MOT Epifluorescent microscope (Carl Zeiss, Inc) with Hamamatsu ORCA ER black/white CCD camera.. For Ring1/Bmi1 and Pc2K191me2 immunofluorescence, rabbit anti-Ring1/Bmi1 and anti-Pc2K191me2 IgGs were labeled by Zenon® Alexa Fluor® 594 and 488 Rabbit IgG Labeling Kits, respectively (Molecular Probe). Quantification of colocalization was determined by Pearson’s correlation coefficient using commercially available Colocalizer Pro software and the analysis was performed on at least 100 cells per condition with three independent experiments.
Immuno-Fluorescence in situ Hybridization
Cell nuclei isolation and Immuno-FISH was carried out according to a method previously described (Lavrov et al., 2004) and the probes were commercially labeled by Empire Genomics and listed in Extended Experimental Procedures. At least 100 cells were counted in each experiment for statistical analysis.
In Vivo Cross-Linking and ImmunoPrecipitation (CLIP) Assay
CLIP analyses of methylated and unmethylated Pc2 were performed as previously described (Ule et al., 2005). Briefly, cells were ultraviolet (UV) irradiated to covalently crosslink RNA-protein complexes. After UV irradiation, cells were lysed and the RNA was partially digested, allowing a small fragment to remain attached to protein. RNA-protein complexes of interest were then partially purified by immunoprecipitation and non-covalently associated RNAs were removed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). These purified RNA-protein complexes were isolated and treated with proteinase K, which removes protein but leaves intact RNAs. The recovered RNAs were subjected to RNA linker ligation, RT-PCR amplification, and sequencing.
Molecular Beacon and RNA FISH
Custom designed molecular beacon targeting TUG1 was synthesized by IDT. RNA FISH was performed using Stellaris™ RNA FISH technology according to manufacturer’s instruction (Biosearch Technologies, Inc.). Stellaris™ RNA FISH probes targeting TUG1 and NEAT2 were designed at http://www.singlemoleculefish.com/designer.html.
Data Analysis and Statistics
Relative quantities of cell cycle gene expression level were normalized to Actin. The relative quantities of ChIP samples were normalized by individual inputs respectively. Results are reported as mean ± SEM of three independent experiments. Comparisons were performed using two-tailed paired Student t-test. *p<0.05, **p<0.01 and ***p<0.001.
Supplementary Material
Acknowledgments
We thank C. Nelson for assistance with cell culture; J. Hightower for artwork; D. Benson, M. Fisher and R. Pardee for assistance with the manuscript; Dr. B. Tanasa for providing E2F1 target sites from previous published ChIP-Seq data; Dr. P. Sun and Dr. R. Liao for providing BJ cells and technical supports for senescence study; Dr. R. June for assistance with flow cytometry analysis; and Dr. M. Ghassemian for mass spectrometry analysis. Michael G. Rosenfeld is a Howard Hughes Medical Institute Investigator. This study was funded by grants from DK018477, DK74868, DK39949, CA97134, NS34934, W81XWH-08-1-0665 and the Prostate Cancer Foundation to M. G. R. L. Yang is the recipient of a DoD Era of Hope Postdoctoral Award (GRANT00325108); and C. Lin is the recipient of a Susan G. Komen for the Cure Fellowship (KG080247).
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- Bagchi S, Weinmann R, Raychaudhuri P. The retinoblastoma protein copurifies with E2F-I, an E1A-regulated inhibitor of the transcription factor E2F. Cell. 1991;65:1063–1072. doi: 10.1016/0092-8674(91)90558-g. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanero MR, Flemington EK. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci U S A. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Czvitkovich S, Sauer S, Peters AH, Deiner E, Wolf A, Laible G, Opravil S, Beug H, Jenuwein T. Over-expression of the SUV39H1 histone methyltransferase induces altered proliferation and differentiation in transgenic mice. Mech Dev. 2001;107:141–153. doi: 10.1016/s0925-4773(01)00464-6. [DOI] [PubMed] [Google Scholar]
- de la Luna S, Burden MJ, Lee CW, La Thangue NB. Nuclear accumulation of the E2F heterodimer regulated by subunit composition and alternative splicing of a nuclear localization signal. J Cell Sci. 1996;109(Pt 10):2443–2452. doi: 10.1242/jcs.109.10.2443. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Ho CS, Ponzielli R, Barsyte-Lovejoy D, Bouffet E, Picard D, Hawkins CE, Penn LZ. Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res. 2005;65:5607–5619. doi: 10.1158/0008-5472.CAN-05-0500. [DOI] [PubMed] [Google Scholar]
- Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, et al. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lavrov S, Dejardin J, Cavalli G. Combined immunostaining and FISH analysis of polytene chromosomes. Methods Mol Biol. 2004;247:289–303. doi: 10.1385/1-59259-665-7:289. [DOI] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AH, van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, Zhang MQ, Lazebnik Y, Bar-Sagi D, Lowe SW. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- Nevins JR. Transcriptional regulation. A closer look at E2F. Nature. 1992;358:375–376. doi: 10.1038/358375a0. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Olson DJ, van der Vlag J, Hamer KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R, Otte AP. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt RG, Lachner M, Vargas M, Hamer KM, den Blaauwen JL, Hendrix T, Melcher M, Schweizer D, Jenuwein T, Otte AP. Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of Polycomb group proteins. Mol Cell Biol. 2002;22:5539–5553. doi: 10.1128/MCB.22.15.5539-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczerbal I, Bridger JM. Association of adipogenic genes with SC-35 domains during porcine adipogenesis. Chromosome Res. 2010;18:887–895. doi: 10.1007/s10577-010-9176-1. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hoang S, Mayo MW, Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics. 2010;11:396. doi: 10.1186/1471-2105-11-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.