Abstract
Protonated tryptic peptides, somatostatin-14, and oxytocin have been subjected to reactions with doubly deprotonated 4-formyl-1,3-benzenedisulfonic acid (FBDSA) in the gas phase. The major product is a negatively-charged complex comprised of the peptide and the reagent. Upon dehydration of the complex, all peptides show evidence for Schiff base formation involving a primary amine of the peptide. Some peptides also show evidence for the formation of a relatively strong electrostatic interaction without Schiff base formation (i.e., a mixture of isomeric precursor ions is generated upon dehydration of the complex). Ion trap collision-induced dissociation of the dehydration products from all peptides examined gave distinct product ion spectra relative to the deprotonated and protonated forms of the peptides. The distinct behavior of the modified ions is attributed to the highly stable charge carrying sulfonate group, which tends to inhibit intramolecular proton transfer in negatively charged species. Modified anions of the peptides with an intramolecular disulfide linkage show evidence for cleavage of both the disulfide linkage and an amide bond in the loop defined by the disulfide bond. Modification of protonated peptides via charge inversion with FBDSA is a useful means for generating novel and distinct ion-types that can provide complementary structural information upon subsequent activation to that obtained from dissociation of protonated or deprotonated forms of the peptide.
Keywords: Ion/ion reactions, peptide ion modification, peptide anion CID
1. Introduction
Primary structure characterization by mass spectrometry is generally dependent upon fragmentation of ions derived from the molecule of interest. The two main options for maximizing the structural information of interest are selection of ion-type (e.g., protonated molecule versus radical cation versus deprotonated molecule, etc.) and ion activation conditions [1]. The ion-type is usually determined by the means for ionization. It is also common to subject analyte species, such as peptides or proteins, to a derivatization reaction in solution to facilitate ionization [2], quantification [3], or structural characterization [4] via mass spectrometry or tandem mass spectrometry. Alteration of the ion-type can also take place in the gas phase via ion/electron [5], ion/molecule [6], or ion/ion reactions [7]. Most gas phase reactions have involved the gain or loss of protons or electrons by the analyte ions. Examples of selective covalent modification of polypeptide ions in the gas-phase are restricted to ion-molecule [8] and ion-ion reactions and are still rare. However, several recent findings suggest that the selective modification of primary amines in peptide ions via ion/ion chemistry is straightforward via either Schiff base formation [9] using aldehyde containing reagents or via amide bond formation [10] using N-hydroxysuccinimide ester containing reagents. The ability to form covalent bonds selectively in the gas-phase opens up new possibilities for probing the structures of gaseous polypeptides. For example, the cross-linking of peptide ions in the gas-phase has recently been demonstrated using bi-functional N-hydroxysuccimide ester reagent ions [11].
Schiff base formation involving either the N-terminus or the ε -NH2 group of lysine residues has been effected by reacting peptide cations with either singly or doubly deprotonated 4-formyl-1,3-benzenedisulfonic acid (FBDSA). A sulfonic acid group in the reagent anion can interact strongly with a protonated site on the peptide resulting in a long-lived complex. Upon collisional activation of the complex, the aldehyde group can undergo nucleophilic attack by an available unprotonated primary amine resulting in loss of water and formation of a Schiff base. Water loss from the complex is therefore consistent with Schiff base formation, although water loss is a common process from peptide ions. Strong electrostatic interactions between the reagent anion and the peptide cation can occur, particularly between the sulfonate group and protonated arginine. Hence, either covalent or non-covalent modification of the peptide may alter the fragmentation behavior of a peptide ion. The identities of the fragmentation products arising from further activation of the water loss product to give cleavages along the peptide backbone can provide clues regarding the nature of the modification. Loss of intact FBDSA, for example, from the water loss product is a clear indication of an electrostatic interaction for those ions that undergo this loss.
Singly deprotonated FBDSA is useful for modifying multiply-protonated peptides [9], as shown in process (1):
| (1) |
Note that the diamond symbol (◆) corresponds to the addition of FBDSA to M minus a water molecule. Doubly deprotonated FBDSA can be used to modify either singly protonated peptides or peptides with three or more excess charges in a single step. In the case of singly protonated peptides, reaction with doubly deprotonated FBDSA results in the conversion of the peptide cation to an anionic product, as indicated in process (2): [12]
| (2) |
Doubly protonated peptides can also undergo charge inversion in reactions with FBDSA di-anions but this entails two consecutive ion/ion reactions with the first involving single proton transfer (process (3)) to yield the singly protonated peptide, which can then undergo process (2):
| (3) |
Single proton transfer, as represented by process (3), competes with long-lived complex formation, which constitutes the first step of processes (1) and (2) [13]. Complex formation is the dominant process for the species relevant to this work but single proton transfer typically occurs as a competitive process.
All work to date using ion/ion reactions involving FBDSA anions with peptide cations has indicated that the modified products upon collisional activation often yield sequence information that is complementary to that derived from unmodified ions [9,12]. However, the work has been restricted to a limited number of model peptide ions. In this work, we have extended the range of observations to include peptides with an intramolecular disulfide linkage as well as ions derived from tryptic digestion of a protein. Tryptic peptides are of interest because of their relevance to bottom-up protein identification [14]. Peptides with intramolecular disulfide linkages often present a challenge for deriving sequence information, particularly for protonated peptides, due to the fact that the disulfide linkage must be cleaved along with a backbone bond [15, 16]. In this work, somatostatin-14 and oxytocin, two peptide hormones, were chosen as model cyclic peptides.
2. Experimental
Materials
Methanol, glacial acetic acid, and ammonium hydroxide were purchased from Mallinckrodt (Phillipsburg, NJ). Ubiquitin from bovine erythrocytes, TPCK treated trypsin from bovine pancreas, somatostatin-14, 4-formyl-1,3-benzenedisulfonic acid, tris (2-carboxyethyl) phosphine hydrochloride, and ammonium bicarbonate were purchased from Sigma-Aldrich (St. Louis, MO). Peptides MQIFVK, TITLEVEPSDTIENVK, EGIPPDQQR, LIFAGK, TLSDYNIQK, and ESTLHLVLR were generated from the digestion of ubiquitin by trypsin. Oxytocin was obtained from Bachem Bioscience Inc. (Torrance, CA). Peptide analytes were prepared in ~100 μM aqueous solution prior to positive nanoelectrospray ionization (nESI). The anion reagent was prepared at a concentration of 3.5 mM in solution of 49.5/49.5/1 (v/v/v) water/methanol/ammonium hydroxide for negative nESI.
Tryptic Digest
The procedure for the tryptic digestion of ubiquitin has been described previously [17]. Separation of the tryptic peptides was performed using a reverse-phase HPLC (Agilent 1100, Palo Alto, CA) equipped with an Aquapore RP-300 (7μm pore size, 100 × 4.6 mm i.d.) column (Perkin-elmer, Wellesley, MA). The gradient for the HPLC separation has been described previously [17]. Following separation, collected fractions were lyophilized and dissolved in 250 μL of water.
Disulfide Reduction
The procedure for disulfide reduction has been described previously [18]. Reduced cyclic peptides were filtered through a disposable PD-10 desalting column (GE Healthcare) using a 1% acetic acid solution. Collected fractions were lyophilized and dissolved in 250 μL of water.
Mass Spectrometry
Experiments were performed on QqQ tandem mass spectrometers (QTRAP 2000 and QTRAP 4000, AB Sciex, Concord, ON, Canada), which have been modified for ion/ion reactions [19]. The QTRAP 4000 was used in cases in which the Schiff base product ions were of m/z greater than 1700 due to its higher mass range. Alternately pulsed ESI emitters allowed for sequential ion injection into the q2 reaction cell [20, 21]. First, doubly or singly deprotonated FBDSA was accumulated in the q2 reaction cell. Next, the peptide cations were generated and transferred to the q2 cell to undergo mutual storage with the FBDSA anions for 500-1200 milliseconds. The product ions were then transferred to the Q3 ion trap for subsequent mass analysis via mass-selective axial ejection (MSAE) [22] or for further interrogation via MSn.
3. Results and Discussion
Tryptic Peptides
Ubiquitin was subjected to digestion with trypsin and cations derived from the following peptides were observed in the positive nESI mass spectrum: MQIFVK, LIFAGK, TITLEVEPSDTIENVK, EGIPPDQQR, TLSDYNIQK, and ESTLHLVLR. Cations of each of these peptides were subjected to ion/ion reactions with anions derived from FBDSA and the results are provided below and in Supplementary Information. In all cases, comparisons were made of the CID behavior of the unmodified [M-H]− peptide as well as that of the singly or doubly protonated species.
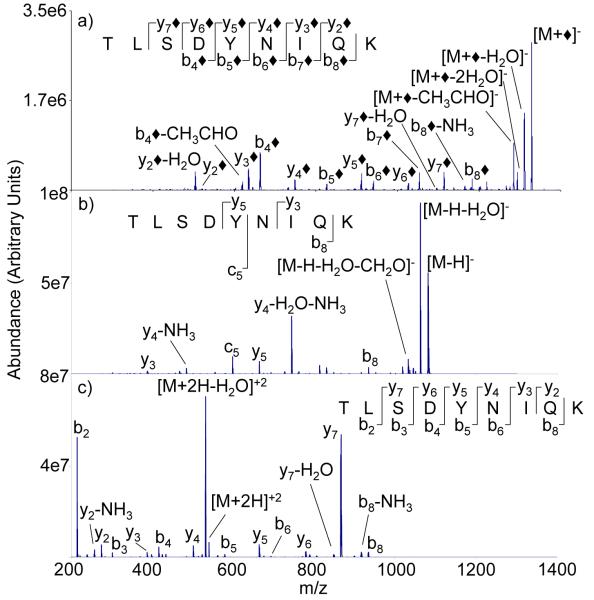
The ion trap CID product ion spectrum of the modified TLSDYNIQK anion is compared to those of the singly deprotonated and doubly protonated forms of the peptide in Figure 1. In this case, cations of TLSDYNIQK were reacted with [FBDSA-2H]2- to yield the negatively charged [TLSDYNIQK+FBDSA-H]− complex. Ion trap collisional activation of the complex gave rise to a highly abundant water loss product, which is indicated as [M+◆]− in Figure 1(a). The diamond symbol (◆) associated with some products indicates a fragment that is consistent with the indicated product with the expected mass shift associated with the covalent modification. We note, however, that the labeled fragments can also, in principle, arise from non-covalent FBDSA adduction with water loss arising elsewhere in the peptide ion. In this particular case, no clear evidence for electrostatic binding, such as the appearance of [FBDSA-H]− or the attachment of FBDSA to a product ion (see below) is apparent. The most abundant products are consistent with modified y-type and b-type ions, using the nomenclature that is adapted [23] from the protonated peptide literature [24], which would correspond to α-ions and β-ions, respectively, in the nomenclature employed by Bowie [25]. When compared to the unmodified anion (i.e., the [M-H]− ion), the CID of the modified peptide in this case generates more sequence information (compare Figures 1(a) and 1(b)). Figure 1(c) shows the CID product ion spectrum of doubly protonated TLSDYNIQK, as this is the most abundant cation in the nESI mass spectrum. The modified peptide and the unmodified peptide cation of TLSDYNIQK produce similar sequence information (Figures 1(a) and 1(c)). Both spectra reflect cleavages of the same peptide bonds, although the relative contributions differ markedly. For example, the complementary y7/b2 pair dominates the cation spectrum whereas less selectivity in cleavage among amide bonds is apparent in the CID of the modified anion.
Figure 1.
Ion trap CID product ion spectra of a) modified product, [M+◆]−, b) [M-H]−, c) [M+2H]2+ derived from M = TLSDYNIQK
The results of TLSDYNIQK are illustrative for tryptic peptides with a C-terminal lysine residue. The results for ions derived from MQIFVK (Figure S-1), LIFAGK (Figure S-2), and TITLEVEPSDTIENVK (Figure S-3) are provided as Supplementary Information. In all three cases, the major fragmentation products from the [M+◆]− ions correspond to y◆- or b◆-type ions. The CID product ion spectra of the [M-H]− ions, on the other hand, provide fewer sequence-informative fragment ions. The results reported here are consistent with previous work involving condensed-phase covalent derivatization of the C-terminus of peptides with 4-aminonaphthalenesulphonic acid [26]. The resulting anions showed abundant modified y-type ions, the origin of which was attributed to a charge-remote mechanism [27, 28]. The sulfonate anionic site is highly stable and is expected to inhibit intramolecular proton transfer relative to a carboxylate site, thereby reducing the contributions from at least some charge-mediated reactions.
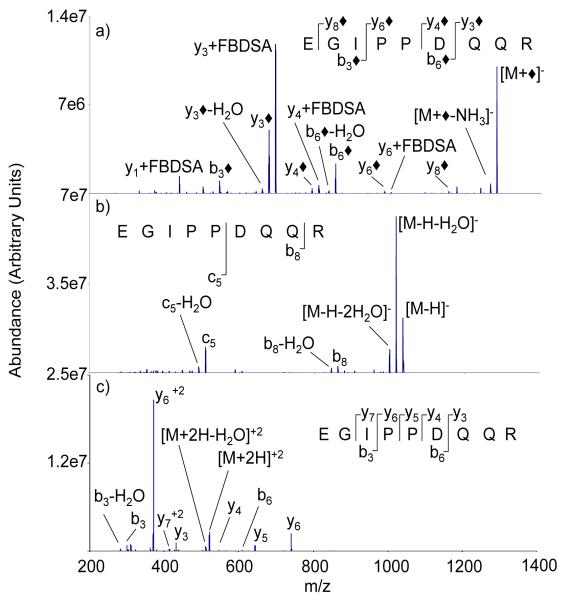
Fragmentation spectra of the modified tryptic peptides with a C-terminal arginine residue shared commonalities with those of the C-terminal lysine peptides but also showed some unique products. As an example, the CID product ion spectra for ions derived from EGIPPDQQR are compared in Figure 2. In the spectrum of the [M+◆]− ion, both b◆- and y◆-ions are observed (Figure 2(a)), which suggests that the precursor ion population is comprised of a mixture of species with the attachment at either end of the peptide. However, the N-terminus is the only site with an available primary amine for Schiff base formation. The y◆-ions are interpreted to arise from precursor ions with strong electrostatic attachment of a sulfonate group to the guanidinium side-chain of the C-terminal arginine residue, such interactions are known to be strong in the condensed phase [29, 30], or via formation of a negatively charged proton-bound dimer with the C-terminus. Ion/ion charge inversion studies have shown that the C-terminus can play an important role in adduct formation [13, 31]. The water loss resulting from activation of the [EGIPPDQQR+FBDSA-H]− complex presumably can arise from several sites on the peptide (e.g., the Glu and Asp residues). The appearance of abundant [y+FBDSA]− products in Figure 2(a) is consistent with this interpretation, as the site from which water is lost is not expected to be dependent upon the site of electrostatic attachment. On the other hand, the water lost upon Schiff base formation must come from the aldehyde group of FBDSA. If all peptide ions were to undergo Schiff base formation, no products with an FBDSA adduct should appear.
Figure 2.
Ion trap CID product ion spectra of a) [M+◆]−, b) [M-H]−, and c) [M+2H]2+ derived from M = ESTLHLVLR
Ion trap CID spectra of the [M-H]− and [M+2H]2+ ions of EGIPPDQQR are shown in Figures 2(b) and 2(c), respectively. The [M-H]− ion provides relatively few sequence-related product ions, the most prominent of which is the c5-ion, presumably arising from the previously noted cleavage associated with the aspartic acid side-chain [25]. The doubly protonated species generates a series of complementary b- and y-type ions with cleavage N-terminal to the proline at the fourth residue accounting for a large fraction of the observed fragmentation. Clearly, each precursor ion reflected in Figure 2 gives distinct fragmentation pattern. It is particularly noteworthy that the apparent non-covalent attachment of FBDSA can affect cleavage of the peptide backbone, rather than lose FBDSA, at least for the arginine-containing peptides. A similar set of comparisons for ESTLHLVLR is shown in Figure S-4.
Intramolecular Disulfide-Linked Peptides
Cations of two peptides with an intramolecular disulfide linkage, somatostatin-14 and oxytocin, were subjected to ion/ion reactions with anions of FBDSA to explore the effect of modification on fragmentation of the cyclic portions of the peptide ions. Polypeptide ions with intramolecular disulfide linkages, particularly protonated forms, often show little evidence for cleavage within the peptide loop defined by the disulfide bridge [15]. For this reason, the peptides are often reduced prior to tandem mass spectrometry [32]. However, gas-phase means for the selective cleavage of disulfide linkages have also been explored, such as the CID of deprotonated species [33, 34], CID of alkali or alkaline earth cationized ions [35], CID of coinage metal cationized ions [15, 36, 37], photodissociation [38], electron induced dissociation [39], electron capture dissociation [40], and electron transfer dissociation [15]. Some approaches rely on altering the ionization conditions while ECD/ETD require the reactant polypeptide to be multiply-charged. The use of [FBDSA-2H]2- allows for the modification of singly-protonated species.
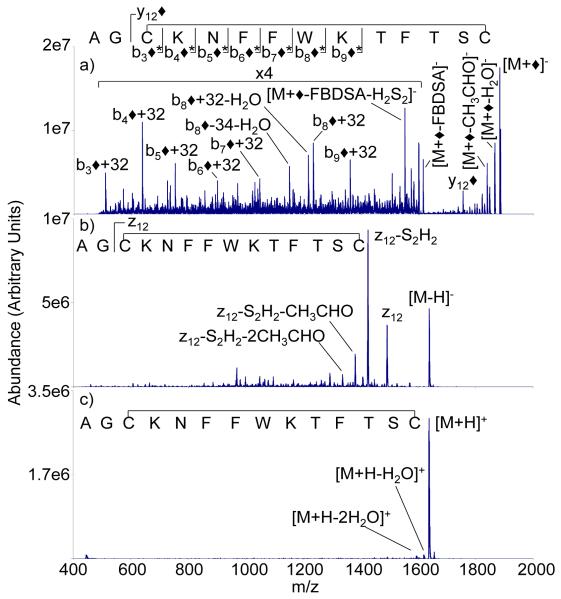
Figure 3 compares the ion trap CID product ion spectra of the modified form of somatostatin-14 (i.e., the [M+◆]− species) (Figure 3(a)) with those of the [M-H]− and [M+H]+ ions (Figures 3(b) and 3(c), respectively). Ion trap CID of the [M+H]+ species yields essentially no sequence information. As previously reported in the CID of deprotonated somatostatin [15], the z12-ion and an associated loss of H2S2 dominate the product ion spectrum (Figure 3(b)). The formation of the z12-ion is a two-step process that begins with cleavage of the disulfide linkage at the CH2-S bond of Cys3. Some internal fragments that contain neither the N-terminus nor the C-terminus are also noted, although much of the low level signal is not readily assigned [15]. CID of the modified somatostatin ions produces a major loss of FBDSA, which suggests that a significant fraction of the precursor ions are comprised of a non-covalently bound FBDSA. However, a series of bn◆+32 ions, where n=3-9, is noted. This series of ions is consistent with Schiff base formation N-terminal to the backbone cleavage (e.g., the b3◆+32 and b4◆+32 ions suggest Schiff base formation at the N-terminus while the b9+32 can be modified at the N-terminus, at Lys4, or at Lys9) as well as cleavage of the CH2-S bond of Cys14. Ion trap CID of disulfide linked peptide anions have shown cleavage at all three bonds along the disulfide linkage [30] (i.e., at the S-S bond or at either of the CH2-S bonds of R-CH2-S-S-CH2-R). Evidence for a few bn◆-ions that follow cleavage either at the S-S bond or at the CH2-S bond of Cys3 was noted but the abundances were very low relative to the bn◆+32 ions.
Figure 3.
Ion trap CID product ion spectra of A) [M+◆]−, B) [M-H]−, C) [M+H]+ ions derived from M = AGCKNFFWKTFTSC (somatostatin). The b ◆* n symbols in the somatostatin sequence at the top of the figure represent modified b-ions that originate from the loop defined by the disulfide linkage. These ions may contain different numbers of sulfur atoms depending upon which of the bonds of the disulfide linkage was cleaved.
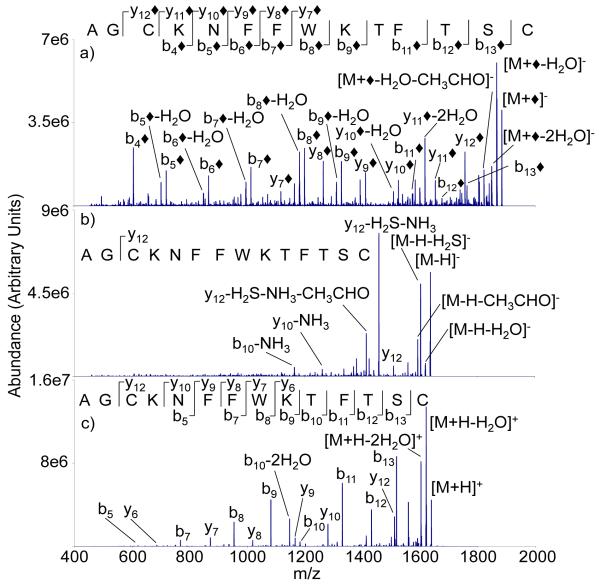
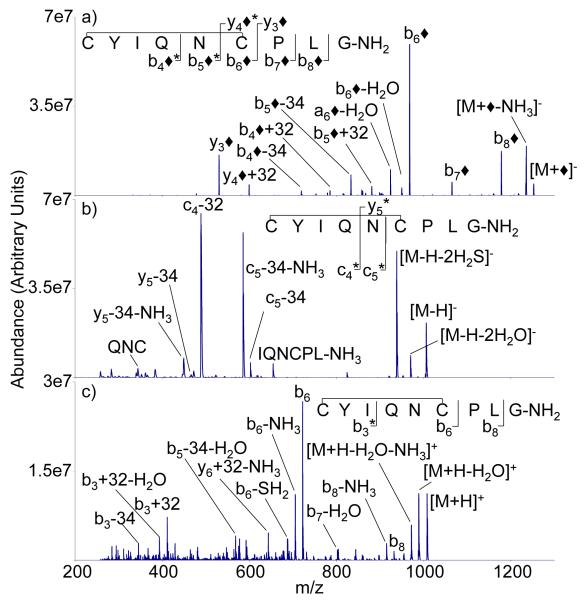
The presence of the disulfide linkage in somtatostatin has a profound effect on the ion trap CID of all three ion types, as reflected in the respective CID spectra shown in Figure 4. Modification of reduced somatostatin via reaction with FBDSA dianions leads to a spectrum rich in sequence-related ions (Figure 4(a)) that is comprised primarily of b◆- and y◆-type ions. As there is no need to break a disulfide linkage in order to observe cleavage products between the cysteine residues, no product ions with either missing or additional sulfur atoms are observed. As expected, the ion trap CID spectrum of the reduced [M-H]− ion shows no evidence for a z12-ion, as this ion is generated via a sequential fragmentation that begins with cleavage of the Cys3 CH2-S bond (see Figure 4(b)). Rather, the spectrum is dominated by losses of small neutral molecules, such as H2S, and one major product from an apparent backbone cleavage that also involved small molecules losses (i.e., the y12-H2S-NH3 ion). Ion trap CID of the protonated peptide (Figure 4(c)) provides a similar range of backbone cleavages to that observed from the modified anion, although the relative abundances are markedly different. There is a small degree of complementarity associated with the data of Figures 4(a) and 4(c), however, as the modified anion shows unique evidence for cleavage of the Cys3-Lys4 amide bond, while the protonated molecule shows unique evidence for cleavage of the Thr10-Phe11 amide bond.
Figure 4.
Ion trap CID product ion spectra of a) [M+◆]−, b) [M-H]−, c) [M+H]+ derived from M = AGCKNFFWKTFTSC (reduced somatostatin-14)
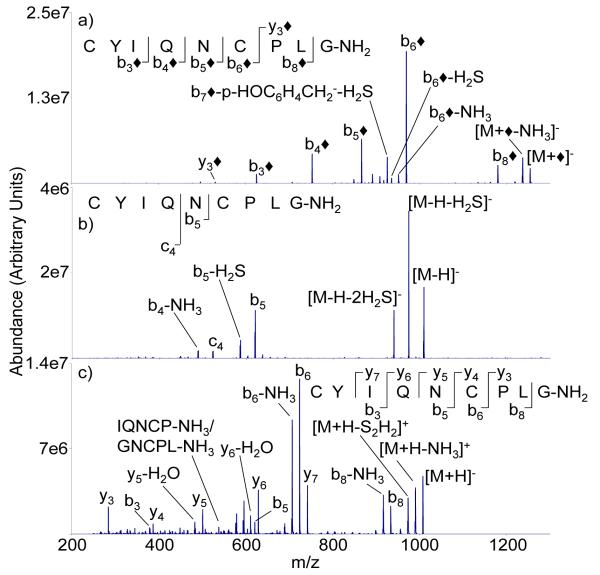
Oxytocin represents another peptide hormone with a cyclic portion defined by a disulfide linkage, in this case between Cys1 and Cys6. Figure 5 provides the ion trap CID product ion spectra from the modified anion (i.e., the [M+◆]− species, Figure 5(a)), the deprotonated molecule (Figure 5(b)), and the protonated molecule (Figure 5(c)). Similar to the case for somatostatin, CID of the [M+H]+ and [M-H]− ions of oxytocin produces limited sequence information. The unmodified peptide anion and cation produce products from cleavages between two and three of eight interresidue linkages, respectively. Activation of the [M-H]− ion produces a few prominent c-ions, which are commonly observed in CID of deprotonated peptides with an asparagine residue [25]. Dissociation of the protonated molecule shows a dominant N-terminal proline cleavage that falls outside of the loop defined by the disulfide linkage. A cleavage at the same amide bond is also highly abundant for the modified anion. Both the protonated molecule and the modified anion show evidence for cleavage of the disulfide bond in oxytocin by virtue of the appearance of b- or y-type ions from within the loop defined by the disulfide bond. Ions that contain both sulfur atoms or no sulfur atoms are clear indicators of cleavages of the disulfide linkage at CH2-S bonds. Unlike the somatostatin case, no clear preference for cleavage of either of the two CH2-S bonds is noted for the oxytocin ions. The [M+◆]− ions show evidence for cleavages of five of the eight amide linkages, which yields primary structure information that largely overlaps with that derived from the protonated molecule. Taken together, the [M+H]+ and [M+◆]− ions show cleavages at six of eight interresidue linkages. For comparison, the ion trap CID product ion spectra of the reduced forms of the three ion types are shown in Figure 6. Fragmentation of the [M-H]− ion (Figure 6b) proceeds largely through loss of one or two molecules of H2S. The [M+H]+ (Figure 6a) and [M+◆]− (Figure 6c) ions of the reduced form of oxytocin show evidence for cleavages at six and five interresidue linkages, respectively. However, the spectrum of the [M+H]+ ion tends to show more extensive losses of small molecules, such as water and ammonia, than does the [M+◆]− ion.
Figure 5.
Ion trap CID product ion spectra of a) [M+◆]−, b) [M-H]−, and c) [M+H]+ derived from M = CYIQNCPLG-NH2 (oxytocin)
Figure 6.
Ion trap CID product ion spectra of a) [M+◆]−, b) [M-H]−, and c) [M+H]+ derived from M = CYIQNCPLG-NH2 (reduced oxytocin)
Doubly deprotonated 4-formyl-1,3-benzenedisulfonic acid reacts with protonated peptides largely via complex formation. Schiff base formation involving a primary amine of the peptide and the aldehyde group of the reagent anion can occur in conjunction with dehydration of the complex. However, water loss that is unrelated to the reagent can also occur. An electrostatic (or dipole/dipole) interaction between a sulfonate of the reagent anion and a protonated site on the peptide can be sufficiently strong that covalent bond cleavages can compete with simple detachment of the reagent. This work provides evidence for Schiff base formation associated with many of the ion/ion reactions studied, while, in some cases, the formation of complexes with a strong electrostatic interaction without Schiff base formation is indicated as well. This work suggests that Schiff base formation can generally be expected for tryptic peptides and is also demonstrated for the peptide hormones somatostatin-14 and oxytocin, which both contain an intramolecular disulfide linkage. In all cases, the modified anions (i.e., those that undergo Schiff base formation as well as those that undergo non-covalent anion attachment) show fragmentation behavior that is distinct from either protonated or deprotonated forms of the peptide. Significantly more structural information, in particular, was derived from the modified anions than from the deprotonated species. The highly stable sulfonate group is expected to inhibit intramolecular proton transfer, which likely accounts for the markedly different fragmentation behavior of the modified anions relative to the deprotonated species. The structural information available from the dissociation of protonated forms of the various peptides overlapped with that from the modified anions but some complementarity was noted in most cases. Modification of peptide ions in the gas-phase via ion/ion reactions is an attractive means for increasing structural information from dissociation because it does not require chemical modification in solution. Schiff base formation and strong electrostatic binding are two possible means for making modifications. Future work will be devoted to understanding when these phenomena compete, which may point to novel reagents that react exclusively by one means or the other.
Supplementary Material
Acknowledgments
This research was supported by AB Sciex and the National Institutes of Health under Grant GM 45372.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].McLuckey SA, Mentinova M. Ion/Neutral, Ion/Electron, Ion/Photon, and Ion/Ion Interactions in Tandem Mass Spectrometry: Do we need them all? Are they enough? J. Am. Soc. Mass Spectrom. 2011;22:3–12. doi: 10.1007/s13361-010-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stewart NA, Pham VT, Choma CT, Kaplan H. Improved Peptide Detection with Matriz-assisted Laser Desorption Ionization Mass Spectrometry by Trimethylation of Amino Groups. Rapid Commun. Mass Spectrom. 2002;16:1448–1453. doi: 10.1002/rcm.726. [DOI] [PubMed] [Google Scholar]
- [3].Ross PL, Huang YLN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed Protein Quantitation in Saccharomyces Cerevisiae Using Amine-reactive Isobaric Tagging Reagents. Molec. Cellular Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- [4].Mendoza VL, Vachet RW. Probing Protein Structure by Amino Acid-Specific Covalent Labeling and Mass Spectrometry. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zubarev RA. Reactions of Polypeptide Ions with Electrons in the Gas Phase. Mass Spectrom. Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- [6].Brodbelt JS. Analytical Applications of Ion-Molecule Reactions. Mass Spectrom. Rev. 1997;16:91–110. [Google Scholar]
- [7].Huang T-Y, McLuckey SA. Gas-Phase Chemistry of Multiply-Charged Bio-ions in Analytical Mass Spectrometry. Ann. Rev. Anal. Chem. 2010;3:365–285. doi: 10.1146/annurev.anchem.111808.073725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].O’Hair RAJ, Reid GE. Derivatization of Protonated Peptides via Gas Phase Ion-Molecule Reactions with Acetone. J. Am. Soc. Mass Spectrom. 2000;11:244–256. doi: 10.1016/S1044-0305(99)00142-7. [DOI] [PubMed] [Google Scholar]
- [9].Han H, McLuckey SA. Selective Covalent Bond Formation in Polypeptide Ions via Gas-Phase Ion/Ion Reaction Chemistry. J. Am. Chem. Soc. 2009;131:12884–12884. doi: 10.1021/ja904812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mentinova M, McLuckey SA. Covalent Modification of Gaseous Peptide Ions with N-Hydroxysuccinimide Ester Reagent Ions. J. Am. Chem. Soc. 2010;132:18248–18257. doi: 10.1021/ja107286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mentinova M, McLuckey SA. Intra- and Inter-Molecular Cross-Linking of Peptide Ions in the Gas-Phase: Reagents and Conditions. J. Am. Soc. Mass Spectrom. 2011;22:912–921. doi: 10.1007/s13361-011-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hassell KM, Stutzman JR, McLuckey SA. Gas-Phase Bioconjugation of Peptides via Ion/Ion Reactions Charge Inversion: Schiff Base Formation on the Conversion of Cations to Anions. Anal. Chem. 2010;82:1594–1597. doi: 10.1021/ac902732v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Emory JF, McLuckey SA. The Role of Amino Acid Composition on the Charge Inversion of Deprotonated Peptides via Gas-Phase Ion/Ion Reactions. J. Am. Soc. Mass Spectrom. 2009;20:180–187. doi: 10.1016/j.jasms.2008.08.015. [DOI] [PubMed] [Google Scholar]
- [14].Aebersold R, Mann M. Mass Spectrometry-based Proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- [15].Mentinova M, Han H, McLuckey SA. Dissociation of Disulfide-Intact Somatostatin Ions: The Roles of Ion Type and Dissociation Methods. Rapid Commun. Mass Spectrom. 2009;23:2647–2655. doi: 10.1002/rcm.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chrisman PA, Pitteri SJ, Hogan JM, McLuckey SA. SO2−* Electron Transfer Ion/Ion Reactions with Disulfide Linked Polypeptide Ions. J. Am. Soc. Mass Spectrom. 2005;16:1020–1030. doi: 10.1016/j.jasms.2005.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Han H, Xia Y, McLuckey SA. Ion Trap Collisional Activation of c and z Ions Formed via Gas-Phase Ion/Ion Electron Transfer Dissociation. J. Proteome. Res. 2007;6:3062–3069. doi: 10.1021/pr070177t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hogan JM, McLuckey SA. Charge State Dependent Collison-induced Dissociation of Native and Reduced Porcine Elastase. J. Mass Spectrom. 2003;38:245–256. doi: 10.1002/jms.458. [DOI] [PubMed] [Google Scholar]
- [19].Xia Y, Wu J, Londry FA, Hager JW, McLuckey SA. Mutual Storage Mode Ion/Ion Reactions in Hybrid Linear Ion Trap. J. Am. Soc. Mass Spectrom. 2005;16:71–81. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- [20].Liang X, Xia Y, McLuckey SA. Alternately Pulsed Nano-electrospray Ionization/Atmospheric Pressure Chemical Ionization for Ion/Ion Reactions in an Electrodynamic Ion Trap. Anal.Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liang X, Han H, Xia Y, McLuckey SA. A Pulsed Triple Ionization Source for Sequential Ion/Ion Reactions in an Electrodynamic Ion Trap. J. Am. Soc. Mass Spectrom. 2007;18:369–376. doi: 10.1016/j.jasms.2006.10.004. [DOI] [PubMed] [Google Scholar]
- [22].Londry FA, Hager JW. Mass Selective Axial Ion Ejection from a Linear Quadrupole Ion Trap. J. Am. Soc. Mass Spectrom. 2003;14:1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- [23].Harrison AG, Young AB. Fragmentation Reactions of Deprotonated Peptides Containing Proline. The Proline Effect. J. Mass Spectrom. 2005;40:1176–1183. doi: 10.1002/jms.891. [DOI] [PubMed] [Google Scholar]
- [24].Roepstorff P, Fohlman J. Proposal for a Common Nomenclature for Sequence Ions in Mass Spectra of Peptides. Biomed. Mass Spectrom. 1984;11:601–602. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- [25].Bowie JH, Brinkworth CS, Dua S. Collision-induced Fragmentations of the (M - H)- Parent Anions of Underivatized Peptides: an Aid to Structure Determination and Some Unusual Negative Ion Cleavages. Mass Spectrom. Rev. 2002;21:87–107. doi: 10.1002/mas.10022. [DOI] [PubMed] [Google Scholar]
- [26].Lindh I, Griffiths WJ, Bergman T, Sjovall J. Charge-Remote Fragmentation of Peptides Derivatized with 4-Aminonapthlenesulphonic Acid. Rapid Commun. Mass Spectrom. 1994;8:797–803. doi: 10.1002/rcm.1290081002. [DOI] [PubMed] [Google Scholar]
- [27].Gross ML. Charge-remote Fragmentations: Method, Mechanism and Applications. Int. J. Mass Spectrom. 1992;118/119:137–165. [Google Scholar]
- [28].Cheng C, Gross ML. Applications and Mechanism of Charge-Remote Fragmentation. Mass Spectrom. Rev. 2000;19:398–420. doi: 10.1002/1098-2787(2000)19:6<398::AID-MAS3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- [29].Schug K, Lindner W. Noncovalent Binding between Guanidinium and Anionic Groups: Focus on Biological- and Synthetic-Based Arginine/Guanidinium Interactions with Phosph(on)ate and Sulf(on)ate Residues. Chem. Rev. 2005;105:67–113. doi: 10.1021/cr040603j. [DOI] [PubMed] [Google Scholar]
- [30].Ojala WH, Sudbeck EA, Lu LK, Richardson TI, Lovrien RE, Gleason WB. Complexes of Lysine, Histidine, and Arginine with Sulfonated Azo Dyes: Model Systems for Understanding the Biomolecular Recognition of Glycosaminoglycans by Proteins. J. Am. Chem. Soc. 1996;118:2131–2142. [Google Scholar]
- [31].He M, Emory JF, McLuckey SA. Reagent Anions for Charge Inversion of Polypeptide/Protein Cations in the Gas Phase. Anal. Chem. 2005;77:3173–3182. doi: 10.1021/ac0482312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Stephenson JL, Jr., Cargile BJ, McLuckey SA. Ion Trap Collisional Activation of Disulfide Linkage Intact and Reduced Multiply Protonated Polypeptides. Rapid Commun. Mass Spectrom. 1999;13:2040–2048. doi: 10.1002/(SICI)1097-0231(19991030)13:20<2040::AID-RCM754>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- [33].Chrisman PA, McLuckey SA. Dissociations of Disulfide-Linked Gaseous Polypeptide/Protein Anions: Ion Chemistry with Implications for Protein Identification and Characterization Dissociations of Disulfide-Linked Gaseous Polypeptide/Protein Anions: Ion Chemistry with Implications for Protein Identification and Characterization. J. Proteome Res. 2002;1:549–557. doi: 10.1021/pr025561z. [DOI] [PubMed] [Google Scholar]
- [34].Bilusich D, Bowie JH. Fragmentations of (M-H)- Anions of Underivatised Peptides. Part 2: Characteristic Cleavages of Ser and Cys and of Disulfides and Other Post-translational Modifications, Together with Some Unusual Internal Processes. Mass Spectrom. Rev. 2009;28:20–34. doi: 10.1002/mas.20206. [DOI] [PubMed] [Google Scholar]
- [35].Kim HI, Beauchamp JL. Mapping Disulfide Bonds in Insulin with the Route 66 Method: Selective Cleavage of S-C bonds using Alkali and Alkaline Earth Metal Enolate Complexes. J. Am. Soc. Mass Spectrom. 2009;20:157–166. doi: 10.1016/j.jasms.2008.10.003. [DOI] [PubMed] [Google Scholar]
- [36].Gunawardena HP, O’Hair RAJ, McLuckey SA. Selective Disulfide Bond Cleavage in Gold (I) Cationized Polypeptide Ions formed via Gas Phase Ion/ion Cation Switching. J. Proteome Res. 2006;5:2087–2092. doi: 10.1021/pr0602794. [DOI] [PubMed] [Google Scholar]
- [37].Lioe H, Duan M, O’Hair RAJ. Can metal ions be used as gas-phase disulfide bond cleavage reagents? A survey of coinage metal complexes of model peptides containing an intramolecular disulfide bond. Rapid Commun. Mass Spectrom. 2007;21:2727–2733. doi: 10.1002/rcm.3122. [DOI] [PubMed] [Google Scholar]
- [38].Fung YME, Kjeldsen F, Silivra OA, Chan TWD, Zubarev RA. Facile Disulfide Bond Cleavage in Gaseous Peptide and Protein Cations by Ultraviolet Photodissiation at 157 nm. Angew. Chem. Int. Ed. 2005;44:6399–6403. doi: 10.1002/anie.200501533. [DOI] [PubMed] [Google Scholar]
- [39].Lioe H, O’Hair AJ. Comparison of collision-induced dissociation and electron-induced dissociation of singly protonated aromatic amino acids, cystine and related simple peptides using a hybrid linear ion trap-FT-ICR mass spectrometer. Anal. Bioanal. Chem. 2007;389:1429–1437. doi: 10.1007/s00216-007-1535-1. [DOI] [PubMed] [Google Scholar]
- [40].Zubarev RA, Kruger NA, Fridriksson MA, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. Electron Capture Dissociation of Gaseous Multiply-charged Proteins is Favored at Disulfide Bonds and Other Sites of High Hydrogen Atom Affinity. J. Am. Chem. Soc. 1999;121:2857–2862. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.