Abstract
The asymmetric unit of the title compound, {[Sm2(C8H2NO6)2(H2O)5]·H2O}n, contains two independent SmIII ions, two pyridine-2,4,6-tricarboxylate (ptc) ligands, five aqua ligands and one lattice water molecule. One SmIII ion is nine-coordinated by one N and five O atoms from the three ptc ligands and three aqua ligands in a distorted monocapped square antiprismatic geometry, and the other is eight-coordinated by one N and five O atoms from three ptc ligands and two aqua ligands in a 4,4′-bicapped trigonal antiprismatic geometry. The ptc ligands brigde the SmIII ions into a three-dimensional polymeric framework. Extensive O—H⋯O hydrogen bonding is observed in the crystal structure.
Related literature
For related compounds, see: Gao et al. (2006 ▶); Ghosh & Bharadwaj (2005 ▶); Li et al. (2008 ▶); Wang et al. (2007 ▶).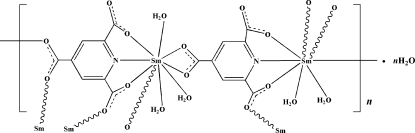
Experimental
Crystal data
[Sm2(C8H2NO6)2(H2O)5]·H2O
M r = 825.01
Monoclinic,

a = 18.426 (4) Å
b = 6.9082 (14) Å
c = 18.583 (4) Å
β = 111.98 (3)°
V = 2193.6 (8) Å3
Z = 4
Mo Kα radiation
μ = 5.40 mm−1
T = 293 K
0.43 × 0.28 × 0.21 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▶) T min = 0.177, T max = 0.321
20300 measured reflections
4963 independent reflections
4776 reflections with I > 2σ(I)
R int = 0.035
Refinement
R[F 2 > 2σ(F 2)] = 0.018
wR(F 2) = 0.041
S = 1.19
4963 reflections
344 parameters
H-atom parameters constrained
Δρmax = 0.69 e Å−3
Δρmin = −0.84 e Å−3
Data collection: RAPID-AUTO (Rigaku, 1998 ▶); cell refinement: RAPID-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2004 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812002462/cv5233sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812002462/cv5233Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O7—H7A⋯O14i | 0.82 | 1.87 | 2.686 (3) | 172 |
| O7—H7B⋯O2ii | 0.86 | 1.80 | 2.645 (3) | 165 |
| O8—H8A⋯O12iii | 0.89 | 1.83 | 2.727 (3) | 177 |
| O8—H8B⋯O9iv | 0.87 | 2.38 | 2.831 (3) | 113 |
| O9—H9A⋯O6i | 0.87 | 1.84 | 2.698 (3) | 171 |
| O9—H9B⋯O5i | 0.93 | 2.55 | 3.052 (3) | 114 |
| O16—H16A⋯O4i | 0.82 | 2.31 | 3.075 (3) | 157 |
| O16—H16B⋯O15v | 0.79 | 1.97 | 2.747 (3) | 171 |
| O17—H17A⋯O18vi | 0.84 | 1.86 | 2.697 (3) | 176 |
| O17—H17B⋯O5vii | 0.80 | 1.96 | 2.731 (3) | 162 |
| O18—H18A⋯O14viii | 0.85 | 2.06 | 2.905 (3) | 174 |
| O18—H18B⋯O13 | 0.82 | 1.93 | 2.736 (3) | 168 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  ; (vii)
; (vii)  ; (viii)
; (viii)  .
.
Acknowledgments
This project was supported by the Scientific Research Fund of Ningbo University (grant Nos. XKL11058 and XYL11005). Sincere thanks are also extended to the K. C. Wong Magna Fund of Ningbo University.
supplementary crystallographic information
Comment
In recent years, research of lanthanide pyridine-2,4,6-tricarboxylate coordination polymers has been of great interest in the fields of molecule adsorption, host-guest interaction and luminescence materials (Gao et al., 2006; Li et al., 2008; Wang et al., 2007;) etc. The compound {[Pr(H2O)2(ptc)].2H2O} reported by Ghosh et al. (2005) represents an example of three-dimensional MOF, which could potentially be utilized as an adsorption material. In this contribution, we report the structure of the title compound.
The asymmetric unit of title compound contains two SmIII ions (Sm1, Sm2), two ptc ligands (denoted as ptc1 and ptc2 ligands, which contain N1 and N2 atoms, respectively) (H3ptc = pyridine-2,4,6-tricarboxylate), five aqua ligands and one lattice water molecule as illustrated in Fig. 1. The Sm1 and Sm2 are respectively 9- and 8-coordinated with the ligating atoms occupying the corners of distorted monocapped square anti-prism and 4,4'-bicapped trigonal anti-prism, respectively. The bond distances about the Sm1 and Sm2 atoms fall in the regions 2.406-2.587 Å and 2.359-2.529 Å, the corresponding bond angles are in the regions 50.9-147.8° and 63.3-159.7°, respectively.
Each ptc1 ligand links four SmIII centers with the 4-carboxylate bonded to two metal ions in syn-syn bridging fashion and the 2-carboxylate coordinated to two metal ions in anti-syn bridging fashion, while each ptc2 ligand connects three SmIII centers with the 4-carboxylate chelating one metal ion and the 2-carboxylate bridging two metal ions in anti-anti bridging fashions.
Both ptc1 and ptc2 ligands coordinate Sm1 atoms to form a linear [Sm(ptc)2] metallo-ligand, which, in turn, bridges the Sm2 atoms to generate one-dimensional ribbon-like chains with rectangular and 8-membered rhombic rings alternating (Fig. 2). Within the rectangular ring, the two adjacent ptc1 and ptc2 ligands orientate approximately parallelly to each other with a dihedral angle of 7.4° and the mean interplanar distance of 3.33 Å suggests significant intrachain π···π stacking interactions. The resulting one-dimensional chains donate carboxylate O atoms (O1 and O10) to coordinate with Sm atoms from two neighboring chains to construct a three-dimensional framework (Fig. 3).
Experimental
All chemicals were obtained from commerical sources and were used as obtained. A mixture of SmCl3.nH2O (0.60 mmol), pyridine-2,4,6-tricarboxylic acid (0.0537 g, 0.25 mmol), Malonic acid (0.0261 g, 0.25 mmol), NaOH (1 ml, 1 M) and H2O (20 ml) was sealed into a 23 ml Teflon-lined stainless autoclave, which was heated up to 180°C, at which temperature the reactor was held for 3 days, and then cooled to room temperature. A mixture of brownish deposit and a small amount of colourless block-liked crystals were obtained.
Refinement
H atoms bonded to C atoms were palced in geometrically calculated position, and were refined using a riding model, with Uiso(H) = 1.2 Ueq(C). H atoms attached to O atoms were found in a difference Fourier synthesis and were refined using a riding model, with the O–H distances fixed as initially found and with Uiso(H) values set at 1.2 Ueq(O).
Figures
Fig. 1.
A portion of the crystal structure showing the atomic numbering and 45% probability dispalcement ellipsoids [symmetry codes: (i) 1/2-x, -1/2+y, 1/2-z; (ii) x, y, 1+z; (iii) 1-x, 1-y, 1-z; (iv) 1/2-x, -1/2+y, 3/2-z].
Fig. 2.
The one-dimensional chain with the rectangular and 8-membered rhombic rings.
Fig. 3.
The three-dimensional metal-organic framework in the title compound.
Crystal data
| [Sm2(C8H2NO6)2(H2O)5]·H2O | F(000) = 1576 |
| Mr = 825.01 | Dx = 2.498 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 19369 reflections |
| a = 18.426 (4) Å | θ = 3.2–27.5° |
| b = 6.9082 (14) Å | µ = 5.40 mm−1 |
| c = 18.583 (4) Å | T = 293 K |
| β = 111.98 (3)° | Block, colorless |
| V = 2193.6 (8) Å3 | 0.43 × 0.28 × 0.21 mm |
| Z = 4 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 4963 independent reflections |
| Radiation source: fine-focus sealed tube | 4776 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.035 |
| Detector resolution: 0 pixels mm-1 | θmax = 27.5°, θmin = 3.2° |
| ω scans | h = −23→23 |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | k = −8→8 |
| Tmin = 0.177, Tmax = 0.321 | l = −24→22 |
| 20300 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.018 | H-atom parameters constrained |
| wR(F2) = 0.041 | w = 1/[σ2(Fo2) + (0.0066P)2 + 3.5019P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.19 | (Δ/σ)max = 0.002 |
| 4963 reflections | Δρmax = 0.69 e Å−3 |
| 344 parameters | Δρmin = −0.84 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.00244 (6) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Sm1 | 0.373309 (7) | 0.222784 (18) | 0.378119 (7) | 0.01049 (4) | |
| Sm2 | 0.374965 (8) | 0.250031 (17) | 0.896720 (7) | 0.01111 (5) | |
| N1 | 0.42996 (13) | 0.3322 (3) | 0.27802 (12) | 0.0125 (4) | |
| C1 | 0.38355 (15) | 0.4105 (3) | 0.21147 (15) | 0.0127 (5) | |
| C2 | 0.40694 (15) | 0.4323 (3) | 0.14877 (15) | 0.0139 (5) | |
| H2A | 0.3735 | 0.4861 | 0.1023 | 0.017* | |
| C3 | 0.48176 (15) | 0.3710 (3) | 0.15774 (15) | 0.0125 (5) | |
| C4 | 0.53246 (15) | 0.3029 (4) | 0.22948 (15) | 0.0136 (5) | |
| H4A | 0.5840 | 0.2707 | 0.2377 | 0.016* | |
| C5 | 0.50357 (15) | 0.2846 (3) | 0.28859 (15) | 0.0119 (5) | |
| C6 | 0.30319 (15) | 0.4640 (4) | 0.20964 (15) | 0.0145 (5) | |
| O1 | 0.26044 (11) | 0.5719 (3) | 0.15807 (11) | 0.0185 (4) | |
| O2 | 0.28618 (11) | 0.3912 (3) | 0.26427 (11) | 0.0206 (4) | |
| C7 | 0.50797 (15) | 0.3719 (3) | 0.08959 (14) | 0.0124 (5) | |
| O3 | 0.45938 (12) | 0.3160 (3) | 0.02671 (11) | 0.0219 (4) | |
| O4 | 0.57737 (11) | 0.4236 (3) | 0.10278 (11) | 0.0172 (4) | |
| C8 | 0.55086 (16) | 0.2082 (4) | 0.36836 (16) | 0.0146 (5) | |
| O5 | 0.62134 (12) | 0.1852 (3) | 0.38807 (12) | 0.0278 (5) | |
| O6 | 0.51110 (11) | 0.1716 (3) | 0.41069 (11) | 0.0183 (4) | |
| O7 | 0.36682 (12) | −0.0503 (3) | 0.29113 (11) | 0.0220 (4) | |
| H7B | 0.3178 | −0.0743 | 0.2654 | 0.026* | |
| H7A | 0.3869 | −0.0502 | 0.2585 | 0.026* | |
| O8 | 0.40849 (13) | 0.5640 (3) | 0.39790 (13) | 0.0267 (5) | |
| H8A | 0.4563 | 0.6135 | 0.4218 | 0.032* | |
| H8B | 0.3682 | 0.5986 | 0.4083 | 0.032* | |
| O9 | 0.37653 (13) | −0.0753 (3) | 0.45208 (12) | 0.0232 (4) | |
| H9A | 0.4159 | −0.0992 | 0.4946 | 0.028* | |
| H9B | 0.3363 | −0.1166 | 0.4672 | 0.028* | |
| N2 | 0.38147 (13) | 0.2822 (3) | 0.76366 (13) | 0.0126 (4) | |
| C9 | 0.31935 (15) | 0.3494 (3) | 0.70511 (15) | 0.0136 (5) | |
| C10 | 0.31717 (16) | 0.3588 (4) | 0.62931 (15) | 0.0164 (5) | |
| H10A | 0.2726 | 0.4017 | 0.5888 | 0.020* | |
| C11 | 0.38335 (16) | 0.3023 (4) | 0.61614 (15) | 0.0144 (5) | |
| C12 | 0.44856 (17) | 0.2360 (3) | 0.67751 (16) | 0.0142 (5) | |
| H12A | 0.4938 | 0.2004 | 0.6699 | 0.017* | |
| C13 | 0.44461 (16) | 0.2242 (3) | 0.75073 (16) | 0.0135 (5) | |
| C14 | 0.25231 (16) | 0.4146 (4) | 0.72805 (15) | 0.0148 (5) | |
| O10 | 0.19320 (12) | 0.4815 (3) | 0.67583 (11) | 0.0210 (4) | |
| O11 | 0.26175 (12) | 0.3950 (3) | 0.79795 (11) | 0.0232 (4) | |
| C15 | 0.38312 (17) | 0.3038 (4) | 0.53467 (15) | 0.0157 (5) | |
| O12 | 0.44658 (13) | 0.2832 (3) | 0.52504 (12) | 0.0221 (4) | |
| O13 | 0.31898 (12) | 0.3173 (3) | 0.47848 (11) | 0.0212 (4) | |
| C16 | 0.50960 (15) | 0.1461 (3) | 0.82233 (15) | 0.0134 (5) | |
| O14 | 0.57112 (12) | 0.0880 (3) | 0.81788 (11) | 0.0206 (4) | |
| O15 | 0.49458 (12) | 0.1467 (3) | 0.88390 (11) | 0.0198 (4) | |
| O16 | 0.39568 (13) | −0.0498 (3) | 0.97353 (11) | 0.0244 (4) | |
| H16B | 0.4251 | −0.0676 | 1.0162 | 0.029* | |
| H16A | 0.3898 | −0.1564 | 0.9530 | 0.029* | |
| O17 | 0.28096 (12) | 0.3100 (3) | 0.95460 (13) | 0.0283 (5) | |
| H17B | 0.2358 | 0.3132 | 0.9261 | 0.034* | |
| H17A | 0.2806 | 0.2465 | 0.9929 | 0.034* | |
| O18 | 0.21204 (15) | 0.6072 (3) | 0.41898 (14) | 0.0374 (6) | |
| H18B | 0.2389 | 0.5106 | 0.4367 | 0.045* | |
| H18A | 0.1688 | 0.5550 | 0.3908 | 0.045* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Sm1 | 0.00985 (8) | 0.01369 (7) | 0.00821 (7) | 0.00008 (4) | 0.00368 (6) | 0.00015 (4) |
| Sm2 | 0.00839 (8) | 0.01720 (7) | 0.00731 (7) | −0.00064 (4) | 0.00243 (6) | −0.00005 (4) |
| N1 | 0.0109 (11) | 0.0167 (10) | 0.0101 (10) | 0.0002 (8) | 0.0040 (9) | 0.0003 (8) |
| C1 | 0.0095 (12) | 0.0143 (11) | 0.0133 (12) | 0.0001 (9) | 0.0032 (10) | 0.0017 (9) |
| C2 | 0.0132 (13) | 0.0169 (11) | 0.0101 (12) | −0.0002 (9) | 0.0026 (10) | 0.0028 (9) |
| C3 | 0.0128 (13) | 0.0136 (11) | 0.0118 (12) | −0.0032 (9) | 0.0055 (10) | −0.0006 (9) |
| C4 | 0.0108 (13) | 0.0162 (11) | 0.0144 (13) | −0.0015 (9) | 0.0055 (11) | −0.0011 (10) |
| C5 | 0.0115 (13) | 0.0128 (10) | 0.0111 (12) | −0.0005 (9) | 0.0041 (11) | 0.0002 (9) |
| C6 | 0.0113 (13) | 0.0184 (12) | 0.0133 (12) | 0.0010 (9) | 0.0042 (10) | −0.0015 (10) |
| O1 | 0.0137 (10) | 0.0243 (9) | 0.0164 (10) | 0.0060 (7) | 0.0045 (8) | 0.0061 (8) |
| O2 | 0.0128 (10) | 0.0339 (10) | 0.0171 (10) | 0.0053 (8) | 0.0079 (8) | 0.0105 (8) |
| C7 | 0.0137 (13) | 0.0127 (10) | 0.0105 (12) | 0.0009 (9) | 0.0044 (10) | 0.0024 (9) |
| O3 | 0.0208 (11) | 0.0313 (10) | 0.0112 (10) | −0.0046 (8) | 0.0031 (9) | −0.0038 (8) |
| O4 | 0.0150 (10) | 0.0224 (9) | 0.0158 (9) | −0.0015 (7) | 0.0077 (8) | 0.0010 (7) |
| C8 | 0.0127 (13) | 0.0158 (11) | 0.0140 (13) | 0.0001 (9) | 0.0036 (11) | −0.0002 (10) |
| O5 | 0.0108 (11) | 0.0466 (13) | 0.0235 (12) | 0.0054 (9) | 0.0035 (9) | 0.0103 (10) |
| O6 | 0.0137 (10) | 0.0295 (10) | 0.0121 (9) | 0.0051 (8) | 0.0053 (8) | 0.0073 (8) |
| O7 | 0.0143 (10) | 0.0353 (11) | 0.0188 (10) | −0.0037 (8) | 0.0090 (9) | −0.0115 (9) |
| O8 | 0.0268 (12) | 0.0190 (9) | 0.0329 (12) | −0.0045 (8) | 0.0098 (10) | −0.0050 (8) |
| O9 | 0.0273 (12) | 0.0248 (10) | 0.0169 (10) | 0.0035 (8) | 0.0074 (9) | 0.0062 (8) |
| N2 | 0.0114 (11) | 0.0163 (9) | 0.0095 (11) | 0.0000 (8) | 0.0033 (9) | 0.0004 (8) |
| C9 | 0.0136 (13) | 0.0157 (11) | 0.0113 (12) | 0.0018 (9) | 0.0044 (10) | 0.0009 (9) |
| C10 | 0.0168 (14) | 0.0197 (12) | 0.0111 (13) | 0.0039 (10) | 0.0034 (11) | 0.0030 (10) |
| C11 | 0.0173 (14) | 0.0152 (11) | 0.0114 (13) | 0.0001 (9) | 0.0062 (11) | −0.0017 (10) |
| C12 | 0.0137 (14) | 0.0169 (12) | 0.0128 (13) | 0.0007 (9) | 0.0058 (12) | −0.0007 (9) |
| C13 | 0.0131 (14) | 0.0140 (11) | 0.0133 (13) | −0.0002 (9) | 0.0049 (11) | −0.0013 (9) |
| C14 | 0.0139 (13) | 0.0171 (11) | 0.0126 (12) | 0.0028 (9) | 0.0039 (10) | 0.0006 (10) |
| O10 | 0.0170 (11) | 0.0284 (10) | 0.0149 (10) | 0.0110 (8) | 0.0029 (8) | 0.0043 (8) |
| O11 | 0.0167 (11) | 0.0417 (12) | 0.0124 (10) | 0.0096 (9) | 0.0067 (9) | 0.0040 (8) |
| C15 | 0.0216 (14) | 0.0164 (11) | 0.0105 (13) | 0.0019 (10) | 0.0076 (11) | −0.0003 (10) |
| O12 | 0.0175 (11) | 0.0370 (11) | 0.0136 (10) | 0.0003 (8) | 0.0077 (9) | −0.0027 (8) |
| O13 | 0.0180 (11) | 0.0347 (11) | 0.0099 (9) | 0.0068 (8) | 0.0042 (8) | −0.0007 (8) |
| C16 | 0.0129 (13) | 0.0135 (11) | 0.0132 (12) | −0.0016 (9) | 0.0042 (10) | 0.0001 (9) |
| O14 | 0.0137 (10) | 0.0303 (10) | 0.0194 (10) | 0.0067 (8) | 0.0079 (9) | 0.0014 (8) |
| O15 | 0.0152 (10) | 0.0336 (10) | 0.0113 (9) | 0.0068 (8) | 0.0057 (8) | 0.0043 (8) |
| O16 | 0.0297 (12) | 0.0225 (10) | 0.0140 (10) | 0.0053 (8) | 0.0002 (9) | 0.0034 (8) |
| O17 | 0.0111 (11) | 0.0532 (13) | 0.0215 (11) | 0.0062 (9) | 0.0071 (9) | 0.0058 (10) |
| O18 | 0.0347 (15) | 0.0383 (13) | 0.0323 (13) | 0.0133 (10) | 0.0046 (11) | −0.0015 (10) |
Geometric parameters (Å, º)
| Sm1—O6 | 2.406 (2) | O3—Sm2vi | 2.377 (2) |
| Sm1—O2 | 2.422 (2) | O4—Sm2iv | 2.4185 (18) |
| Sm1—O8 | 2.436 (2) | C8—O5 | 1.221 (3) |
| Sm1—O7 | 2.4582 (19) | C8—O6 | 1.285 (3) |
| Sm1—O9 | 2.4642 (19) | O7—H7B | 0.8654 |
| Sm1—O13 | 2.5110 (18) | O7—H7A | 0.8193 |
| Sm1—O1i | 2.5243 (19) | O8—H8A | 0.8930 |
| Sm1—N1 | 2.564 (2) | O8—H8B | 0.8667 |
| Sm1—O12 | 2.587 (2) | O9—H9A | 0.8662 |
| Sm1—C15 | 2.901 (3) | O9—H9B | 0.9304 |
| Sm2—O10ii | 2.3600 (19) | N2—C9 | 1.333 (3) |
| Sm2—O3iii | 2.377 (2) | N2—C13 | 1.335 (3) |
| Sm2—O17 | 2.3925 (19) | C9—C10 | 1.396 (3) |
| Sm2—O15 | 2.4120 (19) | C9—C14 | 1.518 (3) |
| Sm2—O4iv | 2.4185 (19) | C10—C11 | 1.387 (3) |
| Sm2—O11 | 2.420 (2) | C10—H10A | 0.9300 |
| Sm2—O16 | 2.463 (2) | C11—C12 | 1.389 (4) |
| Sm2—N2 | 2.530 (2) | C11—C15 | 1.512 (3) |
| N1—C1 | 1.327 (3) | C12—C13 | 1.392 (4) |
| N1—C5 | 1.337 (3) | C12—H12A | 0.9300 |
| C1—C2 | 1.394 (3) | C13—C16 | 1.518 (4) |
| C1—C6 | 1.514 (3) | C14—O10 | 1.245 (3) |
| C2—C3 | 1.391 (4) | C14—O11 | 1.252 (3) |
| C2—H2A | 0.9300 | O10—Sm2vii | 2.3600 (19) |
| C3—C4 | 1.393 (4) | C15—O13 | 1.254 (3) |
| C3—C7 | 1.515 (3) | C15—O12 | 1.256 (3) |
| C4—C5 | 1.394 (3) | C16—O14 | 1.234 (3) |
| C4—H4A | 0.9300 | C16—O15 | 1.274 (3) |
| C5—C8 | 1.506 (4) | O16—H16B | 0.7852 |
| C6—O1 | 1.237 (3) | O16—H16A | 0.8179 |
| C6—O2 | 1.273 (3) | O17—H17B | 0.8023 |
| O1—Sm1v | 2.5243 (19) | O17—H17A | 0.8383 |
| C7—O3 | 1.238 (3) | O18—H18B | 0.8223 |
| C7—O4 | 1.260 (3) | O18—H18A | 0.8524 |
| O6—Sm1—O2 | 125.63 (6) | C2—C1—C6 | 124.1 (2) |
| O6—Sm1—O8 | 84.72 (7) | C3—C2—C1 | 118.0 (2) |
| O2—Sm1—O8 | 73.68 (8) | C3—C2—H2A | 121.0 |
| O6—Sm1—O7 | 80.78 (7) | C1—C2—H2A | 121.0 |
| O2—Sm1—O7 | 86.62 (7) | C2—C3—C4 | 119.8 (2) |
| O8—Sm1—O7 | 141.88 (7) | C2—C3—C7 | 120.7 (2) |
| O6—Sm1—O9 | 86.22 (7) | C4—C3—C7 | 119.5 (2) |
| O2—Sm1—O9 | 139.32 (7) | C3—C4—C5 | 117.9 (2) |
| O8—Sm1—O9 | 140.81 (7) | C3—C4—H4A | 121.1 |
| O7—Sm1—O9 | 73.17 (7) | C5—C4—H4A | 121.1 |
| O6—Sm1—O13 | 121.87 (7) | N1—C5—C4 | 121.8 (2) |
| O2—Sm1—O13 | 101.87 (6) | N1—C5—C8 | 114.3 (2) |
| O8—Sm1—O13 | 78.10 (7) | C4—C5—C8 | 123.8 (2) |
| O7—Sm1—O13 | 138.87 (7) | O1—C6—O2 | 125.5 (2) |
| O9—Sm1—O13 | 74.70 (7) | O1—C6—C1 | 119.9 (2) |
| O6—Sm1—O1i | 147.10 (6) | O2—C6—C1 | 114.6 (2) |
| O2—Sm1—O1i | 72.76 (7) | C6—O1—Sm1v | 136.93 (16) |
| O8—Sm1—O1i | 128.16 (7) | C6—O2—Sm1 | 127.30 (17) |
| O7—Sm1—O1i | 72.87 (6) | O3—C7—O4 | 126.5 (2) |
| O9—Sm1—O1i | 67.72 (7) | O3—C7—C3 | 116.3 (2) |
| O13—Sm1—O1i | 71.60 (7) | O4—C7—C3 | 117.1 (2) |
| O6—Sm1—N1 | 63.12 (7) | C7—O3—Sm2vi | 170.38 (18) |
| O2—Sm1—N1 | 62.70 (7) | C7—O4—Sm2iv | 127.27 (16) |
| O8—Sm1—N1 | 70.53 (7) | O5—C8—O6 | 125.2 (3) |
| O7—Sm1—N1 | 71.46 (6) | O5—C8—C5 | 120.0 (2) |
| O9—Sm1—N1 | 136.02 (6) | O6—C8—C5 | 114.8 (2) |
| O13—Sm1—N1 | 147.78 (7) | C8—O6—Sm1 | 127.38 (17) |
| O1i—Sm1—N1 | 123.39 (7) | Sm1—O7—H7B | 107.1 |
| O6—Sm1—O12 | 70.93 (7) | Sm1—O7—H7A | 124.1 |
| O2—Sm1—O12 | 139.30 (7) | H7B—O7—H7A | 104.9 |
| O8—Sm1—O12 | 71.36 (7) | Sm1—O8—H8A | 127.1 |
| O7—Sm1—O12 | 134.07 (7) | Sm1—O8—H8B | 95.7 |
| O9—Sm1—O12 | 69.64 (7) | H8A—O8—H8B | 123.4 |
| O13—Sm1—O12 | 50.94 (7) | Sm1—O9—H9A | 120.4 |
| O1i—Sm1—O12 | 114.82 (7) | Sm1—O9—H9B | 124.7 |
| N1—Sm1—O12 | 121.65 (7) | H9A—O9—H9B | 99.0 |
| O6—Sm1—C15 | 96.45 (8) | C9—N2—C13 | 119.8 (2) |
| O2—Sm1—C15 | 123.75 (7) | C9—N2—Sm2 | 119.21 (16) |
| O8—Sm1—C15 | 75.45 (7) | C13—N2—Sm2 | 120.87 (18) |
| O7—Sm1—C15 | 140.98 (7) | N2—C9—C10 | 122.1 (2) |
| O9—Sm1—C15 | 67.82 (7) | N2—C9—C14 | 114.5 (2) |
| O13—Sm1—C15 | 25.51 (7) | C10—C9—C14 | 123.4 (2) |
| O1i—Sm1—C15 | 92.01 (7) | C11—C10—C9 | 118.0 (2) |
| N1—Sm1—C15 | 141.37 (7) | C11—C10—H10A | 121.0 |
| O12—Sm1—C15 | 25.64 (7) | C9—C10—H10A | 121.0 |
| O10ii—Sm2—O3iii | 137.50 (7) | C10—C11—C12 | 119.7 (2) |
| O10ii—Sm2—O17 | 94.18 (8) | C10—C11—C15 | 120.1 (2) |
| O3iii—Sm2—O17 | 79.54 (8) | C12—C11—C15 | 120.1 (2) |
| O10ii—Sm2—O15 | 91.23 (7) | C11—C12—C13 | 118.5 (2) |
| O3iii—Sm2—O15 | 83.25 (7) | C11—C12—H12A | 120.8 |
| O17—Sm2—O15 | 159.69 (7) | C13—C12—H12A | 120.8 |
| O10ii—Sm2—O4iv | 148.20 (7) | N2—C13—C12 | 121.8 (3) |
| O3iii—Sm2—O4iv | 73.67 (7) | N2—C13—C16 | 113.8 (2) |
| O17—Sm2—O4iv | 99.20 (7) | C12—C13—C16 | 124.4 (2) |
| O15—Sm2—O4iv | 86.13 (7) | O10—C14—O11 | 126.2 (2) |
| O10ii—Sm2—O11 | 76.61 (8) | O10—C14—C9 | 117.2 (2) |
| O3iii—Sm2—O11 | 137.89 (7) | O11—C14—C9 | 116.6 (2) |
| O17—Sm2—O11 | 72.89 (7) | C14—O10—Sm2vii | 149.34 (19) |
| O15—Sm2—O11 | 127.43 (6) | C14—O11—Sm2 | 125.26 (16) |
| O4iv—Sm2—O11 | 79.98 (7) | O13—C15—O12 | 121.9 (2) |
| O10ii—Sm2—O16 | 66.67 (7) | O13—C15—C11 | 118.9 (2) |
| O3iii—Sm2—O16 | 70.83 (7) | O12—C15—C11 | 119.2 (3) |
| O17—Sm2—O16 | 82.31 (7) | O13—C15—Sm1 | 59.59 (13) |
| O15—Sm2—O16 | 81.90 (7) | O12—C15—Sm1 | 63.06 (14) |
| O4iv—Sm2—O16 | 143.57 (7) | C11—C15—Sm1 | 168.04 (17) |
| O11—Sm2—O16 | 133.64 (7) | C15—O12—Sm1 | 91.30 (17) |
| O10ii—Sm2—N2 | 73.77 (7) | C15—O13—Sm1 | 94.91 (15) |
| O3iii—Sm2—N2 | 136.39 (7) | O14—C16—O15 | 125.1 (3) |
| O17—Sm2—N2 | 137.00 (8) | O14—C16—C13 | 120.0 (2) |
| O15—Sm2—N2 | 63.27 (7) | O15—C16—C13 | 114.9 (2) |
| O4iv—Sm2—N2 | 76.81 (6) | C16—O15—Sm2 | 127.11 (17) |
| O11—Sm2—N2 | 64.22 (7) | Sm2—O16—H16B | 128.1 |
| O16—Sm2—N2 | 125.92 (7) | Sm2—O16—H16A | 121.5 |
| C1—N1—C5 | 120.1 (2) | H16B—O16—H16A | 104.5 |
| C1—N1—Sm1 | 120.05 (16) | Sm2—O17—H17B | 117.1 |
| C5—N1—Sm1 | 119.15 (16) | Sm2—O17—H17A | 121.9 |
| N1—C1—C2 | 122.0 (2) | H17B—O17—H17A | 103.8 |
| N1—C1—C6 | 113.8 (2) | H18B—O18—H18A | 100.8 |
Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) −x+1/2, y−1/2, −z+3/2; (iii) x, y, z+1; (iv) −x+1, −y+1, −z+1; (v) −x+1/2, y+1/2, −z+1/2; (vi) x, y, z−1; (vii) −x+1/2, y+1/2, −z+3/2.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7—H7A···O14viii | 0.82 | 1.87 | 2.686 (3) | 172 |
| O7—H7B···O2i | 0.86 | 1.80 | 2.645 (3) | 165 |
| O8—H8A···O12iv | 0.89 | 1.83 | 2.727 (3) | 177 |
| O8—H8B···O9ix | 0.87 | 2.38 | 2.831 (3) | 113 |
| O9—H9A···O6viii | 0.87 | 1.84 | 2.698 (3) | 171 |
| O9—H9B···O5viii | 0.93 | 2.55 | 3.052 (3) | 114 |
| O16—H16A···O4viii | 0.82 | 2.31 | 3.075 (3) | 157 |
| O16—H16B···O15x | 0.79 | 1.97 | 2.747 (3) | 171 |
| O17—H17A···O18ii | 0.84 | 1.86 | 2.697 (3) | 176 |
| O17—H17B···O5xi | 0.80 | 1.96 | 2.731 (3) | 162 |
| O18—H18A···O14xii | 0.85 | 2.06 | 2.905 (3) | 174 |
| O18—H18B···O13 | 0.82 | 1.93 | 2.736 (3) | 168 |
Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) −x+1/2, y−1/2, −z+3/2; (iv) −x+1, −y+1, −z+1; (viii) −x+1, −y, −z+1; (ix) x, y+1, z; (x) −x+1, −y, −z+2; (xi) x−1/2, −y+1/2, z+1/2; (xii) x−1/2, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CV5233).
References
- Gao, H.-L., Yi, L., Ding, B., Wang, H.-S., Cheng, P., Liao, D.-Z. & Yan, S.-P. (2006). Inorg. Chem. 45, 481–483. [DOI] [PubMed]
- Ghosh, S. K. & Bharadwaj, P. K. (2005). Eur. J. Inorg. Chem. pp. 4886–4889.
- Higashi, T. (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Li, C.-J., Peng, M.-X., Leng, J.-D., Yang, M.-M., Lin, Z.-J. & Tong, M.-L. (2008). CrystEngComm, 10, 1645–1652.
- Rigaku (1998). RAPID-AUTO Rigaku Corporation, Tokyo, Japan.
- Rigaku/MSC (2004). CrystalStructure Rigaku/MSC Inc., The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, H.-S., Zhao, B., Zhai, B., Shi, W., Cheng, P., Liao, D.-Z. & Yan, S.-P. (2007). Cryst. Growth Des. 7, 1851–1857.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812002462/cv5233sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812002462/cv5233Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





