Abstract
The title compound, C10H8O6, a promising raw material to obtain colorless polyimides which are applied to microelectronic and optoelectronic devices, adopts a folded conformation in which the dihedral angle between the two anhydro rings is 55.15 (8)°. The central six-membered ring assumes a conformation intermediate between boat and twist-boat. In the crystal, molecules are linked by weak C—H⋯O interactions, forming a layer parallel to the bc plane.
Related literature
For microelectronic applications of the present compound, see: Ando et al. (2010 ▶). For background to polyimides, see: Hasegawa et al. (2007 ▶, 2008 ▶); Hasegawa & Horie (2001 ▶). For a related structure, see: Uchida et al. (2003 ▶).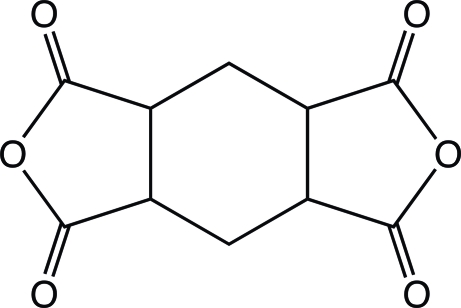
Experimental
Crystal data
C10H8O6
M r = 224.16
Monoclinic,
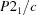
a = 12.167 (2) Å
b = 7.1380 (14) Å
c = 10.626 (2) Å
β = 90.12 (3)°
V = 922.8 (3) Å3
Z = 4
Mo Kα radiation
μ = 0.14 mm−1
T = 296 K
0.51 × 0.42 × 0.42 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.934, T max = 0.945
6648 measured reflections
2285 independent reflections
2015 reflections with I > 2σ(I)
R int = 0.026
Refinement
R[F 2 > 2σ(F 2)] = 0.045
wR(F 2) = 0.140
S = 1.00
2285 reflections
146 parameters
H-atom parameters constrained
Δρmax = 0.30 e Å−3
Δρmin = −0.18 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT-Plus (Bruker, 2007 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶) and ORTEPIII (Burnett & Johnson, 1996 ▶); software used to prepare material for publication: SHELXL97 and PLATON.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812003571/is5043sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812003571/is5043Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812003571/is5043Isup3.mol
Supplementary material file. DOI: 10.1107/S1600536812003571/is5043Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1⋯O3i | 0.98 | 2.40 | 3.3384 (19) | 159 |
| C3—H3B⋯O6ii | 0.97 | 2.58 | 3.429 (2) | 146 |
Symmetry codes: (i) 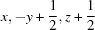 ; (ii)
; (ii) 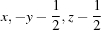 .
.
supplementary crystallographic information
Comment
Aromatic polyimides (PI) have been widely applied to microelectronic and optoelectronic devices for their reliable combined properties: considerably high glass transition temperatures (Tg), non-flammability, and good dielectric and mechanical properties (Ando et al., 2010). Conventional aromatic PI films are intensively colored on the basis of charge-transfer (CT) interactions (Hasegawa & Horie, 2001). However, the coloration often disturbs optical applications of PIs. Recently, there is a strong demand that further lightens the total weights of flat panel displays by replacing fragile inorganic glass substrates (~400 µm thick) by plastic substrates (~100 µm thick). However, it is not easy to develop the practically useful plastic substrates simultaneously possessing excellent optical transparency and sufficient heat resistance (Tg's > 250 °C) for the device fabrication processes such as inorganic transparent electrode deposition. The most effective strategy for completely erasing the significant PI film coloration is to use non-aromatic (cycloaliphatic) monomers either in tetracarboxylic dianhydride or diamine, thereby the CT interactions are inhibited. Our previous work illustrated that the equimolar polyaddition of cis, cis, cis-1,2,4,5-cyclohexanetetracarboxylic dianhydride (H-PMDA), synthesized by hydrogenation of pyromellitic dianhydride (PMDA), and some diamines indeed led to colorless PIs with very high Tg's (Hasegawa et al., 2007). However, the obtained PI films were very brittle in some cases owing to poor chain entanglement caused by insufficient molecular weights of the resultant PIs, which come from the insufficient reactivity of H-PMDA with diamines. The low reactivity of H-PMDA can be explained in terms of its steric structure (Uchida et al., 2003). In order to solve this crucial problem, we developed another H-PMDA isomer, i.e., 1R*, 2S*, 4S*, 5R*-cyclohexanetetracarboxylic dianhydride (H"-PMDA) (Hasegawa et al., 2008). The present work reports the crystal structure of this compound.
Experimental
H"-PMDA was synthesized as follows (Hasegawa et al., 2008); PMDA was hydrolyzed with a NaOH aqueous solution. The pyromellitic acid tetrasodium salt formed was hydrogenated in a high-pressure hydrogen atmosphere in the presence of a ruthenium catalyst, and neutralized with conc. HCl. The tetracarboxylic acid obtained was isomerized by dehydrating with acetic anhydride at a precisely controlled temperature. Crystals of the title compound suitable for X-ray analysis were obtained from an acetic anhydride solution.
Refinement
All H atoms were placed in geometrical positions (C—H = 0.98 and 0.97 Å for CH and CH2, respectively) and constrained to ride on their parent atoms with Uiso(H) = 1.2Ueq(C).
Figures
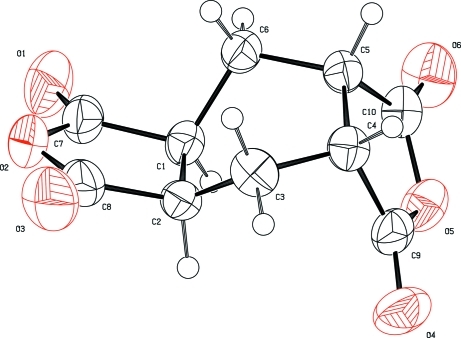
Fig. 1.
The molecular structure of the title compound, showing displacement ellipsoids at the 50% probability level.
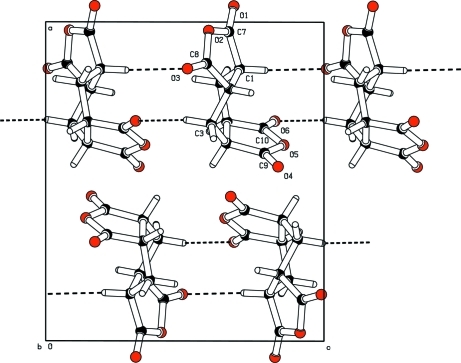
Fig. 2.
The crystal packing of the title compound viewed along the b axis. The dashed lines indicate C—H···O intermolecular interactions.
Crystal data
| C10H8O6 | F(000) = 464 |
| Mr = 224.16 | Dx = 1.613 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3549 reflections |
| a = 12.167 (2) Å | θ = 2.5–28.3° |
| b = 7.1380 (14) Å | µ = 0.14 mm−1 |
| c = 10.626 (2) Å | T = 296 K |
| β = 90.12 (3)° | Block, colorless |
| V = 922.8 (3) Å3 | 0.51 × 0.42 × 0.42 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD area-detector diffractometer | 2285 independent reflections |
| Radiation source: fine-focus sealed tube | 2015 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.026 |
| φ and ω scans | θmax = 28.3°, θmin = 3.3° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −16→16 |
| Tmin = 0.934, Tmax = 0.945 | k = −9→9 |
| 6648 measured reflections | l = −7→14 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.045 | H-atom parameters constrained |
| wR(F2) = 0.140 | w = 1/[σ2(Fo2) + (0.0954P)2 + 0.121P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max < 0.001 |
| 2285 reflections | Δρmax = 0.30 e Å−3 |
| 146 parameters | Δρmin = −0.18 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.057 (8) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 1.05049 (8) | −0.0274 (2) | 0.67601 (13) | 0.0712 (4) | |
| O2 | 0.97350 (9) | 0.22073 (16) | 0.58580 (11) | 0.0583 (3) | |
| O3 | 0.85376 (13) | 0.43198 (17) | 0.50962 (13) | 0.0773 (4) | |
| O4 | 0.54696 (9) | 0.16073 (17) | 0.83493 (12) | 0.0642 (3) | |
| O5 | 0.61479 (8) | −0.12795 (16) | 0.85526 (9) | 0.0504 (3) | |
| O6 | 0.68964 (10) | −0.40914 (17) | 0.82316 (12) | 0.0662 (4) | |
| C1 | 0.85268 (9) | 0.02100 (17) | 0.69704 (11) | 0.0371 (3) | |
| H1 | 0.8492 | 0.0003 | 0.7881 | 0.045* | |
| C2 | 0.79051 (10) | 0.20009 (16) | 0.66127 (11) | 0.0382 (3) | |
| H2 | 0.7781 | 0.2748 | 0.7373 | 0.046* | |
| C3 | 0.68030 (11) | 0.16836 (17) | 0.59459 (13) | 0.0439 (3) | |
| H3A | 0.6350 | 0.2794 | 0.6032 | 0.053* | |
| H3B | 0.6930 | 0.1473 | 0.5056 | 0.053* | |
| C4 | 0.62021 (9) | 0.00055 (17) | 0.65005 (11) | 0.0374 (3) | |
| H4 | 0.5530 | −0.0204 | 0.6010 | 0.045* | |
| C5 | 0.68715 (10) | −0.18238 (16) | 0.65113 (11) | 0.0363 (3) | |
| H5 | 0.6573 | −0.2689 | 0.5881 | 0.044* | |
| C6 | 0.80993 (10) | −0.15105 (16) | 0.62629 (12) | 0.0401 (3) | |
| H6A | 0.8509 | −0.2608 | 0.6528 | 0.048* | |
| H6B | 0.8216 | −0.1342 | 0.5367 | 0.048* | |
| C7 | 0.96943 (10) | 0.0603 (2) | 0.65718 (13) | 0.0479 (3) | |
| C8 | 0.86994 (13) | 0.3023 (2) | 0.57707 (14) | 0.0508 (4) | |
| C9 | 0.58864 (9) | 0.0285 (2) | 0.78582 (13) | 0.0431 (3) | |
| C10 | 0.66715 (10) | −0.2597 (2) | 0.78074 (13) | 0.0434 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0325 (5) | 0.0985 (10) | 0.0825 (9) | 0.0007 (5) | −0.0011 (5) | 0.0044 (7) |
| O2 | 0.0524 (6) | 0.0621 (7) | 0.0605 (7) | −0.0161 (5) | 0.0189 (5) | −0.0008 (5) |
| O3 | 0.1034 (10) | 0.0551 (7) | 0.0736 (8) | −0.0151 (7) | 0.0130 (7) | 0.0214 (6) |
| O4 | 0.0493 (6) | 0.0756 (8) | 0.0679 (7) | 0.0162 (5) | 0.0054 (5) | −0.0189 (6) |
| O5 | 0.0465 (5) | 0.0679 (7) | 0.0369 (5) | 0.0015 (4) | 0.0045 (4) | 0.0062 (4) |
| O6 | 0.0716 (7) | 0.0576 (7) | 0.0694 (7) | 0.0034 (5) | 0.0005 (6) | 0.0278 (6) |
| C1 | 0.0332 (5) | 0.0456 (6) | 0.0326 (5) | −0.0024 (4) | 0.0015 (4) | 0.0029 (5) |
| C2 | 0.0437 (6) | 0.0349 (6) | 0.0359 (6) | −0.0044 (4) | 0.0042 (5) | −0.0022 (4) |
| C3 | 0.0494 (7) | 0.0388 (6) | 0.0436 (7) | 0.0024 (5) | −0.0066 (5) | 0.0077 (5) |
| C4 | 0.0318 (5) | 0.0427 (6) | 0.0377 (6) | 0.0014 (4) | −0.0066 (4) | 0.0019 (5) |
| C5 | 0.0359 (6) | 0.0361 (6) | 0.0368 (6) | −0.0030 (4) | 0.0003 (4) | 0.0013 (4) |
| C6 | 0.0365 (6) | 0.0379 (6) | 0.0461 (7) | 0.0009 (4) | 0.0073 (5) | −0.0005 (5) |
| C7 | 0.0383 (6) | 0.0595 (8) | 0.0460 (7) | −0.0094 (6) | 0.0031 (5) | −0.0051 (6) |
| C8 | 0.0627 (9) | 0.0427 (7) | 0.0471 (7) | −0.0126 (6) | 0.0090 (6) | 0.0006 (6) |
| C9 | 0.0282 (5) | 0.0548 (7) | 0.0464 (7) | 0.0024 (5) | −0.0005 (5) | −0.0042 (5) |
| C10 | 0.0368 (6) | 0.0487 (7) | 0.0447 (7) | −0.0040 (5) | −0.0011 (5) | 0.0091 (5) |
Geometric parameters (Å, º)
| O1—C7 | 1.1850 (19) | C2—C3 | 1.5322 (18) |
| O2—C7 | 1.3745 (19) | C2—H2 | 0.9800 |
| O2—C8 | 1.391 (2) | C3—C4 | 1.5226 (17) |
| O3—C8 | 1.1869 (19) | C3—H3A | 0.9700 |
| O4—C9 | 1.1923 (17) | C3—H3B | 0.9700 |
| O5—C9 | 1.3754 (18) | C4—C9 | 1.5070 (18) |
| O5—C10 | 1.3853 (18) | C4—C5 | 1.5390 (16) |
| O6—C10 | 1.1898 (18) | C4—H4 | 0.9800 |
| C1—C7 | 1.5095 (17) | C5—C10 | 1.5039 (17) |
| C1—C6 | 1.5304 (17) | C5—C6 | 1.5341 (17) |
| C1—C2 | 1.5328 (17) | C5—H5 | 0.9800 |
| C1—H1 | 0.9800 | C6—H6A | 0.9700 |
| C2—C8 | 1.5068 (18) | C6—H6B | 0.9700 |
| C7—O2—C8 | 110.60 (10) | C5—C4—H4 | 108.4 |
| C9—O5—C10 | 110.53 (10) | C10—C5—C6 | 111.75 (10) |
| C7—C1—C6 | 109.29 (10) | C10—C5—C4 | 103.39 (10) |
| C7—C1—C2 | 103.86 (10) | C6—C5—C4 | 112.99 (9) |
| C6—C1—C2 | 112.36 (10) | C10—C5—H5 | 109.5 |
| C7—C1—H1 | 110.4 | C6—C5—H5 | 109.5 |
| C6—C1—H1 | 110.4 | C4—C5—H5 | 109.5 |
| C2—C1—H1 | 110.4 | C1—C6—C5 | 111.25 (10) |
| C8—C2—C3 | 111.02 (11) | C1—C6—H6A | 109.4 |
| C8—C2—C1 | 103.55 (10) | C5—C6—H6A | 109.4 |
| C3—C2—C1 | 114.98 (10) | C1—C6—H6B | 109.4 |
| C8—C2—H2 | 109.0 | C5—C6—H6B | 109.4 |
| C3—C2—H2 | 109.0 | H6A—C6—H6B | 108.0 |
| C1—C2—H2 | 109.0 | O1—C7—O2 | 120.18 (13) |
| C4—C3—C2 | 110.96 (9) | O1—C7—C1 | 129.62 (14) |
| C4—C3—H3A | 109.4 | O2—C7—C1 | 110.16 (12) |
| C2—C3—H3A | 109.4 | O3—C8—O2 | 121.05 (14) |
| C4—C3—H3B | 109.4 | O3—C8—C2 | 129.08 (16) |
| C2—C3—H3B | 109.4 | O2—C8—C2 | 109.86 (12) |
| H3A—C3—H3B | 108.0 | O4—C9—O5 | 120.41 (13) |
| C9—C4—C3 | 112.95 (11) | O4—C9—C4 | 129.29 (13) |
| C9—C4—C5 | 103.95 (10) | O5—C9—C4 | 110.30 (10) |
| C3—C4—C5 | 114.57 (10) | O6—C10—O5 | 119.85 (13) |
| C9—C4—H4 | 108.4 | O6—C10—C5 | 129.67 (14) |
| C3—C4—H4 | 108.4 | O5—C10—C5 | 110.48 (11) |
| C7—C1—C2—C8 | 13.48 (13) | C6—C1—C7—O2 | 109.53 (12) |
| C6—C1—C2—C8 | −104.52 (11) | C2—C1—C7—O2 | −10.57 (13) |
| C7—C1—C2—C3 | 134.77 (11) | C7—O2—C8—O3 | −172.68 (15) |
| C6—C1—C2—C3 | 16.77 (14) | C7—O2—C8—C2 | 6.61 (15) |
| C8—C2—C3—C4 | 155.23 (11) | C3—C2—C8—O3 | 42.5 (2) |
| C1—C2—C3—C4 | 38.11 (15) | C1—C2—C8—O3 | 166.44 (16) |
| C2—C3—C4—C9 | 63.98 (13) | C3—C2—C8—O2 | −136.70 (12) |
| C2—C3—C4—C5 | −54.81 (14) | C1—C2—C8—O2 | −12.78 (14) |
| C9—C4—C5—C10 | 11.25 (11) | C10—O5—C9—O4 | −176.99 (12) |
| C3—C4—C5—C10 | 134.99 (11) | C10—O5—C9—C4 | 3.01 (13) |
| C9—C4—C5—C6 | −109.73 (11) | C3—C4—C9—O4 | 45.97 (18) |
| C3—C4—C5—C6 | 14.02 (14) | C5—C4—C9—O4 | 170.76 (14) |
| C7—C1—C6—C5 | −172.96 (10) | C3—C4—C9—O5 | −134.04 (11) |
| C2—C1—C6—C5 | −58.22 (13) | C5—C4—C9—O5 | −9.25 (12) |
| C10—C5—C6—C1 | −74.30 (13) | C9—O5—C10—O6 | −175.24 (13) |
| C4—C5—C6—C1 | 41.81 (13) | C9—O5—C10—C5 | 4.90 (14) |
| C8—O2—C7—O1 | −179.25 (14) | C6—C5—C10—O6 | −68.34 (18) |
| C8—O2—C7—C1 | 2.77 (15) | C4—C5—C10—O6 | 169.85 (14) |
| C6—C1—C7—O1 | −68.20 (19) | C6—C5—C10—O5 | 111.50 (11) |
| C2—C1—C7—O1 | 171.70 (15) | C4—C5—C10—O5 | −10.31 (13) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1···O3i | 0.98 | 2.40 | 3.3384 (19) | 159 |
| C3—H3B···O6ii | 0.97 | 2.58 | 3.429 (2) | 146 |
Symmetry codes: (i) x, −y+1/2, z+1/2; (ii) x, −y−1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS5043).
References
- Ando, S., Ueda, M., Kakimoto, M., Kochi, M., Takeichi, T., Hasegawa, M. & Yokota, R. (2010). The Latest Polyimides: Fundamentals and Applications, 2nd ed. Tokyo: NTS.
- Bruker (2007). APEX2 and SAINT-Plus Bruker AXS Inc., Madison, Wisconsin, USA.
- Burnett, M. N. & Johnson, C. K. (1996). ORTEPIII Report ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.
- Hasegawa, M., Fujii, M., Uchida, A., Hirano, D., Yamaguchi, S., Takezawa, E. & Ishikawa, A. (2008). Polym. Prep. Jpn, 57, 4031–4032.
- Hasegawa, M. & Horie, K. (2001). Prog. Polym. Sci. 26, 259–335.
- Hasegawa, M., Horiuchi, M. & Wada, Y. (2007). High Perform. Polym. 19, 175–193.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Uchida, A., Hasegawa, M. & Manami, H. (2003). Acta Cryst. C59, o435–o438. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812003571/is5043sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812003571/is5043Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812003571/is5043Isup3.mol
Supplementary material file. DOI: 10.1107/S1600536812003571/is5043Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report