Abstract
In the title compound, C7H11NO4, prepared via a Morita–Baylis–Hillman adduct, the five-membered ring bearing three O atoms approximates to a twisted conformation, whereas the other ring is close to an envelope, with a C atom in the flap position. The dihedral angle between their mean planes (all atoms) is 23.11 (9)°. The new stereocenters are created in a trans-diaxial configuration. In the crystal, O—H⋯O and O—H⋯(O,O) hydrogen bonds link the molecules, generating a three-dimensional network. A weak C—H⋯O interaction also occurs.
Related literature
For the utilization of this type of pyrrolizidinone as an inihibitor of glicosidase, see: D’Alanzo et al. (2009) ▶; Ayad et al. (2004 ▶) and for their huge therapeutical potential for the treatment of a number of diseases such as cancer, diabetes, and lysosomal storage disorders, see: Baumann (2007 ▶). For related literature concerning preparation of the title compound, see: Freire et al. (2007 ▶). Analysis of the absolute structure was also performed using likelihood methods, see: Hooft et al. (2008 ▶).
Experimental
Crystal data
C7H11NO4
M r = 173.17
Monoclinic,

a = 4.6983 (3) Å
b = 14.5424 (10) Å
c = 5.5271 (4) Å
β = 99.663 (3)°
V = 372.28 (4) Å3
Z = 2
Cu Kα radiation
μ = 1.09 mm−1
T = 100 K
0.31 × 0.27 × 0.25 mm
Data collection
Bruker Kappa APEXII DUO diffractometer
3697 measured reflections
1229 independent reflections
1228 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.029
wR(F 2) = 0.073
S = 1.14
1229 reflections
112 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.27 e Å−3
Δρmin = −0.41 e Å−3
Absolute structure: Flack (1983 ▶), 537 Friedel pairs
Flack parameter: 0.20 (17)
Data collection: APEX2 (Bruker, 2010) ▶; cell refinement: SAINT (Bruker, 2010 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: WinGX (Farrugia,1999 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812002292/hb6566sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812002292/hb6566Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812002292/hb6566Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯O2i | 0.84 | 1.98 | 2.8190 (15) | 174 |
| O2—H2⋯O1ii | 0.84 | 2.50 | 3.1745 (15) | 138 |
| O2—H2⋯O4iii | 0.84 | 2.25 | 2.8589 (15) | 129 |
| O4—H4⋯O3iv | 0.84 | 1.84 | 2.6636 (15) | 167 |
| C4—H4A⋯O4ii | 1.00 | 2.41 | 3.3057 (18) | 148 |
Symmetry codes: (i) 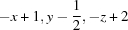 ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Acknowledgments
The authors acknowledge the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for financial support. FLO and KRLF were supported by fellowships from CAPES and FAPESP, respectively. RA and FC are recipients of research fellowships from CNPq.
supplementary crystallographic information
Comment
Crystallographic data of the title polyhydroxylated pyrrolizidinone are disclosed. Compounds of this class can be used as glycosidase inhibitors and present a huge therapeutical potential for the treatment of a number of diseases such as cancer, diabetes, and lysosomal storage disorders (Baumann, 2007). The title compound has been prepared, for the first time, using a synthetic strategy based on a Morita-Baylis-Hillman adduct, easily obtained from a reaction between N-Boc-4(R)-hydroxy-2(S)-prolinal and methyl acrylate in 70% yield, as a mixture of diastereoisomers. After chromatographic separation, the minor isomer was transformed into the title compound. This compound was synthesized in five steps and 5.2% overall yield.
The asymmetric pyrrolizidinone, C7H11NO4, a new molecule with four stereocenters from a Morita-Baylis-Hillman adduct is shown in Fig. 1. The crystal packing (Fig. 2) is stabilized by hydrogen bonds. The dihedral angles of H7—C7—C6—H6 = 153.8° and H1A—C1—C7—H7 = 161.6° show that the H atoms 1A, 7 and 6 of the two new stereocenters are created in the trans-diaxial configuration. These values agree with the coupling constants values obtained for these protons in the 1H NMR analysis, 3JH6,H7 = 7.2 Hz e 3JH1A,H7 = 8.8 Hz. The crystallography parameters for this new molecule confirm its absolute configuration.
Experimental
A solution of pyrrolizidinone (II) (0.10 g, 0.59 mmol) in MeOH/CH2Cl2 (3:7, 15 mL) was cooled to -72°C. After that a stream of oxygen/ozone was bubbled into it for 8–10 min (the reaction evolution was followed by TLC). Then, NaBH4 (0.112 g, 4.45 mmol) was added at -72°C and the resulting mixture was stirred for 6 h at room temperature. The reaction medium was initially acidified to pH 2–3 with a solution of HCl in methanol, then it was neutralized to pH 6–7 with solid Na2CO3. The resulting mixture was filtered over a pad of Celite(R) and the solid was washed with methanol. The filtrates were combined and the solvents were removed under reduced pressure. The residue was purified by flash silica gel column chromatography (CH2Cl2:MeOH 95:05) to afford pyrrolizidinone I (0.08 g), as a white solid, in 80% yield. The title compound was recrystallized by using the liquid-vapor saturation method. The compound was dissolved with ethanol and crystallized with a vapor pressure of a second less polar liquid (chloroform), in a closed camera, providing the slow formation of crystals. [α]D20 + 3 (c 1, MeOH); M. p. 150–152°C; IR (KBr, vmax): 3499, 3374, 2993, 2910, 1681, 1446, 1362, 1327, 1262, 1129, 1111, 1014 cm-1; 1H NMR (400 MHz, D2O) δ 1.78 (ddd, J 13.4, 5.0, 4.9 Hz, 1H, H-5B); 2.28 (dd, J 13.4, 5.7 Hz, 1H, H-5 A); 3.10 (d, J 12.8 Hz, 1H, H-3B; 3.77 (dd, J 12.9, 4.9 Hz, 1H, H-3 A); 3.91 (m, 1H, H-6); 3.99 (dd, JH7,H1A 8.8, JH6,H7 7.2 Hz, 1H, H-7); 4.60 (d, JH7,H1A 8.8 Hz, 1H, H-1 A); 4.70 (t, J 4.9 Hz, 1H, H-4 A); 13C NMR (62.5 MHz, MeOD) δ 40.7, 52.1, 63.0, 73.2, 80.1, 83.3, 174.0; HRMS (ESI-TOF) Calcd. for C7H12NO4 [M + H]+ 174.0766, Found 174.0754.
Refinement
The calculated Flack parameter was F=0.20 (17) (Flack, 1983). Analysis of the absolute structure was also performed using likelihood methods (Hooft et al., 2008) as implemented in PLATON (Spek, 2009). The resulting value for the Hooft parameter was y=0.12 (4), with a corresponding probability for an inverted structure smaller than 1 × 10-100. Taken togheter, these results indicate that the absolute structure has been determined correctly.
Figures
Fig. 1.
Molecular view of the title compound showing displacement ellipsoids drawn at the 50% probability level. H atoms are presented as a small spheres of arbitrary radius.
Fig. 2.
Title compound involved into hydrogen bonds. The presence of several hydroxyl groups in its structure leads this compound to behave as a sugar.
Fig. 3.
The conversion of (I) to pyrrolizidinone (II).
Crystal data
| C7H11NO4 | F(000) = 184 |
| Mr = 173.17 | Dx = 1.545 Mg m−3 |
| Monoclinic, P21 | Cu Kα radiation, λ = 1.54178 Å |
| a = 4.6983 (3) Å | Cell parameters from 1229 reflections |
| b = 14.5424 (10) Å | θ = 6.1–66.8° |
| c = 5.5271 (4) Å | µ = 1.09 mm−1 |
| β = 99.663 (3)° | T = 100 K |
| V = 372.28 (4) Å3 | Rectangular block, colorless |
| Z = 2 | 0.31 × 0.27 × 0.25 mm |
Data collection
| Bruker Kappa APEXII DUO diffractometer | 1228 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.027 |
| Graphite monochromator | θmax = 66.8°, θmin = 6.1° |
| Bruker APEX CCD area–detector scans | h = −5→5 |
| 3697 measured reflections | k = −16→16 |
| 1229 independent reflections | l = −6→6 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.029 | H-atom parameters constrained |
| wR(F2) = 0.073 | w = 1/[σ2(Fo2) + (0.0498P)2 + 0.0546P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.14 | (Δ/σ)max = 0.012 |
| 1229 reflections | Δρmax = 0.27 e Å−3 |
| 112 parameters | Δρmin = −0.41 e Å−3 |
| 1 restraint | Absolute structure: Flack (1983), 537 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.20 (17) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.6971 (2) | −0.03528 (7) | 0.86162 (19) | 0.0163 (3) | |
| H1 | 0.7222 | −0.0594 | 1.0016 | 0.024* | |
| O2 | 0.2633 (2) | 0.38620 (8) | 0.6676 (2) | 0.0167 (3) | |
| H2 | 0.1802 | 0.4123 | 0.5394 | 0.025* | |
| O3 | 0.9071 (2) | 0.13035 (8) | 1.12983 (19) | 0.0194 (3) | |
| O4 | 0.1530 (2) | 0.02523 (7) | 0.5015 (2) | 0.0167 (3) | |
| H4 | 0.0715 | 0.0504 | 0.3714 | 0.025* | |
| N1 | 0.5577 (3) | 0.20182 (9) | 0.8573 (2) | 0.0128 (3) | |
| C1 | 0.5216 (3) | 0.04341 (11) | 0.8610 (3) | 0.0134 (3) | |
| H1A | 0.3599 | 0.0303 | 0.9528 | 0.016* | |
| C2 | 0.6890 (3) | 0.12859 (11) | 0.9697 (3) | 0.0141 (3) | |
| C3 | 0.6663 (3) | 0.29584 (11) | 0.8577 (3) | 0.0143 (3) | |
| H3A | 0.8780 | 0.2966 | 0.8633 | 0.017* | |
| H3B | 0.6191 | 0.3310 | 0.9992 | 0.017* | |
| C4 | 0.5078 (3) | 0.33520 (10) | 0.6142 (3) | 0.0136 (3) | |
| H4A | 0.6384 | 0.3753 | 0.5346 | 0.016* | |
| C5 | 0.4128 (3) | 0.25050 (10) | 0.4564 (3) | 0.0142 (3) | |
| H5A | 0.2409 | 0.2644 | 0.3318 | 0.017* | |
| H5B | 0.5701 | 0.2286 | 0.3721 | 0.017* | |
| C6 | 0.3420 (3) | 0.17927 (10) | 0.6401 (2) | 0.0124 (3) | |
| H6 | 0.1420 | 0.1887 | 0.6752 | 0.015* | |
| C7 | 0.3995 (3) | 0.07635 (12) | 0.6019 (3) | 0.0132 (3) | |
| H7 | 0.5501 | 0.0697 | 0.4949 | 0.016* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0237 (5) | 0.0097 (6) | 0.0144 (5) | 0.0043 (5) | 0.0003 (4) | 0.0025 (5) |
| O2 | 0.0219 (5) | 0.0113 (6) | 0.0152 (5) | 0.0040 (4) | −0.0017 (4) | 0.0004 (4) |
| O3 | 0.0229 (6) | 0.0171 (6) | 0.0149 (5) | 0.0001 (5) | −0.0062 (4) | 0.0016 (4) |
| O4 | 0.0209 (5) | 0.0118 (6) | 0.0141 (5) | −0.0016 (4) | −0.0065 (4) | 0.0014 (4) |
| N1 | 0.0192 (6) | 0.0101 (6) | 0.0077 (6) | 0.0004 (5) | −0.0019 (5) | −0.0002 (5) |
| C1 | 0.0175 (7) | 0.0110 (7) | 0.0111 (8) | 0.0019 (6) | 0.0007 (6) | 0.0014 (5) |
| C2 | 0.0196 (7) | 0.0140 (8) | 0.0086 (7) | −0.0004 (6) | 0.0021 (6) | −0.0007 (6) |
| C3 | 0.0172 (7) | 0.0116 (8) | 0.0130 (7) | −0.0010 (6) | −0.0009 (6) | −0.0009 (6) |
| C4 | 0.0185 (8) | 0.0093 (7) | 0.0124 (7) | 0.0001 (6) | 0.0007 (6) | 0.0006 (5) |
| C5 | 0.0213 (7) | 0.0106 (8) | 0.0097 (7) | 0.0001 (6) | −0.0005 (6) | 0.0014 (6) |
| C6 | 0.0152 (7) | 0.0113 (8) | 0.0098 (7) | 0.0010 (6) | −0.0005 (6) | 0.0003 (6) |
| C7 | 0.0151 (7) | 0.0120 (7) | 0.0114 (7) | 0.0002 (6) | −0.0007 (6) | 0.0020 (6) |
Geometric parameters (Å, º)
| O1—C1 | 1.410 (2) | C1—H1A | 1.0000 |
| O1—H1 | 0.8400 | C3—C4 | 1.535 (2) |
| O2—C4 | 1.439 (2) | C3—H3A | 0.9900 |
| O2—H2 | 0.8400 | C3—H3B | 0.9900 |
| O3—C2 | 1.2368 (19) | C4—C5 | 1.532 (2) |
| O4—C7 | 1.409 (2) | C4—H4A | 1.0000 |
| O4—H4 | 0.8400 | C5—C6 | 1.526 (2) |
| N1—C2 | 1.331 (2) | C5—H5A | 0.9900 |
| N1—C3 | 1.459 (2) | C5—H5B | 0.9900 |
| N1—C6 | 1.4721 (18) | C6—C7 | 1.542 (2) |
| C1—C7 | 1.528 (2) | C6—H6 | 1.0000 |
| C1—C2 | 1.535 (2) | C7—H7 | 1.0000 |
| C1—O1—H1 | 109.5 | C5—C4—C3 | 104.56 (12) |
| C4—O2—H2 | 109.5 | O2—C4—H4A | 111.1 |
| C7—O4—H4 | 109.5 | C5—C4—H4A | 111.1 |
| C2—N1—C3 | 127.91 (12) | C3—C4—H4A | 111.1 |
| C2—N1—C6 | 113.83 (12) | C6—C5—C4 | 104.01 (12) |
| C3—N1—C6 | 113.74 (12) | C6—C5—H5A | 111.0 |
| O1—C1—C7 | 112.59 (13) | C4—C5—H5A | 111.0 |
| O1—C1—C2 | 113.15 (12) | C6—C5—H5B | 111.0 |
| C7—C1—C2 | 101.60 (12) | C4—C5—H5B | 111.0 |
| O1—C1—H1A | 109.7 | H5A—C5—H5B | 109.0 |
| C7—C1—H1A | 109.7 | N1—C6—C5 | 101.20 (12) |
| C2—C1—H1A | 109.7 | N1—C6—C7 | 102.44 (12) |
| O3—C2—N1 | 125.51 (15) | C5—C6—C7 | 120.32 (12) |
| O3—C2—C1 | 127.26 (14) | N1—C6—H6 | 110.6 |
| N1—C2—C1 | 107.22 (11) | C5—C6—H6 | 110.6 |
| N1—C3—C4 | 103.30 (12) | C7—C6—H6 | 110.6 |
| N1—C3—H3A | 111.1 | O4—C7—C1 | 110.98 (13) |
| C4—C3—H3A | 111.1 | O4—C7—C6 | 114.49 (13) |
| N1—C3—H3B | 111.1 | C1—C7—C6 | 102.87 (12) |
| C4—C3—H3B | 111.1 | O4—C7—H7 | 109.4 |
| H3A—C3—H3B | 109.1 | C1—C7—H7 | 109.4 |
| O2—C4—C5 | 111.40 (12) | C6—C7—H7 | 109.4 |
| O2—C4—C3 | 107.38 (12) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···O2i | 0.84 | 1.98 | 2.8190 (15) | 174 |
| O2—H2···O1ii | 0.84 | 2.50 | 3.1745 (15) | 138 |
| O2—H2···O4iii | 0.84 | 2.25 | 2.8589 (15) | 129 |
| O4—H4···O3iv | 0.84 | 1.84 | 2.6636 (15) | 167 |
| C4—H4A···O4ii | 1.00 | 2.41 | 3.3057 (18) | 148 |
Symmetry codes: (i) −x+1, y−1/2, −z+2; (ii) −x+1, y+1/2, −z+1; (iii) −x, y+1/2, −z+1; (iv) x−1, y, z−1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6566).
References
- Ayad, T., Génisson, A. & Baltas, M. (2004). Curr. Med. Chem. 8, 1211–1233.
- Baumann, K. (2007). WO Patent 2007039286; Chem. Abstr. 146, 421836.
- Bruker (2010). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- D’Alanzo, D., Guaragna, A. & Palumbo, G. (2009). Curr. Med. Chem. 16, 473–505. [DOI] [PubMed]
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Freire, K. R. L., Tormena, C. F. & Coelho, F. (2011). Synlett, pp. 2059–2063.
- Hooft, R. W. W., Straver, L. H. & Spek, A. L. (2008). J. Appl. Cryst. 41, 96–103. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812002292/hb6566sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812002292/hb6566Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812002292/hb6566Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





