Abstract
Aims
DNA methylation is increasingly proposed as a mechanism for underlying asthma-related inflammation. However, epigenetic studies are constrained by uncertainties on whether samples that can be easily collected in human individuals can provide informative results.
Methods
Two nasal cell DNA samples were collected on different days by nasal brushings from 35 asthmatic children aged between 8 and 11 years old. We correlated DNA methylation of IL-6, iNOS, Alu and LINE-1 with fractional exhaled nitric oxide, forced expiratory volume in 1 s and wheezing.
Results
Fractional exhaled nitric oxide increased in association with lower promoter methylation of both IL-6 (+29.0%; p = 0.004) and iNOS (+41.0%; p = 0.002). Lower IL-6 methylation was nonsignificantly associated with wheezing during the week of the study (odds ratio = 2.3; p = 0.063).
Conclusion
Our findings support the use of nasal cell DNA for human epigenetic studies of asthma.
Keywords: airway obstruction, asthma, children, DNA methylation, epigenetics, inflammation
Asthma is the most common chronic disease of childhood in developed countries, affecting nearly 6.5 million children in the USA [1], and 234.9 million individuals worldwide [2]. Airway inflammation is a key feature in the pathogenesis of childhood asthma [3], and is characterized by the presence of inflammatory cells and release of inflammatory mediators in the airways [4]. Growing evidence shows that expression and responsiveness of several inflammatory mediators are programmed through epigenetic mechanisms, such as DNA methylation [5–7]. In humans, DNA methylation contributes to silencing gene expression through the addition of methyl groups to cytosine to form 5-methyl-cytosine (5mC) [8]. DNA methylation is largely established in utero or during early life, but also shows changes thereafter in response to environmental stressors [9–12].
As a feature of the asthma-associated eosinophilic inflammation, asthma patients exhibit an increase in nitric oxide (NO) production [4], predominantly due to overexpression in the airway epithelium of the inducible nitric oxide synthase (iNOS) [13]. Studies of iNOS activation have shown that lower DNA methylation in the gene promoter is associated with increased expression [7]. Among inflammatory mediators that are relevant to asthma, consistent evidence has shown that IL-6 expression is associated with reduced DNA methylation of its gene promoter [5,6]. IL-6 is central to inflammatory processes underlying chronic inflammatory diseases, including allergic asthma, and have been shown to induce the expression of other genes that might contribute to the asthma phenotype [14].
Although inflammation-related processes have been associated with changes in DNA methylation of promoters in specific genes, including IL-6 and iNOS, the bulk of DNA methylation in the human genome is located in intergenic DNA [15,16]. In particular, Alu and long interspersed nucleotide element 1 (LINE-1) repetitive elements, which are sequences of intergenic DNA repeated in up to 1 million copies per haploid genome, represent approximately 30% of the human genome and are heavily methylated [17]. Alu and LINE-1 methylation have been shown to correlate with the global amount of DNA methylation in the genome based on studies with cancer tissues [15,16], has been shown to decrease in response to inflammation and oxidative stress [18–20].
DNA methylation patterns are tissue specific, and one critical limitation for human epigenetic studies is that tissues that are relevant for disease etiology cannot be easily obtained in most cases from patients and study participants [21]. Human in vivo studies of DNA methylation have often used blood [10,20,22–28] or buccal cells [11,29,30] as easily obtainable biospecimens in patients as well as in healthy individuals. Nasal epithelial cells have been proposed as surrogates for bronchial epithelial cells in airway inflammation studies [31]. However, to the best of our knowledge, nasal cell DNA methylation has never been evaluated in relation to asthma. In the present work, we sought proof-of-principle as to whether the levels of methylation of the IL-6 and iNOS gene promoters, and of Alu and LINE-1 repetitive elements – measured in nasal cells – were correlated with fractional exhaled nitric oxide (FENO), forced expiratory volume in 1 second (FEV1), and wheezing in a small panel study of children with current asthma.
Materials & methods
Study subjects
Between December 2007 and April 2008, we performed a panel study in children with asthma, identified during a cross-sectional investigation conducted in the area of Milazzo-Valle del Mela (Sicily, Italy). The cross-sectional screening was originally prompted by concerns due to the presence of a major petrochemical plant and an oil-powered thermal plant in the area and was conducted on all the 2506 resident children (8–11 years old) attending the local primary schools (response rate: 89.5%), in order to provide data on their respiratory health.
We used the International Study of Asthma and Allergy in Childhood (ISAAC) core questionnaire [32], to ascertain lifetime and past year prevalence of asthma and wheezing, and added questions about child’s respiratory health and risk factors for asthma derived from the Italian Studies on Respiratory Diseases in Childhood and the Environment (SIDRIA) Phase II study [33], the Italian section of ISAAC. Questionnaires were completed at home by the parents. For the panel study, we selected all the children who: had a physician diagnosis of asthma; reported wheezing symptoms in the previous 12 months; and had chest tightness and/or use of bronchodilators in the last 12 months (n = 50).
Written informed consent to participate in the panel study was obtained from the parents of 35 of the 50 children, and therefore comprised our study population of 35 participants. The reason for refusal was the concern for the invasiveness of the nasal brushing procedure. The children who did not participate in the study were not different in asthma severity from those who participated (data not shown). Each child was followed-up for 7 consecutive days. A diary on daily respiratory symptoms (e.g., symptoms of cold to rule out acute respiratory infections; wheezing symptoms and chest tightness); and on bronchodilators, inhaled steroids and antileukotrienes use, was completed by the parents of study subjects. The protocol of the study was approved by the Ethics Committee of the University of Cagliari, Italy.
Nasal mucosa cell collection & DNA extraction from nasal cell pellets
In the afternoon (4–6 pm) on days 4 and 7 (Tuesday and Friday) of the study, each child went to a dedicated out-patient clinic to undergo nasal brushing to collect nasal cells for DNA methylation analysis. Nasal brushing was performed on day 4 in the right nostril and on day 7 in the left nostril by a trained nurse, following standardized procedures. Each of the children was first asked to blow his/her nose into nonscented tissue paper to clear any mucus discharge. In case of mucus the nurse washed the nasal cavity with 2–4 ml saline solution (0.9% NaCl) inserted via a syringe (with no needle inserted) and cleaned it with a cotton swab. Nasal cells were collected by soft brushing of the inferior turbinate with a cytobrush Plus® (CooperSurgical, CT, USA). Nasal cells were gently rinsed from the cytobrush into a tube containing 6 ml of saline solution (0.9% NaCl) and 10% acetylcysteine (200 mg/ml) and then incubated for 30 min at room temperature (100 cycles/min shaking frequency). After centrifugation, the cell pellets were kept at −20°C until shipment from the study site to the central laboratory. All specimens were blind coded. DNA was extracted with the Maxwell Automatic Extractor® (Promega, Madison, WI, USA) using tissue cartridges and eluted in a 300-μl final volume.
DNA methylation analyses by bisulfite pyrosequencing
We used quantitative analysis by means of PCR-pyrosequencing on bisulfite-treated DNA to measure DNA methylation in the promoter regions of IL-6 and iNOS, as well as in Alu and LINE-1 repetitive elements [10,25]. EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) was used to treat 0.5 μg DNA (concentration 50 ng/μl), according to the manufacturer’s protocol. Final elution was performed with 30 μl M-elution buffer. The within-sample coefficients of variation between the pyrosequencing duplicates for these markers were 0.7% for LINE-1, 1.6% for Alu, 0.7% for iNOS and 4.0% for IL-6. Primers and conditions for each of the assays are reported in Table 1. Methods and primers for measuring iNOS promoter DNA methylation, as well as analyses of Alu and LINE-1 methylation that allow for the amplification of a representative pool of repetitive elements, were recently described [10]. We developed a new assay for IL-6 promoter methylation, by locating the IL-6 promoter using the Genomatix Software (Genomatix Software Inc., Ann Arbor, MI, USA) on chromosome 7 (start = 22732791, end = 22733685), and amplified the sequence between 22733756 and 22733893. A 50-μL PCR was carried out in 25 μl GoTaq® Green Master mix (Promega), 10 pmol forward primer, 10 pmol reverse primer, 50 ng bisulfite-treated genomic DNA and water. PCR cycling conditions were 95°C for 60 s, 57°C for 60 s and 72°C for 60 s for 45 cycles. PCR products were purified and sequenced by pyrosequencing as previously described using a 0.3-μM sequencing primer [25].
Table 1.
Primers and conditions for DNA methylation analysis.
| Sequence ID | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Sequencing primer (5′ to 3′) | Sequence analyzed† |
|---|---|---|---|---|
| Gene-specific methylation analysis | ||||
| iNOS | AATGAGAGTTGTTGGGAAGTGTTT | Biotin-CCACCAAACCCAACCAAACT | TAAAGGTATTTTTGTTTTAA | C/TGATTTTC/TGGGTTTTTTTTTATTTTG |
| IL-6 | Biotin-TATTTTAGTTTTGAGAAAGGAGGTG | CAATACTCTAAAACCCAACAAAAAC | TCCTAATACAAACAACCCC | CG/AACCACACG/ACAAAAAC |
| Global methylation analysis | ||||
| Alu | Biotin-TTTTTATTAAAAATATAAAAATT | CCCAAACTAAAATACAATAA | AATAACTAAAATTACAAAC | G/AC/TG/AC/TG/ACCACCA |
| LINE-1 | TTTTGAGTTAGGTGTGGGATATA | Biotin-AAAATCAAAAAATTCCCTTTC | AGTTAGGTGTGGGATATAGT | TTC/TGTGGTGC/TGTC/TG |
Nucleotides at which DNA methylation was measured are underlined.
LINE-1: Long interspersed nucleotide element 1.
For all assays we used built-in controls to verify bisulfite conversion efficiency. Compared with other common methods of DNA methylation analysis, pyrosequencing-based assays have the advantage of producing individual measures of methylation at more than one CpG dinucleotide, thus reflecting more accurately DNA methylation in the region [34,35]. In the Alu or LINE-1 assays, we measured the percentage of 5mC (%5mC) at each of three CpG dinucleotide positions that are repeated over the human genome with the sequence of interest. In the iNOS promoter assay, we measured %5mC at each of two individual CpG dinucleotides within a CpG island located in the gene promoter. In the IL-6 promoter assay, we measured %5mC at two individual CpG dinucleotides within a CpG island located downstream in the proximity of the gene promoter. %5mC levels of individual CpG dinucleotides were averaged to obtain a mean measure for each of the assays. The basis for using the average was the observation that methylation values in adjacent CpG sites over a short amplicon such as that used in pyrosequencing analysis are usually highly correlated [36]. In our previous work, we observed that averages of DNA methylation within each amplicon provide more robust methylation measures compared with analyses of individual CpGs [37]. Every sample was tested twice for each assay to confirm reproducibility, and the average of the two measurements was used in the statistical analysis.
FENO & FEV1 measurements & wheezing symptoms
FENO and FEV1 measurements were performed during the same visits when nasal cell DNA was collected. The children were trained by a nurse on the day before the beginning of the 7-day follow-up, and collection of exhaled air for FENO measurement and the forced expiratory maneuver were supervised by the same nurse during the study week. Ambient NO and FENO concentrations were assessed by chemiluminescence using the NIOX analyzer (Aerocrine, Stockholm, Sweden) and following the American Thoracic Society/European Respiratory Society recommended procedures (American Thoracic Society/European Respiratory Society) [38]. FEV1 was recorded using the PiKo®1 electronic FEV1 meter (Ferraris Respiratory Europe, Hereford, UK), a monitoring device that uses a patented pressure/flow sensor technology, and which has been demonstrated to be accurate at measuring the FEV1 in similar conditions [39]. The occurrence of wheezing (during the day, night or exercise) was recorded by the daily diary.
Statistical methods
Means and standard deviations (SDs) were used to summarize the data. In our study, each child participated in two visits in which FEV1, FENO and methylation data were collected. Therefore, our data may lack independence. To evaluate the association of DNA methylation with FEV1 or FENO, we fitted generalized estimating equation (GEE) population averaged models. GEE were fitted by specifying a generalized linear model (GLM) extension to each of the visits at which FENO, FEV1, and DNA methylation were measured. We used the GEE extension of GLM. We specified a γ family for the continuous responses (FEV1 or FENO), because both FEV1 and FENO exhibited slightly asymmetrical distributions with constant coefficients of variation [40]. We used a log-link, which also allows reporting the results as easily interpretable percentage variation for unit increase in the predictor. These choices are common in GEE implementation and supported in standard textbooks [40,41]. We specified an independent working correlation, which is robust in the presence of noisy data, and used the Huber–White estimator of standard errors [40]. The adoption of the Huber–White estimator ensures that standard errors are unbiased even if the correlation structure is misspecified.
DNA methylation, FENO and FEV1 data were used as continuous variables and results were expressed as percent variation (%v) of FENO or FEV1 in relation to interquartile-range (IQR) changes in DNA methylation (i.e., IL-6: IQR = 16.1 %5mC; IQR = iNOS: 9.4 %5mC; Alu: IQR = 0.9 %5mC; LINE: IQR = 4.9 %5mC). We fitted GEE regression models in which all four DNA methylation markers (i.e., iNOS, IL-6, Alu and LINE-1) were included as independent explanatory variables. We also determined the association of DNA methylation with wheezing symptoms (yes/no, evaluated on the day of nasal brushing) by fitting a multivariable GEE regression model for binary data (i.e., binomial family with logit link). Again, we specified an independent working correlation and used the Huber–White estimator of standard errors [40].
All observations with FENO levels below the detection limit of 5 parts per billion (five measurements) were excluded from the analysis. In addition, to avoid interference from ambient NO on the FENO readings, we excluded readings (two measurements) taken when ambient NO was >5 parts per billion. One additional observation was excluded due to missing FEV1 data. Therefore, out of the total 70 observations (35 subjects × two measurements), we conducted statistical analyses on 62 observations without missing data for any of the variables mentioned above.
Because the main purpose of the present observational study was to explore associations between DNA methylation and respiratory outcomes, we interpreted our data, based on the strength of the associations, as often done in observational studies [42], within the context of the available evidence on biological functions [43]. Accordingly, we reported confidence intervals using 90% levels, as indicated in the Sterne and Davey-Smith’s suggested guidelines for the reporting of results of statistical analyses in medical journals [43]. However, to conform to standard practice in biological reports, in the text we indicated findings with two-sided p < 0.05 as statistically significant. The study protocol included power calculations on the primary out-come (FEV1). We used the formula for panel studies reported in [40]. The sample size required ranged between 30 and 60 for within-subject correlation from 0.5–0.0, power 80% α = 0.05 two-sided. All the statistical analyses were performed using Stata 11 (Stata Corporation, College Station, TX, USA).
Results
Study population
The panel study consisted of 35 Caucasian children, 27 males and 8 females whose characteristics are reported in Table 2.
Table 2.
Characteristics of study participants.
| Characteristics | n (%) |
|---|---|
| Age, year (mean ± standard deviation) | 8.9 ± 0.8 |
|
| |
| Male | 27 (77) |
|
| |
| Parental schooling: | |
| – University | 8 (23) |
| – High school (13 year) | 22 (63) |
| – <13 year | 5 (14) |
|
| |
| Maternal tobacco smoke exposure | 8 (23) |
| Paternal tobacco smoke exposure | 11 (31) |
|
| |
| Damp or mould in child’s bedroom | 5 (14) |
|
| |
| Traffic intensity on street of residence (high) | 3 (9) |
|
| |
| Rhino conjunctivitis symptoms (past 12 months) | 12 (34) |
|
| |
| Chest tightness (past 12 months) | 16 (48) |
|
| |
| Prescription of bronchodilators (past 12 months) | 22 (63) |
|
| |
| Chest tightness in the study week | 6 (17) |
|
| |
| Respiratory infections in the study week and in the previous week | 7 (20) |
|
| |
| Bronchodilators in the study week | 10 (29) |
|
| |
| Antileukotrienes in the study week | 6 (17) |
|
| |
| Inhaled steroids in the study week | 2 (6) |
|
| |
| Antihistamines in the study week | 5 (14) |
DNA methylation & its relationship with FENO, FEV1 & wheezing
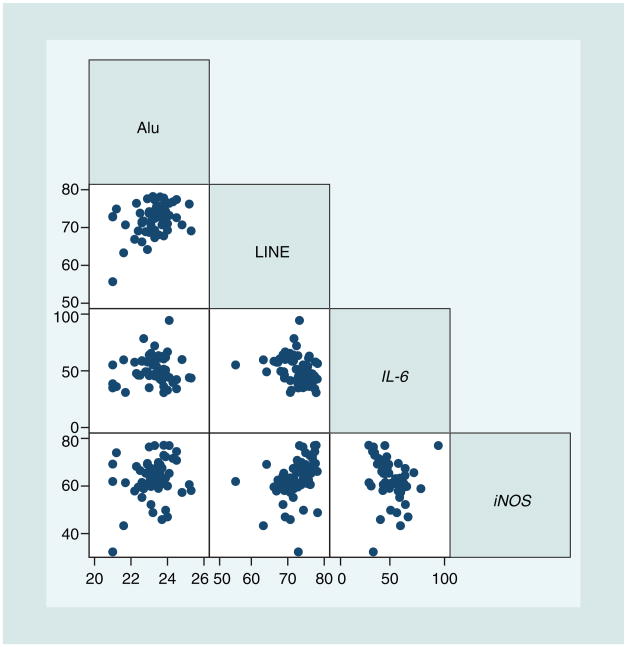
Table 3 reports descriptive statistics for FENO, FEV1 and DNA methylation of the IL-6 and iNOS promoters, as well as of Alu and LINE-1 repetitive elements. The correlation matrix of IL-6, iNOS, Alu and LINE-1 methylation showed significant correlations among the methylation markers (Figure 1). Consequently, we fitted multivariable GEE regression models to estimate the net association of each methylation markers (IL-6, iNOS, Alu or LINE-1) with FENO or FEV1 taking into account the methylation levels of the other methylation markers. In these models (Table 4), FENO was negatively associated with both IL-6 (−29.0 %v per interquartile change in IL-6 methylation; 90% CI: −44.0 to −12.9 %v; p = 0.004) and iNOS methylation (−41.0 %v per interquartile change in iNOS methylation; 90% CI: −61.6 to −20.4 %v; p = 0.002). FEV1 was not associated with IL-6 (5.6 %v per interquartile change in IL-6 methylation, 90% CI: −1.5–12.7 %v; p = 0.20) and iNOS methylation (0.05 %v per interquartile change in iNOS methylation, 90% CI: −3.5–3.6 %v; p = 0.98). Alu and LINE-1 methylation showed no associations with FENO and FEV1 (Table 4). When we evaluated the DNA methylation markers in relation to wheezing (yes/no) during the week of the study, we found a moderate nonsignificant association with lower methylation of the IL-6 promoter (odds ratio = 2.3 for wheezing; 90% CI: 1.1–4.9; p = 0.063 per interquartile decrease in IL-6 methylation).
Table 3.
Fractional exhaled nitric oxide, forced expiratory volume in 1 s and levels of DNA methylation of the IL-6 and iNOS promoters and Alu and long interspersed nucleotide element 1 repetitive elements.
| Variable (ppb) | Mean | SD | IQR |
|---|---|---|---|
| FENO | 36.30 | 29.80 | 36.00 |
| FEV1(l) | 2.17 | 0.47 | 0.68 |
| IL-6 (%5mC) | 51.40 | 12.00 | 16.10 |
| iNOS (%5mC) | 63.00 | 8.40 | 9.40 |
| Alu (%5mC) | 23.30 | 0.90 | 0.90 |
| LINE-1 (%5mC) | 72.30 | 4.10 | 4.90 |
%5mC: Percentage of 5-methyl-cytosine; FENO: Fractional exhaled nitric oxide; FEV1: Forced expiratory volume in 1 s; IQR: Interquartile range; ppb: Parts per billion; SD: Standard deviation.
Figure 1. Correlation among DNA methylation of IL-6 and iNOS promoters, and in Alu and LINE-1 repetitive elements (n = 62).
Significant correlations were found between LINE-1 and Alu methylation (r = 0.37; p = 0.003), LINE-1 and IL-6 methylation (r = −0.29; p = 0.02), and LINE-1 and iNOS methylation (r = 0.36; p = 0.004).
LINE: Long interspersed nucleotide element.
Table 4.
Association of fractional exhaled nitric oxide and forced expiratory volume in 1 s with DNA methylation of IL-6 and iNOS promoters, and Alu and long interspersed nucleotide element 1 repetitive elements.†
| Gene | FENO |
FEV1 |
||||
|---|---|---|---|---|---|---|
| %v‡ | (90% CI) | p-value | %v‡ | (90% CI) | p-value | |
| IL-6 | −29.0 | (−44.0 to −12.9) | 0.004 | 5.6 | (−1.5–12.7) | 0.198 |
|
| ||||||
| iNOS | −41.0 | (−61.6 to −20.4) | 0.002 | 0.05 | (−3.5–3.6) | 0.983 |
|
| ||||||
| Alu | 7.0 | (−10.3–20.4) | 0.503 | 2.6 | (−0.8–6.0) | 0.205 |
|
| ||||||
| LINE-1 | −7.1 | (−32.9–18.7) | 0.648 | −1.4 | (−6.2–3.3) | 0.617 |
Shows results from generalized estimating equation models including IL-6, iNOS, Alu and LINE-1 methylation as independent variables. Because correlations were found among IL-6, iNOS, Alu and LINE-1 methylation, all four methylation variables were fitted in the same model to estimate the net association of each methylation marker independently of the other markers. Results for FENO are adjusted for age and gender. Results for FEV1 are adjusted for age, gender and height.
%v per interquartile range (IQR) of methylation (IL-6: IQR = 16.1 %5mC; iNOS: IQR = 9.4 %5mC; Alu: IQR = 0.9 %5mC; LINE: IQR = 4.9 %5mC).
%5mC: Percentage of 5-methyl-cytosine; %v: Percent variation; 5mC: 5-methyl-cytosine; FENO: Fractional exhaled nitric oxide; FEV1: Forced expiratory volume in 1 s; LINE-1: Long interspersed nucleotide element 1.
We also explored whether DNA methylation was affected by exposure to environmental tobacco smoke (ETS), using information on maternal smoking as proxy of exposure. Neither current nor past maternal smoking was associated with IL-6 methylation (p = 0.64, and p = 0.73 respectively in analysis adjusted for child age and gender) or iNOS methylation (p = 0.19 and p = 0.49, respectively, in analysis adjusted for child age and gender). Most of the children included in our study were boys (n = 27). We performed a separate set of analyses on boys only, which did not show relevant differences from the analyses conducted on all children. For instance, the age- and gender-adjusted associations of IL-6 and iNOS methylation with FENO in boys only were −28.4 %v per interquartile change in IL-6 methylation (90% CI: −43.2 to −13.6 %v; p = 0.002) and −38.5 %v per interquartile change iNOS methylation (90% CI: −56.4 to −20.6 %v; p = 0.001).
Sensitivity analyses
All models reported above were adjusted for age (as a continuous variable) and gender. Because FEV1 is highly dependent on height, all modelsused to evaluate FEV1 were also adjusted by height (as a continuous variable). In sensitivity statistical analyses, we fitted multivariable models that also included as independent variables subject’s height and weight, parental education, ETS exposure, mould or damp in the childs room, symptoms of rhino-conjunctivitis (as a proxy of atopy), traffic intensity in the street of residence (as measured in [44]), recent respiratory infections, use of inhaled steroid for asthma, day of the week, outdoor temperature and relative humidity. The regression coefficients expressing the associations of the DNA methylation variables with the response variables in the set of models adjusted for the potential confounders mentioned above did not show major differences from coefficients adjusted for the smaller set of confounders (data not shown).
Discussion
Implications for human epigenetics of asthma
In this study of a small sample of asthmatic children, we found that individuals with lower DNA methylation of the IL-6 and iNOS promoters in nasal cells had higher airway inflammation, as measured by increased FENO. Promoter hypomethylation has been demonstrated as a mechanism through which both iNOS [7] and IL-6 [5] are derepressed or brought at a higher set point. The levels of FENO in patients with asthma are predominantly due to iNOS expression and activity in the airway epithelium [13], and vary depending on the inflammatory status of the airways [4]. We also found that lower IL-6 promoter methylation was associated with higher FENO, and that – as shown in multivariable regression – this association was independent from the methylation status of the iNOS promoter. Hypomethylation-related expression of IL-6 might therefore increase FENO by inducing iNOS expression either through mechanisms of gene expression control alternative to iNOS promoter demethylation or through activation at the post-transcriptional level.
Taken together, our findings indicate that demethylation of the IL-6 and iNOS promoters is a feature of increased airway inflammation that can be detected in nasal cell DNA. Based on our findings, we surmise that DNA methylation measured in nasal cell DNA may help capture epigenetic modifications that extend to the lower airways. McDougall et al. have previously reported correlations between nasal and lower airway epithelial cells in the expression of a number of mediators, including IL-6 [31]. Also, Sridhar et al. showed strong correlations between the nasal and bronchial epithelia in the mRNA levels of a subset of genes from a microarray platform that were expressed in the bronchial airways of healthy nonsmoking adult subjects [30]. However, these genes did not include those evaluated in our study. Additional research is warranted to determine whether DNA methylation in nasal cells collected by brushing is correlated with DNA methylation measured in other respiratory tissues. Whereas our results suggest that nasal cell DNA analysis is informative in the investigation of childhood asthma, and potentially of other respiratory conditions, previous large human investigations have mostly collected buccal and/or blood DNA. The present study was specifically focused on nasal cell DNA and did not include any collection of buccal or blood DNA. Therefore, we are unable to evaluate whether the DNA methylation associations that we found in nasal cell DNA can be also captured by using other frequently collected cell types.
Although we selected IL-6 and iNOS because of the established role of DNA methylation in programming their expression and their potential roles in relation with FENO and airways obstruction, inflammatory states in asthma depend on a much higher number of genes across different related pathways, and several of them would also represent strong candidates for DNA methylation studies. For instance, mouse studies have shown that inhaled diesel exposure and intranasal Aspergillus fumigatus allergen-induced hypermethylation at multiple sites of the IFN-γ promoter and hypomethylation at the CpG 408 site of the IL-4 promoter [45]. Altered methylation of both gene promoters was correlated significantly with changes in IgE levels [45]. Perera et al. recently reported that DNA methylation of the ACSL3 gene measured in umbilical cord white blood cell DNA from a subset of children in a cohort with high prevalence of asthma was related with polycyclic aromatic hydrocarbon exposure and with asthma symptoms in children prior to the age of 5 years [28]. In a mouse study, Hollingsworth et al. showed that a maternal diet rich in methyl donors was associated with greater levels of airway hyperactivity, airway eosinophilic inflammation, and IgE production in the F1 progeny, and partially in the F2 generation mice [46]. That same work discovered loci differentially methylated in lung tissues after in utero supplementation with a methyl-rich diet, including Runx3, a gene known to negatively regulate allergic airway inflammation. Future studies are therefore warranted to expand our findings on the IL-6 and iNOS promoters and evaluate DNA methylation of a larger number of genes.
Hypomethylation of Alu and LINE-1 repetitive elements has been previously found in peripheral blood leukocytes in response to risk factors for asthma and respiratory disease, such as exposure to airborne particulate matter [10,20] and pollutants from vehicular traffic [25]. In the present study, we found that nasal cell LINE-1 methylation showed a negative correlation with IL-6 and a positive correlation with iNOS methylation. Nonetheless, LINE-1 methylation did not show any association with FENO, FEV1 nd wheezing. The biological background underlying the associations of LINE-1 methylation with IL-6 and iNOS methylation is unclear. Alu methylation showed a moderate positive correlation with LINE-1 methylation, but not with IL-6 and iNOS methylation. Also, Alu methylation did not show any association with FENO, FEV1 and wheezing. It is worth noting that although Alu and LINE-1 methylation analyses have been proposed as surrogate measures of DNA methylation based on findings on cancer tissues [15,16], no evidence is available to suggest that Alu and LINE-1 methylation measures are associated with the global DNA methylation content in noncancerous DNA sources. Conversely, Choi et al. did not find any correlation between LINE-1 methylation and 5mC levels in blood leukocyte DNA from healthy individuals [47]. In addition, most Alu and LINE-1 methylation sites are extragenic and would therefore represent methylation levels in the vast proportion of the human genome composed by noncoding sequences, rather than that in the smaller proportion of gene-coding DNA [48].
Strengths & limitations
Our study was based on analysis of DNA methylation using pyrosequencing, which is highly reproducible and accurate at quantifying DNA methylation [15,25]. Whereas other DNA methylation techniques are only qualitative (i.e., produce a binary call of DNA methylation as negative or positive methylation), pyrosequencing analysis is quantitative and generates a measure of the proportion of cytosines that are methylated at a given CpG dinucleotide position within a sequence of interest. Therefore, the measure obtained, expressed as %5mC, reflects how many of the cells that contributed to the tested DNA were methylated in the specific CpG dinucleotides evaluated. To that extent, data from pyrosequencing analysis more adequately represent the control of gene expression, which would not be appropriately captured by qualitative methods. In addition, each of the DNA methylation assays we used in our study measured multiple individual CpG dinucleotides, which were averaged to obtain a mean measure for each marker, and were repeated twice on each sample to minimize the assay variability. We also designed our assays to evaluate DNA methylation in the promoters of IL-6 and iNOS. Recently, gene silencing by DNA methylation has been suggested to be associated also with methylation in regions up to 2 kb from the islands (i.e., on the island edges), termed CpG island shores [49,50]. Future studies are warranted to evaluate CpG shore methylation in children with asthma.
DNA methylation in the present study was limited to analyses on nasal cell DNA. We did not collect additional tissues, such as, for example, buccal cells or blood, that have been extensively used in studies on children and adults. Therefore, the correlation of nasal cell DNA methylation with those DNA sources could not be explored in our study. Also, we could not collect nasal cell samples using protocols that would have allowed for mRNA expression studies.
Our study was designed to evaluate whether DNA methylation was associated with FENO, FEV1 and wheezing within a group of children with asthma and did not include a group of healthy controls. Therefore, we were unable to identify differences in DNA methylation that are associated with asthma. Also, the present work did not consider potential effects from nearby sources of exposures. The presence of a major petrochemical plant and an oil-powered thermal plant in the area might have contributed to exacerbate asthmatic symptoms in these children. The study group included children between 8–11 years of age, who were Caucasians and predominantly males. The characteristics of the study participants limit the generalizability of our findings to other age ranges and ethnical groups, as well as to the female gender. In consideration of the small sample size we elected to report unadjusted analyses, although we performed sensitivity analyses that confirmed the results of the study. This is an additional limitation related to the small sample size of the study.
Conclusion
This study provides proof of principle that DNA methylation in nasal cell DNA is related with airway inflammation in a small group of children with current asthma. Whether our findings can be extended to children with different degrees of asthma severity remains to be determined. Our findings may establish a new model to obtain noninvasive measures of epigenetic marks using nasal cell DNA, which might prove useful for research and clinical purposes. Future studies are warranted to evaluate the determinants of DNA methylation changes that are associated with asthma inflammation and severity, as well as to further explore the role of DNA methylation in the natural history of asthma.
Executive summary.
DNA methylation is increasingly being proposed as a mechanism for underlying asthma-related inflammation.
Whether altered DNA methylation contributes to airway inflammation and obstruction in humans is largely unexplored.
In this work, we tested the use of DNA from nasal cells obtained noninvasevely through nasal brushing in 8–11 year old children with asthma.
We measured DNA methylation of candidate biomarkers using highly quantitative bisulfite-PCR-pyrosequencing.
Levels of exhaled nitric oxide, a measure of lower-airway inflammation, increased in association with lower promoter methylation of IL-6 and iNOS in nasal cell DNA.
Lower IL-6 methylation in nasal cell DNA was nonsignificantly associated with wheezing during the week of the study.
Alu and LINE-1 methylation levels were not associated with any of the outcomes.
Our findings support the use of nasal cell DNA for human epigenetic studies of asthma.
Footnotes
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
Funding was recieved from project ‘Environment and Health in Sicily’ – WHO Europe, Health Impact Assessment, Methods and Strategies, Rome Office and Regione Siciliana; PRIN20072S2HT8 Grant – Italian Ministry of Education and Scientific Research; ESTHER2007-5469 Grant – non-profit CARIPLO Foundation; and HSPH–NIEHS Center for Environmental Health New Investigator Fund (P30ES000002). The authors have no other relevant affilitions or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.USEPA. America’s Children and the Environment. EPA’s National Service Center for Environmental Publications; Research Triangle Park, NC, USA: 2007. [Google Scholar]
- 2.WHO. The Global Burden of Disease: 2004 Update. WHO; Geneva, Switzerland: 2008. [Google Scholar]
- 3.Holt PG, Sly PD. Non-atopic intrinsic asthma and the ‘family tree’ of chronic respiratory disease syndromes. Clin Exp Allergy. 2009;39(6):807–811. doi: 10.1111/j.1365-2222.2009.03258.x. [DOI] [PubMed] [Google Scholar]
- 4.Pijnenburg MW, De Jongste JC. Exhaled nitric oxide in childhood asthma: a review. Clin Exp Allergy. 2008;38(2):246–259. doi: 10.1111/j.1365-2222.2007.02897.x. [DOI] [PubMed] [Google Scholar]
- 5.Armenante F, Merola M, Furia A, Palmieri M. Repression of the Il-6 gene is associated with hypermethylation. Biochem Biophys Res Commun. 1999;258(3):644–647. doi: 10.1006/bbrc.1999.0566. [DOI] [PubMed] [Google Scholar]
- 6.Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthrit Rheum. 2008;58(9):2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 7.Chan GC, Fish JE, Mawji IA, Leung DD, Rachlis AC, Marsden PA. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J Immunol. 2005;175(6):3846–3861. doi: 10.4049/jimmunol.175.6.3846. [DOI] [PubMed] [Google Scholar]
- 8.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21(1):243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adcock IM, Ford P, Ito K, Barnes PJ. Epigenetics and airways disease. Respir Res. 2006;7:21. doi: 10.1186/1465-9921-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarantini L, Bonzini M, Apostoli P, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–222. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11▪.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180(5):462–467. doi: 10.1164/rccm.200901-0135OC. Identifies gene-specific methylation alterations associated with exposure to prenatal tobacco smoke. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.London SJ, Romieu I. Gene by environment interaction in asthma. Ann Rev Pub Health. 2009;30:55–80. doi: 10.1146/annurev.publhealth.031308.100151. [DOI] [PubMed] [Google Scholar]
- 13.Lane C, Knight D, Burgess S, et al. Epithelial inducible nitric oxide synthase activity is the major determinant of nitric oxide concentration in exhaled breath. Thorax. 2004;59(9):757–760. doi: 10.1136/thx.2003.014894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheller J, Ohnesorge N, Rose-John S. Interleukin-6 trans-signalling in chronic inflammation and cancer. Scand J Immunol. 2006;63(5):321–329. doi: 10.1111/j.1365-3083.2006.01750.x. [DOI] [PubMed] [Google Scholar]
- 15.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):E38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by methylight. Nucleic Acids Res. 2005;33(21):6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28(6):527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 18.Teneng I, Stribinskis V, Ramos KS. Context-specific regulation of LINE-1. Genes Cells. 2007;12(10):1101–1110. doi: 10.1111/j.1365-2443.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 19.Thomson SJ, Goh FG, Banks H, et al. The role of transposable elements in the regulation of IFN-λ1 gene expression. Proc Natl Acad Sci USA. 2009;106(28):11564–11569. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179(7):572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baccarelli A, Rienstra M, Benjamin EJ. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ Cardiovasc Genet. 2010;3(6):567–573. doi: 10.1161/CIRCGENETICS.110.958744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou L, Wang H, Sartori S, et al. Blood leukocyte DNA hypomethylation and gastric cancer risk in a high-risk polish population. Int J Cancer. 2010;127(8):1866–1874. doi: 10.1002/ijc.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright RO, Schwartz J, Wright RJ, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118(6):790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavanello S, Bollati V, Pesatori AC, et al. Global and gene-specific promoter methylation changes are related to anti-B[a] PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. 2009;125(7):1692–1697. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- 25.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67(3):876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 26.Baccarelli A, Tarantini L, Wright RO, et al. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics. 2010;5(3):222–228. doi: 10.4161/epi.5.3.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu ZZ, Hou L, Bollati V, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2010 doi: 10.1093/ije/dyq154. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28▪▪.Perera F, Tang WY, Herbstman J, et al. Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS ONE. 2009;4(2):E4488. doi: 10.1371/journal.pone.0004488. Identifies gene-specific methylation differences in cord blood DNA associated with child asthma phenotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talens RP, Boomsma DI, Tobi EW, et al. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J. 2010;24(9):3135–3144. doi: 10.1096/fj.09-150490. [DOI] [PubMed] [Google Scholar]
- 30.Sridhar S, Schembri F, Zeskind J, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomicss. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDougall CM, Blaylock MG, Douglas JG, Brooker RJ, Helms PJ, Walsh GM. Nasal epithelial cells as surrogates for bronchial epithelial cells in airway inflammation studies. Am J Respir Cell Mol Biol. 2008;39(5):560–568. doi: 10.1165/rcmb.2007-0325OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ISAAC. The International Study of Asthma and Allergies in Childhood: Phase 1 Manual. 2. ISAAC; Auckland, New Zealand: 1993. [Google Scholar]
- 33.Galassi C, Forastiere F, Biggeri A, et al. Sidria second phase: objectives, study design and methods. Epidemiol Prev. 2005;29(Suppl 2):S9–S13. [PubMed] [Google Scholar]
- 34▪.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. Biotechniques. 2003;35(1):152–156. doi: 10.2144/03351md02. Illustrates the use of pyrosequencing in DNA methylation analysis. [DOI] [PubMed] [Google Scholar]
- 35.Aparicio A, North B, Barske L, et al. LINE-1 methylation in plasma DNA as a biomarker of activity of DNA methylation inhibitors in patients with solid tumors. Epigenetics. 2009;4(3):176–184. doi: 10.4161/epi.4.3.8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nautiyal S, Carlton VE, Lu Y, et al. High-throughput method for analyzing methylation of CpGs in targeted genomic regions. Proc Natl Acad Sci USA. 2010;107(28):12587–12592. doi: 10.1073/pnas.1005173107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou L, Zhang X, Tarantini L, et al. Ambient PM exposure and DNA methylation in tumor suppressor genes: a cross-sectional study. Part Fibre Toxicol. 2011;8:25. doi: 10.1186/1743-8977-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.American Thoracic Society; European Respiratory Society. ATS/ERS recommendations for standardized procedures for the online and offline measurement of exhaled lower respiratory nitric oxide and nasal nitric oxide, 2005. Am J Respir Crit Care Med. 2005;171(8):912–930. doi: 10.1164/rccm.200406-710ST. [DOI] [PubMed] [Google Scholar]
- 39.Fonseca JA, Costa-Pereira A, Delgado L, et al. Pulmonary function electronic monitoring devices: a randomized agreement study. Chest. 2005;128(3):1258–1265. doi: 10.1378/chest.128.3.1258. [DOI] [PubMed] [Google Scholar]
- 40.Diggle P, Heagerty P, Liang KY, Zeger SL. Analysis of Longitudinal Data. 2. Oxford University; Oxford, UK: 2002. [Google Scholar]
- 41.Mccullagh P, Nelder J. Generalized Linear Models. 2. Chapman and Hall/CRC; FL, USA: 1989. [Google Scholar]
- 42.Goodman SN. Of p-values and bayes: a modest proposal. Epidemiology. 2001;12(3):295–297. doi: 10.1097/00001648-200105000-00006. [DOI] [PubMed] [Google Scholar]
- 43.Sterne JA, Davey Smith G. Sifting the evidence – what’s wrong with significance tests? BMJ. 2001;322(7280):226–231. doi: 10.1136/bmj.322.7280.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciccone G, Forastiere F, Agabiti N, et al. Road traffic and adverse respiratory effects in children. SIDRIA collaborative group. Occup Environ Med. 1998;55(11):771–778. doi: 10.1136/oem.55.11.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Ballaney M, Al-Alem U, et al. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicol Sci. 2008;102(1):76–81. doi: 10.1093/toxsci/kfm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hollingsworth JW, Maruoka S, Boon K, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118(10):3462–3469. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Choi JY, James SR, Link PA, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30(11):1889–1897. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48▪.Bollati V, Baccarelli A. Environmental epigenetics. Heredity. 2010;105(1):105–112. doi: 10.1038/hdy.2010.2. Discusses concepts related to epigenetic mechanisms as intermediates between acquired risk factors and human disease outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49▪▪.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. Describes CpG shores, newly identified regions that suggested to have particular relevance for gene expression control by DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roger T, Ding X, Chanson AL, Renner P, Calandra T. Regulation of constitutive and microbial pathogen-induced human macrophage migration inhibitory factor (MIF) gene expression. Eur J Immunol. 2007;37(12):3509–3521. doi: 10.1002/eji.200737357. [DOI] [PubMed] [Google Scholar]