Abstract
Glycopeptide antibiotics, including vancomycin, form complexes via a set of five hydrogen bonds with the acyl-l-Lys-d-Ala-d-Ala portion of the peptidyl stems of the bacterial cell wall peptidoglycan. This complexation deprives the organism from the ability to cross-link peptidyl stems of the peptidoglycan, leading to bacterial cell death. Four synthetic fragments as surrogates of the components of the bacterial cell wall have been prepared in our lab in multistep syntheses. These synthetic samples were used in investigations of the thermodynamics properties (ΔG°, ΔH°, and TΔS°) for the complexation with vancomycin by isothermal titration calorimetry (ITC). Complexation with the glycopeptide analogues are largely enthalpy-driven (formation of five hydrogen bonds) and in the analogues with a single peptidyl stem the complexation is 1:1. The complexation is more complicated with an approximately 2-kDa cell wall surrogate (compound 4), which possesses two peptidyl stems. The data were suggestive of interactions between the two vancomycin molecules, with an entropic penalty attributable to restriction of molecular movements within the complex due to restriction of motion of the highly mobile acyl-d-Ala-d-Ala moiety of the peptidyl stems. These data were reconciled with the recently determined NMR solution structure for the peptidoglycan fragment 4 and its implications for the larger cell wall.
Vancomycin, a representative member of the glycopeptide family of antibiotics, has become an important agent in treatment of infections caused by Gram-positive bacteria resistant to many antibiotics, of which methicillin-resistant Staphylococcus aureus (“MRSA”) is one important example.1,2 Understanding of the causes of antibiotic resistance and the mechanisms of action of the antibiotics are central in our ability to treat infections by these organisms.
The cell wall is critical for survival of bacteria. As such, the cell wall itself and its biosynthetic enzymes are targets of antibiotics. The backbone of the cell wall is made up of alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). The NAM residues are incorporated with a pentapeptidyl stem, terminating in acyl-d-Ala-d-Ala. This terminal acyl-d-Ala-d-Ala is the site of the critical cross-linking of individual peptidoglycan residues to each other.3,4 Both β-lactam and glycopeptide antibiotics interfere with the cross-linking step. β-Lactams are mechanism-based inhibitors of transpeptidases, enzymes that perform the cross-linking reaction.3–5 Glycopeptides form a non-covalent complex involving a network of five critical hydrogen bonds to the acyl-d-Ala-d-Ala portion of the individual cell-wall peptidoglycan, preventing the formation of the cross-linked product.6,7 Some glycopeptide antibiotics also inhibit transglycosylases.4,8
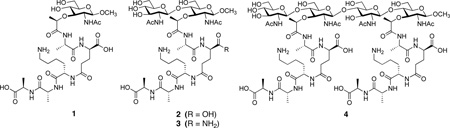
In our efforts toward understanding of the processes that involve the bacterial cell wall, we have undertaken the syntheses of its various components. Toward this goal, we have reported the preparation of compounds 1–4.9,10 Compound 4 has four alternating NAG and NAM. The NAM have been appended with the pentapeptidyl stem (NAM-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala) seen in many bacteria. The synthetic compounds 1 and 2 are smaller fragments of compound 4. In some S. aureus, the γ-d-Glu, is substituted with γ-d-Gln, and compound 3 was prepared to mimic this possibility. In this report we have investigated the thermodynamics of interactions of these mimics of cell wall structure (compounds 1–4) with vancomycin. The data indicate that enthalpy is the primary driving force in complexation between these cell wall surrogates with vancomycin. However, the larger peptidoglycan 4 interacts with vancomycin in a more complicated manner, involving cooperativity in binding of a second antibiotic, which is explained in light of the recently solved structure for the bacterial peptidoglycan.11
The earlier studies of the interactions of glycopeptide antibiotics with cell wall fragments have been limited to the use of diacetyl-l-Lys-d-Ala-d-Ala ((Ac)2KAA; see reference 7 and the citations therein). Compounds 1–4 have the full pentapeptidyl stem in their structures, and they each contain from one to four of the sugar residues. Each compound was prepared for this study in multistep syntheses, as reported.9,10 Preparation of 4 was the most challenging, a task accomplished in 37 synthetic steps.10
Affinity of (Ac)2KAA toward vancomycin in aqueous acetate buffer was determined previously,12 and thus it was used as a reference compound in our isothermal titration calorimetry (ITC) experiments. The values for the association constant (K) from earlier work and from our experimental ITC data (Table 1) are in a good agreement. Association of (Ac)2KAA with vancomycin is driven exclusively by the enthalpy change with some entropic drawback (Table 1).
Table 1.
Thermodynamic parameters for complexation of vancomycin with cell wall fragments in sodium acetate buffer, pH 4.7 (298 K). Results of microcalorimetric experiments are in a good agreement with 1:1 a complexation model for the first four entries and with 2:1 complexation model for compound 4.
| ligand | K (M−1) | ΔG° (kJ mol−1) |
ΔH° (kJ mol−1) |
TΔS (kJ mol−1) |
|---|---|---|---|---|
| (Ac)2KAA | (4.8±0.7) × 105 | −32.4 ± 0.4 | −40.1 ± 1.0 | −7.7 ± 1.0 |
| 1 | (6.6±1.0) × 105 | −33.2 ± 0.4 | −40.2 ± 1.0 | −7.0 ± 1.0 |
| 2 | (5.4±0.8) × 105 | −32.7 ± 0.4 | −39.2 ± 1.0 | −6.5 ± 1.0 |
| 3 | (2.5±0.3) × 105 | −30.8 ± 0.3 | −40.2 ± 1.0 | −9.4 ± 1.0 |
| 4 (1st) | (1.1±0.2) × 106 | −34.5 ± 0.5 | −41.2 ± 1.0 | −6.7 ± 1.0 |
| 4 (2nd) | (4.0±0.7) × 105 | −32.0 ± 0.5 | −45.4 ± 1.0 | −13.4 ± 1.0 |
Peptidoglycans 1 and 2 exhibit similar thermodynamic parameters for complexation with vancomycin, when compared with (Ac)2KAA, indicating that the saccharide units of 1 and 2 do not participate directly in complex formation. However, conversion of the negatively charged γ-d-Glu (as in 1 and 2) to uncharged γ-d-Gln (as in 3) resulted in reduction in affinity of approximately two-fold, which is due to an entropic contribution. This entropic difference is likely due to differences in solvation of the carboxylate vs the amide. The associations of these peptidoglycan derivatives with vancomycin fit to a 1:1 complex between the two.
Interestingly, the situation with peptidoglycan 4 was quite different. Experimental ITC data indicate two distinct binding events. A 1:1 complex formation between one vancomycin and 4 was followed by binding of an additional vancomycin molecule to the 1:1 complex. A distinctive pattern of experimental data points allowed reliable determination of association constants and thermodynamics parameters for the first and the second binding events from the same titration curve. The association constant for the first vancomycin binding event to 4 increased by slightly over two-fold, compared to the related peptidoglycans 1 and 2, a predominantly enthalpy-driven process. This observation might suggest the existence of certain weak electrostatic interactions of the second peptidyl stem of 4 with vancomycin. Significantly, we observe an additional favorable enthalpic increment and a more unfavorable entropic contribution for the second step events (i.e., complex of 4-vancomycin + vancomycin). These data are suggestive of contact between two vancomycin molecules (gain of enthalpy due to additional electrostatic interactions such as hydrogen-bonding). The entropic penalty on the other hand can be attributed to restriction of molecular movements within the complex due to restriction of motion of the highly mobile acyl-d-Ala-d-Ala moiety of the peptidyl stem (elaborated below).
The pioneering early work has revealed that glycopeptide antibiotics, including vancomycin, are prone to dimerization.7,13 However, the cooperativity that we see in the two binding events of vancomycin to 4 is not due to dimerization. These experiments were done under conditions that did not favor dimer formation (concentrations of vancomycin below the equilibrium constant for the dimerization). Nonetheless, it would appear that the first binding event influences the second. This information on complexation is revealing in light of the solution (NMR) structural information that has recently emerged for peptidoglycan 4 from our laboratory.11
Glycopeptide 4 is a dimer of the repeating unit of the bacterial cell wall (NAG-NAM-peptide) and the three sets of torsion angles that define the rotamers for the glycosidic bonds of 4 were shown to be constant in the structure. The highly regular structure of 4 allowed the construction of a larger model for the peptidoglycan with the preservation of the requisite glycosidic torsion angles.11 The structure of the peptidoglycan model conforms to a right-handed helix with respect to the saccharide backbone. The pentapeptidyl stems project outward from the central helical motif. Whereas the helical motif of the saccharide backbone is stable, in the peptide only the l-Ala-γ-d-Glu portion of the peptidyl stem conforms to some minimal structure; the acyl-l-Lys-d-Ala-d-Ala portion of the peptidyl stem apparently does not conform to any three-dimensional structure (absence of NOE measurements in NMR experiments and dynamics simulations).11 As such, binding of two vancomycins to glycopeptide 4 restricts the motions of both the vancomycin molecules and the acyl-l-Lys-d-Ala-d-Ala moieties in both peptides, consistent with the entropic penalty documented in Table 1.
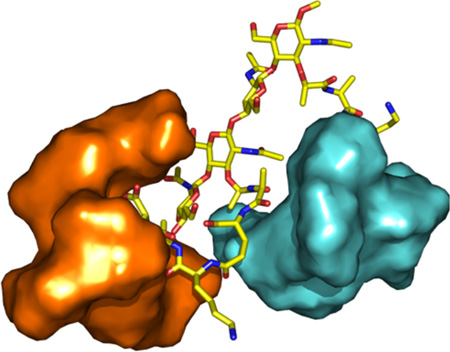
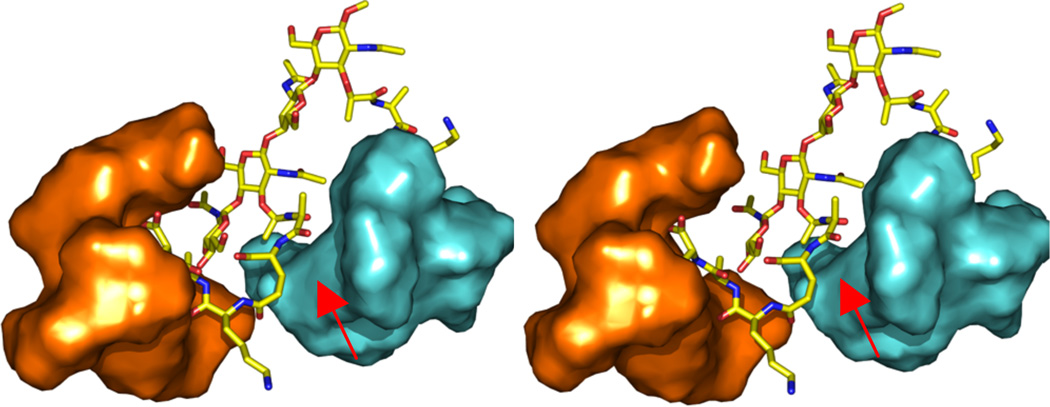
The NMR structure of compound 4 complexed to two vancomycins was constructed and submitted to explicit-solvent molecular dynamics simulations over the course of 12 ns (MPEG animation in Supplemental Information). In the early stages of the trajectory, the vancomycin molecules were distal from each other. At approximately 5 ns the vancomycin molecules were seen approaching each other, culminating in the structure that is shown in Fig. 1 by 6 ns of the simulation. This structure remained stable to the end of the trajectory (12 ns). This complex reveals one vancomycin molecule (cyan) embracing the pentapeptide (from the d-lactyl moiety to d-Glu; see red arrow) that is in complex with the other vancomycin (orange). This interaction would lead to a decrease in motion experienced by the system and hence the entropic penalty evaluated experimentally for the 2:1 complex (−13.4 kJ mol−1), when compared to the 1:1 complex (−6.7 kJ mol−1).
Figure 1.
Stereoview of the complex of compound 4 with two vancomycin molecules. Compound 4 is shown in capped-sticks representation, colored according to atom types (C, N, O in yellow, blue, and red). The vancomycin molecules are shown in Connolly solvent-accessible surfaces, colored in orange and cyan. The red arrow is used to point to the region of interaction between peptide and the second vancomycin.
The data presented here do not exclude the possibility of a single vancomycin interacting with a single peptidoglycan, or a dimer of vancomycin binding two separate peptidoglycans (binding of dimer to two peptide stems in one peptidoglycan is impossible based on the NMR structure). However, the ITC and the molecular dynamics results confirm the cooperative binding ability of two vancomycin molecules on the cell wall mimic, an event that is likely to take place in vivo based on the favorable thermodynamics.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health.
Footnotes
Supporting Information Available: Microcalorimetric titration experiments condition and titration curves and computational procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sakoulas G, Moellering RC, Eliopoulos GM. Clin. Infec. Dis. 2006;42:S40–S50. doi: 10.1086/491713. [DOI] [PubMed] [Google Scholar]; Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chambers HF. Clin. Microbiol. Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vancomycin-resistant Staphylococcus aureus. Morbidity Mortality Weekly Report 2002. Vol. 51. Pennsylvania: Centers for Disease Control and Prevention; 2002. p. 931. [Google Scholar]; Vancomycin-resistant Staphylococcus aureus. Morbidity Mortality Weekly Report 2004. Vol. 53. New York: Centers for Disease Control and Prevention; 2004. pp. 322–323. [PubMed] [Google Scholar]; Bartley J. Infect. Control Hosp. Epidemiol. 2002;23:480. doi: 10.1017/s0195941700082333. [DOI] [PubMed] [Google Scholar]; Walsh TR, Bolmstrom A, Qwarnstrom A, Ho P, Wootton M, Howe RA, MacGowan AP, Diekema D. J. Clin. Microbiol. 2001;39:2439–2444. doi: 10.1128/JCM.39.7.2439-2444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher JF, Meroueh SO, Mobashery S. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]; Lee W, McDonough MA, Kotra LP, Li ZH, Silvaggi NR, Takeda Y, Kelly JA, Mobashery S. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1427–1431. doi: 10.1073/pnas.98.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahne D, Leimkuhler C, Wei L, Walsh C. Chem. Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 5.Tipper DJ, Strominger JL. Proc. Natl. Acad. Sci. U. S. A. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh CT. Science. 1993;261:308–309. doi: 10.1126/science.8392747. [DOI] [PubMed] [Google Scholar]; Williams DH, Stephens E, O'Brien DP, Zhou M. Angew. Chem. Int. Ed. Engl. 2004;43:6596–6616. doi: 10.1002/anie.200300644. [DOI] [PubMed] [Google Scholar]
- 7.Williams DH, Bardsley B. Angew. Chem. Int. Ed. Engl. 1999;38:1173–1193. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1172::AID-ANIE1172>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 8.Silver LL. Curr. Opin. Microbiol. 2003;6:431–438. doi: 10.1016/j.mib.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Fuda C, Hesek D, Lee M, Morio K, Nowak T, Mobashery S. J. Am. Chem. Soc. 2005;127:2056–2057. doi: 10.1021/ja0434376. [DOI] [PubMed] [Google Scholar]; Hesek D, Suvorov M, Morio K, Lee M, Brown S, Vakulenko SB, Mobashery S. J. Org. Chem. 2004;69:778–784. doi: 10.1021/jo035397e. [DOI] [PubMed] [Google Scholar]
- 10.Hesek D, Lee MJ, Morio KI, Mobashery S. J. Org. Chem. 2004;69:2137–2146. doi: 10.1021/jo035583k. [DOI] [PubMed] [Google Scholar]
- 11.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper A, McAuley-Hecht KE. Philo. Trans. R. Soc. London A. 1993;345:23–35. [Google Scholar]; Williamson MP, Williams DH, Hammond SJ. Tetrahedron. 1984;40:569–577. [Google Scholar]; Nieto M, Perkins HR. Biochem. J. 1971;123:773–787. doi: 10.1042/bj1230773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderone CT, Williams DH. J. Am. Chem. Soc. 2001;123:6262–6267. doi: 10.1021/ja003016y. [DOI] [PubMed] [Google Scholar]; Kerns R, Dong SD, Fukuzawa S, Carbeck J, Kohler J, Silver L, Kahne D. J. Am. Chem. Soc. 2000;122:12608–12609. [Google Scholar]; Williams DH, Maguire AJ, Tsuzuki W, Westwell MS. Science. 1998;280:711–714. doi: 10.1126/science.280.5364.711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.