Abstract
Crystal structures of insulin are remarkable for a long-range reorganization among three families of hexamers (designated T6, , and R6). Although these structures are well characterized at atomic resolution, the biological implications of the TR transition remain the subject of speculation. Recent studies indicate that such allostery reflects a structural switch between distinct folding-competent and active conformations. Stereospecific modulation of this switch by corresponding d- and l-amino-acid substitutions yields reciprocal effects on protein stability and receptor-binding activity. Naturally occurring human mutations at the site of conformational change impair the folding of proinsulin and cause permanent neonatal-onset diabetes mellitus. The repertoire of classical structures thus foreshadows the conformational lifecycle of insulin in vivo. By highlighting the richness of information provided by protein crystallography—even in a biological realm far removed from conditions of crystallization—these findings validate the prescient insights of the late D. C. Hodgkin. Future studies of the receptor-bound structure of insulin may enable design of novel agonists for the treatment of diabetes mellitus.
I. Introduction
Insulin is a small globular protein containing two chains, A (21 residues) and B (30 residues) (Fig. 2.1A). Stored in the β cell as a Zn2+-stabilized hexamer, the hormone dissociates in the bloodstream to function as a Zn2+-free monomer (Fig. 2.1B). The structure of the free hormone has been well characterized by X-ray crystallography and NMR spectroscopy (Baker et al., 1988; Hua et al., 1996b; Olsen et al., 1996). Key receptor-binding contacts (Baker et al., 1988; Blundell et al., 1972; Liang et al., 1994; Pullen et al., 1976) have been defined by rare naturally occurring mutations (positions A3, B24, and B25; red in Fig. 2.1A) associated with impaired activity and diabetes mellitus (Shoelson et al., 1983). Classical structure–function relationships were originally inferred from patterns of sequence conservation (Baker et al., 1988) and subsequently tested through systematic studies of insulin analogs (Baker et al., 1988; Hu et al., 1993; Huang et al., 2004; Kitagawa et al., 1984; Liang et al., 1994; Mirmira and Tager, 1989; Mirmira et al., 1991; Nakagawa and Tager, 1986, 1992; Nakagawa et al., 2000; Pullen et al., 1976; Xu et al., 2002b). Despite an abundance of such data and complementary advances in characterization of the insulin receptor (IR) (Lou et al., 2006; McKern et al., 2006), a molecular understanding of how insulin binds to its receptor has remained elusive (Ward et al., 2008).
Figure 2.1.
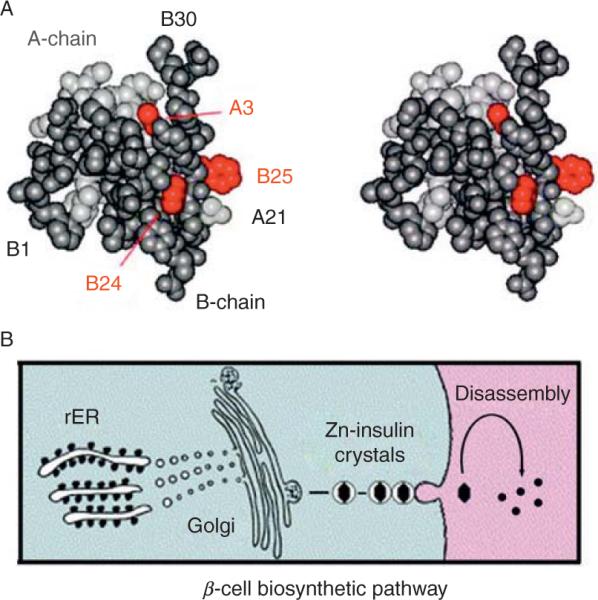
Globular structure of an insulin monomer and pathway of insulin biosyn-thesis. (A) Space-filling model of an insulin monomer highlighting sites of classical diabetes-associated mutations (red): ValA3→Leu, PheB24→ Ser, and PheB25→Leu (Shoelson et al., 1983). The A- and B-chains are otherwise shown in light and dark gray, respectively. Atomic coordinates were obtained from Protein Databank entry 4INS (2-Zn molecule 1). (B) Nascent proinsulin folds as a monomer in ER wherein zinc-ion concentration is low; in Golgi apparatus zinc-stabilized proinsulin hexamer assembles. The prohormone is processed by cleavage of connecting peptide in post-Golgi vescicles to yield mature insulin. Zinc-insulin crystals are observed in secretory granules. Insulin hexamers dissociate in the bloodstream to release active zinc-free monomers.
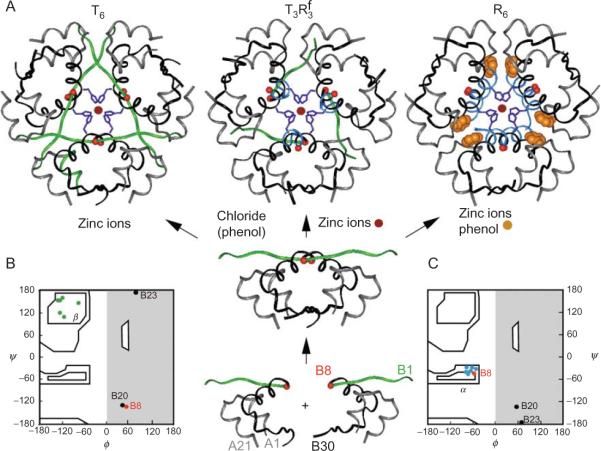
A rigorous foundation for studies of insulin folding, assembly, and dynamics has been provided by an extensive database of crystal structures (Adams et al., 1969; Bentley et al., 1976; Brader and Dunn, 1991; Ciszak et al., 1995; Derewenda et al., 1989). Such studies have spanned more than four decades and have played a central role in the history of structural biology. The structure of porcine insulin as a zinc-stabilized hexamer was first determined by the late Dorothy C. Hodgkin and coworkers in the 2-Zn form (Adams et al., 1969). This celebrated structure, stabilized by two axial zinc ions, is now designated T6. A second crystal form, known as 4-Zn insulin, was obtained in high concentrations of chloride ion by Schlichtkrull (1958). Now designated , the structure of this variant zinc hexamer was described by the Hodgkin laboratory (Bentley et al., 1976) and extended to the corresponding structure of human insulin (Smith et al., 1984). These two rhombohedral crystal forms exhibit extensive structural differences, designated the TR transition. (This nomenclature was adopted in honor of Max Perutz but is unrelated to the cooperative mechanism of oxygen binding to hemoglobin.) In 1989 this transition was observed in complete form by Dodson and coworkers in an R6 phenol-stabilized zinc insulin hexamer (Derewenda et al., 1989). The TR transition can be observed in crystals (Bentley et al., 1978) and monitored in solution by spectroscopy (Roy et al., 1989; Thomas and Wollmer, 1989).
The three families of insulin hexamers (T6, , and R6) are shown in Fig. 2.2A (top row). In the T-state conformation, the A-chain contains an N-terminal α-helix (residues A1–A8) followed by a noncanonical turn, second helix (A12–A18), and C-terminal segment (A19–A21); the B-chain contains an N-terminal segment (residues B1–B6), type II′ β-turn (B7–10), central α-helix (B9–B19), type I β-turn (B20–B23), and C-terminal β-strand (B24–B28), extended by less well-ordered terminal residues B29 and B30. In the R-state, the N-terminal portion of the B-chain participates in a single long α-helix. The resulting B1–B19 α-helix (or B3–B19 in the frayed Rf state) projects from the globular core of the protomer to make extensive hexamer contacts, including formation of specific phenol-binding pockets at symmetry-related trimer interfaces (Derewenda et al., 1989). This change in the secondary structure of the B-chain is highlighted in Fig. 2.2A: the T-specific conformation of the B1–B8 is shown in green, the R-specific conformation in powder blue. Interconversion among the three families of hexamers (T6, , and R6) is regulated by ionic strength (Bentley et al., 1976; Blundell et al., 1971) and the binding of small cyclic alcohols (Derewenda et al., 1989; Krebs et al., 2005) (arrows in Fig. 2.2). Crystal structures of zinc-free insulin dimers and monomeric fragments exhibit T-family features (Bi et al., 1983, 1984; Whittingham et al., 2006). Together, this set of structures has established a pathway of insulin assembly (central panel of Fig. 2.2).
Figure 2.2.
Structural families of insulin hexamers. (A) Left to right, ribbon models of T6, , and R6 zinc insulin hexamers. Central panel indicates pathway of insulin assembly and regulators of conformational reorganization (arrows). The Cα positions of GlyB8 are indicated by red balls; the variable secondary structures of the N-terminal segment of the B-chain (residues B1–B7) are shown highlighted in green (extended in T-state) or powder blue (α-helical in R-state). The B-chain is otherwise shown in black, and A-chain in gray. The side chains of HisB10 in the metal-ion binding sites are shown in dark blue; zinc ions in magenta; and phenol in burnt amber. (B and C) The TR transition is associated with a conformational change of GlyB8 from right to left in the Ramachandran plot. Main-chain dihedral conformations of the three glycines in the B-chain (residues B8, B20, and B23) and residues B2–B8 in a representative T-state (B) or R-state (C) protomer. The conformation of GlyB8 is indicated by red; black circles indicate GlyB20 and GlyB23. Residues B2–B7 are shown in green (in β-region in T-state) or blue (within α-helical island in R-state).
Structural variation among crystal structures of insulin has provided an important model of conformational change and protein flexibility (Fig. 2.3) (Chothia et al., 1983). Yet the specific biological relevance of the TR transition, if any, has remained a matter of speculation. On the one hand, NMR studies have established that an insulin monomer in solution resembles the T-state (Hua et al., 1991, 1996a; Olsen et al., 1996). On the other hand, a variety of evidence suggests that the hormone undergoes a conformational change on receptor binding (Derewenda et al., 1991; Hua et al., 1991). In this chapter, we explore the possible biological relevance of the TR transition. Evidence is reviewed that the choreography of conformational changes in the crystalline state, in fact, identifies a functional switch (Hua et al., 2006b; Nakagawa et al., 2005; Wan et al., 2008). We envisage that this switch operates to enable both the productive folding of proinsulin in the endoplasmic reticulum (ER) of pancreatic β-cells and binding of the mature hormone to the IR. Remarkably, naturally occurring mutations in the human insulin gene have recently been identified that lead to impairment of this switch and in turn to permanent neonatal-onset diabetes mellitus (Stoy et al., 2007). These findings suggest that the lessons of protein structures can extend far beyond the immediate conditions of crystallization to inform a broad range of biological phenomena.
Figure 2.3.
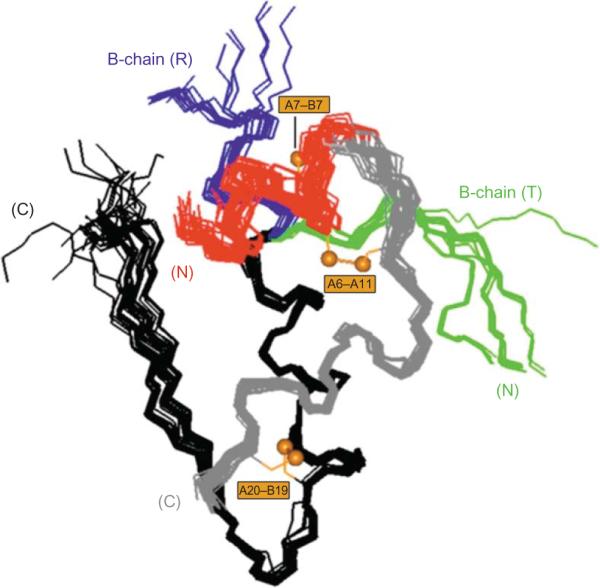
Structural variation among crystallographic protomers. Superposition of crystallographic protomers (15 T states and 15 R states). The structures were aligned according the main-chain atoms of residues B9–B24 and A12–A21. The A1–A8 α-helix in each protomer is shown in red; the variable secondary structure of the N-terminal segment of the B-chain is shown in green (extended in T state; right) or blue (extended α-helix in R state; left). Structures were obtained from the following entries in the Protein Data Bank: (T states), 4INS, 1APH, 1BPH, 1CPH, 1DPH, 1TRZ, 1TYL, 1TYM, 2INS, 1ZNI, 1LPH, 1G7A, 1MSO; (R states), 1EV6, 1ZNJ, 1TRZ, 1ZNI, 1LPH.
II. Structure–Activity Relationships
A. Chiral mutagenesis of insulin
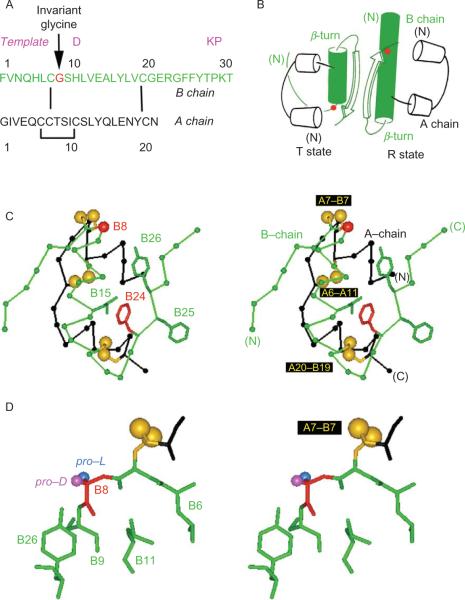
The crux of the TR transition occurs at the junction between the N-terminal segment of the B-chain and its central α-helix. In the T-state, this junction forms a type II′ β-turn comprising residues B7–B10. The local structure of this turn requires a glycine at position B8 (arrow in Fig. 2.4A) whose main-chain (ϕ, ψ) dihedral angles lie on the right-hand side of the Ramachandran plane (ϕ > 0; red circle in Fig. 2.2B). This conformation is ordinarily “forbidden” to l-amino acids. By contrast, in the R-state GlyB8 adopts a negative ϕ angle as expected within the interior of an α-helix (Fig. 2.2C). In either protomer, GlyB8 lies on the protein surface adjacent to CysB7 (part of the canonical cystine A7–B7; this disulfide bridge provides a link between the junctional B-chain element and the A-chain (Fig. 2.4C)). The contrasting structural environments of GlyB8 in the T- and R-states are shown in Fig. 2.4B (red circles). Glycine is invariant at this site among mammalian insulins, insulin-like growth factors, and relaxin-like polypeptides (Blundell and Humbel, 1980).
Figure 2.4.
Conformation of GlyB8 in a T-state-specific α-turn. (A) Sequence of B chain (top) and A chain (bottom); arrow indicates invariant GlyB8 (red). Shown above B chain in magenta are the three substitutions in the monomeric DKP template. (B) Cylinder models of TR dimer based on crystal structure of zinc insulin hexamers (PDB ID: 1TRZ). The T state is at left and R state at right. B-chain α-helices are shown in green; the α-carbons of GlyB8 are shown as red circles. Three families of hexamers have been characterized, designated T6, , and R6. The R-state conformation has only been observed within hexamers. (C) Structure of insulin T state (stereo pair) showing positions of selected side chains (labeled at left) relative to GlyB8 Cα (red) and disulfide bridges (gold; labeled at right). The B chain is shown in green, and A chain in black. (D) Structure of T-state-specific B7–B10 β-turn (stereo pair). Main chain of GlyB8 is shown in red; its pro-l and pro-d Hα atoms are highlighted in blue and magenta, respectively. This figure is reprinted from Hua et al. (2006b).
In a recent series of studies “chiral mutagenesis”—comparison of corresponding d- and l amino-acid substitutions—has been employed to probe the functional importance of GlyB8 (Nakagawa et al., 2005). The essential idea is that d or l substitutions impose a reciprocal bias on the setting of the B8 ϕ angle and may hence modulate the conformational equilibrium between T- and R states. Respective sites of l and d substituents are indicated in blue (the pro-l Hα of GlyB8) or magenta (pro-d Hα) (Fig. 2.4D). Substitution of GlyB8 by l-amino acids was observed to impair the folding of a single-chain insulin precursor (Guo et al., 2005) and to impede disulfide pairing in insulin chain combination (Nakagawa et al., 2005). Substitution of this turn-specific “d-glycine” by a d-amino acid by contrast was observed to augment the stability of the native T-like fold. Remarkably, whereas unstable l-analogs can be highly active, d-analogs exhibit reduced binding to the IR. The extent of impairment caused by d-amino-acid substitutions at B8 (between 102- and 103-fold) is more marked than is usually observed on single amino-acid substitutions at other sites. Such loss of activity is associated with impairment of the TR transition in crystallographic and NMR studies of hexameric assemblies of d-AlaB8-insulin (Nakagawa et al., 2005). Crystal structures of zinc insulin hexamers containing representative d- or l amino-acid substitutions at B8 nonetheless demonstrate that either chirality can be accommodated within such assemblies (Weiss and Dodson, unpublished results).
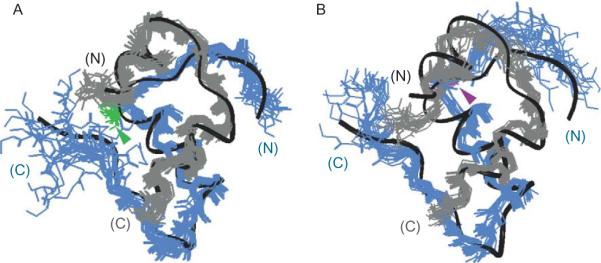
The structural basis of reciprocal stereospecific effects of B8 substitutions was investigated through comparative studies of diastereomeric analogs containing either d- or l-SerB8 within an engineered T-like monomer (Hua et al., 2006b). (Such engineering is generally required to avoid dimerization and higher-order protein assembly (Brange et al., 1988), which would otherwise limit the feasibility of NMR analysis (Weiss et al., 1991)). Although the NMR-derived structures of the analogs each resemble wild-type insulin, the unstable but active l-SerB8 variant exhibits greater dynamic flexibility (Fig. 2.5). Enhanced flexibility or imprecision in the ensemble of l-specific structures is associated with an attenuated far-ultraviolet circular dichroism (CD) spectrum (Hua et al., 2006b). Since the CD spectrum of insulin primarily reflects its α-helical subdomain and since the B8 site of substitution in a T-like monomer is peripheral to this subdomain, the attenuated CD spectrum of the l-SerB8 analog indicates a global change in protein dynamics. The CD spectrum of the d-SerB8 analog is by contrast essentially identical to that of the parent T-like monomer (Hua et al., 2006b). Studies of amide proton exchange in D2O suggest that D substitutions damp local conformational fluctuations near the site of substitution (Hua and Weiss, unpublished results).
Figure 2.5.
NMR-derived solution structures of d-SerB8 and l-SerB8 analogs of an engineered insulin monomer. Front and back views of (A) d-SerB8-DKP-insulin and (B) l-SerB8-DKP-insulin. In each case the A chain is shown in gray, and B chain in blue. The d- and l-SerB8 side chains are shown in green and purple, respectively (arrow-heads). Ribbons indicate mean structure of DKP-insulin. This figure is reprinted from Hua et al. (2006b).
Together, the above studies highlighted the dual importance of GlyB8. On the one hand, a T-like conformation (with positive ϕ angle) is required for initial disulfide pairing and for thermodynamic and dynamic stability of the T-like monomer once folding is achieved. On the other hand, stereo-specific enhancement of such stability by d-amino-acid substitutions impedes receptor binding. Further, whereas d-amino-acid substitutions at B8 impede disulfide pairing and impair the stability of the T-like monomer, the high activity of such unstable analogs suggests that a local R-like conformation (with negative ϕ angle) is required for biological activity. We, therefore, suggest that GlyB8 recapitulates local aspects of the TR transition to function a switch between folding-competent and active conformations.
B. Uncoupling activity from allostery
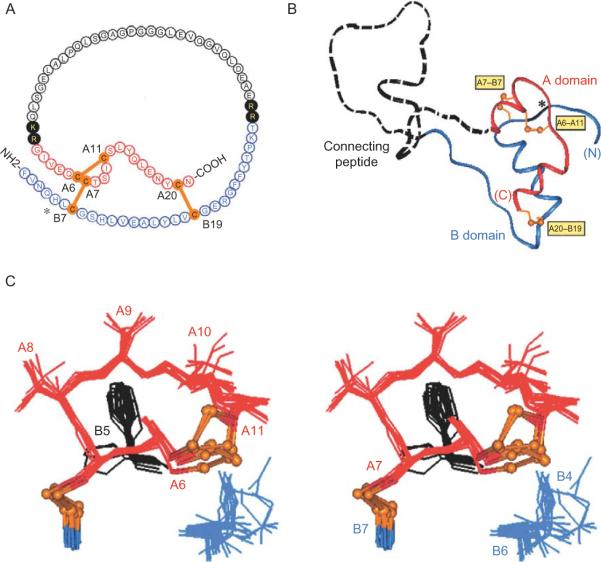
Interpretation of crystallographic studies of insulin is limited by its state of assembly. Whereas crystal structures feature zinc-free dimers or zinc-stabilized hexamers, the hormone circulates in the bloodstream and binds to target cells as a monomer (Fig. 2.1B). Local modulation of the B8 dihedral angle by stereospecific substitutions (discussed earlier) does not address whether global aspects of the TR transition may pertain to the insulin monomer. As a further test of the relationship between the TR transition and biological activity, a recent study has exploited a species variant at position B5 (Wan et al., 2008). In human insulin as in other eutherian mammals residue B5 is conserved as histidine (Fig. 2.6A). Attention was focused on its substitution by Arg (as observed in some hystricomorph mammals, birds, and reptiles) because of the distinctive structural environments of HisB5 in wild-type insulin hexamers. In the T-state the imidazole ring packs within a solvated crevice at the edge of the protein surface (Fig. 2.6C; Baker et al., 1988) whereas in the R-state the side chain packs at an internal interface between dimers. Although this interface is in part solvated, NMR studies of the pKa of HisB5 suggested that an R-specific interface would be destabilized by the uncompensated positive charge of an ArgB5 substituent. In accord with this prediction, spectroscopic results demonstrated that substitution of HisB5 by Arg impedes the TR transition in solution. Further, crystals of ArgB5-insulin grown under conditions leading to stabilization of wild-type R6 hexamers (i.e., in the presence of phenolic ligands) were observed to contain only T6 hexamers. The crystal structure of ArgB5-insulin as a variant T6 hexamer, determined at a resolution of 1.3 Å, was found to be essentially identical to that of the wild-type T6 hexamer (Fig. 2.7A and B), including analogous interactions by the wild-type variant B5 side chain (HisB5 and ArgB5) (Wan et al., 2008). The corresponding packing of HisB5 and ArgB5 in similar inter-chain pockets is illustrated in Fig. 2.7C and D, respectively. Because ArgB5-human insulin binds well to the IR (with affinity ca. 40% relative to wild-type human insulin), these results collectively demonstrate that competence to undergo the TR transition in a hexamer is not required for the biological activity of an insulin monomer. These findings are restricted to zinc insulin hexamers and so do not address whether competence of an insulin monomer to adopt an R-like conformation is required for high-affinity receptor binding. Whereas interfacial substitutions such as ArgB5 may be regarded as extrinsic probes of protein allostery, d and l substitutions at B8 modulate the intrinsic conformational repertoire of the monomer in relation to both the TR transition and receptor binding.
Figure 2.6.
Structure of proinsulin and T-state-specific environment of HisB5. (A) Sequence of human proinsulin: insulin moiety is shown in red (A chain) and blue (B chain). The connecting region is shown in black: flanking dibasic cleavage sites (filled circles) and C-peptide (open circles). (B) Structural model of insulin-like moiety and disordered connecting peptide (dashed line). Cystines are labeled in yellow boxes. (C) Structural environment of HisB5 within A-chain-related crevice. Structures are drawn from T-state coordinates given by PDB code in legend to Figure 2.3. This figure is reprinted from Hua et al. (2006a).
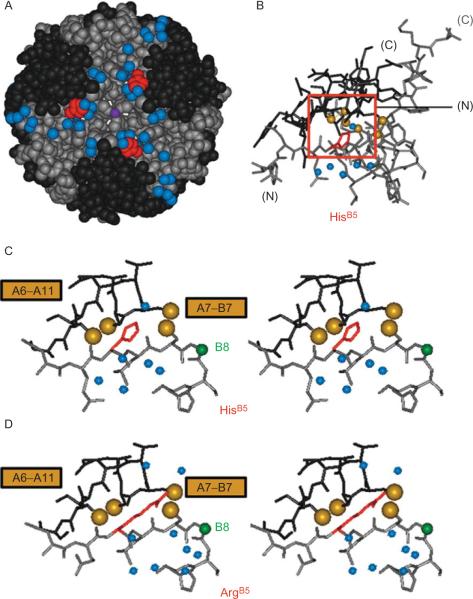
Figure 2.7.
Structural environment of residue B5 in T6 hexamer and T-state protomers. (A) Spacing filling model of T6 hexamer showing side chain of HisB5 (red) lying along protein surface near bound water molecules (blue). (B) Stick model of single T-state protomer with B-chain in black and A-chain in gray. Red box encloses environment of HisB5 (red) in inter-chain crevice near A7–B7 and A6–A11 disulfide bridges (sulfur atoms shown in gold). (C) Expansion of boxed region in panel B providing stereo view of packing of HisB5. (D) Corresponding stereo view of ArgB5 (red) in crystallographic T-state protomer. Analogous side-chain NH functions of HisB5 and ArgB5 are near the main-chain of the A-chain. Cystine A7–B7 in each case lies on the protein surface whereas cystine A6–A11 packs within the core of the protomer. Bound water molecules near respective B5-related crevices are shown as blue spheres. This figure is reprinted from Wan et al. (2008).
III. Implications for the Genetics of Diabetes Mellitus
Insulin chain combination, developed more than four decades ago by Katsoyannis and colleagues (with independent and important contributions from the laboratories of Brandenburg and Zahn in Aachen, Germany, and from the laboratories of Cao and Gong in Shanghai, China) has provided a robust synthetic route to the preparation of insulin analogs (for review, see Katsoyannis, 1966). Despite its general success, chain combination fails in the case of selected substitutions in the A- or B-chains (Hu et al., 1993; Hua et al., 2002). Examples of a block to disulfide pairing are provided by substitutions at positions B5 and B8. Very low yields were encountered on substitution of HisB5 by Ala or Met (Hua et al., 2006a; Wan et al., 2008) and on substitution of GlyB8 by any l-amino acid (Hua et al., 2006b; Nakagawa et al., 2005). The origin of these synthetic blocks is not well understood. Because efficient syntheses of unstable insulin analogs have previously been reported (Hua et al., 2002; Weiss et al., 2000; Xu et al., 2002a), we have hypothesized that unfavorable substitutions at B5 or B8 imposes a kinetic barrier to disulfide pairing. It is possible that in a reaction intermediate a productive orientation of CysB7 (and in turn its alignment with CysA7) requires (i) nascent interactions between the B5 side chain and the A-chain (resembling those observed in the native T-state; Figs. 2.6C and 2.7C) and (ii) a positive dihedral angle at B8 (as in the native T-state or as stabilized by d-amino-acid substitutions; Fig. 2.1B). The crystal structure of ArgB5-insulin (discussed earlier) may thus have implications beyond the TR transition: HisB5 and ArgB5 both contain nitrogenous side chains able to form specific hydrogen bonds to the main-chain of the A-chain (Fig. 2.7C and D). The biological relevance of these chemical findings is supported by their extension to corresponding studies of the biosynthetic expression of proinsulin variants and single-chain insulin analogs in eukaryotic cells (Guo et al., 2005; Hua et al., 2006a).
Recent advances in human genetics have identified a large collection of clinical mutations in the insulin gene causing permanent neonatal-onset diabetes mellitus (Colombo et al., 2008; Edghill et al., 2008; Molven et al., 2008; Polak et al., 2008; Stoy et al., 2007). The majority of such mutations add or remove a cysteine from proinsulin, leading to an odd number of thiol groups and hence protein misfolding in the ER. Although the crystal structure of proinsulin has not been determined, a variety of spectroscopic evidence indicates that it consists of a folded insulin-like domain and unfolded connecting peptide (Fig. 2.6B). Misfolding of proinsulin in the ER presumably activates the unfolded protein response (UPR), leading to chronic ER stress and apoptosis of β-cells. Such a mechanism would rationalize the onset of apparent type 1 diabetes mellitus prior to maturation of an immune system capable of mounting an autoimmune attack (Stoy et al., 2007). This hypothesis is supported by the observation of a human mutation associated with neonatal diabetes mellitus that recapitulates a classical diabetes-associated mutation in the mouse (the Akita mouse; Araki et al., 2003; Oyadomari et al., 2002; Ron, 2002) in which CysA7 is substituted by Tyr. Affected patients are heterozygous, implying that misfolding of the variant proinsulin perturbs the folding and trafficking of the wild-type polypeptide. A subset of mutations may lead to less severe folding defects, leading to the onset of diabetes mellitus later in life (Edghill et al., 2008; Molven et al., 2008).
Remarkably, among patients with permanent neonatal-onset diabetes mellitus, residues B5 and B8 provide “hot spots” for mutations not involving cysteine directly. The clinical database includes (l) SerB5, providing an unusual example in which physical studies of a mutant protein preceded and anticipated a clinical phenotype (Guo et al., 2005; Hua et al., 2006b; Nakagawa et al., 2005). This clinical correlation strongly suggests that insulin chain combination provides an informative peptide model for the physiological folding of nascent proinsulin in the ER of human β-cells as originally envisaged by Katsoyannis (1966). We propose that the conservation of HisB5 and the invariance of GlyB8 are enjoined by the requirement to ensure the foldability of proinsulin—and conversely to avoid the severe pathological consequences of proteotoxity following its misfolding (Araki et al., 2003; Liu et al., 2007). This perspective thus highlights the achirality of glycine as a uniquely “ambidextrous” residue strategically placed at a site of conformational change.
IV. Concluding Remarks
Structural studies of insulin analogs strongly suggest that the classical insulin T-state represents an inactive conformation of the hormone. By identifying a critical hinge point at GlyB8, studies of mirror-image d- and l substitutions in insulin have provided evidence for induced fit on receptor binding. The receptor-bound conformation of insulin may in part resemble the crystallographic R-state, but these processes may readily be uncoupled. We suggest that the classical allosteric reorganization of zinc insulin hexamers, so elegantly characterized by the late D. C. Hodgkin and colleagues, identifies sites of flexibility utilized by the insulin monomer on binding to its receptor. Remarkably, structural relationships observed in the inactive T-state are nonetheless of key biological importance: analogous interactions are required—presumably within the nascent structure of oxidative folding intermediates (Hua et al., 2001, 2002; Weiss et al., 2000)—for native disulfide pairing. The evolution of insulin sequences has thus been constrained by the dual and interlocking requirements of function and foldability. An understanding of structure–function relationships in insulin is of both basic and translational interest. Whereas analysis of ER-directed disulfide pairing is central to the emerging genetics of diabetes mellitus (Stoy et al., 2007), characterization of the receptor-bound structure of insulin promises to enable design of novel agonists for the treatment of diabetes mellitus.
ACKNOWLEDGMENTS
The author thanks Profs. P. Arvan, G. G. Dodson, Q. Hua, P. Katsoyannis, D. F. Steiner, and J. Whittaker for helpful discussion and is grateful to members of his laboratory for all aspects of the work cited. Figure 2.2 was designed with the assistance of G. G. Dodson; the present studies have been greatly aided by his collaboration and deep historical understanding of insulin crystallography. Studies of insulin analogs at CWRU were supported in part by grants from the National Institutes of Health (DK40949 and DK069764) and American Diabetes Association. This chapter represents a contribution from the Cleveland Center for Membrane and Structural Biology.
REFERENCES
- Adams MJ, Blundell TL, Dodson EJ, Dodson GG, Vijayan M, Baker EN, Hardine MM, Hodgkin DC, Rimer B, Sheet S. Structure of rhombohedral 2 zinc insulin crystals. Nature. 1969;224:491–495. [Google Scholar]
- Araki E, Oyadomari S, Mori M. Endoplasmic reticulum stress and diabetes mellitus. Intern. Med. 2003;42:7–14. doi: 10.2169/internalmedicine.42.7. [DOI] [PubMed] [Google Scholar]
- Baker EN, Blundell TL, Cutfield JF, Cutfield SM, Dodson EJ, Dodson GG, Hodgkin DM, Hubbard RE, Isaacs NW, Reynolds CD. The structure of 2Zn pig insulin crystals at 1.5Å resolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1988;319:369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- Bentley G, Dodson E, Dodson G, Hodgkin D, Mercola D. Structure of insulin in 4-zinc insulin. Nature. 1976;261:166–168. doi: 10.1038/261166a0. [DOI] [PubMed] [Google Scholar]
- Bentley G, Dodson G, Lewitova A. Rhombohedral insulin crystal transformation. J. Mol. Biol. 1978;126:871–875. doi: 10.1016/0022-2836(78)90026-8. [DOI] [PubMed] [Google Scholar]
- Bi RC, Cutfield SM, Dodson EJ, Dodson GG, Giorgino F, Reynolds CD, Tolley SP. Molecular-replacement studies on crystal forms of despentapeptide insulin. Acta Crystallogr. B. 1983;39:90–98. [Google Scholar]
- Bi RC, Dauter Z, Dodson E, Dodson G, Giordano F, Reynolds C. Insulin structure as a modified and monomeric molecule. Biopolymers. 1984;23:391–395. [Google Scholar]
- Blundell TL, Cutfield JF, Cutfield SM, Dodson EJ, Dodson GG, Hodgkin DC, Mercola DA, Vijayan M. Atomic positions in rhombohedral 2-zinc insulin crystals. Nature. 1971;231:506–511. doi: 10.1038/231506a0. [DOI] [PubMed] [Google Scholar]
- Blundell TL, Dodson GG, Hodgkin DC, Mercola DA. Insulin: The structure in the crystal and its reflection in chemistry and biology. Adv. Protein Chem. 1972;26:279–402. [Google Scholar]
- Blundell TL, Humbel RE. Hormone families: Pancreatic hormones and homologous growth factors. Nature. 1980;287:781–787. doi: 10.1038/287781a0. [DOI] [PubMed] [Google Scholar]
- Brader ML, Dunn MF. Insulin hexamers: New conformations and applications. Trends Biochem. Sci. 1991;16:341–345. doi: 10.1016/0968-0004(91)90140-q. [DOI] [PubMed] [Google Scholar]
- Brange J, Ribel U, Hansen JF, Dodson G, Hansen MT, Havelund S, Melberg SG, Norris F, Norris K, Snel L. Monomeric insulins obtained by protein engineering and their medical implications. Nature. 1988;333:679–682. doi: 10.1038/333679a0. [DOI] [PubMed] [Google Scholar]
- Chothia C, Lesk AM, Dodson GG, Hodgkin DC. Transmission of conformational change in insulin. Nature. 1983;302:500–505. doi: 10.1038/302500a0. [DOI] [PubMed] [Google Scholar]
- Ciszak E, Beals JM, Frank BH, Baker JC, Carter ND, Smith GD. Role of C-terminal B-chain residues in insulin assembly: The structure of hexameric LysB28ProB29-human insulin. Structure (Lond.) 1995;3:615–622. doi: 10.1016/s0969-2126(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Colombo C, Porzio O, Liu M, Massa O, Vasta M, Salardi S, Beccaria L, Monciotti C, Toni S, Pedersen O, Hansen T, Federici L, Early Onset Diabetes Study Group of the Italian Society of Pediatric Endocrinology and Diabetes Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. J. Clin. Invest. 2008;118:2148–2156. doi: 10.1172/JCI33777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda U, Derewenda Z, Dodson EJ, Dodson GG, Bing X, Markussen J. X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue. J. Mol. Biol. 1991;220:425–433. doi: 10.1016/0022-2836(91)90022-x. [DOI] [PubMed] [Google Scholar]
- Derewenda U, Derewenda Z, Dodson EJ, Dodson GG, Reynolds CD, Smith GD, Sparks C, Swenson D. Phenol stabilizes more helix in a new symmetrical zinc insulin hexamer. Nature. 1989;338:594–596. doi: 10.1038/338594a0. [DOI] [PubMed] [Google Scholar]
- Edghill EL, Flanagan SE, Patch AM, Boustred C, Parrish A, Shields B, Shepherd MH, Hussain K, Kapoor RR, Malecki M, MacDonald MJ, Stoy J, Neonatal Diabetes International Collaborative Group Insulin mutation screening in 1044 patients with diabetes: Mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes. 2008;57:1034–1042. doi: 10.2337/db07-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZY, Zhang Z, Jia XY, Tang YH, Feng YM. Mutational analysis of the absolutely conserved B8Gly: Consequence on foldability and activity of insulin. Acta Biochim. Biophys. Sin. (Shanghai) 2005;10:673–679. doi: 10.1111/j.1745-7270.2005.00093.x. [DOI] [PubMed] [Google Scholar]
- Hu SQ, Burke GT, Schwartz GP, Ferderigos N, Ross JB, Katsoyannis PG. Steric requirements at position B12 for high biological activity in insulin. Biochemistry. 1993;32:2631–2635. doi: 10.1021/bi00061a022. [DOI] [PubMed] [Google Scholar]
- Hua QX, Chu YC, Jia W, Phillips NB, Wang RY, Katsoyannis PG, Weiss MA. Mechanism of insulin chain combination. Asymmetric roles of A-chain α-helices in disulfide pairing. J. Biol. Chem. 2002;277:43443–43453. doi: 10.1074/jbc.M206107200. [DOI] [PubMed] [Google Scholar]
- Hua QX, Hu SQ, Frank BH, Jia W, Chu YC, Wang SH, Burke GT, Katsoyannis PG, Weiss MA. Mapping the functional surface of insulin by design: Structure and function of a novel A-chain analogue. J. Mol. Biol. 1996a;264:390–403. doi: 10.1006/jmbi.1996.0648. [DOI] [PubMed] [Google Scholar]
- Hua QX, Liu M, Hu SQ, Jia W, Arvan P, Weiss MA. A conserved histidine in insulin is required for the foldability of human proinsulin. Structure and function of an AlaB5 analog. J. Biol. Chem. 2006a;281:24889–24899. doi: 10.1074/jbc.M602617200. [DOI] [PubMed] [Google Scholar]
- Hua QX, Nakagawa SH, Hu SQ, Jia W, Wang S, Weiss MA. Toward the active conformation of insulin. Stereospecific modulation of a structural switch in the B chain. J. Biol. Chem. 2006b;281:24900–24909. doi: 10.1074/jbc.M602691200. [DOI] [PubMed] [Google Scholar]
- Hua QX, Nakagawa SH, Jia W, Hu SQ, Chu YC, Katsoyannis PG, Weiss MA. Hierarchical protein folding: Asymmetric unfolding of an insulin analogue lacking the A7–B7 interchain disulfide bridge. Biochemistry. 2001;40:12299–12311. doi: 10.1021/bi011021o. [DOI] [PubMed] [Google Scholar]
- Hua QX, Narhi L, Jia W, Arakawa T, Rosenfeld R, Hawkins N, Miller JA, Weiss MA. Native and non-native structure in a protein-folding intermediate: Spectroscopic studies of partially reduced IGF-I and an engineered alanine model. J. Mol. Biol. 1996b;259:297–313. doi: 10.1006/jmbi.1996.0320. [DOI] [PubMed] [Google Scholar]
- Hua QX, Shoelson SE, Kochoyan M, Weiss MA. Receptor binding redefined by a structural switch in a mutant human insulin. Nature. 1991;354:238–241. doi: 10.1038/354238a0. [DOI] [PubMed] [Google Scholar]
- Huang K, Xu B, Hu SQ, Chu YC, Hua QX, Qu Y, Li B, Wang S, Wang RY, Nakagawa SH, Theede AM, Whittaker J, et al. How insulin binds: The B-chain α-helix contacts the L1 β-helix of the insulin receptor. J. Mol. Biol. 2004;341:529–550. doi: 10.1016/j.jmb.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Katsoyannis PG. Synthesis of insulin. Science. 1966;154:1509–1514. doi: 10.1126/science.154.3756.1509. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Ogawa H, Burke GT, Chanley JD, Katsoyannis PG. Critical role of the A2 amino acid residue in the biological activity of insulin: [2-glycine-A]- and [2-alanine-A]insulins. Biochemistry. 1984;23:1405–1413. doi: 10.1021/bi00302a011. [DOI] [PubMed] [Google Scholar]
- Krebs MR, Bromley EH, Donald AM. The binding of thioflavin-T to amyloid fibrils: Localisation and implications. J. Struct. Biol. 2005;149:30–37. doi: 10.1016/j.jsb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Liang DC, Chang WR, Wan ZL. A proposed interaction model of the insulin molecule with its receptor. Biophys. Chem. 1994;50:63–71. doi: 10.1016/0301-4622(94)85020-8. [DOI] [PubMed] [Google Scholar]
- Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin maturation, misfolding, and proteotoxicity. Proc. Natl. Acad. Sci. USA. 2007;104:15841–15846. doi: 10.1073/pnas.0702697104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou M, Garrett TP, McKern NM, Hoyne PA, Epa VC, Bentley JD, Lovrecz GO, Cosgrove LJ, Frenkel MJ, Ward CW. The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proc. Natl. Acad. Sci. USA. 2006;103:12429–12434. doi: 10.1073/pnas.0605395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKern NM, Lawrence MC, Streltsov VA, Lou MZ, Adams TE, Lovrecz GO, Elleman TC, Richards KM, Bentley JD, Pilling PA, Hoyne PA, Cartledge KA, et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature. 2006;443:218–221. doi: 10.1038/nature05106. [DOI] [PubMed] [Google Scholar]
- Mirmira RG, Nakagawa SH, Tager HS. Importance of the character and configuration of residues B24, B25, and B26 in insulin-receptor interactions. J. Biol. Chem. 1991;266:1428–1436. [PubMed] [Google Scholar]
- Mirmira RG, Tager HS. Role of the phenylalanine B24 side chain in directing insulin interaction with its receptor: Importance of main chain conformation. J. Biol. Chem. 1989;264:6349–6354. [PubMed] [Google Scholar]
- Molven A, Ringdal M, Nordbo AM, Raeder H, Stoy J, Lipkind GM, Steiner DF, Philipson LH, Bergmann I, Aarskog D, Undlien DE, Joner G, et al. Mutations in the insulin gene can cause MODY and autoantibody-negative type 1 diabetes. Diabetes. 2008 doi: 10.2337/db07-1467. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Nakagawa SH, Tager HS. Role of the phenylalanine B25 side chain in directing insulin interaction with its receptor. Steric and conformational effects. J. Biol. Chem. 1986;261:7332–7341. [PubMed] [Google Scholar]
- Nakagawa SH, Tager HS. Importance of aliphatic side-chain structure at positions 2 and 3 of the insulin A chain in insulin-receptor interactions. Biochemistry. 1992;31:3204–3214. doi: 10.1021/bi00127a023. [DOI] [PubMed] [Google Scholar]
- Nakagawa SH, Tager HS, Steiner DF. Mutational analysis of invariant valine B12 in insulin: Implications for receptor binding. Biochemistry. 2000;39:15826–15835. doi: 10.1021/bi001802+. [DOI] [PubMed] [Google Scholar]
- Nakagawa SH, Zhao M, Hua QX, Hu SQ, Wan ZL, Jia W, Weiss MA. Chiral mutagenesis of insulin. Foldability and function are inversely regulated by a stereospecific switch in the B chain. Biochemistry. 2005;44:4984–4999. doi: 10.1021/bi048025o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen HB, Ludvigsen S, Kaarsholm NC. Solution structure of an engineered insulin monomer at neutral pH. Biochemistry. 1996;35:8836–8845. doi: 10.1021/bi960292+. [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J. Clin. Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak M, Dechaume A, Cave H, Nimri R, Crosnier H, Sulmont V, de Kerdanet M, Scharfmann R, Lebenthal Y, Froguel P, Vaxillaire M. Heterozygous missense mutations in the insulin gene are linked to permanent diabetes appearing in the neonatal period or in early-infancy: A report from the French ND Study Group. Diabetes. 2008;57:1115–1119. doi: 10.2337/db07-1358. [DOI] [PubMed] [Google Scholar]
- Pullen RA, Lindsay DG, Wood SP, Tickle IJ, Blundell TL, Wollmer A, Krail G, Brandenburg D, Zahn H, Gliemann J, Gammeltoft S. Receptor-binding region of insulin. Nature. 1976;259:369–373. doi: 10.1038/259369a0. [DOI] [PubMed] [Google Scholar]
- Ron D. Proteotoxicity in the endoplasmic reticulum: Lessons from the Akita diabetic mouse. J. Clin. Invest. 2002;109:443–445. doi: 10.1172/JCI15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M, Brader ML, Lee RW, Kaarsholm NC, Hansen JF, Dunn MF. Spectroscopic signatures of the T to R conformational transition in the insulin hexamer. J. Biol. Chem. 1989;264:19081–19085. [PubMed] [Google Scholar]
- Schlichtkrull J. Insulin Crystals: Chemical and Biological Studies on Insulin Crystals and Insulin Zinc Suspensions. Kobenhavns universitet; Copenhagen, Munksgaard: 1958. p. 139. [Google Scholar]
- Shoelson S, Haneda M, Blix P, Nanjo A, Sanke T, Inouye K, Steiner D, Rubenstein A, Tager H. Three mutant insulins in man. Nature. 1983;302:540–543. doi: 10.1038/302540a0. [DOI] [PubMed] [Google Scholar]
- Smith GD, Swenson DC, Dodson EJ, Dodson GG, Reynolds CD. Structural stability in the 4-zinc human insulin hexamer. Proc. Natl. Acad. Sci. USA. 1984;81:7093–7097. doi: 10.1073/pnas.81.22.7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoy J, Edghill EL, Flanagan SE, Ye H, Paz VP, Pluzhnikov A, Below JE, Hayes MG, Cox NJ, Lipkind GM, Lipton RB, Greeley SA, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc. Natl. Acad. Sci. USA. 2007;104:15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B, Wollmer A. Cobalt probing of structural alternatives for insulin in solution. Biol. Chem. Hoppe Seyler. 1989;370:1235–1244. doi: 10.1515/bchm3.1989.370.2.1235. [DOI] [PubMed] [Google Scholar]
- Wan Z, Huang K, Whittaker J, Weiss MA. The structure of a mutant insulin uncouples receptor binding from protein allostery. An electrostatic block to the TR transition. J. Biol. Chem. 2008;283:21198–21210. doi: 10.1074/jbc.M800235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C, Lawrence MC, Streltsov V, Garrett T, McKern N, Lou MZ, Lovrecz G, Adams T. Structural insights into ligand-induced activation of the insulin receptor. Acta Physiol. (Oxf.) 2008;192:3–9. doi: 10.1111/j.1748-1716.2007.01781.x. [DOI] [PubMed] [Google Scholar]
- Weiss MA, Hua QX, Jia W, Chu YC, Wang RY, Katsoyannis PG. Hierarchiacal protein “un-design”: Insulin's intrachain disulfide bridge tethers a recognition α-helix. Biochemistry. 2000;39:15429–15440. doi: 10.1021/bi001905s. [DOI] [PubMed] [Google Scholar]
- Weiss MA, Hua QX, Lynch CS, Frank BH, Shoelson SE. Heteronuclear 2D NMR studies of an engineered insulin monomer: Assignment and characterization of the receptor-binding surface by selective 2H and 13C labeling with application to protein design. Biochemistry. 1991;30:7373–7389. doi: 10.1021/bi00244a004. [DOI] [PubMed] [Google Scholar]
- Whittingham JL, Youshang Z, Zakova L, Dodson EJ, Turkenburg JP, Brange J, Dodson GG. I222 crystal form of despentapeptide (B26–B30) insulin provides new insights into the properties of monomeric insulin. Acta Crystallogr. D Biol. Crystallogr. 2006;62:505–511. doi: 10.1107/S0907444906006871. [DOI] [PubMed] [Google Scholar]
- Xu B, Hua QX, Nakagawa SH, Jia W, Chu YC, Katsoyannis PG, Weiss MA. A cavity-forming mutation in insulin induces segmental unfolding of a surrounding α-helix. Protein Sci. 2002a;11:104–116. doi: 10.1110/ps.32102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Hua QX, Nakagawa SH, Jia W, Chu YC, Katsoyannis PG, Weiss MA. Chiral mutagensis of insulin's hidden receptor-binding surface: Structure of an allo-IleA2 analogue. J. Mol. Biol. 2002b;316:435–441. doi: 10.1006/jmbi.2001.5377. [DOI] [PubMed] [Google Scholar]