Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common form of human lymphoma. While a number of structural alterations have been associated with the pathogenesis of this malignancy, the full spectrum of genetic lesions that are present in the DLBCL genome, and therefore the identity of dysregulated cellular pathways, remains unknown. By combining next-generation sequencing and copy number analysis, we show that the DLBCL coding genome contains on average more than 30 clonally represented gene alterations/case. This analysis also revealed mutations in genes not previously implicated in DLBCL pathogenesis, including those regulating chromatin methylation (MLL2, 24% of cases) and immune recognition by T cells. These results provide initial data on the complexity of the DLBCL coding genome and identify novel dysregulated pathways underlying its pathogenesis.
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most common lymphoid malignancy in adulthood, is considered a curable disease in only ~50% of cases1,2. This incomplete success indicates the need for further studies aimed at elucidating its pathogenesis through the identification of genes and cellular pathways that are structurally altered in this disease and may represent targets for novel therapeutic approaches.
Current understanding of the biology of DLBCL indicates the existence of several distinct subgroups reflecting the derivation from B-cells at discrete stages of differentiation, namely germinal-center B-cell-like (GCB) DLBCL, activated B-cell-like (ABC) DLBCL, and primary mediastinal B-cell lymphoma (PMBCL)3–5. These subtypes are associated in part with distinct genetic lesions, indicating the involvement of separate oncogenic pathways6,7. In particular, chromosomal translocations involving the BCL2 and MYC oncogenes and mutations of the EZH2 methyltransferase are almost exclusively observed in GCB-DLBCL7–9, while the less curable ABC-DLBCL is preferentially associated with structural alterations disrupting terminal B-cell differentiation (PRDM1 inactivation)10–12 and with a variety of lesions that result in the constitutive activation of the NF-κB transcription complex (most commonly, mutations of TNFAIP3, CARD11, CD79B and MYD88)13–16. More recently, chromosomal translocations involving CIITA and amplifications/rearrangements of the genes encoding for the CD274/PDL1 and CD273/PDL2 ligands have been reported as recurrent events in PMBCL17–19. Additional lesions are common to GCB- and ABC-DLBCL, including BCL6 translocations20 and inactivation of the acetyltransferase genes CREBBP and EP30021. Overall, these alterations only affect a fraction of cases, while the full spectrum of genomic lesions that contribute to malignant transformation remains unknown.
Major improvements in sequencing technologies have now provided an unprecedented opportunity to examine the cancer genome for large-scale identification of genomic alterations in a comprehensive and unbiased manner. The present study was aimed at expanding our current knowledge of the number and type of genetic alterations that are present in the DLBCL coding genome by integrating massively parallel whole-exome sequencing (WES) and genome-wide high-density single nucleotide polymorphism (SNP) array analysis. This combined approach allowed the definition of the degree of complexity characterizing the DLBCL coding genome, and revealed the involvement of several previously unrecognized dysregulated genes/pathways.
Results
Mutational load by exome sequencing analysis
In order to determine the load of somatic, non-silent mutations that are clonally represented in the DLBCL genome, we performed massively parallel sequencing of paired tumor and normal DNAs from six untreated, de novo DLBCL patients (3 ABC, 1 GCB, 1 non-GCB, 1 unclassified; Supplementary Table 1) by using a hybridization-capture method for the enrichment of non-repetitive protein-coding genes (~85% of the CCDS database) followed by next-generation sequencing using the 454 Genome Sequencer FLX instrument (Supplementary Fig. 1). We also analyzed by direct sequencing the coding exons of selected genes that have been previously implicated in other hematologic cancers, but are not represented in the capture array (e.g., NOTCH1, MLL2 and CYLD)22–27. The WES approach produced 11.8 billion base pairs of mapped sequences (~2,3 million reads/case, of average length 305bp), with a mean depth of 9.8× (range: 8.9×–12.8× per case), and on average 77.7% of the target sequence being covered by at least 5 reads (range: 70.8%–81.4%)(Supplementary Table 2). This level of resolution allowed us to provide an initial estimate of the order of magnitude of the mutation load in the disease, and to identify genes that are relevant to its pathogenesis because recurrently mutated across different subtypes (probability of detecting mutations in genes that are affected at ~30% prevalence, 70%)(Methods and Supplementary Note).
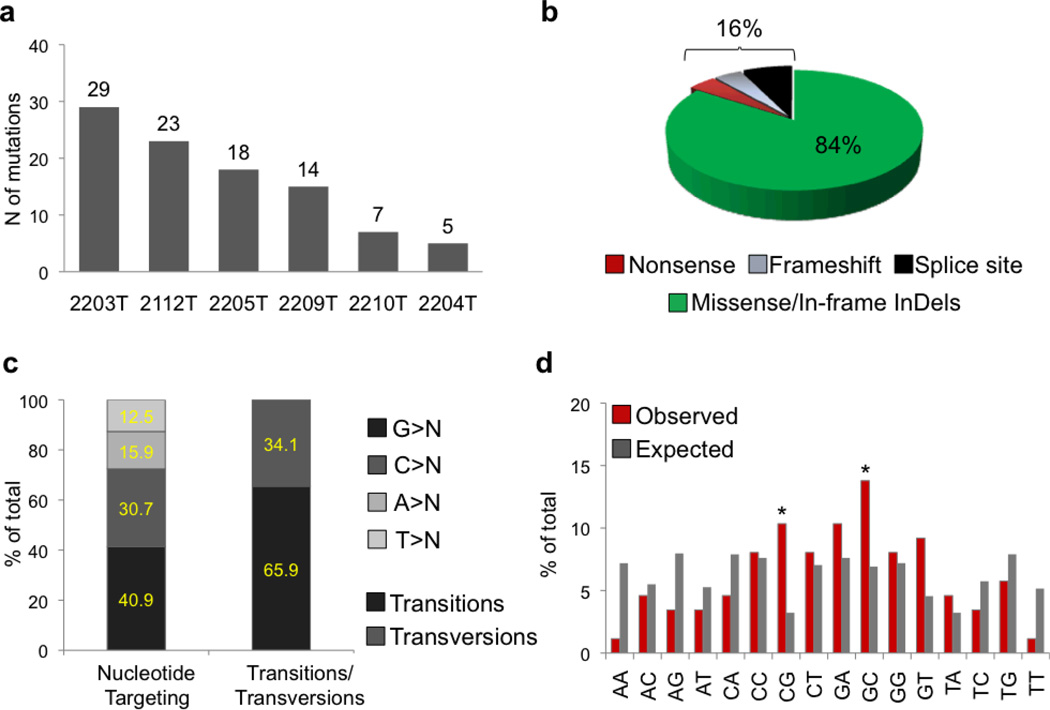
Sanger-based re-sequencing of candidate variants in the matched tumor-normal DNAs validated the presence of 96 somatic, non-silent mutations involving 93 distinct genes and present in a major clone (Fig. 1a and Supplementary Tables 3 and 4). The overall mutation load/case varied significantly across samples (mean, 16; range, 5–29) (Figure 1). Mutations were largely represented by single nucleotide substitutions leading to amino acid changes (n=78; 82.2%), but also included in-frame insertions/deletions (n=3), nonsense mutations (n=4), alterations in canonical splice sites (n=7), and frameshift deletions of short nucleotide stretches (n=4), collectively accounting for ~16% of the events (Fig. 1b and Supplementary Table 3). Analogous to the spectrum reported in other cancer types26,28, we observed a predominance of transitions over transversions (n=58:30; ratio, 1.9) and a preferential targeting of G and C nucleotides (71.6% vs 28.4% affecting A/T nucleotides; P value after correction for target exome sequence composition: 0.004) (Fig. 1c); moreover, there was a significant bias toward alterations at 5’-CpG-3’ dinucleotides, which account for 10% of all non-synonymous changes (frequency of CpG nucleotides in the target exome, ~3%; p<0.0001)(Fig. 1d) (see Discussion).
Figure 1. DLBCL non-silent mutation load.
Number (a) and type (b) of Sanger-validated, non-silent mutations identified in the 6 DLBCL discovery patients. c, Nucleotide targeting of the DLBCL-associated point mutations (nucleotide composition of the target exome: G, 26.2%; C, 25.7%; T, 22.0%; A, 25.9%). d, Observed mutation frequencies at specific dinucleotides (red bars). The expected frequencies (grey bars) correpond to the dinucleotide sequence composition of the target exome. Asterisks denote statistically significant differences in overrepresented changes, as assessed by a Poisson distribution after correction for multiple hypotheses.
The 93 mutated genes identified include most of the ones previously implicated in the pathogenesis of this disease, namely PRDM1, TNFAIP3, CARD11, CD79B and MYD88, as well as the acetyltransferase gene CREBBP (Supplementary Table 3). Of the remaining 87 genes, 26 have never been implicated in cancer to date, while 60 are reported in the COSMIC database and 4 are listed in the Cancer Gene Census database as causally related to cancer (Supplementary Table 3)21. Although the functional significance of the mutations found in these 87 genes is largely unknown, 71 of them (81.6%) are expected to alter the function of the encoded protein, based on two distinct prediction algorithms (Supplementary Table 3).
Copy Number analysis
The same six DLBCL samples (and paired normal DNAs) were then analyzed for the presence of copy number changes by using the Affymetrix SNP6.0 platform, which interrogates ~1.8 million markers, including SNPs and copy number probes. This analysis identified 90 somatically-acquired genomic alterations (66 deletions and 24 gains), with significant variability across individual samples (range: 1–31) (Fig. 2 and Supplementary Table 5). Chromosomes 1, 2, 3 and 6 comprised the highest number of lesions, in agreement with previous studies using chromosomal and array-based CGH6,29, and ten of the changes (3 losses and 7 gains) involved whole chromosomes or chromosome arms. The SNP array approach correctly identified two deletions of the PRDM1 gene on chromosome 6q21 (case 2204 and 2210) and a 17p deletion (case 2204), which had been previously detected by FISH analysis. Additional known DLBCL-associated alterations include a focal homozygous deletion of CDKN2A/CDKN2B and a 6q23.3 deletion spanning the TNFAIP3 tumor suppressor. Of the remaining lesions, nine encompassed single genes that likely represent the target of the aberration (Supplementary Table 5). Overall, the detection of 90 copy number alterations, with over 30-fold differences across the 6 patients studied, indicates that the genetic landscape of DLBCL is remarkably heterogeneous, and that largely distinct types of genes are affected in individual cases.
Figure 2. Copy number analysis of the 6 DLBCL discovery cases.

a, dChipSNP heatmap showing median-smoothed log2 copy number ratio of all chromosomes in the 6 index DLBCL biopsies and their paired normal DNAs. In the red-blue scale, white corresponds to a normal (diploid) copy number log-ratio, blue is deletion and red is gain. b, Curated segmentation data for the 6 DLBCL samples shown in panel a, obtained as described in Methods and visualized using the Integrative Genomics Viewer (IGV) software (http://www.broadinstitute.org/igv). b, Overall number and frequency of copy number alterations.
Overall complexity of the DLBCL coding genome
The combination of whole-exome sequencing and copy number data from the 6 index patients, together with FISH analysis for three common chromosomal translocations in lymphoma (BCL6, BCL2 and MYC), provided an integrated snapshot on the complexity of alterations affecting the DLBCL coding genome. In addition to the described mutations and copy number aberrations, chromosomal translocations of BCL6 were present in two cases, while no alterations were observed at the MYC and BCL2 loci. Few genes were biallelically inactivated by a combination of truncating mutations and/or deletions, as typical of tumor suppressor genes (e.g. TNFAIP3, PRDM1, CDKN2A/CDKN2B, TMEM30A, CD58 and IGSF3). Interestingly, the relative representation of point mutations versus copy number changes varied significantly within individual cases (Fig. 3). Thus, the overall load of tumor-acquired lesions that are present in the major clone was relatively heterogeneous across the 6 cases studied (average load 31.5, range 16–49).
Figure 3. DLBCL harbors a heterogeneous load of numerical and structural genomic aberrations.
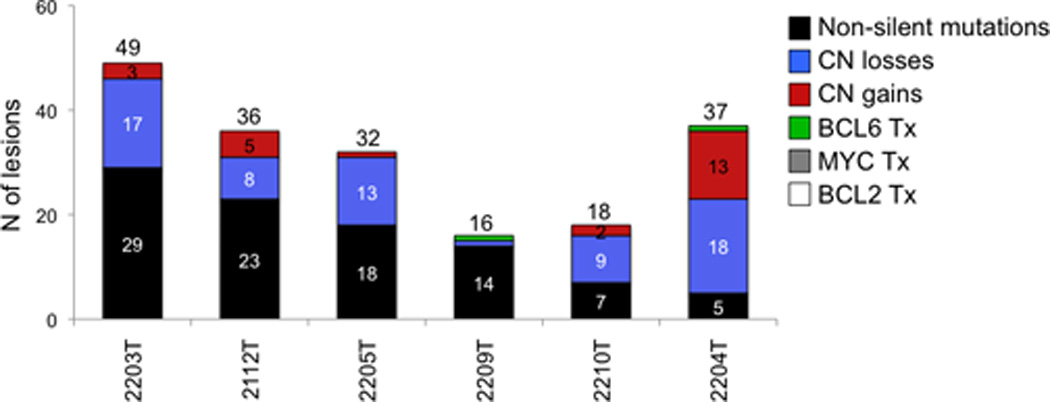
Combined load of genetic lesions that were present in the major tumor clone of the 6 DLBCL discovery cases (including point mutations, copy number aberrations, and chromosomal translocations at three common target loci).
Identification of recurrent targets of point mutations and copy number changes
One criterion to assess the pathogenetic relevance of candidate genetic lesions is to examine their recurrence in the disease, which can involve different modes of alteration, including mutations and copy number changes. To increase our ability to detect relevant tumor-associated genomic alterations across and within different DLBCL subgroups, and to identify additional candidate genes for downstream mutation screening in a larger DLBCL dataset, we first extended the SNP array analysis to 73 DLBCL biopsies representative of the two major subtypes. Consistent with the results obtained from the discovery panel, this screening confirmed the high degree of complexity of the DLBCL genome, which displayed an average of 24 acquired copy number aberrations/case, with great variability across individual samples (range: 0 to 92), regardless of their DLBCL subtype. Losses were more common than gains (n=1108 and 765, respectively). We identified a total of 325 minimal common regions (MCR) of aberration measuring <1Mb in size and encompassing 1–3 genes, which most likely represent relevant selected targets. Of these, 241 were commonly deleted, with 154 spanning to a single gene, and 84 were gained, and including 20 focal high-level amplifications (in total 474 involved genes).
Gene-annotation clustering analysis using the DAVID algorithm and the list of 560 genes obtained by combining the WES-candidates (n=93) and the MCRs candidates (n=474, 7 of which are in common between the two groups) revealed a significant enrichment in specific functional categories, including regulation of transcription, lymphocyte activation/differentiation, chromatin modification/DNA methylation, and antigen processing and presentation (Supplementary Table 6), suggesting that dysregulation of these biological processes plays a central role in DLBCL pathogenesis.
Out of this large array of candidates, genes were prioritized for further analysis based on one or more of the following criteria: i) mutation frequency in the 6 discovery genomes; ii) participation in both point mutations and focal MCRs of aberration; iii) significance scores, as assessed by two independent statistical approaches for the analysis of copy number data, including GISTIC (which is based on the amplitude and the frequency of occurrence of copy number changes)30 (Supplementary Fig. 2, Supplementary Tables 7 and 8) and ComFocal, a newly developed algorithm based on the size, amplitude and frequency of the copy number aberration (Supplementary Note and Supplementary Fig. 3); iv) functional annotation. Genes that had been previously implicated in DLBCL or in other hematologic malignancies were also included in the analysis. Based on these criteria, 56 genes were subjected to Sanger-based resequencing of their complete coding exons and consensus splice sites in an independent panel of at least 48 (up to 105) biopsies representative of the main DLBCL subtypes.
Figure 4 shows the prevalence of mutated cases in each of the 56 genes analyzed. Of these, sixteen were never found mutated in the 48 “screening” cases (overall frequency, including the discovery cases: 1/54, 1.8%). The remaining 40 genes harbored mutations in at least one additional patient, and 30 of them (including 14 not previously reported in the disease) were altered in >5% of cases, indicating that our selection criteria had effectively enriched the list for recurrent, thus presumably relevant targets (see Supplementary Tables 9, 10, 12, 14 and 18 for details on the mutations found). These genes point to the involvement of specific biological programs, as described in the following sections.
Figure 4. Recurrent mutations in DLBCL.

Percentage of DLBCL primary cases harboring mutations in 56 candidate genes after targeted-resequencing of an expanded screening dataset. The final number of mutated cases over total analyzed (including the 6 DLBCL discovery cases) is given for each gene; this number includes validated somatic mutations as well as variants that are not reported in any available SNP databases (see Methods). The somatic origin of the mutations was confirmed by analysis of paired normal DNA in at least one case/gene. Red asterisks denote putative tumor suppressor genes, as suggested by the presence of predominantly inactivating mutations (stop codons, frameshift mutations and experimentally validated missense mutations). Green asterisks denote genes where the oncogenic activity of the mutation was functionally proven. Black asterisks indicate genes harboring mutations of unclear pathogenic role, but predicted to truncate the encoded protein in at least one case.
Frequent alterations in genes controlling chromatin methylation
A prominent feature of the DLBCL genome was the presence of multiple lesions targeting histone/chromatin modification genes. In addition to the recurrent inactivation of the histone acetyltransferases (HATs) CREBBP and EP30021, we discovered alterations of several genes involved in the regulation of histone methylation, with mixed-lineage leukemia 2 (MLL2) being the most frequently mutated. MLL2 encodes a histone methyltransferase (HMT) that controls gene transcription by modifying the lysine-4 position of histone 3 (H3K4) and by promoting PolII-dependent activation of target genes31. Targeted re-sequencing of the MLL2 coding exons in 115 DLBCLs (58 ABC-DLBCLs and 57 GCB-DLBCLs) revealed a total of 33 sequence variants distributed in 28 samples, including 21/92 biopsies and 7/23 cell lines (Fig. 5a). In most cases, the mutations were clearly inactivating events represented by nonsense mutations (n=10), frameshift insertions/deletions (n=11), and a consensus splice site mutation (Fig. 5a,b and Supplementary Table 10). As a consequence, the corresponding MLL2 alleles are predicted to generate truncated proteins lacking the entire C-terminal cluster of conserved domains (including the SET domain) or significant portions of it (Fig. 5a). Eleven additional missense mutations were distributed along the MLL2 protein, with no apparent clustering (Fig. 5a and Supplementary Table 10). Where available (n=3 patients), analysis of paired normal DNA confirmed their somatic origin, which is strongly suggested for the remaining variants based on their absence in public and our own SNP databases (see Methods). While the functional consequences of these amino acid changes will have to be tested experimentally, five of them were located within or in close proximity to the conserved PHD, FYRN and SET domains, which are central to the MLL2 protein function.
Figure 5. The MLL2 gene is mutated in a large fraction of DLBCL.

a, Schematic diagram of the MLL2 gene (top) and protein (bottom), with its conserved functional domains (PHD, FYRN, FYRC, SET). In black, untranslated exons. Arrows, color-coded as in (b), indicate the position of the mutations found (cell lines on top, primary cases at the bottom). b, Overall percentage of DLBCL cases carrying MLL2 mutations, according to mutation type (cell lines and biopsies). c, Prevalence of MLL2 mutated cases in major DLBCL phenotypic subtypes; numbers on top indicate the actual number of mutated samples over total analyzed. d, Allelic distribution of the mutations in 26 of 28 affected samples for which this data could be obtained. Mutations are color-coded as in (b).
In cell lines, MLL2 mutations were exclusively associated with a GCB-DLBCL phenotype, where they account for ~50% of samples (n=7/16, versus 0/7 ABC-DLBCL), while their distribution in primary biopsies was not significantly different between the two subgroups (GCB-DLBCL, 27%; ABC-DLBCL, ~20%)(Fig. 5c). With one exception, MLL2 mutations affected a single allele in all evaluable cases, and were not accompanied by deletion of the second copy (Fig. 5d). This pattern of monoallelic inactivation suggests a role for MLL2 as a haploinsufficient tumor suppressor, mutated in ~23% of primary DLBCLs (24.3% including cell lines)(see Discussion).
In addition to MLL2, several genes involved in histone methylation were targeted by genomic alterations. Our analysis confirmed the occurrence of EZH2 mutations in 6/107 (5.6%) biopsies and 4/23 lines, almost exclusively of the GCB-type (n=9/63, 14.2%)(Figs. 4 and 6a); these variants all target a specific hotspot residue that has been associated with increased H3K27 trimethylation activity8,32,33. In 11 patients, we identified somatic mutations (n=4) or deletions (n=7) of KDM2B, a gene encoding for a H3K36 histone demethylase (Fig. 6a, Supplementary Tables 9 and 11). Additionally, individual patients harbored genomic deletions or rearrangements of MLL3 (n=6), MLL5 (n=2) and MLL (Fig. 6b and Supplementary Table 11).
Figure 6. Disruption of histone/chromatin modification genes is a major feature of DLBCL.
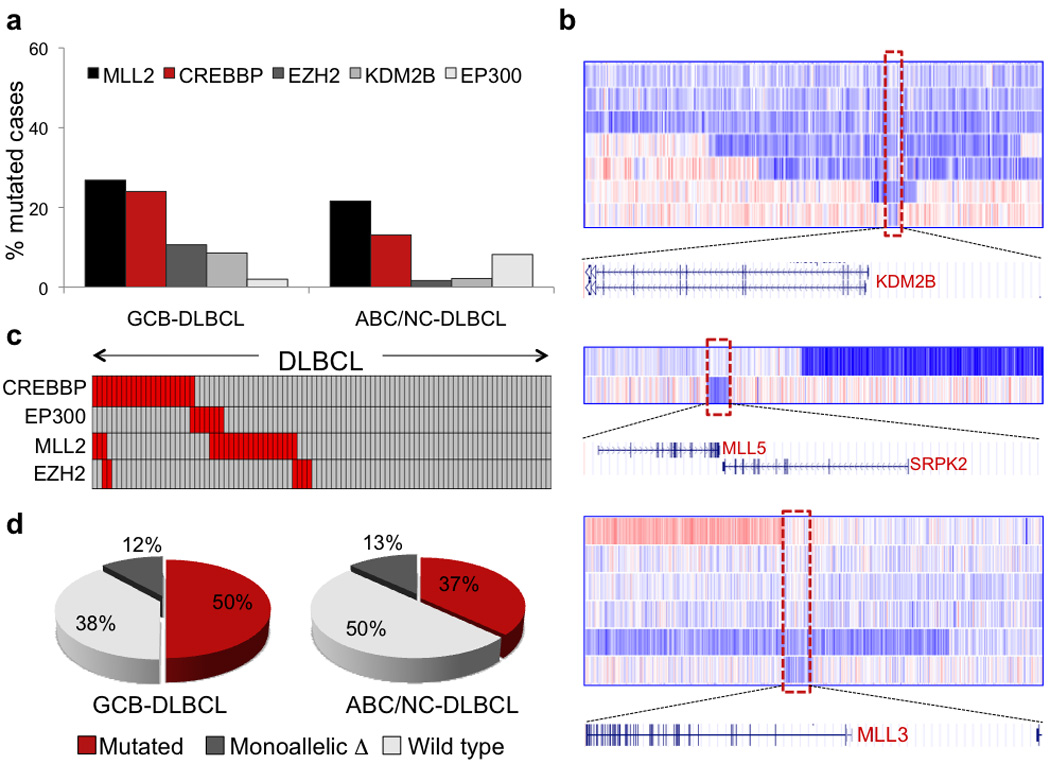
a, Mutation frequency of five genes encoding for histone-acetyltransferases and histone-methylation modifiers in GCB- and ABC-DLBCL (primary cases only). b, dChipSNP heatmap showing median-smoothed log2 copy number ratio for DLBCL biopsies harboring KDM2B, MLL3 and MLL5 deletions; in the red-blue scale, white corresponds to a normal (diploid) copy number, blue is deletion and red is gain. Note that, due to the presence of non-tumor cells infiltrating the biopsies, the inferred copy number, and the corresponding color intensity, may vary across samples (see Supplementary Table 11). The boxed area corresponds to the commonly deleted region, which is expanded below the heatmap to show the included genes. c, Relationship between MLL2 mutations and mutations of HATs. In the heatmap, rows correspond to the indicated genes and columns represent individual samples, color-coded according to the gene status (grey, unmutated; red, mutated or deleted). Of the 3 MLL2-mutated cases showing simultaneous lesions at CREBBP, one harbored an amino acid substitution whose functional significance is presently unclear. Thus, it is possible that this change represents a passenger event or a private germline variant not annotated in currently available databases; alternatively, these mutations may have milder effects, requiring additional cooperating lesions in the same pathway. d, Overall fraction of DLBCL biopsies carrying mutations at one or more of the four histone modification genes shown in (c).
Since H3K4 methylation has been linked to other chromatin-modifiers, such as HATs and chromatin remodelers34–38, we looked at the relationship between genetic lesions affecting MLL2 and CREBBP/EP300, which were recently shown to be monoallelically inactivated in up to 39% of DLBCL21. Although the number of cases studied does not allow robust statistical analyses, most patients displayed a mutually exclusive involvement of these genes (Fig. 6c), suggesting that alterations at these chromatin modifiers may represent alternative mechanisms converging on a common transcriptional program. Together, these data suggest that disruption of histone modification/chromatin remodeling genes plays a central role in DLBCL pathogenesis (Fig. 6d).
Mutations of genes controlling immune recognition by T cells
A second set of lesions recurrently observed in DLBCL involve immune recognition and antigen presenting functions. Expanding on previous observations from isolated cases, we found frequent inactivating mutations and deletions in the β2-microglobulin (B2M) gene. B2M encodes a polypeptide found in association with the major histocompatibility complex (MHC) class I on the surface of nearly all nucleated cells39. In sixteen samples (13/111 biopsies and 3/23 cell lines) both B2M alleles were lost due to homozygous deletions (8 cases), biallelic mutations (4 cases) and hemizygous deletions with inactivating mutation of the second allele (4 cases)(Supplementary Fig. 4 and Supplementary Tables 12 and 13). Nine additional patients harbored monoallelic nonsense or frameshift mutations; however, the relatively low density of probe coverage for this small gene in the array may have prevented the identification of submicroscopic deletions in the second allele. Most mutations are predicted to generate truncated proteins as the result of premature stop codons (n=3), splice site mutations (n=2), out-of-frame indels (n=6) or mutations at the translation-initiating methionine codon (n=6); moreover, 4 missense mutations were found associated with inactivation of the second allele in 4 cases, as documented by sequencing analysis of cloned PCR products, suggesting that they may also be functionally relevant (Supplementary Table 12). Taken together, these lesions predict the loss of B2M expression, which is required for cell surface expression of HLA class I molecules and recognition by cytotoxic T lymphocytes39.
Focal homozygous deletions (n=4, including one cell line), truncating mutations (n=7, distributed in 5 cases and 1 cell line), in-frame deletions (n=1) and hemizygous deletions (n=10) were also recurrently detected in the CD58 gene, a member of the immunoglobulin superfamily that functions as a ligand of the CD2 protein on T lymphocytes, participating in their adhesion and activation40 (Supplementary Fig. 4, Supplementary Tables 14 and 15). Although no copy number data were available to assess the status of the second allele in 3 of the mutated cases, and one patient apparently retained a normal allele, the detection of frequent biallelic inactivation (n=6 samples) suggests that CD58 may play a tumor suppressor function in DLBCL.
A novel finding was the presence of focal deletions in TNFSF9, the gene encoding for a transmembrane cytokine that belongs to the tumor necrosis factor (TNF) ligand family (n=10/79 cases, of which 3 homozygous) (Supplementary Table 16). TNFSF9 interacts with a costimulatory receptor molecule in T lymphocytes and follicular dendritic cells (TNFRSF9), and is involved in antigen presentation and in the generation of cytotoxic T cells41. Interestingly, TNFSF9 knock-out mice develop GC-derived lymphoma42.
Other lesions affecting regulators of immune responses include mutations and genomic breakpoints of the MHC class II transactivator gene CIITA, as well as amplifications and breaks in the genes encoding for the receptor immunomodulatory proteins PDL2 and PDL1, often occurring simultaneously (Supplementary Fig. 4a,d). In PMBCL, rearrangements of CIITA have been recently shown to cause downregulation of surface HLA class II expression19, which is associated with reduced tumor cell immunogenicity, while amplifications of the PDL1 locus (also found in PMBCL and Hodgkin lymphoma) have been linked to impaired anti-tumor immune responses in several cancers17,18. While no clear relationship was observed between alterations at these five loci, their ability to interfere with the interaction between tumor cells and the microenvironment suggests that they may facilitate lymphomagenesis by allowing escape of immuno-surveillance mechanisms.
Alterations in pathways of post-GC differentiation
Our results extend previous reports on the high frequency of genetic lesions affecting a variety of signaling pathways that share their ability to induce the NF-κB transcription complex, including the B-cell receptor (BCR), CD40 and Toll-like receptor pathways13–16,43, Overall, components of these pathways were found structurally altered in 63% (36/59) ABC- and ~31% (12/41) GCB-DLBCL biopsies (Supplementary Fig. 5a), consistent with the observation that >90% of ABC-DLBCL and ~30% of GCB-DLBCL display nuclear NF-κB, as direct evidence of its constitutive activation13,44; these comprise previously identified genes (e.g., the negative regulator TNFAIP3 and the positive regulators CARD11, MYD88 and CD79B), but also novel targets, such as ITPKB and TRAF3 (Supplementary Fig. 5b,c). NF-κB alterations showed a largely mutually exclusive pattern of distribution (Supplementary Fig. 5d), implicating activation of NF-κB as a major downstream effect shared by the above lesions. Nonetheless, additional consequences of individual lesions may include deregulation of distinct signaling pathways, such as PI3K and MAPK (mutations of CD79B and CARD11) and/or JAK/STAT (MYD88) (Supplementary Fig. 5e).14,16
NF-κB alterations were frequently associated with a set of genetic lesions in downstream components of the signaling pathway that regulates GC exit and the generation of plasma cells, ultimately causing a block in terminal B-cell differentiation. Specifically, we confirmed the common biallelic loss of PRDM1/BLIMP1 in ABC-DLBCL and its mutually exclusive relationship with chromosomal translocations of BCL6, which substitute its promoter region making it constitutively active and resistant to IRF4-mediated suppression45 (Supplementary Fig. 6). Since PRDM1 is a direct target of BCL6, these translocations will also result in the block of plasma cell differentiation (Supplementary Fig. 5e). However, deregulation of BCL6 has broader consequences, including suppression of DNA damage responses by its direct targets (e.g. p53 and p21)46,47, which may be provided by other presently unknown lesions in the remaining cases.
Other transcription factors involved in B-cell differentiation emerged as common targets of various numerical or structural aberrations, including PAX5, IRF4, ETS1 (Supplementary Table 17) and MEF2B, a gene highly expressed in the GC48 which was found mutated in 8/96 biopsies (Fig. 4 and Supplementary Table 18) and rearranged in one case (not shown). Although the functional consequences of the above lesions remain to be defined, these findings indicate that, collectively, over half of all DLBCL harbor defects at one or more genes involved in GC differentiation.
Discussion
The main findings of this study are the initial elucidation of the complexity of the DLBCL coding genome, and the discovery of a novel set of recurrent lesions that may be of relevance for the understanding of the pathogenesis of this malignancy.
The combined set of mutations and CNAs detected in the six DLBCL discovery cases does not allow a final assessment of the precise number of genomic alterations affecting coding genes, but provides information about the order of magnitude of the lesions associated with this malignancy. The estimate of >30 alterations/case emerging from this study reflects only those events with abundant to complete clonal representation, i.e. changes that were likely present during the initial phases of tumor expansion, and thus promoted malignant transformation. Furthermore, this estimate does not include an additional 40% of mutations that may have been missed by the WES approach due to the relatively low depth of coverage (see Methods), as well as chromosomal translocations other than those affecting BCL6, BCL2 and MYC (not detectable by the two methodologies used). Thus, it can be concluded that the coding genome of DLBCL contains <100 lesions on average. Although very approximate, this figure is informative for future studies and may serve as an initial database for the determination of recurrence in additional panels of cases.
When compared to other malignancies, the order of magnitude of lesions detected in DLBCL appears lower than that reported for certain epithelial cancers49,50. Among hematologic malignancies, the complexity of the DLBCL genome is not significantly different from multiple myeloma51, while it appears much more complex than acute myeloid leukemia52–54 and chronic lymphocytic leukemia23,27 in terms of both CNAs and mutational load. On the other hand, the observed predominance of transitions over transversions, the preferential targeting of C:G and G:C basepairs and the significant bias toward alterations at CpG dinucleotides is emerging as a common feature shared by most malignancies studied so far, including those of epithelial derivation. This pattern of alterations is generally derived from endogenous biochemical processes, such as the spontaneous deamination of 5-methylcytosine residues. However, it cannot be excluded that the abnormal and/or ectopic activity of activation-induced cytidine deaminase (AICDA) has a role in the generation of DNA lesions affecting these residues; indeed, AICDA has been shown to target the 5’ portion of multiple genes –mostly noncoding– in DLBCL, and some of the mutations detected in this study may in fact reflect the activity of aberrant somatic hypermutation (for example, PIM1)55.
Among the vast array of genetic lesions identified in the DLBCL genome, alterations of MLL family members, and in particular MLL2, appear especially frequent. MLL2 encodes a trimethyltransferase with well-documented influence on the expression of a large number of genes, including homeobox genes. The pattern of monoallelic somatic inactivation observed in DLBCL suggests a role for MLL2 as a haploinsufficient tumor suppressor, consistent with the observation that monoallelic MLL2 truncating mutations leading to descreased gene dosage have a pathogenic effect in Kabuki syndrome, a congenital disorder characterized by developmental and intellectual abnormalities56. A role for MLL genes in malignant transformation is supported by the involvement of MLL in the pathogenesis of acute leukemia57, although by a distinct and partially unclear mechanism, and by the recent finding of somatic inactivating mutations in both MLL2 and MLL3 in various cancers26,51,58. Collectively, alterations of chromatin modifying enzymes, including HMT and HAT, emerge as the most frequent alterations associated with DLBCL pathogenesis, being present in over one third of cases independent of disease subtype (GCB-DLBCL, ~50%; ABC-DLBCL, ~30%).
A second novel finding in the DLBCL coding genome is the high frequency of alterations in genes that are involved in immune recognition by T cells. In particular, the observation that bi-allelic inactivation of B2M is more common than originally implicated based on only a few cases analyzed59 suggests a major role for these lesions in causing the loss of HLA class I expression, which constitutes a frequent event in DLBCL60. The findings herein provide a mechanistic explanation for a fraction of these cases, and suggest the existence of additional genetic or epigenetic mechanisms preventing the expression of HLA class I molecules. Since it is well established that the lack of these molecules makes cells insensitive to cytotoxic T cell-mediated killing, loss of B2M may represent a major mechanism of tumor escape from immune surveillance.
The distribution of recurrent lesions among DLBCL subtypes defined by cell of origin supports the existence of both common and subtype-specific genetic lesions. The former are represented by those inactivating chromatin-modifying functions, including HAT and HMT genes, as well as the newly discovered alterations of immune recognition functions. These alterations may represent common pathways necessary for the development of the DLBCL phenotype, independent of cell of origin. Conversely our results confirm the preferential association of BCL2 and MYC alterations with GCB-DLBCL7, and of alterations in the NF-κB and BCL6/BLIMP1 axis with ABC-DLBCL10. The observed distribution has immediate clinical implications since it suggests the development of therapies that combine drugs targeting commonly altered pathways with those targeting pathways selectively disrupted in DLBCL subtypes.
Supplementary Material
Acknowledgements
We would like to thank T. Palomero, E. Tzilianos, V. Miljkovic and the Genomics Technologies Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University; D. Burgess at Roche NimbleGen (Madison, WI), and B. Boese at 454 Life Sciences (Branford, CT) for assistance with the whole exome capture and sequencing procedure; K. Basso and A. Holmes for help with the manual curation and analysis of the copy number data; M. Malladi and Y.K. Lieu for help with the sequencing analysis of B2M and CD58; J. Zhang for filtering the list of candidate mutations against the database of germline variants discovered from the St Jude Children's Research Hospital - Washington University Pediatric Cancer Genome Project. The Affymetrix SNP6.0 array experiments were processed in part at the Affymetrix Research Services Laboratory. Automated DNA sequencing was performed at Genewiz.Inc. This work was supported by N.I.H. Grants PO1-CA092625 and RO1-CA37295 (to R.D.-F.), a Specialized Center of Research grant from the Leukemia & Lymphoma Society (to R.D.-F.), N.I.H. Grant CA121852-05, the Northeast Biodefence Center (U54-AI057158) and the National Library of Medicine (1R01LM010140-01)(to RR), and the AIRC Special Program Molecular Clinical Oncology – 5 per mille (Contract No. 10007, Milan, Italy). L.P. is on leave from the Institute of Hematology, University of Perugia Medical School, Perugia, Italy.
Footnotes
Accession codes. The SNP Array 6.0 data and the whole exome sequencing data reported in this paper have been deposited in dbGaP under accession no. phs000328.v1.p1.
Authors Contributions. L.P and R.D.-F. designed the study and wrote the manuscript. L.P. conducted experiments, analyzed data and supervised the study. G.F., A.C., A.G., V.A.W. and M.M. performed PCR amplification and sequencing analysis. C.G.M. and J.M. developed methods for analysis of high-density SNP array data, which was conducted by C.G.M., J.M, L.P. and G.F. D.R., G.G., G.B. and A.Chadburn provided pathologically characterized patient samples. V.T. and R.R. analyzed high throughput sequencing data and developed the ComFocal algorithm for analysis of copy number data, with the help of O.E. and J.C. All authors read and approved the manuscript.
The Authors declare no competing financial interests.
References
- 1.Abramson JS, Shipp MA. Advances in the biology and therapy of diffuse large B-cell lymphoma: moving toward a molecularly targeted approach. Blood. 2005;106:1164–1174. doi: 10.1182/blood-2005-02-0687. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, et al. Lyon: International Agency for Research on Cancer (IARC); 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 3.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage KJ, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- 6.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenwald A, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 8.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savage KJ, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–3537. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 10.Mandelbaum J, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasqualucci L, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam W, et al. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- 13.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 16.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2010 doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green MR, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rui L, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steidl C, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471:377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iqbal J, et al. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia. 2007;21:2332–2343. doi: 10.1038/sj.leu.2404856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Annunziata CM, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabbri G, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011 doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keats JJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendez-Lago M, Morin R, Andrew J, Mungall A, Chan S, Marra M. Mutations In MLL2 and MEF2B Genes In Follicular Lymphoma and Diffuse Large B-Cell Lymphoma. Blood (ASH Annual Meeting Abstracts) 2010:116–473. [Google Scholar]
- 26.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011 doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cigudosa JC, et al. Cytogenetic analysis of 363 consecutively ascertained diffuse large B- cell lymphomas. Genes Chromosomes Cancer. 1999;25:123–133. [PubMed] [Google Scholar]
- 30.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad R, Zhadanov AB, Sedkov Y, Bullrich F, Druck T, Rallapalli R, Yano T, Alder H, Croce CM, Huebner K, Mazo A, Canaani E. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene. 1997 Jul 31;15(5):549–560. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- 32.Sneeringer CJ, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 35.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 37.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 39.Cresswell P, Ackerman AL, Giodini A, Peaper DR, Wearsch PA. Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol Rev. 2005;207:145–157. doi: 10.1111/j.0105-2896.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 40.Moingeon P, et al. CD2-mediated adhesion facilitates T lymphocyte antigen recognition function. Nature. 1989;339:312–314. doi: 10.1038/339312a0. [DOI] [PubMed] [Google Scholar]
- 41.Wang C, Lin GH, McPherson AJ, Watts TH. Immune regulation by 4-1BB and 4-1BBL: complexities and challenges. Immunol Rev. 2009;229:192–215. doi: 10.1111/j.1600-065X.2009.00765.x. [DOI] [PubMed] [Google Scholar]
- 42.Middendorp S, et al. Mice deficient for CD137 ligand are predisposed to develop germinal center-derived B-cell lymphoma. Blood. 2009;114:2280–2289. doi: 10.1182/blood-2009-03-208215. [DOI] [PubMed] [Google Scholar]
- 43.Lenz G, Staudt LM. Mechanisms of Disease: Aggressive Lymphomas. New England Journal of Medicine. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito M, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 47.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 48.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 49.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radtke I, et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106:12944–12949. doi: 10.1073/pnas.0903142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walter MJ, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A. 2009;106:12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 56.Ng SB, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 58.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jordanova ES, Riemersma SA, Philippo K, Schuuring E, Kluin PM. Beta2-microglobulin aberrations in diffuse large B-cell lymphoma of the testis and the central nervous system. Int J Cancer. 2003 Jan 20;103(3):393–398. doi: 10.1002/ijc.10824. [DOI] [PubMed] [Google Scholar]
- 60.Riemersma SA, et al. Extensive genetic alterations of the HLA region, including homozygous deletions of HLA class II genes in B-cell lymphomas arising in immune-privileged sites. Blood. 2000;96:3569–3577. [PubMed] [Google Scholar]
- 61.Tchernitchko D, Goossens M, Wajcman H. In silico prediction of the deleterious effect of a mutation: proceed with caution in clinical genetics. Clin Chem. 2004;50:1974–1978. doi: 10.1373/clinchem.2004.036053. [DOI] [PubMed] [Google Scholar]
- 62.Lin M, et al. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 63.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 64.Pounds S, et al. Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics. 2009;25:315–321. doi: 10.1093/bioinformatics/btn624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.