Abstract
Here we describe an automated method, which we call serial two-photon (STP) tomography, that achieves high-throughput fluorescence imaging of mouse brains by integrating two-photon microscopy and tissue sectioning. STP tomography generates high-resolution datasets that are free of distortions and can be readily warped in 3D, for example, for comparing multiple anatomical tracings. This method opens the door to routine systematic studies of neuroanatomy in mouse models of human brain disorders.
Since the pioneering work of Ramón y Cajal, advances in light microscopy have been central to many discoveries in neuroanatomy. The recent focus on systematic generation of whole-brain datasets, for example the Allen Mouse Brain Atlas for gene expression1 and the ongoing Mouse Brain Architecture Project for mesoscopic connectivity2, has created a pressing need for the development of new instrumentation for high-throughput whole-brain imaging. Here we describe a microscopy method, which we call serial two-photon (STP; pronounced “step”) tomography, that achieves automated high-throughput imaging of fluorescently labeled mouse brains. This method uses whole-mount two-photon microscopy3, 4 and thus generates datasets of well aligned, high-resolution serial optical sections. We tested the versatility of STP tomography by imaging four mouse brains with cell type-specific fluorescent protein expression and systematically mapping input and output connections of mouse somatosensory cortex. These experiments show that STP tomography is a robust imaging method that can transform the emerging field of systematic whole-brain anatomy, until now limited to dedicated atlasing initiatives1, 2, into a routine methodology applicable, for example, to the study of mouse models of human brain disorders in standard laboratory settings.
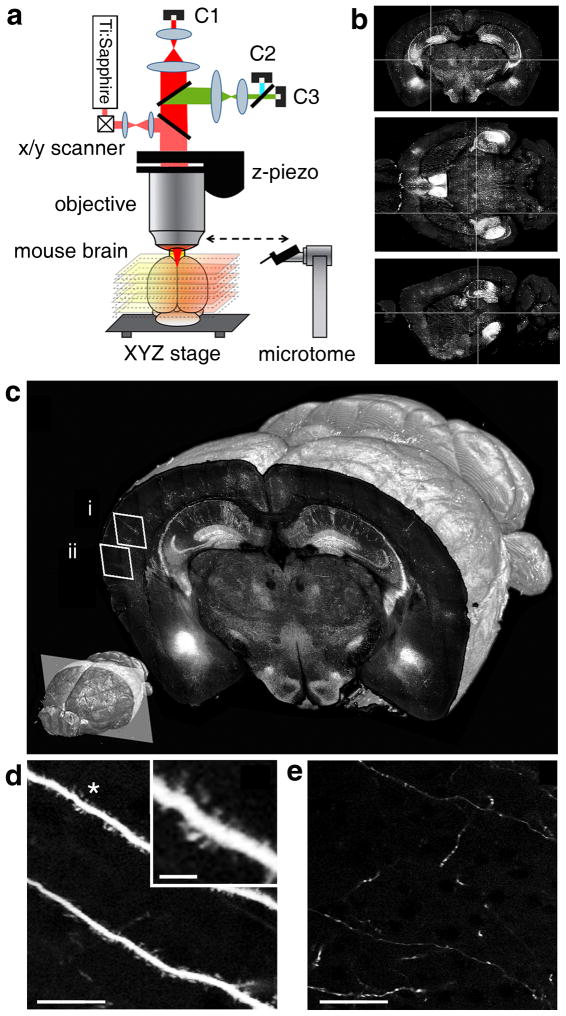
STP tomography works as follows (Fig. 1; Supplementary Video 1). First, a fixed agar-embedded mouse brain is placed in a water bath on integrated XYZ stages under the objective of a two-photon microscope and imaging parameters are entered in the operating software (Online Methods). Once these are set, the instrument works fully automatically: 1) the XYZ stages move the brain under the objective so that an optical section (or an optical Z-stack) is imaged as a mosaic of fields of view (FOVs); 2) a builtin vibrating blade microtome mechanically cuts off a tissue section from the top; and 3) the steps of overlapping optical and mechanical sectioning are repeated until the dataset is collected. The instrument is based on a previous prototype4 that was redesigned for imaging of fluorescently labeled mouse brains, including the integration of a custom-build vibrating blade microtome instead of a milling machine and the use of high-speed galvanometric scanners instead of a rotating polygonal scanner (Online Methods). Sectioning by vibrating blade microtome allows the use of brains prepared by simple procedures of formaldehyde fixation and agar embedding, which have minimal detrimental effects on fluorescence and brain morphology. High-speed galvanometric scanning enables fast imaging at different sampling resolutions.
Figure 1.
STP tomography. (a) Schema of the method. Computer-controlled XYZ stages move the brain under the objective of a two-photon microscope, so that the top view is imaged as a mosaic. The stages also deliver the brain to a built-in vibrating blade microtome for sectioning (Supplementary Video 1). (b) Coronal (top), horizontal (mid) and sagittal (bottom) views of a GFPM dataset of 260 coronal sections (Supplementary Fig. 1) after 3D reconstruction. (c) 3D view of a coronal section of the GFPM brain imaged with a 20x objective at 0.5 μm XY sampling. Lower left: position of the coronal plane in the imaged mouse brain (approximately −2.5 mm from Bregma). (d-e) Enlarged views from regions (i) and (ii) in panel (c), respectively, demonstrating visualization of dendritic spines (d) and fine axon fibers (e); scale bar = 25 μm (d-e) and 5 μm (insert in d). See Supplementary Figs. 2 and 3 for a comparison of different imaging conditions.
In the first set of experiments, we used Thy1-GFPM mice, which express green fluorescent protein (GFP) mainly in hippocampal and cortical pyramidal neurons, to determine the optimal conditions for imaging mouse brains at different sampling resolutions. We imaged the GFPM brain as a dataset of 260 coronal sections, evenly spaced by 50 μm, with 10x and 20x objectives at XY imaging resolution 2.0, 1.0 and 0.5 μm (Fig. 1; Supplementary Figs. 1–3; Supplementary Video 2). The 10x objective (0.6 NA) allows fast imaging at a resolution sufficient to visualize the distribution and morphology of GFP-labeled neurons, including their dendrites and axons (Supplementary Figs. 2–3). The data collection times for a 10×-objective dataset of 260 coronal sections are ~6 1/2 and 8 1/2 hrs at x-y sampling of 2 and 1 μm, respectively (Supplementary Table 1). The 20x objective (1.0 NA) enables visualization of dendritic spines and fine axonal arborizations (Fig. 1 and Supplementary Figs. 2–3; note that in this application axons are detected within single XY optical sections, but not traced in Z, because of the spacing of 50 μm between each section). The data collection times for a 260-section dataset using the 20x objective are ~15 1/2 and 24 hrs at x-y sampling of 1 and 0.5 μm, respectively (Supplementary Table 1). Taken together, these experiments show that STP tomography can be used as an automated method for collection of high resolution datasets of fluorescently labeled mouse brains.
Transgenic mice with cell type-specific fluorescent protein expression allow easy identification of different types of neurons and glia5. Next, we show examples of whole-brain mapping of different cell types in two BAC transgenic mice and one gene-targeted (knockin) mouse. The Mobp-GFP5 mouse reveals a pattern of whole-brain myelination as a result of GFP expression in oligodendrocytes from the promoter of myelin-associated oligodendrocyte basic protein (Mobp) (Supplementary Video 3). The ChAT-GFP5 mouse allows visualization of whole-brain cholinergic innervation as a result of GFP expression in cholinergic neurons from the choline acetyltransferase (ChAT) promoter (Supplementary Video 4). The SST-ires-Cre::Ai96 mouse reveals brain-wide distribution of somatostatin-expressing interneurons as a result of Cre recombinase expression from the somatostatin (SST) gene6 (Supplementary Video 5). These experiments show the ease of generating brain atlas-like datasets of cell-type distribution and innervation by STP tomography in transgenic mice. In addition, a more complete visualization of a specific cell-type distribution can be achieved by imaging Z-stack volumes, instead of single optical sections, between the steps of mechanical tissue sectioning. As an example of this application, we include a dataset of 800 optical sections (2.5 μm Z-spacing) revealing the distribution of all somatostatin-expressing interneurons in the olfactory bulbs of the SST-ires-Cre::Ai96 mouse (Supplementary Video 6 and 7). Imaging with high Z resolution, of course, increases the acquisition time and currently it would take about 7 days to image a whole mouse brain at the same resolution. However, increasing the imaging speed (at present 0.4 μs pixel residence time) by, for example, integration of resonant scanners, should make high Z-resolution imaging of whole mouse brains by STP tomography more practical in the future.
In the final set of experiments, we demonstrate the use of STP tomography for mapping brain connectivity by imaging mouse brains injected with anatomical tracers in the somatosensory barrel cortex, a brain region with projections well documented by both retro- and anterograde tracing7, 8. We imaged brains injected with CTB-Alexa-488 for retrograde tracing and adeno-associated virus expressing GFP (AAV-GFP) for anterograde tracing at 1 μm XY resolution (20x objective). As expected, Alexa-488-labeled neurons were found in brain areas known to project to the mouse barrel cortex7, 8 and GFP-labeled axons were detected in brain areas known to receive barrel cortex projections7 (Figs. 2 and 3, Supplementary Figs. 4 and 5). The experiments also revealed two brain regions with sparse labeling that were not previously reported in the literature: retrogradely labeled contralateral orbital cortex (Supplementary Fig. 4b panel 2) and anterogradely labeled contralateral motor cortex (Supplementary Fig. 5b panel 2). Taken together, the replication of the previously described pattern of connectivity and the detection of putative new connections in the contralateral cortical areas demonstrate that STP tomography is both a high-throughput and highly sensitive method for whole mouse brain anatomy. The 3D alignment of the datasets, in addition, facilitates direct comparison between different samples. We demonstrate this by warping the AAV-GFP brain onto the CTB Alexa-488 brain for direct comparison of anterograde and retrograde tracings (Fig. 3; Online Methods; Supplementary Video 7). We estimated the precision of co-registration of anatomical landmarks between two brains to be approximately 100 μm (Supplementary Fig. 7). Warping of multiple brains to one space thus provides a simple alternative to multiple tracer injections and can be extended to include many brains in a virtual brainbow9-like tracing.
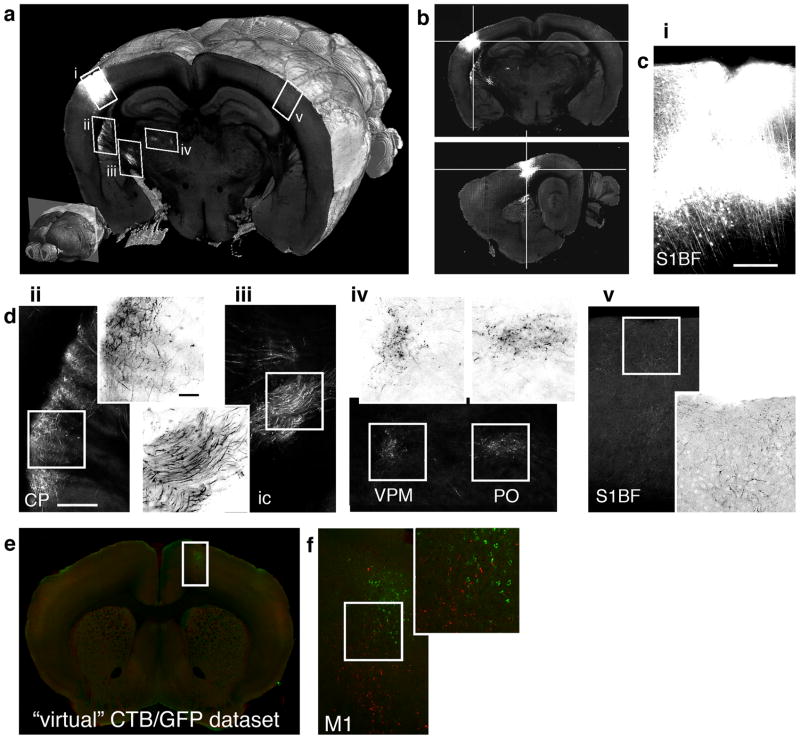
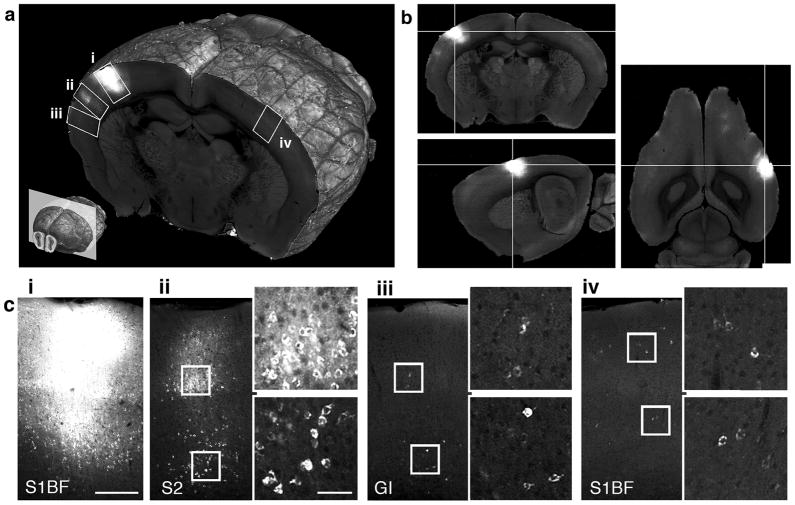
Figure 2.
Retrograde tracing by CTB-Alexa-488. (a) 3D view of a coronal section comprising the injection site (i) and several retrogradely labeled regions (ii-iv). Lower left: position of the section in the whole brain (approximately −1.15 mm from Bregma). (b) Coronal (top), sagittal (bottom), and horizontal views of the injection site. (c) Cortical regions marked up in (a), comprising: (i) the injection site in the barrel field of the primary somatosensory cortex (S1BF), (ii) ipsilateral secondary somatosensory cortex (S2), (iii) granular insular cortex (GI), and (iv) contralateral S1BF. The panels (ii-iv) are shown with enlarged regions from supragranular and infragranular cortical layers comprising CTB labeled cells. The scale bar is 250 μm in panel (i) and 50 μm in the enlarged view of panel (ii).
Figure 3.
Anterograde tracing by AAV-GFP and brain warping. (a) 3D view of a coronal section comprising the injection site (i) and several anterogradely labeled regions (ii-v). Lower left: position of the section in the whole brain (approximately −1.9 mm from Bregma). (b) Coronal (top) and sagittal (bottom) views of the injection site. (c) The injection site (S1BF). (d) Brain regions (ii-v) marked up in (a), comprising: (ii) ipsilateral caudoputamen (CP); (iii) axon fibers in the internal capsule (ic) (iv) ventral posteromedial thalamus (VPM) and posterior thalamus (PO), and (v) contralateral barrel cortex (S1BF). The enlarged views show inverted grayscale images for better visualization of axon fibers and varicosities. The scale bar in (c) and (d-ii) and the enlarged view of (d-ii) is 250 μm. (e) One section from a combined “virtual” two-tracer dataset generated by warping AAV-GFP brain onto CTB-Alexa-488 brain (see Supplementary Video 8). (f) Brain region marked up in (e) comprising motor cortex (M1) with overlapping anterograde (AAV-GFP, red color) and retrograde (CTB-Alexa-488, green color) labeling.
STP tomography is part of a growing field of automated methods for ex-vivo imaging of animal (mainly mouse) brain tissue1, 3, 10–14. The combination of serial imaging and sectioning was pioneered by block-face imaging in light microscopy15 and more recently adapted for two-photon microscopy3, 4, confocal microscopy16, and electron microscopy10. The use of two-photon microscopy allows deep tissue imaging, which is advantageous for focusing below the surface to obtain undisturbed optical sections and to collect high-resolution Z-stacks between sectioning steps, as pioneered in all-optical histology3.
What are the advantages of STP tomography compared to other methods for whole mouse brain imaging? One comparison is with robotized wide-field fluorescence microscopy of mounted serial brain sections1. This approach, however, is more laborious and prone to error, because it requires brain sectioning and mounting of the sections on microscopic slides. In addition, enhancements in scanning speed of STP tomography should allow the use of our method for complete whole-brain anatomical tracings and cell counts, whereas imaging of cut tissue sections will always result in some loss of data.
A second comparison is with light-sheet fluorescence microscopy (LSFM; also known as selective-plane illumination microscopy (SPIM)17 and ultramicroscopy11), which can image mouse brains without mechanical sectioning, but at a lower resolution11. Until now, the use of LSFM for whole mouse brain imaging has also been limited by the need for chemical clearing that quenches fluorescent proteins, but this may be overcome by the development of improved clearing reagents18. Finally, a third comparison is with micro-optical sectioning tomography (MOST14; also known as knife-edge scanning microscopy13), which combines bright-field line-scan imaging and ultra-microtome cutting and was used to image Golgi-stained mouse brains in two studies13, 14. This method, however, has yet to be implemented for fluorescence microscopy and since the tissue preparation requires dehydration and plastic embedding, which decrease GFP intensity, it is not clear how MOST will perform for anatomical tracing and other applications described in our study.
In summary, we conclude that STP tomography can be used to generate high-resolution anatomical datasets that can be readily warped for comparison of multiple brains. STP tomography is particularly well-suited for systematic studies of brain anatomy in genetic mouse models of cognitive disorders, such as autism and schizophrenia. To provide quantitative measurements for such studies, we are currently focusing on anatomical registration19 and the development of computational methods for detection of fluorescence signals in whole-brain datasets generated by STP tomography.
Online Methods
Tissue Preparation
The following mouse strains were used: GENSAT ChAT-GFP Tg(Chat-EGFP)GH293Gsat/Mmcd (#000296-UCD) and Mobp-GFP Tg(Mobp-EGFP)103Gsat/Mmcd (#030483-UCD)5 (MMRRC; www.mmrrc.org); GFPM20; SST-ires-Cre::Ai96; and wild type C57BL/6J (Jax mice; # 000664). As anatomical tracers, we used CTB Alexa Fluor-488 (Invitrogen; #C22841; 0.5 % wt/vol in phosphate buffer) and AAV-GFP with synapsin promoter21, 22. AAV was produced at the Salk vector core (http://vectorcore.salk.edu) as a chimeric 1/2 serotype23, purified by iodoxinal gradient and concentrated to 5.3 × 1011 genomic copy per ml. Stereotaxic injections of the tracers were done as described24. Briefly, the mice were anaesthetized by 1% isoflurane inhalation. A small craniotomy (approximately 300 × 300 μm) was opened over the left primary somatosensory cortex and ~50 nl of virus or 50 nl of 0.05 % CTB Alexa FluorR 488 was injected into layer 2/3 barrel cortex at stereotaxic coordinates: caudal 1.6, lateral 3.2, ventral 0.3 mm relative to bregma. The skin incision was then closed with silk sutures, and the mice were allowed to recover with free access to food and water (meloxicam was given at 1 mg/kg, s.c. for analgesia). The brains were prepared for imaging 10–14 days later (see below).
The mouse brains were prepared for STP tomography as follows. The mice were deeply anesthetized by intraperitoneal (i.p.) injection of the mixture of ketamine (60 mg/kg) and medetomidine (0.5 mg/kg) and transcardially perfused with ~15 ml cold saline (0.9 % NaCl) followed by ~30 ml cold neutral buffered formaldehyde (NBF, 4% w/v in phosphate buffer, pH 7.4). The brains were dissected out and post-fixed in 4% NBF overnight at 4 °C. In order to decrease formaldehyde-induced autofluorescence, the brains were incubated in 0.1 M glycine (adjusted to pH 7. 4 with 1M Tris base) at 4 °C for 2–5 days. The brains were then washed in phosphate buffer (PB) and embedded in 3–5% oxidized agarose as described25, 26. Briefly, agarose (Sigma, cat.# A6013) was oxidized by stirring in 10 mM sodium periodate (NaIO4, Sigma cat.# S1878) solution for 2 hrs at RT, washed 3x and re-suspended in PB to bring the final concentration to 3–5 %. The mouse brain was pat-dried and embedded in melted oxidized agarose using a cube-shaped mold. Covalent crosslinking between brain surface and agarose was activated by equilibrating in excess of 0.5–1 % sodium borohydrate (NaBH4, Sigma cat.# 452882) in 0.05 M sodium borate buffer (pH = 9.0–9.5), gently shaking for 2–4 hrs at RT (or overnight at 4 °C) (note that after rinsing, activated agarose can be stored in PB at 4 °C for up to one week; sodium borohydrate buffer should be prepared fresh). Covalent crosslinking of the agar-brain interface is important to keep the brain firmly embedded during sectioning and to limit shadowing artifacts by insufficiently cut meninges (see Troubleshooting, below).
The Instrument and Software
The experiments were performed on a high speed multiphoton microscope27 with integrated vibratome sectioning (TissueCyte 1000, TissueVision, Inc.; Supplementary Video 1 shows the instrument schema). Laser light from a titanium sapphire laser (Chameleon Ultra, Coherent) is directed through a tube and scan lens assembly towards a pair of galvanometer mirrors (6210H, Cambridge Technology) and reflected by a short pass dichroic towards a microscope objective (either a 20x Olympus XLUMPLFLN20XW lens, NA 1.0, or a 10x Olympus XLUMPLFL10XW-SP lens, NA 0.6). The fluorescent signal from the sample is collected by the same objective, passes through the dichroic and is directed by a series of mirrors and lens onto a photomultiplier tube detection system (PMT, Hamamatsu, R3896). In two- and three-channel multicolor configuration the emission light is split by dichroic mirror(s) onto, respectively, two and three PMTs to allow for simultaneous multichannel data acquisition. 3D scanning of Z-volume stacks is achieved via a microscope objective piezo (PI E-665 LVPZT amplifier, P-725 PIFOC long-travel objective scanner), which translates the microscope objective with respect to the sample. Laser light intensity can be varied by liquid crystal controller (Thorlabs, LCC25) for shuttering purposes and as a function of imaging depth into the sample.
Robust mechanical sectioning is achieved by a vibrating blade microtome that is integrated into the imaging system. It is based on a novel dual flexure design. Flexures are compliant mechanisms consisting of a series of rigid bodes connected by compliant elements that are designed to produce geometrically well defined motion upon application of force. Flexures can achieve smooth displacements down to the sub-micron level with little parasitic motion. The microtome consists of a primary flexure to which the blade is mounted and a secondary flexure which connects the primary flexure to the actuator. The actuator consists of a DC motor with an off-center cam attached to the shaft. The secondary flexure is designed to be rigid in the direction of the cut and compliant in all other directions. In this way, only a force along the direction of the cut is transmitted to the primary flexure which holds the microtome blade and reduces any potential parasitic motion along unwanted axis of motion. For this design, we experimentally verified that the parasitic Z-vertical deflection was less than 2 μm RMS by measuring the motion directly with capacitive sensors. Additional details of the microtome design will be presented in a forthcoming manuscript by Drs. Shih-Chi Chen and Martin Culpepper (MIT, Cambridge, MA). The vibration frequency can be set between 0–60 Hz and the blade angle between 5–30 degrees. By the use of different cams, the amplitude can be adjusted from 0.8 mm to 2 mm. We empirically determined the sectioning parameters for brain tissue to be 0.8 mm amplitude at 60 Hz and at a blade angle of 11 degrees (the blade was Vibroslice from WPI). The reliability of sectioning was verified by measurements of the brain surface and overlapping Z-planes before and after sectioning during a whole brain dataset (Supplementary Fig. 6). As emphasized in the Troubleshooting section below, to achieve reliable sectioning it is essential to use brains covalently crosslinked in oxidized agarose (see Tissue Preparation, above).
The instrument is controlled by custom software, written in C++ and C#. It handles the scanning, stage motion, microtome control, and data acquisition. The software is comprised of several discrete services, each of which controls a particular hardware component or function of the instrument. Sequences of events are coordinated by a master orchestrator service. For instance, in order to scan a section, a command is sent from the orchestrator service to the galvanometer scanner service commanding it to unshutter the laser and scan an image. The orchestrator service waits until the scanner service reports that the image acquisition has been completed, and then sends a command to the XY stages to move the sample to the next position. Once the XY stages completes the requested motion, a command is sent back to the orchestrator service, which in turn issues a command to the scanner service to acquire a second image. During the imaging, background services handle the data acquisition and saving of the 16 bit TIFF images to a local or network attached storage device. The process continues until an entire section has been acquired. Similarly, to acquire a whole-brain dataset, at the end of each mosaic section acquisition the orchestrator service commands the Z-stage service to move the sample upwards by the desired slice thickness. Simultaneously, the sample is directed towards the microtome by the XY stage service. Once in position, the microtome is turned on and the sample is translated through the microtome and a tissue section is cut. The sample is then translated back underneath the objective, and the next section is imaged. This process repeats until all sections are imaged. The software is highly modular and additional services can be introduced or specific hardware can be exchanged with minimal changes to higher level routines. For instance, services to automate additional features, such as the capture of the slices after sectioning, can be added in the future.
In comparison to an earlier prototype4 there are a substantial number of improvements in the design of the current instrument. The previous version used a milling machine to machine the surface of a paraffin embedded tissue. Because paraffin quenches fluorescence, an integrated vibrating blade microtome (Supplementary Video 1) is now used. This allows imaging of formaldehyde fixed brains embedded in agar, a histological procedure with low quenching. As an additional advantage, the sections can be used for further histochemical analysis as they are no longer destroyed by the milling process (currently, the sections sink to the bottom of the water bath and can be collected and sorted at the end of the experiment). The incorporation of low-magnification (10–20X) high-numerical aperture (NA 0.6–1.0) lenses has increased fluorescence collection compared to a standard 60× objective, without compromising the resolution at large imaging depths28. The combination of a low-magnification lens with large aperture optics have increased the image field of view that can be scanned with even illumination from ~200 to 1400 μm. High speed galvanometric scanning has replaced a polygonal scanning approach. Galvanometric scanners are far more flexible than polygonal scanners and allow a wide range of pixel sizes and residence times to be set depending on the requirements of the sample. Finally, high speed XYZ stages were constructed to allow positioning of the sample over centimeters of travel with sub-micron accuracy. The custom Z-stage was designed to hold two commercial X and Y stages (Polytech, PI) and be rotationally rigid with a pitch and yaw of less than 1 micron over the entire travel range of the X and Y stage assembly. The X and Y axes have a 0.1 μm positional accuracy, a settling time of 0.1 ms and a speed up to 50 mm/s. The high speed and small settling time allows for rapid positioning of the sample and minimizes acquisition time of a section, while the positional accuracy decreases post-processing registration time. The Z-axis has a precision of 0.15 μm, maximum velocity of 1 mm/s and maximum Z travel distance 5 cm. Since this stage is only used to raise the sample to the microtome blade and objective, its speed has negligible impact on the imaging time.
The Instrument Operation
Once the brain is positioned under the objective and the imaging and sectioning parameters are chosen (see below), the instrument operates in a fully automated mode. The brain is mounted in saline (we used 50 mM PB, pH 7.4) in a water bath positioned on the computer controlled XYZ stages. After identifying Z-position of the brain surface under the objective, the following parameters are set in the software: FOV size, FOV mosaic size, pixel size, pixel residence time, laser power, sectioning speed, sectioning frequency, Z-step for each sectioning cycle and a number of Z sections. The imaging plane is set below the brain surface to ensure an undisturbed optical section throughout. We typically use 50 μm below surface, but a comparable image resolution can be obtained down to about 100 μm below surface with small adjustments in laser power. The laser power is set constant for imaging of single optical sections between each sectioning steps. For collection of Z-volumes between sectioning steps, such as the dataset of SST-ires-Cre::Ai9 olfactory bulb imaged at Z-resolution 2.5 μm (Supplementary Video 6), the laser power is adjusted based on the Z depth to compensate for increased light scattering with increased depth.
The number of FOV tiles per mosaic is set to cover the extent of the sample and allow for a small overlap between the FOV tiles for post-processing stitching (see below). The experiments with the 10X objective employ 6 × 8 overlapping mosaic of 1.66 × 1.66 mm FOV, the XY stage movement is 1.5 mm, pixel size 1 or 2 μm and pixel residence time between 0.4 to 1.0 μs. The experiments with the 20x objective employ 11 × 17 mosaic of 0.83 × 0.83 mm FOV, the XY stage movement is 0.7 mm, pixel size 0.5 or 1μm and pixel residence time between 0.4 to 1 μs. Once a mosaic is completed, the same XYZ stages used for the mosaic imaging move the sample from the microscope objective towards a vibrating blade microtome to section the uppermost portion of the tissue. The times for imaging of 260 section mouse brain datasets are given Table 1.
Image processing
The images are constructed from the PMT signal, with the tile and pixel size set by a combination of the scan angle and pixel sampling rate. The tiles are saved as tif files (named as Tile_Z{zzz}_Y{yyy}_X{xxx}.tif) and processed in the following way. First, each tile is cropped to remove illumination artifacts near the edges (the number of pixels cropped is determined empirically based on the objective used and FOV; e.g. we crop 15 and 10 pixels at each side of X and Y direction, respectively, for 832 × 832 pixel FOV). Second, all tiles from one brain dataset (for example 52,360 tiles for 11×17 mosaic of 280 sections) are loaded in Fiji (ImageJ-based image-processing software; http://fiji.sc/wiki/index.php/Fiji) and used to generate an average-intensity image for illumination correction by a Z-project function. Third, all tiles are divided by the average-intensity image to correct for uneven illumination (Plugins > TissueVision > Divide sequence by image). Fourth, illumination-corrected tiles are used to stitch the sequence of mosaic images (Plugins > Stitching > Stitch Sequence of Grids of Images; fusion method = linear blending, fusion alpha = 1.5, regression threshold = 0.3, max/avg displacement = 2.5, absolute displacement = 3.5; select “compute overlap”). The transformation between the tiles is modeled as a translation transform. For each section, the X and Y translations are determined by cross correlation29 between the tiles. At the overlapping regions, the pixels are blended linearly30, 31. The overlapping regions may show some photobleaching when large power (>150 mW) is used for samples with low fluorescence. In such case, since bleaching occurs mainly for the second overlapping tile, it is better to display the image from the first tile and use the second tile only for XY registration. This can be achieved by rendering the tiles into the mosaic in the reverse order they where scanned by the microscope: the pixels of the first scanned tile overwrite the same pixels scanned later in the second. The whole brain dataset of 11×17 mosaic of 280 sections of raw tiles scanned at a 16-bit depth occupies ~40GB. The final stitched slices occupy ~25GB with LZW compression on the final stitched TIFF slices. All image processing was run on Mac/Linux desktop machines with at least 8 GB on RAM.
Image Warping
The warping was done by an affine registration followed by an elastic B-spline-based transformation32 using autofluorescence signal from STP tomography datasets downsampled by factor of 20 (resolution 20 × 20 × 50 μm). The registration was done in a multi-resolution approach for a more efficient and robust alignment33. The affine transform was calculated using 4 resolution levels while the elastic step uses 6 resolution steps. Advanced Mattes mutual information34 was used as the metric to measure the similarity of registration. In this parametric registration method, Mattes Mutual information is used as the similarity measure between the moving and fixed images. The registration problem is posed as an optimization problem, where the image discrepancy/similarity function is minimized for a set of transformation parameters. The transformation parameters are then estimated in multi-resolution approach, which ensures a more robust approach compared to a single resolution approach. The image similarity function is estimated and minimized for a set of randomly chosen samples with the images at each resolution in a iterative way. On a 8 core CPU with 16 GB RAM, the registration takes 12 hrs on 650×450×300 sized image with 20×20×50 micron pixel spacing. The entire image warping experiment is setup using elastix32, an image registration tool based on Kitware’s ITK with Parameters setup according to our dataset. To determine the effectiveness of the warping procedure, we compared the displacement of 42 anatomical manually identified landmark points of interest in two mouse brain scans before and after warping one dataset onto the other (Supplementary Fig. 7). The mean (± SEM) distance between the corresponding points in the two brains was 749.5 ± 52.1 and 102.5 ± 45 μm before and after warping, respectively.
Troubleshooting
We chose to establish STP tomography for imaging formaldehyde-fixed brains embedded in agar, because this provides maximal preservation of GFP fluorescence. However, vibrating microtome-based sectioning of a relatively soft agar-brain block is mechanically more challenging than sectioning of hard resin-embedded tissue and can lead to imaging artifacts. First, the brain may become “loose” in the agar block after many hours of imaging and this can cause the remaining part of the brain to fall out of the agar or the brain edges to shift out of the imaging plane. These problems can be completely prevented by covalent crosslinking of brain-agar after embedding (see above). Second, brain meninges sometime fail to be completely cut with every sectioning step and the remaining small stripes of meninges can cause shadowing artifacts during imaging. This problem is again largely solved by brain-agar crosslinking, but small uncut meninges sometimes remain in the regions between hemispheres that are not in contact with agar (this is seen in a few sections in our datasets; an example from the Thy1-GFPM dataset is shown in Supplementary Fig. 8a). These artifacts are infrequent and only near the brain surface and thus are unlikely to have a substantial effect on data interpretation in most experimental applications. Carefully removing the meninges under a dissecting microscope will help to limit this problem. Third, in the current configuration of the instrument, cut brain sections freely sink to the bottom of the water bath. However, a floating section may sometimes remain between the objective and the brain block, causing substantial shadowing artifacts (an example from the Thy1-GFPM dataset is shown in Supplementary Fig. 8b). This happens infrequently (typically once per several samples) and can be largely avoided by setting a longer waiting time after each sectioning step in the instrument software (we used only a 1 sec waiting time for fast data collection). Recent software update also added a lateral movement of the stage at the end of sectioning, which helps to direct the sinking section away from the objective. In addition, we are working on integrating a section retrieval module, which will allow an automated capture of each brain section and eliminate the problem with floating sections. Finally, meninges between the frontal cortex and the accessory olfactory bulb sometimes show very bright autofluorescence (an example from the Chat-GFP is shown in Supplementary Fig. 8c). This only occurs in the rostral brain areas and since it is on the brain surface it does not affect data interpretation.
Supplementary Material
Supplementary Figure 1. 2D montage of a GFPM STP-tomography dataset.
Supplementary Figure 2. XY sampling resolutions for imaging dendritic spines.
Supplementary Figure 3. XY sampling resolutions for imaging axons.
Supplementary Figure 4. Retrograde tracing by CTB-Alexa-488.
Supplementary Figure 5. Anterograde tracing by AAV-GFP.
Supplementary Figure 6. Evaluation of Z-plane consistency before and after sectioning.
Supplementary Figure 7. Quantification of warping accuracy.
Supplementary Figure 8. Troubleshooting.
Supplementary Table 1. Imaging conditions for STP tomography.
Supplementary Video 1. STP tomography. The schema shows a representation of the instrument, including the X, Y, and Z stages, water bath with brain sample, and vibratome. Optical excitation path from Ti:sapphire laser is drawn as a dashed red line; the laser beam is scanned using galvanometric X- and Y-axis mirrors; fluorescence from the sample is collected via an optical detection path, drawn as a dashed green light, and detected with photomultiplier tube (PMT). The video (after pressing “play” button) shows a zoom-in view of the water bath during imaging and cutting of a brain sample, with a simultaneous mosaicing of a coronal GFPM brain section.
Supplementary Video 2. 260-coronal section datasets showing 3D reconstruction of the GFPM mouse brain. The STP tomography 2D brain images were downsampled by factor of 5.
Supplementary Video 3. 260-coronal section datasets showing 3D reconstruction of the Mobp-GFP mouse brain. The STP tomography 2D brain images were downsampled by factor of 5.
Supplementary Video 4. 260-coronal section datasets showing 3D reconstruction of the ChAT-GFP mouse brain. The STP tomography 2D brain images were downsampled by factor of 5.
Supplementary Video 5. 260-coronal section datasets showing 3D reconstruction of SST-ires-Cre::Ai9 mouse brain. The STP tomography 2D brain images were downsampled by factor of 5.
Supplementary Video 6. 800-coronal section dataset showing 3D reconstruction of olfactory bulbs from the SST-ires-Cre::Ai9 mouse brain, imaged at 2.5 μm Z-resolution. The STP tomography 2D images were downsampled by factor of 5.
Supplementary Video 7. The same SST-ires-Cre::Ai9 olfactory bulb dataset as shown in Supplementary Video 6 after optical re-slicing in YZ at isotropic 2.5 × 2.5 × 2.5 μm.
Supplementary Video 8. AAV-GFP brain was warped onto CTB-Alexa-488 brain and the datasets are shown as overlays in red and green color, respectively. The 2D brain images were downsampled by factor of 20 before warping.
Acknowledgments
We thank Y. Bao and R. Palaniswamy for expert technical assistance, S.C. Chen and M. Culpepper (MIT, Boston) for design of the flexure-based microtome, Z. J. Huang (CSHL, CSH) for the SST-ires-Cre::Ai9 mouse, W. Denk (Max Planck Institute for Medical Research, Heidelberg), K. Rockland (MIT, Boston), and F. Scalia (State University of New York, Brooklyn) for critical reading of the manuscript, and J. Kuhl for art graphics and animation. This work was supported by Simons Foundation grant (137480) to P.O., McKnight Technological Innovations in Neuroscience Award to P.O., HHMI Collaborative Innovation Award #43667 to H.S.S., and NIH grant 1 R43 HL093897-01 to T.R.
Footnotes
Conflict of Interest
Karsten Bahlmann and Timothy Ragan are shareholders and employees of TissueVision, Inc., Jason Sutin is an employee of TissueVision Inc., Pavel Osten is a consultant of TissueVision Inc. and a shareholder and consultant of Certerra Inc.
Author Contribution
J.S., T.R., K.B. and P.O. planned the redesign of the instrument, T.R., J.S., and K.B. engineered and built the instrument, T.R and J.S. wrote the operating software, L.R.K did all imaging experiments, J.T. contributed to early imaging experiments, Y.K. validated warping accuracy, K.U.V. and I.A-C. set up all image processing methods, H.S.S. supervised image processing, T.R and K.B. contributed to writing instrument portions of the paper, P.O. supervised imaging, coordinated all work, and wrote the paper.
References
- 1.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 2.Bohland JW, et al. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS computational biology. 2009;5:e1000334. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai PS, et al. All-optical histology using ultrashort laser pulses. Neuron. 2003;39:27–41. doi: 10.1016/s0896-6273(03)00370-2. [DOI] [PubMed] [Google Scholar]
- 4.Ragan T, et al. High-resolution whole organ imaging using two-photon tissue cytometry. Journal of biomedical optics. 2007;12:014015. doi: 10.1117/1.2435626. [DOI] [PubMed] [Google Scholar]
- 5.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 6.Taniguchi H, et al. A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronoff R, et al. Long-range connectivity of mouse primary somatosensory barrel cortex. The European journal of neuroscience. 2010;31:2221–2233. doi: 10.1111/j.1460-9568.2010.07264.x. [DOI] [PubMed] [Google Scholar]
- 8.Hoffer ZS, Arantes HB, Roth RL, Alloway KD. Functional circuits mediating sensorimotor integration: quantitative comparisons of projections from rodent barrel cortex to primary motor cortex, neostriatum, superior colliculus, and the pons. The Journal of comparative neurology. 2005;488:82–100. doi: 10.1002/cne.20579. [DOI] [PubMed] [Google Scholar]
- 9.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 10.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS biology. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodt HU, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nature methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 12.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayerich D, Abbott L, McCormick B. Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. Journal of microscopy. 2008;231:134–143. doi: 10.1111/j.1365-2818.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- 14.Li A, et al. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science. 2011;330:1404–1408. doi: 10.1126/science.1191776. [DOI] [PubMed] [Google Scholar]
- 15.Odgaard A, Andersen K, Melsen F, Gundersen HJ. A direct method for fast three-dimensional serial reconstruction. Journal of microscopy. 1990;159:335–342. doi: 10.1111/j.1365-2818.1990.tb03038.x. [DOI] [PubMed] [Google Scholar]
- 16.Sands GB, et al. Automated imaging of extended tissue volumes using confocal microscopy. Microscopy research and technique. 2005;67:227–239. doi: 10.1002/jemt.20200. [DOI] [PubMed] [Google Scholar]
- 17.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 18.Hama H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of. Nature neuroscience. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 19.Hawrylycz M, et al. Digital atlasing and standardization in the mouse brain. PLoS computational biology. 2011;7:e1001065. doi: 10.1371/journal.pcbi.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 21.Kugler S, Lingor P, Scholl U, Zolotukhin S, Bahr M. Differential transgene expression in brain cells in vivo and in vitro from AAV-2 vectors with small transcriptional control units. Virology. 2003;311:89–95. doi: 10.1016/s0042-6822(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 22.Dittgen T, et al. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:18206–18211. doi: 10.1073/pnas.0407976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauck B, Chen L, Xiao W. Generation and characterization of chimeric recombinant AAV vectors. Mol Ther. 2003;7:419–425. doi: 10.1016/s1525-0016(03)00012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cetin A, Komai S, Eliava M, Seeburg PH, Osten P. Stereotaxic gene delivery in the rodent brain. Nat Protocols. 2007;1:3166–3173. doi: 10.1038/nprot.2006.450. [DOI] [PubMed] [Google Scholar]
- 25.Shainoff JR. Glyoxal agarose and zonal immobilization of proteins therewith. The Clevelend Clinic Foundation; US: 1982. [Google Scholar]
- 26.Sallee CJ, Russell DF. Embedding of neural tissue in agarose or glyoxyl agarose for vibratome sectioning. Biotech Histochem. 1993;68:360–368. doi: 10.3109/10520299309105642. [DOI] [PubMed] [Google Scholar]
- 27.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 28.Oheim M, Beaurepaire E, Chaigneau E, Mertz J, Charpak S. Two-photon microscopy in brain tissue: parameters influencing the imaging depth. Journal of neuroscience methods. 2001;111:29–37. doi: 10.1016/s0165-0270(01)00438-1. [DOI] [PubMed] [Google Scholar]
- 29.Kuo D, Girod B. Direct estimation of displacement histograms. Proceedings of the Optical Society of America Meeting on Understanding and Machine Vision. 1989;7376 [Google Scholar]
- 30.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardona A, et al. Identifying Neuronal Lineages of Drosophila by Sequence Analysis of Axon Tracts. The Journal of Neuroscience. 2010;30:7538–7553. doi: 10.1523/JNEUROSCI.0186-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. Elastix: a toolbox for intensity based medical image registration. IEEE Transactions on Medical Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 33.Lester H, Arridge SR. A survey of hierarchical non-linear medical image registration. Pattern Recognition. 1999;32:129–149. [Google Scholar]
- 34.Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. PET-CT image registration in the chest using free-form deformations. IEEE Transactions in Medical Imaging. 2003;22:120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. 2D montage of a GFPM STP-tomography dataset.
Supplementary Figure 2. XY sampling resolutions for imaging dendritic spines.
Supplementary Figure 3. XY sampling resolutions for imaging axons.
Supplementary Figure 4. Retrograde tracing by CTB-Alexa-488.
Supplementary Figure 5. Anterograde tracing by AAV-GFP.
Supplementary Figure 6. Evaluation of Z-plane consistency before and after sectioning.
Supplementary Figure 7. Quantification of warping accuracy.
Supplementary Figure 8. Troubleshooting.
Supplementary Table 1. Imaging conditions for STP tomography.
Supplementary Video 1. STP tomography. The schema shows a representation of the instrument, including the X, Y, and Z stages, water bath with brain sample, and vibratome. Optical excitation path from Ti:sapphire laser is drawn as a dashed red line; the laser beam is scanned using galvanometric X- and Y-axis mirrors; fluorescence from the sample is collected via an optical detection path, drawn as a dashed green light, and detected with photomultiplier tube (PMT). The video (after pressing “play” button) shows a zoom-in view of the water bath during imaging and cutting of a brain sample, with a simultaneous mosaicing of a coronal GFPM brain section.
Supplementary Video 2. 260-coronal section datasets showing 3D reconstruction of the GFPM mouse brain. The STP tomography 2D brain images were downsampled by factor of 5.
Supplementary Video 3. 260-coronal section datasets showing 3D reconstruction of the Mobp-GFP mouse brain. The STP tomography 2D brain images were downsampled by factor of 5.
Supplementary Video 4. 260-coronal section datasets showing 3D reconstruction of the ChAT-GFP mouse brain. The STP tomography 2D brain images were downsampled by factor of 5.
Supplementary Video 5. 260-coronal section datasets showing 3D reconstruction of SST-ires-Cre::Ai9 mouse brain. The STP tomography 2D brain images were downsampled by factor of 5.
Supplementary Video 6. 800-coronal section dataset showing 3D reconstruction of olfactory bulbs from the SST-ires-Cre::Ai9 mouse brain, imaged at 2.5 μm Z-resolution. The STP tomography 2D images were downsampled by factor of 5.
Supplementary Video 7. The same SST-ires-Cre::Ai9 olfactory bulb dataset as shown in Supplementary Video 6 after optical re-slicing in YZ at isotropic 2.5 × 2.5 × 2.5 μm.
Supplementary Video 8. AAV-GFP brain was warped onto CTB-Alexa-488 brain and the datasets are shown as overlays in red and green color, respectively. The 2D brain images were downsampled by factor of 20 before warping.