Abstract
Twin-arginine translocation (Tat) denotes a protein transport pathway in bacteria, archaea and plant chloroplasts, which is specific for precursor proteins harbouring a characteristic twin-arginine pair in their signal sequences. Many Tat substrates receive cofactors and fold prior to translocation. For a subset of them, proofreading chaperones coordinate maturation and membrane-targeting. Tat translocases comprise two kinds of membrane proteins, a hexahelical TatC-type protein and one or two members of the single-spanning TatA protein family, called TatA and TatB. TatC- and TatA-type proteins form homo- and hetero-oligomeric complexes. The subunits of TatABC translocases are predominantly recovered from two separate complexes, a TatBC complex that might contain some TatA, and a homomeric TatA complex. TatB and TatC coordinately recognize twin-arginine signal peptides and accommodate them in membrane-embedded binding pockets. Advanced binding of the signal sequence to the Tat translocase requires the proton-motive force (PMF) across the membranes and might involve a first recruitment of TatA. When targeted in this manner, folded twin-arginine precursors induce homo-oligomerization of TatB and TatA. Ultimately, this leads to the formation of a transmembrane protein conduit that possibly consists of a pore-like TatA structure. The translocation step again is dependent on the PMF.
Keywords: TatA pore, TatBC receptor, TatA-type protein, TatC-type protein, proofreading chaperones, twin-arginine translocation (Tat)
1. Introduction
Tat translocases are found in bacteria, archaea and plant chloroplasts. They are distinguished from other protein translocation systems, like the Sec system, by their ability to translocate fully folded proteins across lipid bilayers. The most prominent representatives of bacterial Tat substrates are redox proteins involved in anaerobic respiration, proteins required for biogenesis and remodelling of the cell envelope, and virulence proteins [1–3]. Many of these Tat substrates acquire cofactors by complex cytosolic insertion processes; others are composed of several subunits, of which only one is equipped with a Tat-specific signal sequence. Obviously, these Tat substrates can target their Tat translocases only after they have adopted their final ternary or quaternary structure. For other Tat substrates that remain monomeric or do not contain any cofactor, such as for most Tat substrates directed to the thylakoids of chloroplasts, fast folding kinetics have been hypothesized to necessitate export by the Tat pathway. Owing to the folded state of their substrate proteins, Tat translocases are confronted with a large spectrum of substrate sizes and diameters.
The second remarkable characteristic of Tat protein transport systems is the signal sequence of their substrate proteins. These signal sequences carry the consensus motif S-R-R-x-F-L-K with the name-giving arginine pair (twin-arginine (RR) precursors) [4]. Other than that, Tat signal sequences, in general, are cleavable and share the canonical tripartite structure with Sec-targeting signal peptides, i.e. an amino-terminal basic n-region, a central hydrophobic domain and a carboxy-terminal region ending with the recognition sequence for signal peptidase I.
Our current knowledge on how Tat translocases are structured and function is still rather limited owing to the scarcity of high-resolution structural information. Nevertheless, recent biochemical data have led to new insights that seem to allow a more detailed picture at least of the first stages of the Tat pathway. The main purpose of this review is to screen old and new facts for their common ground on which a model for membrane targeting of Tat substrates and the formation of a translocase structure can be built. After a brief compilation of the basic features of Tat signal sequences and Tat substrates, homologues of TatC and TatA are introduced as the building blocks of Tat translocases. Their propensity to form homo- and hetero-oligomeric complexes is discussed and, according to the available data, the Tat pathway is pictured in stepwise events. Further aspects, such as energetization of the transport and proofreading of substrates are viewed. Because of the multitude of data collected for the Tat translocation machinery of Escherichia coli, we will primarily focus on this organism but continuously discuss similarities and dissimilarities with the Tat systems of plant chloroplasts and also Gram-positive bacteria. We refer the interested reader to excellent reviews by our colleagues, each one with an individual main emphasis [1,3,5–7].
2. Tat signal peptides and Tat substrates
(a). Specific features of Tat signal sequences
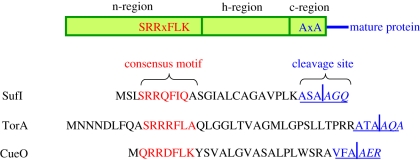
As illustrated in figure 1, the consensus motif S-R-R-x-F-L-K (with x denominating a polar amino acid) is located at the distal end of the n-regions of Tat signal peptides. The RR-pair is almost invariant and a conservative substitution by a KK-pair, in general, abolishes translocation. There seem to be a few natural exceptions having one Arg replaced by a Lys, Asn or Gln [2,8–10] and corresponding mutations introduced into various Tat precursors are partially tolerated [11–14].
Figure 1.
Tat signal sequences. Shown are the general composition of (top) Tat signal sequences and the amino acid sequences of three natural Tat precursor proteins of E. coli. SufI and TorA are the most common Tat substrates used for investigations of the Tat pathway and CueO is a copper oxidase, the RR-signal peptide of which comes closest to the consensus motif.
However, a mere RR-pair is not sufficient to direct a precursor to the Tat route [7]. Other residues of the extended Tat-specific consensus motif S-R-R-x-F-L-K (figure 1) are also important and accordingly are found conserved in more than 50 per cent of cases [15]. Mutations at these positions often affect the translocation efficiency of a particular substrate, but the effect can vary between different Tat signal sequences [12,16]. Mutational analysis of the consensus sequence further revealed that an increase in the overall hydrophobicity of the n-region can direct a Tat substrate to the Sec pathway [17].
The same effect has been described for increasing the hydrophobicity of the h-region of a Tat signal peptide [18]. A statistical analysis revealed that Tat signal peptides of Gram-negative bacteria are, in general, less hydrophobic than Sec signals owing to more Gly and fewer Leu residues [18].
Besides containing the recognition sequence for signal peptidase I [19,20], the c-region of Tat signal peptides is often enriched in basic residues when compared with classical Sec signal sequences [15,17]. In fact, these basic amino acids were shown to function as a Sec avoidance signal [21,22] and together with the overall charge of the adjacent N-terminus of the Tat client they constitute a Tat specificity determinant [23].
(b). Folding and other characteristics of Tat substrates
Using fusions between Tat signal peptides and alkaline phosphatase of E. coli, it could be demonstrated both in vivo and in vitro that Tat-dependent export requires folding of the phosphatase moiety by disulphide-bridge formation [24,25]. Likewise, cofactor insertion into apoproteins proved to be a prerequisite for transport by the Tat translocase [14,26,27]. Attaining a folded state prior to translocation must also occur for hetero-oligomeric protein complexes, of which only a single subunit carries an RR-signal sequence such as in E. coli hydrogenases [28] and dimethyl sulphoxide (DMSO) reductase [29]. This mode of transport, in which one Tat substrate carries signal sequence-less partner proteins across the bacterial plasma membrane has been termed the hitchhiker mechanism [28]. In contrast, monomeric and cofactor-less proteins that use the Tat pathway might exhibit fast folding kinetics. Such a situation seems to prevail in halophilic Archaea, which in fact export the majority of their secreted proteins via the Tat route [30,31]. High salinity seems to determine the need for Tat-dependent secretion also in the Gram-positive bacterium Bacillus subtilis [32].
Most Tat substrates are soluble proteins released after membrane passage, but some remain anchored by either N- or C-terminal transmembrane (TM) domains [33–35]. The number of Tat substrates varies quite dramatically between different organisms and species [3,6,7,31]. Of particular importance are Tat-dependent virulence factors of some pathogenic bacteria because they render the Tat system a potential drug target. Some extracellular Tat substrates involved in the formation and maintenance of the cell envelope are of interest, since their defective export in tat strains determines the tat phenotype. This is characterized by long, non-separated cell chains and outer membrane permeability defects.
3. The components of Tat translocases
(a). Homologues of TatC and TatA
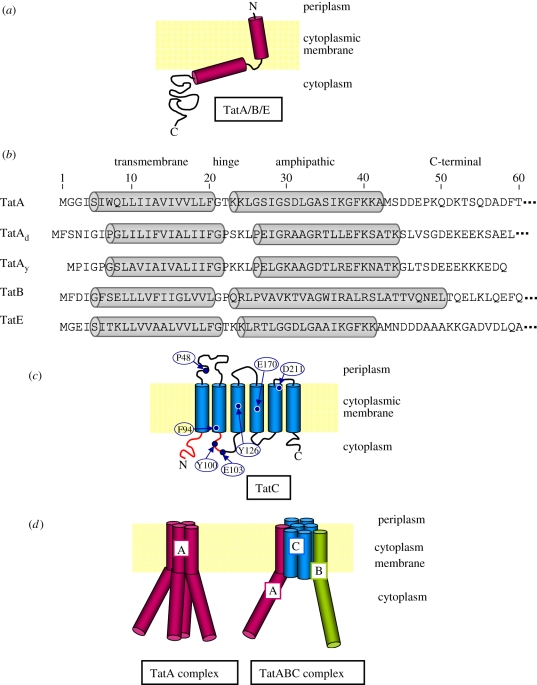
Tat translocases comprise two kinds of membrane proteins, a polytopic TatC-type protein with six predicted TM domains and one or more single-spanning TatA-type proteins denoted TatA, TatB or TatE (figure 2a,c). For a detailed analysis of sequence conservation between TatA- and TatC-type proteins of bacteria, archaea and plant chloroplasts, the reader is referred to a comprehensive review by the Saier group [40].
Figure 2.
The Tat components. (a) The known representatives of the TatA-protein family, TatA, TatB, TatE are depicted according to a recent nuclear magnetic resonance structure solved for an N-terminal fragment of the B. subtilis paralogue TatAd [36]. (b) Sequence alignment of the conserved N-terminal fragments of TatA, TatE and TatB from E. coli and TatAd and TatAy from B. subtilis. Numbering refers to the residues of E. coli TatA. The transmembrane and amphipathic helices and the intervening hinge region are indicated according to references [36–39]. (c) Predicted structure of TatC of E. coli with the approximate positions of residues that influence its activity and the putative substrate binding site highlighted in red. (d) The major Tat complexes recovered from TatABC translocases of Gram-negative bacteria and thylakoids of plant chloroplast. TatA forms homo-oligomeric complexes and is represented here by a tetramer. TatB and TatC form stable complexes independently of TatA. When purified from Gram-negative bacteria, not from chloroplasts, TatBC complexes often contain TatA.
Gram-negative bacteria require a TatA and a TatB protein, because they fulfil diverse functions in these organisms. The same holds true for plant chloroplast, where TatA, TatB, TatC are denominated as Tha4, Hcf106, cpTatC, respectively. In addition to the tatABC genes, Gram-negative bacteria often contain a tatD gene encoding a cytosolic protein, and tatE, which is regarded as a gene duplication of tatA expressing a functional paralogue of TatA. While tatE is monocystronic, tatABCD constitute an operon, the major transcript of which comprises tatABC [41]. Translational fusions to β-galactosidase revealed that in E. coli, TatA is produced at a 20-fold higher level than TatB, at a 50-fold higher level than TatC and at a 100–200-fold higher level than TatE [42]. Although many E. coli Tat substrates are involved in anaerobic respiration, the Tat components are expressed constitutively [42].
In contrast, Gram-positive bacteria express orthologues of TatA and TatC only. The extensively studied B. subtilis expresses two paralogues of TatC—denoted TatCd and TatCy and three paralogues of TatA—denoted TatAd, TatAy, TatAc. The tatAdCd and tatAyCy genes are organized in operon-like arrangements. They code for two substrate-specific Tat translocases, in which the TatA proteins are bifunctional, replacing the individual TatA and TatB orthologues of Gram-negative bacteria [7,43]. Functional Tat translocases that consist only of TatA and TatC subunits are called minimal Tat translocases.
(b). Tat subunits
Homologues of TatA, TatB and TatC are essential components of the Tat translocases of Gram-negative bacteria and plant chloroplasts [44–46].
(i). TatA
TatA of E. coli consists of 89 amino acids (9.6 kDa). Its N-terminal TM helix (figure 2b) is linked via a hinge region to an amphipathic helix and an unstructured C-terminus. Recent solution [47,48] and solid-state [36] NMR structures of TatA orthologues confirmed the previously predicted two α-helices and their L-shaped arrangement in lipid environments that is stabilized by extensive inter-helical contacts. When reconstituted into planar bicelles, the only 14 amino acids long N-terminal TM domain of B. subtilis TatAd was found to be tilted by 13° and deeply inserted in the lipid bilayer. This caused both the hinge region and even the proximal part of the amphipathic helix to be immersed in the lipid bilayer (figure 2a). The amphipathic helix showed a slanted alignment [36] rather than a flat orientation in the lipid–water interphase.
The topology of the TM helix of TatA has remained controversial. Studies probing the accessibility of TatA towards proteases [49] and oxidants [37] suggest a periplasmic location of the N-terminus of TatA. This would also be in line with the removal of an N-terminal extension from the TatA of Providencia stuartii requiring a membrane protease that is oriented towards the periplasm, as discussed in Palmer et al. [3]. On the contrary, using fusions of TatA to topological marker proteins [50] as well as cysteine-accessibility studies [51], an inverse orientation of the TM of TatA was reported. This leaves the possibility that, depending on its functional state, TatA might adopt dual topologies in the membrane.
Another point at issue is whether in vivo cytosolic forms of TatA might occur that represent functional intermediates. Particularly from Gram-positive bacteria, soluble TatA species have been isolated [52–56], and E. coli TatA upon overexpression was reported to form tube-like structures in the cytosol [57]. Notably, a soluble form of TatA present in the chloroplast stroma is functionally active [58].
Amino acid conservation between TatA orthologues is rather low and restricted to the TM and amphipathic helices [38] (figure 2b). Highly conserved residues of the E. coli TatA sequence are F20, G21 and F39 [38]. In agreement with the restricted conservation of TatA homologues, mutagenesis studies mapped residues that are important for the activity of E. coli TatA to the first 42 amino acids (table 1), in particular to the amphipathic helix [37,38,60,62]. Furthermore, truncation of the C-terminal 40 amino acids of E. coli TatA was found not to negatively affect its activity [69]. An F39A mutation completely blocks Tat-dependent transport [61], but the mutant TatA assembles incorrectly in the membrane [70], whereas the K41A mutation destabilizes TatA [38]. Similarly important are residues F20 and G21 in the hinge region (figure 2b) [38,60] and Q8 at the proximal end of the TM helix, where a polar residue seems to be required for helix oligomerization [37]. The acidic D-D-E motif distal of the amphipathic helix of E. coli TatA is required for Tat activity and TatA subunit interactions [59].
Table 1.
Mutagenesis analysis of E.coli TatA, TatB and TatC.
| amino acid | mutation to | conserveda | phenotypes |
references | |||
|---|---|---|---|---|---|---|---|
| growth on |
periplasmic TMAO activity | ||||||
| SDS | TMAO | ||||||
| mutations in TatA | |||||||
| Gln 8 | Ala | − | no influence on TatA oligomerization | n.d.b | n.d. | +++ | [59] |
| Cys | defective TatA oligomerization | n.d. | n.d. | − | [37] | ||
| Ile 12 | Asn | + | ± | n.d. | ++ | [60] | |
| Leu 18 | Pro | + | ± | n.d. | ++ | [60] | |
| Phe 20 | Ala | + | + | + | ++ | [38,61] | |
| Gly 21 | Ala | + | + | + | ++ | [38,60,61] | |
| Cys | n.d. | n.d. | − | [37] | |||
| Thr 22 | Pro | + | + | + | + | [38] | |
| Ala | + | n.d. | ++ | [60] | |||
| Lys 23 | Cys | + | n.d. | n.d. | − | [37] | |
| Lys 24 | Cys | + | n.d. | n.d. | − | [37] | |
| Leu 25 | Cys | + | n.d. | n.d. | − | [37] | |
| His | + | n.d. | ++ | [60] | |||
| Gly 29 | Cys | + | n.d. | n.d. | − | [37] | |
| Asp 31 | Cys | + | n.d. | n.d. | − | [37] | |
| Leu 32 | Cys | + | n.d. | n.d. | − | [37] | |
| Gly 33 | Cys | + | n.d. | n.d. | − | [37] | |
| Asp | ± | n.d. | ++ | [60] | |||
| Ser | − | n.d. | ++ | [60] | |||
| Ala | TorA-GFP transport blocked | n.d. | n.d. | ++ | [62] | ||
| Ile 36 | Cys | + | n.d. | n.d. | − | [37] | |
| Thr | + | n.d. | ++ | [60] | |||
| Lys 37 | Cys | + | n.d. | n.d. | − | [37] | |
| Gly 38 | Cys | + | n.d. | n.d. | − | [37] | |
| Ser | + | n.d. | ++ | [60] | |||
| Phe 39 | Ala | + | dominant mutation | − | − | + | [38,61] |
| Cys | n.d. | n.d. | − | [37] | |||
| Leu | + | n.d. | ++ | [60] | |||
| Lys 40 | Cys | + | n.d. | n.d. | − | [37] | |
| Lys 41 | Ala | + | + | + | +++ | [38] | |
| Ala 42 | Cys | − | n.d. | n.d. | − | [37] | |
| Thr | + | n.d. | ++ | [60] | |||
| DDE 45,46,47 | NNQ | + | n.d. | n.d. | ++ | [59] | |
| DDE 45,46,47 | LLM | + | n.d. | n.d. | − | [59] | |
| Mutations in TatB | |||||||
| Glu 8 | Ala | + | + | + | +++ | [38] | |
| Lys | restores transport of KQ-TorA-MalE | n.d. | n.d. | n.d. | [63] | ||
| Leu 20 | Phe | + | + | + | + | [38] | |
| Gly 21 | Ala | + | phenotype dependent on expression level | + | + | +++ | [38,61] |
| Pro 22 | Leu | + | phenotype dependent on expression level | + | + | + | [38,61] |
| Leu 25 | Ala | + | + | + | +++ | [38] | |
| Pro 26 | Ala | + | phenotype dependent on expression level | + | + | +++ | [38,61,62] |
| TorA-GFP transport blocked | |||||||
| Glu 49 | Ala | + | + | + | +++ | [38] | |
| Glu 53+Glu 58 | Ala | + | + | + | +++ | [38] | |
| Glu 49,53,58 | Ala | + | + | + | + | [38] | |
| Mutations in TatC | |||||||
| Leu 9 | Phe | + | restores transport of KQ-TorA-MalE | n.d. | n.d. | n.d. | [63] |
| His 12 | Ala | + | + | + | + | [64] | |
| Leu 16 | Ala | + | n.d. | + | +++ | [65] | |
| Arg 17 | Ala | + | + | + | + | [61,64,65] | |
| Lys | + | + | +++ | [64] | |||
| Lys 18 | Glu | − | restores transport of KQ-TorA-MalE | n.d. | n.d. | n.d. | [63] |
| Met | restores transport of KQ-TorA-MalE | n.d. | n.d. | n.d. | [63] | ||
| Leu 20 | Ala | − | n.d. | + | + | [65] | |
| Asn 22 | Ile | − | restores transport of KQTorA-MalE | n.d. | n.d. | n.d. | [63] |
| Pro 48 | Ala | + | TatBC complex assembly affected | n.d. | + | n.d. | [61,65] |
| Phe 94 | Ala | + | − | − | + | [61,64,66] | |
| Leu | − | − | + | [64] | |||
| Tyr | + | + | +++ | [64] | |||
| Ser | transports KK TorA-GFP but not RR TorA-GFP | n.d. | n.d. | n.d. | [67] | ||
| Pro 97 | Ala | + | + | + | + | [64] | |
| Leu 99 | Ala | + | ± | ± | + | [64] | |
| Pro | transports KK TorA-GFP but not RR TorA-GFP | n.d. | n.d. | n.d. | [67] | ||
| Tyr 100 | Ala | − | + | + | + | [64] | |
| Glu 103 | Ala | + | phenotype dependent on expression level | − | − | + | [61,64] |
| Asp | − | − | + | [64] | |||
| Gln | − | ± | + | [64] | |||
| Arg | − | − | + | [64] | |||
| Arg 104 | Ala | + | n.d. | + | +++ | [65] | |
| Arg 105 | Ala | + | n.d. | + | +++ | [65] | |
| Try 126 | Ala | + | ± | ± | + | [64] | |
| Pro 142 | Ser | − | transports KQ- and RR TorA-substrates | n.d. | n.d. | n.d. | [67] |
| Tyr 154 | Ser | + | n.d. | + | +++ | [65] | |
| Glu 170 | Ala | + | no phenotype in vitro | + | + | + | [64,68] |
| Asp 211 | Ala | + | phenotype dependent on expression level | + | − | + | [61,64,68] |
| Glu | + | + | +++ | [64] | |||
| Asn | phenotype dependent on expression level | + | + | ++ | [64] | ||
| Gln 215 | Ala | + | + | + | + | [61,64] | |
(ii). TatB
Escherichia coli TatB consists of 171 amino acids with a molecular mass of 18.5 kDa. It shares about 20 per cent sequence identity with TatA [38] (figure 2b). Despite being structural homologues, TatA and TatB are functionally diverse proteins [44]. Highest homologies between prokaryotic orthologues of TatB are found in the predicted TM and amphipathic helices [38]. In accordance with this, the C-terminal part of E. coli TatB beyond the amphipathic helix was found not to be essential for its function [69]. No single mutation has been described that completely inactivates E. coli TatB [38,39,62,71] (table 1).
TatB is virtually essential for the transport of endogenous Tat substrates in E. coli. Nevertheless, artificial Tat substrates made by fusing the RR-signal sequence of E. coli TorA (TMAO reductase) to colicin V [13,72] or maltose-binding protein [73] are slightly exported in the absence of TatB. These findings suggest a low degree of TatB-like activity inherent to E. coli TatA. Such a situation prevails in Gram-positive bacteria that express minimal Tat translocases consisting only of TatA-like homologues and their cognate TatC partners. These organisms did not undergo the early gene duplication event of most Gram-negative bacteria that resulted in the functionally unique TatB paralogues [40]. The TatA homologues of Gram-positive bacteria therefore remained bifunctional. Bifunctionality can be restored to E. coli TatA by discrete mutations in the first N-terminal residues of TatA [73,74].
(iii). TatC
Escherichia coli TatC consists of 258 amino acids with a molecular mass of 28.9 kDa. TatC has six predicted TM (figure 2c), which was experimentally confirmed using reporter fusions [75,76] and thiol-labelling techniques [77].
Mutational analysis (table 1) of conserved residues did not always yield unambiguous results. P48A and F94A (figure 2c) were unanimously described as inactivating TatC [61,64,65,68], which in the case of P48A might be due to a destabilizing effect on the Tat(A)BC complex [70]. Y100A, Y126A and E170A substitutions show decreased activity [64]. E103A and D211A mutants, however, were found to be inactive in one study [64], but active in others [61], which might be due to different expression levels of the mutant proteins. The P48A, F94A, Y100A, E103A and Y126A mutations all interfere with binding of an RR-precursor protein [68].
(iv). TatD
TatD is a 29 kDa cytosolic protein [41]. Escherichia coli expresses two additional paralogues of TatD, and orthologues of TatD exist across all kingdoms, often independently of other Tat proteins. A triple deletion strain of E. coli lacking all TatD paralogues is not impaired in Tat-dependent protein export [41]. Although TatD of E. coli was reported to be required for the degradation of translocation-incompetent, malfolded Tat substrates in vivo [78], this effect was not seen under normal expression levels of the RR-precursor [79]. Consistent with its ubiquitous occurrence, TatD has frequently been characterized as a deoxyribonuclease with no obvious functional link to the Tat pathway [41,80,81].
(v). TatE
Escherichia coli TatE is a protein of 67 amino acids with 53 per cent sequence identity to TatA [45] (figure 2b) that likely arose by a rather late gene duplication event [40]. TatE and TatA are functionally interchangeable, because either homologue can complement the tat phenotype of a tatAtatE double deletion strain [44] and neither protein is by itself essential [45]. TatE seems to be less important than TatA, because more Tat substrates are mislocalized in a ΔtatA mutant than in a ΔtatE strain [45]. On the other hand, a defective TatE strongly impairs the export of some Tat substrates [45]. These genetic findings would be consistent with TatE being an active member of the Tat translocase in E. coli.
(c). Complex formation by Tat subunits
The subunits of Tat translocases have the remarkable property to homo-oligomerize. This has been vastly documented for TatA (see §3e) but was also described for the individually expressed E. coli TatB and TatC subunits [82,83]. When expressed together, the TatC and TatA family members also form hetero-oligomeric complexes both in vitro and in vivo. In E. coli and other Gram-negative bacteria, the TatA, TatB and TatC proteins associate to form TatABC and TatA complexes [84,85] (figure 2d). There are no data available as to the possible association of TatE with either complex. The orthologues Tha4 (TatA), Hcf106 (TatB) and cpTatC in the thylakoidal membrane of chloroplasts show a similar association pattern except that a TatBC complex free of TatA can be isolated [86]. To standardize nomenclature between organisms, we will refer to this complex as Tat(A)BC complex. In Gram-positive bacteria, the sole TatA and TatC subunits can also be recovered from separate TatAC and TatA complexes [87]. Despite the fact that Tat subunits are consistently recovered from these Tat(A)BC and TatA complexes, doubt has been cast on the functional relevance of those isolatable structures. Thus in E. coli, TatA variants, which are able to functionally suppress a TatB deficiency, do so without forming complexes with TatC [74].
(d). Tat(A)BC complex, the site where Tat-dependent translocation starts?
The molecular masses of isolated Tat(A)BC complexes vary between 360 and 700 kDa [85,88–90] depending on the isolation procedure and also owing to some intrinsic heterogeneity [83,91]. Even in the absence of TatA, E. coli TatB and TatC associate into high molecular mass complexes [82,83,88] suggesting that TatB and TatC form the core of the Tat(A)BC complex. TatB and TatC associate in a 1 : 1 stoichiometry and a translational fusion of both proteins retains activity [89]. When purified from TatABC-expressing E. coli cells, this core TatBC complex is usually found associated with TatA [84,85,88,89] and TatA was actually described to stabilize the TatBC complex [92].
In addition to forming hetero-oligomeric complexes, TatB and TatC have the property to self-interact. Homo-oligomeric assemblies of TatB ranging from dimers up to pentamers were detected by chemical cross-linking [93], disulphide mapping [39], bimolecular fluorescence complementation [94] and a two-hybrid approach [71]. Self-association occurs via both the TM and the amphipathic helix of TatB. Homomeric TatB complexes might not only represent resting states, but also have been characterized as multivalent attachment platforms for RR-precursors [95]. Likewise, interaction between two TatC monomers was revealed by cysteine cross-linking [77]. Furthermore, the finding that in E. coli, two inactivating mutations in tatC (F94A and E103A) can be cross-complemented by another tatC mutation (D211A) (figure 2c) suggested that TatC functions as a dimer [64]. This could in fact be confirmed by demonstrating that a genetically fused tandem TatC molecule retains activity, even if one of the fused TatC monomers carries the inactivating point mutation F94A [66]. Based on the above-mentioned findings that TatB and TatC form a 1 : 1 complex and that both have the tendency to homo-oligomerize, a functional TatBC core complex should consist of several TatBC dimers. From single-particle electron microscopy of isolated TatBC complexes that were devoid of TatA, a heptameric composition was estimated [96]. An oligomeric array of TatBC dimers is also strongly supported by the finding that multiple precursor molecules bind to a single TatBC–receptor complex [96,97]. It is further suggested by the cooperative-binding behaviour described for a chloroplast RR-precursor [98]. Because of the self-association of TatB monomers, a model of the TatBC complex was put forward, according to which the TatB protomers would form the central core of the TatBC complex and the TatC protomers a peripheral ring [39] (figure 3a).
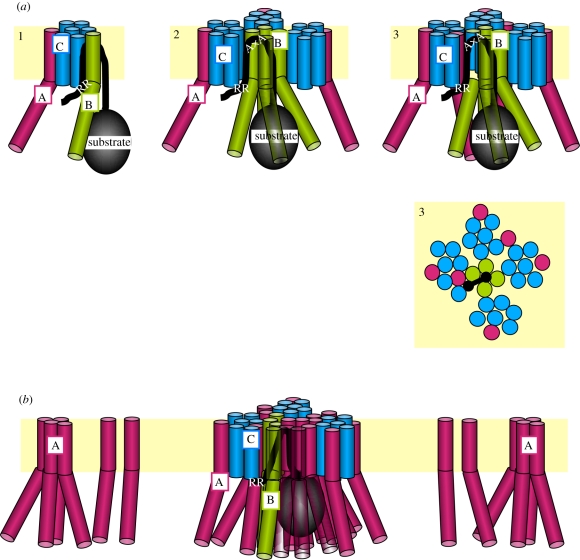
Figure 3.
Model of a substrate-induced formation of a functional Tat translocase. (a) Step 1: superficial binding of a Tat signal sequence to the Tat translocase involves the RR-consensus motif and cytosolic parts of TatC. The coordinate recognition of a Tat signal sequence by TatC and TatB leads to its hairpin-like insertion into the plane of the membrane. Step 2: TatB homo-oligomerizes and encapsulates the folded mature domain of the substrate. Step 3: a proton motive-force-dependent step brings monomeric TatA in close contact to the signal sequence. AxA denotes the C-terminal end of the signal sequence. Step 3 is also shown as top view from the periplasmic side of the membrane. (b) More TatA subunits are recruited from a pool of monomeric TatA or more likely in tetrameric units. Processive oligomerization leads to the pavement of a transmembrane path. This might comprise oligomeric TatA pores, hetero-oligomeric TatA(BC) pores as drawn here or some other undefined TatA structure.
As already mentioned, TatBC complexes, when purified from Gram-negative bacteria, usually contain TatA protomers. A functional association of TatA with the TatBC complex has thus far not been addressed in much detail. Bimolecular fluorescence complementation suggests a close vicinity of TatA to TatC in vivo [94] and recent data from our own laboratory provide evidence for a functional targeting of E. coli RR-precursors not only to TatBC, but also to TatA [99].
The major function of the Tat(A)BC complex is to specifically recognize and bind to RR-precursor proteins. This receptor function was verified by manifold experimental strategies, such as (i) interference of precursor binding to the Tat translocase by antibodies directed against TatB and TatC [86], (ii) co-migration of precursor with a TatBC complex on BN-PAGE [91], (iii) co-purification of precursor, TatB, and TatC upon membrane solubilization [96,100], (iv) chemical and site-directed cross-linking [97,101,102], and (v) identification of TatBC suppressor mutations of export-defective Tat signal peptides [63].
(e). TatA complex, a protein-conducting structure?
The size of isolated TatA complexes varies between about 100 and 700 kDa [70,85,100,103]. Homo-oligomers of TatA clearly exist in vivo as shown by cysteine cross-linking [37] and fluorescent labelling techniques [94,104], even at wild-type expression levels [104]. Oligomerization of TatA occurs via its TM [49,59,93,105,106] and amphipathic domains [37,105], a conserved acidic motif (DDE in E. coli, cf. figure 2b) following the amphipathic helix [59], and the C-terminus (shown for Tha4 of chloroplasts) [105,106]. The finding that on Blue Native-polyacrylamide gel electrophoresis (PAGE) purified TatA resolves into a ladder of bands with a step size of about 40 kDa corresponding to the size of a TatA tetramer [85,103], suggests that a minimal building block of TatA might consist of four protomers (figures 2d and 3b). TatA tetramers might then oligomerize to higher order assemblies [104] of various sizes. Whereas, the formation of TatA dimers and trimers was observed in the absence of TatB and TatC [93], higher order oligomerization usually requires the TatBC subunits [104–106]. Consistent with this, TatA complexes when purified from E. coli cells expressing high levels of Tat proteins also contain TatB [84,107]. TatA complexes are less stable when purified from plant chloroplasts [86] and a recent re-evaluation also indicated that the chloroplast thylakoids of Arabidopsis thaliana seem to contain the TatA-homologue Tha4 in a considerably smaller abundance over TatBC than bacterial organisms [108].
The fact that oligomerization of TatA also requires contact to substrates [105], could reflect the formation of size-fitting pores that allow TM passage of the folded and therefore differently sized Tat substrates. A pore-like structure of TatA complexes purified from E. coli was revealed by single-particle electron microscopy [103]. The size classes of the obtained particles varied between 130 and 390 kDa with estimated subunit numbers of 12–35 and channel diameters of 30–70 Å. This spectrum of diameters would well fit with the size range (20–70 Å) of known folded Tat substrates from bacteria [109]. The walls of the pore-like particles would consist of the TM helices of the TatA protomers, which in fact arrange in a ring-like order as observed by EPR spectroscopy of spin-labelled E. coli TatA variants [110]. TatA particles as seen in the electron microscope possess a lid-like density on their presumed cytosolic face, which Gohlke et al. suggested represented the amphipathic helices of the circularly arranged TatA protomers functioning as a gating device of the TatA pore [103]. The potential of the amphipathic helices of TatA to adopt different orientations was suggested by studies making use of fusions between TatA and topological markers [50,51] as well as by chemical cross-linking [105]. Following these results, the possibility of the TatA amphipathic helices to at least transiently flip into the membrane to line a hydrophilic pore has been proposed [50,51].
The concept of size-fitting pores that consist of an appropriate number of TatA monomers was recently challenged by the finding that TatE, the functional paralogue of TatA in E. coli, also forms pore-like structures which, however, are considerably smaller than those of TatA [111].
4. The Tat pathway
(a). Recognition of an RR-signal sequence by TatBC and TatA
It is an open question whether Tat signal sequences depend on cytosolic chaperones for membran-etargeting. On the one hand, RR-signal sequences are presumably unfolded and unstructured before they contact the membrane [112,113] and as such conceivably might need protection by chaperones. In fact, DnaK, Trigger factor, SlyD and other FK506-binding proteins were reported to interact with Tat signal sequences [114–116]. However, Tat-dependent export was not negatively affected in the absence of any of these chaperones with the notable exception of DnaK, knock-out of which completely abolished transport of the Tat substrate CueO from E. coli [115]. Since the chaperoning effect of DnaK was independent of the Tat signal sequence, it could, however, also be explained by a general stabilizing effect of DnaK on Tat substrates [117]. Finally, Tat-dependent translocation was observed in the bona fide absence of any cytosolic chaperone of E. coli [114].
An overwhelming wealth of results indicates the specific recognition of the RR-signal by the Tat(A)BC–receptor complex (see below). Nevertheless, RR-precursors were found to insert into protein-less lipid bilayers [118–120] and the conversion of membrane-targeted RR-precursors from a lipid-bound into Tat translocase-bound form was described [121]. Furthermore, a TatABC-independent binding of RR-precursors to membrane vesicles was observed using co-sedimentation and filtration assays [121,122], although in these studies interaction with the membrane lipids was not rigorously addressed. On the contrary, co-elution assays using Sepharose columns did not reveal binding of an RR-precursor to membrane vesicles of E. coli [25]. Moreover, in the presence of TatABC-deficient membrane vesicles, RR-signal sequences were found not to cross-link to a non-Tat membrane protein but to a soluble protein exactly as they did in the complete absence of membranes [68]. Thus it is an open question, if lipid-binding is a required step in membrane-targeting of all RR-precursors.
By several experimental approaches, the Tat(A)BC–receptor complex both of chloroplasts and bacteria was identified in vivo and in vitro as the specific binding site for RR-signal sequences [86,100,101]. Cross-linking studies revealed a close proximity between the consensus RR-motif and TatC [25,101,102]. Signal peptide-TatC cross-links are obtained even in the absence of TatB, whereas cross-linking to TatB strictly depends on the presence of TatC and is also quenched by high amounts of TatC [101]. TatC has therefore been regarded as the primary recognition site for RR-signal sequences. Consistent with such a notion, a fusion protein between RR-signal peptides and E. coli alkaline phosphatase (PhoA) was found to mistarget the Sec translocon in the absence of TatC [123]. Further support for TatC specifically recognizing the RR-motif comes from the finding that KK-mutant signal peptides fail to cross-link to TatC [101,102] and do not associate with a TatBC complex resolved by Blue Native-PAGE [91]. In seeming contrast, KK- and other translocation-deficient variants of the RR-consensus motif were demonstrated to still physically interact with Tat translocases [25,98,100]. This discrepancy is explained by assuming that recognition of Tat signal sequences by TatB and TatC extends beyond the RR-pair. In fact, mutational analyses of residues flanking the RR-pair in the S-R-R-x-F-L-K consensus sequence clearly demonstrate that the Ser, Phe and Leu residues are involved in a productive interaction with the Tat translocase [16,124,125]. Moreover, the above-described finding that KK-signal sequences bind to the Tat(A)BC–receptor complex but fail to cross-link to TatC actually suggests that TatB constitutes another half of the precursor-binding site of Tat translocases (as further detailed in §4c). This assumption could experimentally be verified by the finding that an extragenic suppressor mutation restoring transport to a KQ-variant of an RR-precursor mapped to the Glu residue at position 8 of TatB [63]. Fully consistent with a concerted TatBC-binding site for RR-precursor proteins, Tat signal sequences were not only found cross-linked to TatC but also to TatB [25,101,102].
When site-specific cross-linking was performed to identify contacts between RR-precursors and TatB and TatC, a Tat signal sequence was also found to cross-link with TatA [101]. These cross-links disappeared when the TatABC-containing membrane vesicles had been treated with carbonyl cyanide m-chlorophenyl-hydrazone (CCCP) to dissipate their proton-motive force (PMF) [101]. Recently, we obtained evidence that RR-precursors come into contact with the TM helix of TatA before the actual translocation starts. These contacts likely involve the Tat signal sequence after it has been inserted into a TatBC-binding pocket [99]. Such a scenario would invoke a close proximity between the TM parts of TatA and TatC (figure 3a), which was corroborated by site-specific photo cross-linking [99]. Importantly, the observed contacts between RR-precursors and the TatA TM domain were clearly impaired in the absence of a PMF, suggesting that if TatA in fact joins TatBC in recognizing the signal sequence, it might do so only subsequently to TatB and TatC.
(b). Superficial binding sites for RR-precursors
Five single alanine substitutions at residues P48, F94, Y100, E103 and Y126 of E. coli TatC that interfere with translocation both in vivo and in vitro [61,64,65,68] were shown to impair membrane-targeting of RR-precursors [68]. These mutations are spread over the N-terminal half of TatC, including periplasmic, cytosolic and TM sites of the molecule (figure 2c). Therefore, they are less likely constituents of a linear-binding epitope but are rather involved in the formation of a sterical precursor-binding site of TatC. Indirect evidence as to the location of binding site(s) on TatC for Tat signal peptides came from the isolation of suppressor mutants in tatC that partially alleviate a transport block of KQ- and KK-precursor mutants. Suppressors were isolated that mapped to the predicted cytosolic N-terminus and the first cytosolic domain of TatC [63,67]. The interaction of these areas of TatC with RR-precursor proteins was recently confirmed by an extensive cross-linker scanning of the E. coli TatC molecule [126]. Hence the available data suggest that TatC provides a superficially exposed precursor-binding site as highlighted in red in figure 2c. In full agreement with a superficial attachment site, binding of RR-precursors to the thylakoidal Tat translocase still render signal sequences [124] including their N-termini [127] accessible to proteases.
(c). Advanced binding of an RR-signal sequence to the Tat translocase
Whether a first superficial binding of an RR-precursor to cytosolic domains of TatC is immediately followed by a deeper insertion of the signal sequence into the plain of the membrane, is not exactly known. Evidence for a loop-like insertion of an RR-signal peptide into the lipid bilayer of the membrane that is solely executed by TatC was recently obtained [128]. In the following, we summarize the numerous data suggesting a loop-like topology of an RR-signal sequence deeply within a concerted TatBC-binding pocket and discuss the experimental evidence for an advanced step of interaction with the Tat translocase involving the PMF.
As mentioned in §4a, RR-signal peptides seem to be recognized by TatC and TatB in a concerted fashion. Thus, several single-site mutations in tatC were described that simultaneously affected cross-linking of RR-precursors to TatC and TatB [68]. Moreover, suppressors of the translocation defect of KQ-variant precursors were not only isolated in tatC (see §4a) but also in tatB, where they mapped to Glu8 near the trans-sided N-terminus of TatB [63]. The location of this mutation suggests that the RR-signal peptide extends far into the membrane. Consistent with a deep insertion of a Tat signal peptide into a TatBC-binding pocket, the Leu residue at position 9 of TatB next to Glu8 was found to strongly cross-link to the precursors of TorA and SufI [95]. Furthermore, the h-region of Tat signal peptides located distal to the RR-consensus motif also cross-links preferentially to TatB [25,101,102]. If such a deep insertion of an RR-precursor protein into a TatBC-binding pocket occurs in a loop-like fashion, also the stretch distal to the signal peptidase cleavage site might become accommodated next to one of the Tat subunits. This was described to be the case for the precursor of SufI and TatB [101,102]. Furthermore, a tatC suppressor mutant of a KK-variant precursor was isolated that mapped to Pro142 in a predicted periplasmic loop of TatC [67]. The location of this mutation would corroborate again the idea that trans-sided sections of TatB and TatC are involved in the formation of a binding pocket for Tat signal sequences. Finally, preventing the processing of RR-precursors, which per se does not interfere with translocation [129], results in the accumulation of a precursor deep inside the membrane [130].
Binding of RR-precursors to thylakoids [131] and E. coli inner membrane vesicles [25,132] does not depend on the PMF, nor is Tat signal sequence-dependent cross-linking to TatB and TatC affected when the PMF is dissipated [101]. Moreover, a TatC-mediated insertion of Tat signal sequences was observed to occur in the absence of the PMF [128]. All together, these results suggest that insertion of Tat signal sequences into the TatBC-binding pocket does not require energy from the PMF. On the contrary, more advanced binding stages were described that are definitely dependent on the PMF. Thus, a protease-cleavage site that had been engineered into a thylakoid-bound RR-precursor was transferred from an accessible to a membrane-protected environment upon implementation of the PMF [124]. Furthermore, cross-linking of the signal peptide to TatA was observed only in the presence of the PMF [101] and recent data suggest that the TM helix of TatA is involved in this PMF-dependent interaction with the signal sequence [99]. In this context, the TM domain of the chloroplast TatA homologue Tha4 contains a conserved Glu residue, which could become protonated following the trans-sided accumulation of protons. This glutamate was shown to be essential for recruiting Tha4 to the TatBC–precursor complex [133]. Collectively, these results are compatible with a scenario in which RR-precursor proteins, after their primary binding to TatBC and accommodation in a TatBC-binding pocket, undergo a PMF-dependent advanced interaction with the Tat translocase. This advanced binding/insertion step potentially involves a first contact with TatA.
(d). Multivalent binding of TatB to the folded moiety of membrane-targeted RR-precursors
Binding of a whole RR-precursor molecule to the TatBC–receptor complex has been addressed in different studies and thereby revealed multivalent attachment sites. Single-particle electron microscopy of TatA-free TatBC complexes from E. coli produced in vivo and in vitro revealed about 10 nm oval-shaped particles, in which up to seven protomeric TatBC complexes could be fitted [96]. When these complexes were produced in the presence of a natural E. coli RR-precursor, one or two extra densities asymmetrically associated with the surface of the particles were obtained. Into each of these densities, a folded precursor molecule could be modelled. These results suggested that a folded precursor binds to discrete superficial binding sites on an oligomeric TatBC assembly and that a functional TatBC complex might accept more than one substrate at a time. Using chemical cross-linking Ma & Cline [97] recently reported that two or four precursor molecules bind to the chloroplast Tat translocase such that even covalently cross-linked precursor molecules were still efficiently translocated. The assembly of a functional Tat translocase might thus involve recruitment of multiple Tat subunits and substrate molecules. Probing the molecular environment of E. coli RR-precursors after their targeting to the membrane but prior to the actual translocation event unexpectedly revealed the close proximity of several TatB monomers to the folded domains of the precursors [95]. The authors interpreted these findings such that after binding of an RR-precursor to the Tat(A)BC–receptor complex, TatB assembles into an oligomeric structure capable of transiently accommodating folded precursor domains (figure 3a). Encapsulation would most likely be achieved by multiple amphipathic helices of TatB creating a structure that might resemble the ones visualized by electron microscopy of isolated TatBC–precursor particles [96]. This multivalent binding of an RR-precursor to TatB occurs independently of the PMF [95] and would therefore precede the advanced, PMF-dependent interaction of the Tat signal peptide with TatA. Recruitment of several TatB monomers to a folded RR-precursor might also be the starting point for triggering further oligomerization of TatABC subunits, ultimately leading to a functional Tat translocase (figure 3a, step 2).
(e). Membrane translocation
It is not yet understood how the Tat translocase enables the translocation of folded substrates without compromising the ionic seal of the membrane. All models proposed are based on the assumption that TatA plays a major role in this process. As shown for the Tat translocases of chloroplasts, after binding of precursor proteins to the TatBC–receptor complex, TatA (Tha4) subunits are recruited in a PMF-dependent manner [105,134]. Both TatBC and precursor are required to induce oligomerization of TatA [104,105]. These findings would be consistent with TatA forming a protein-conducting channel on the periphery of the TatBC–receptor complex. Thus far, the strongest experimental support for such a scenario comes from the pore-like structures of variable diameters that had been visualized by single-particle imaging of homo-oligomeric TatA particles isolated from E. coli membranes [103]. As discussed in §3e, these and other studies [110] suggest that the TM helices of TatA line a protein-conducting channel. Depending on the number of TatA monomers, they could thus form pores with different diameters to adjust to differently sized Tat substrates. The size of a Tat substrate does, however, not seem to determine the number of recruited TatA protomers, because oligomerization of Tha4 is initiated merely by an RR-signal peptide [106,134]. Whether a cylinder made from the TM helices of TatA would surround an aqueous interior or instead be filled with membrane lipids is not known. An alternative view suggests that the amphipathic helices of TatA fold into the interior of the TM TatA ring in a trap-door-like manner, so that their hydrophilic faces would line the actual protein-conducting channel [6,37,51,105].
Thus far, no translocation intermediate could be generated that was trapped within a ring-like TatA structure, be it made from its TM or its amphipathic helices. A partially membrane-protected translocation intermediate of a folded RR-precursor was, however, described that was dependent on the presence of TatA and accumulated in the immediate vicinity of TatA [25]. In addition, when a membrane-targeted-folded RR-precursor of E. coli was probed for surface contacts by site-specific photo cross-linking, cross-links to TatA that required the PMF were in fact obtained [95]. However, a fusion protein consisting of an RR-precursor, an unstructured linker region and a globular C-terminal domain was found stuck in the thylakoidal membrane without contacting any Tat component [135]. This could, in principle, mean that Tat-dependent translocation does not proceed via pores made from the Tat subunits. Alternatively, pores might quickly disassemble in such a case of a halted translocation event. Since no experimental situation has so far been established that would transiently arrest an RR-precursor in an oligomeric TatA structure, it is also not clear if the presumed pore would exclusively consist of TatA subunits. Because TatB seems to oligomerize around membrane-targeted RR-precursors [95], it cannot be excluded that TatB oligomers constitute a kind of nucleation centre for the pore formation. If so, TatB monomers may even remain part of the final pore. A lateral access of an RR-precursor from the TatBC subunits to the centre of a presumed TatA pore is invoked by the finding that complete translocation can occur when the signal sequence of an RR-precursor is covalently bound to TatC [102]. It is therefore conceivable that TatB and TatC are even integrated in a pore structure (figure 3b).
At variance with the pore-model, Brüser & Sanders suggested that the local accumulation of TatA monomers in the vicinity of the Tat(A)BC–receptor complex could serve to destabilize the ordered lipid bilayer structure, thereby allowing a direct passage of Tat substrates through the membrane lipids [136].
(f). Cleavage of the signal sequence and dissociation of the Tat complex
RR-signal sequences are cleaved by signal peptidase I [19,20]. It is not clear what the exact temporal relationship between translocation and precursor processing is. Obviously, a premature cleavage leading to the detachment of a translocation-competent Tat substrate from the Tat(A)BC–receptor complex would abolish translocation and therefore needs to be prevented. For the Tat system of plant chloroplasts, a situation was described in which a lack of signal sequence processing can lead to the accumulation of non-translocated precursor in the thylakoidal membrane [130]. We have recently obtained experimental evidence pointing to an important function of TatB in controlling a TatC-mediated access of the cleavage site of Tat signal peptides to the peptidase [128]. On the contrary, signal peptide processing is not a prerequisite for a complete Tat-dependent translocation. This has been repeatedly found in vivo and in vitro using mutant RR-precursors harbouring non-functional cleavage sites [25,129,137]. The finding that translocation can occur with a signal peptide covalently linked to TatC [102] could indicate that the processing step might be a rather late event only following complete translocation. Taking this idea one step further, one might speculate that the association of the Tat signal peptides with TatBC has to be maintained during the translocation process in order to keep the protein conduit structurally intact. Only after a complete translocation, would the cleavage site become fully accessible to the signal peptidase, and cleavage of the precursor would then initiate the dissociation of the Tat translocase. In fact, dissociation of the TatABC complex after translocation has been documented for the chloroplast Tat translocase [134]. As discussed in §4e, dissociation of the translocating apparatus would also explain why partially translocated RR-chimeras remaining stuck in the membrane were not found in the vicinity of the Tat subunits, even if they had their signal sequences cleaved off [135].
(g). Energetics
A hallmark of Tat systems is that they do not use nucleotide hydrolysis for energetization but solely the PMF consisting of the pH gradient (ΔpH) and the electrical potential (Δψ) at the membrane [138]. The dependence of Tat-specific translocation on the PMF has frequently been demonstrated using compounds that dissipate the H+-gradient or the electrochemical potential. In addition, the phage-shock protein PspA that is somehow involved in maintaining the PMF under cellular stress conditions [139] was found to improve the efficiency of Tat translocation in bacteria [140,141].
At variance with the preceding statements, processing of natural and model Tat precursor proteins was shown to proceed independently of the PMF in transfected tobacco protoplasts [142]. Although these results do not exclude the possibility that the PMF is involved in completion of translocation, they would at least not be consistent with a PMF-dependent assembly of the Tat translocase. Furthermore, in the green alga Chlamydomonas reinhardtii, the Tat pathway was reported to be unaffected by clamping the ΔpH to zero [143]. This result could be explained by assuming that both components of the PMF, the ΔpH and the Δψ, can contribute equally to the driving force of the Tat pathway as has been experimentally verified [144]. As a consequence of the equivalency of ΔpH and Δψ, a coupling of H+-flow and protein transport would have to be postulated [145], which is in good accordance with previous studies by Theg's group [146,147].
With regard to the involvement of the PMF in individual steps of the Tat pathway, targeting and binding to the TatBC-binding pocket occur independently of the PMF. The same holds true for the oligomerization of TatB around a membrane-bound-folded RR-precursor. On the contrary, advanced binding of Tat signal peptides with the Tat translocase, which might encompass a first interaction with TatA and well precede oligomerization steps of TatA, does require the PMF (see §4c).
In a more recent in vitro study, Bageshwar & Musser [148] came to the conclusion that transport of an E. coli RR-precursor into membrane vesicles requires two Δψ-dependent steps, one that is necessary early and one later during transport.
5. Proof-reading and quality control
(a). Handling of folded and unfolded substrates by Tat translocases
As mentioned earlier, many Tat substrates can exit the cytosol only after they are completely folded, either because cofactor insertion is confined to the cytosol or because hetero-oligomeric substrates have to associate with a single Tat signal sequence-containing subunit. In contrast, the thylakoidal Tat system of plant chloroplasts has long been known also to accept unfolded proteins [149].
The question of whether or not Tat translocases discriminate between the folded and unfolded conformations of a single Tat substrate was experimentally addressed by the use of model Tat substrates. In one case, disulphide-containing proteins were equipped with Tat signal sequences and shown both in vivo and in vitro to be exported in a Tat-dependent manner only when oxidizing conditions allowed disulphide-bridge formation [24,25,91]. Likewise, Tat-mediated export of an RR-signal sequence-containing variant of cytochrome c occurred only after incorporation of haem [26]. In addition, mutations and truncations of Tat substrates, even if they seem to have only a small impact on their conformation, have repeatedly been found to impair Tat-dependent transport both in bacteria and chloroplasts [137,150].
The major question arising from these findings is how an improperly folded Tat substrate is rejected by Tat translocases. Translocation-incompetent Tat substrates can still associate with Tat translocases [25,91,151]. Co-purification of an unfolded Tat substrate with the TatBC complex seems to suggest that neither of those Tat subunits possesses an intrinsic quality control function [91]. Furthermore, overexpression of this translocation-incompatible Tat substrate was found to be toxic for E. coli cells. This has nourished the idea that it is not the Tat translocase that discriminates against malfolded substrates but that an efficient degradation system is required that clears Tat translocases of faulty substrates [91,136]. Degradation of incompletely assembled Fe/S proteins in E. coli was in fact demonstrated to occur, provided that they could interact with the Tat machinery [152].
In contrast, site-specific photo cross-linking revealed a perturbed interaction of the signal sequence of an incorrectly folded RR-precursor with TatB and TatC despite the fact that this precursor was physically associated with the TatABC translocase [25]. Thus, improperly folded Tat substrates do not seem to correctly insert into the TatBC-binding pocket, but it is not understood by which measure this is prevented.
Tat translocases of both chloroplasts and bacteria are capable of transporting unstructured peptides up to a certain length of about 100–120 amino acids, provided that they have a Tat signal peptide and do not contain hydrophobic residues [135,153]. Moreover, improperly folded Tat substrates that the E. coli Tat machinery does not transport, become partially tolerated when they are C-terminally shortened [137]. Why unstructured polypeptides cannot exceed a certain length in order to be Tat-dependently transported is not clear. A likely parameter is the total surface area of an RR-precursor that, when exceeding certain limits, seems to interfere with Tat-dependent translocation [135].
A yet totally speculative idea as to how a Tat-incompatible structure could be recognized on a molecular level, is that oligomerization of TatB following precursor binding (see §4d) would require correctly folded domains. Such a scenario seems to be at odds with the observation that even a translocation-incompetent substrate shows surface contacts to TatB [95], unless one assumes that in this particular case, the Tat substrate retained some tertiary structure [137].
(b). Proofreading of cofactor-containing Tat substrates
Several Tat-dependent redox proteins of bacteria undergo cofactor insertion, folding and even hetero-oligomerization in the cytosol before they become export-competent by the Tat machinery. Because specific chaperones (also termed redox enzyme maturation protein (REMP)) are often involved in these maturation steps, these chaperones serve as proof-reading checkpoints that are independent of the TatABC proteins (reviewed in [3,154,155]).
In E. coli, the best-characterized proof-reading chaperones are TorD, DmsD, HyaE, HybE and NapD, which are specific for the substrates TMAO reductase (TorA), DmsA subunit of DMSO reductase, HyaA subunit of hydrogenase-1, HybO subunit of hydrogenase-2 and NapA subunit of nitrate reductase, respectively. These chaperones, which often dimerize [156], bind to the signal sequences of their cognate substrates [157–161]. A promiscuous interaction was shown for DmsD and the signal peptide of TorA [159], which might be due to some sequence homology between DmsD and TorD [162], but less-specific interactions between REMPs and redox partners were also reported [163]. In addition to the signal sequence, TorD has a second-binding site on the mature domain of its substrate TorA [157], by which it could assist in cofactor insertion. The finding that TorD is a GTP-binding protein and that its affinity is enhanced by the TorA signal peptide suggests that the chaperone–substrate interaction might be regulated by GTP-hydrolysis [154,164]. Importantly, Jack et al. demonstrated that the chaperones TorD and HybE prevent Tat-specific premature targeting and translocation of non-matured substrates [157]. Thus only after complete maturation, would these proof-reading chaperones be released from the RR-signal sequences to allow for a subsequent targeting to the Tat translocase.
6. Perspectives
Although many details recently became manifest of how RR-signal peptides are recognized by Tat translocases, the exact structure and the composition of the signal sequence-binding sites remain to be revealed. In particular, the individual contributions of the TatABC subunits to the binding and insertion of RR-signal sequences that likely occurs in a stepwise manner need to be further substantiated. Undoubtedly, the major challenge for future studies is elucidating the structure, composition and functioning of a leak-proof protein conduit across the membrane. Information relating to the mode of translocation should also settle the issue of whether translocation is paralleled by a topology change of TatA proteins. If future progress should confirm the model of a Tat pore, the mechanism of its assembly from the participating Tat subunits would be a next step of investigation. A pivotal approach towards many of these objectives would be the generation of true translocation intermediates that freeze the molecular environment during TM passage. Understanding how the proton gradient is transduced into the formation of a protein-conducting structure or even directly into the transmembrane movement of Tat substrates will be a milestone in the investigation of the Tat apparatus. Another key question is the fate of translocation-incompetent Tat substrates, be they rejected or cleared from the Tat components. Obviously, any structural information, even on substructures or intermediate assemblies of the Tat subunits, would pave a much more direct way to some of these exciting issues.
Acknowledgements
The authors' work was supported by Sonderforschungsbereich 746 and Forschergruppe 929 of the Deutsche Forschungsgemeinschaft.
References
- 1.Lee P. A., Tullman-Ercek D., Georgiou G. 2006. The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 60, 373–395 10.1146/annurev.micro.60.080805.142212 (doi:10.1146/annurev.micro.60.080805.142212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Widdick D. A., Eijlander R. T., van Dijl J. M., Kuipers O. P., Palmer T. 2008. A facile reporter system for the experimental identification of twin-arginine translocation (Tat) signal peptides from all kingdoms of life. J. Mol. Biol. 375, 595–603 10.1016/j.jmb.2007.11.002 (doi:10.1016/j.jmb.2007.11.002) [DOI] [PubMed] [Google Scholar]
- 3.Palmer T., Sargent F., Berks B. C. 2010. The Tat protein export pathway. In EcoSal—Escherichia coli and Salmonella: cellular and molecular biology (eds Böck A., Curtiss R., III, Kaper J. B., Karp P. D., Neidhardt F. C., Nyström T., Slauch J. M., Squires C. L., Ussery D.). Washington, DC: ASM Press [Google Scholar]
- 4.Berks B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22, 393–404 10.1046/j.1365-2958.1996.00114.x (doi:10.1046/j.1365-2958.1996.00114.x) [DOI] [PubMed] [Google Scholar]
- 5.Robinson C., Matos C. F. R. O., Beck D., Ren C., Lawrence J., Vasisht N., Mendel S. 2011. Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. Biochim. Biophys. Acta 1808, 876–884 10.1016/j.bbamem.2010.11.023 (doi:10.1016/j.bbamem.2010.11.023) [DOI] [PubMed] [Google Scholar]
- 6.Cline K., Theg S. M. 2007. The Sec and Tat protein translocation pathways in chloroplasts. In Molecular machines involved in protein transport across cellular membranes (eds Dalbey R. E., Koehler C. M., Tamanoi F.), pp. 463–492 London, UK: Elsevier [Google Scholar]
- 7.van Dijl J. M., et al. 2002. Functional genomic analysis of the Bacillus subtilis Tat pathway for protein secretion. J. Biotechnol. 98, 243–254 10.1016/S0168-1656(02)00135-9 (doi:10.1016/S0168-1656(02)00135-9) [DOI] [PubMed] [Google Scholar]
- 8.Hinsley A. P., Stanley N. R., Palmer T., Berks B. C. 2001. A naturally occurring bacterial Tat signal peptide lacking one of the ‘invariant’ arginine residues of the consensus targeting motif. FEBS Lett. 497, 45–49 10.1016/S0014-5793(01)02428-0 (doi:10.1016/S0014-5793(01)02428-0) [DOI] [PubMed] [Google Scholar]
- 9.Ignatova Z., Hornle C., Nurk A., Kasche V. 2002. Unusual signal peptide directs penicillin amidase from Escherichia coli to the Tat translocation machinery. Biochem. Biophys. Res. Commun. 291, 146–149 10.1006/bbrc.2002.6420 (doi:10.1006/bbrc.2002.6420) [DOI] [PubMed] [Google Scholar]
- 10.Molik S., Karnauchov I., Weidlich C., Herrmann R. G., Klosgen R. B. 2001. The rieske Fe/S protein of the cytochrome b6/f complex in chloroplasts. Missing link in the evolution of protein transport pathways in chloroplasts? J. Biol. Chem. 276, 42 761–42 766 [DOI] [PubMed] [Google Scholar]
- 11.DeLisa M. P., Samuelson P., Palmer T., Georgiou G. 2002. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J. Biol. Chem. 277, 29 825–29 831 [DOI] [PubMed] [Google Scholar]
- 12.Stanley N. R., Palmer T., Berks B. C. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275, 11 591–11 596 [DOI] [PubMed] [Google Scholar]
- 13.Ize B., Gerard F., Zhang M., Chanal A., Voulhoux R., Palmer T., Filloux A., Wu L. F. 2002. In vivo dissection of the Tat translocation pathway in Escherichia coli. J. Mol. Biol. 317, 327–335 10.1006/jmbi.2002.5431 (doi:10.1006/jmbi.2002.5431) [DOI] [PubMed] [Google Scholar]
- 14.Halbig D., Wiegert T., Blaudeck N., Freudl R., Sprenger G. A. 1999. The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur. J. Biochem. 263, 543–551 10.1046/j.1432-1327.1999.00536.x (doi:10.1046/j.1432-1327.1999.00536.x) [DOI] [PubMed] [Google Scholar]
- 15.Berks B. C., Palmer T., Sargent F. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47, 187–254 10.1016/S0065-2911(03)47004-5 (doi:10.1016/S0065-2911(03)47004-5) [DOI] [PubMed] [Google Scholar]
- 16.Mendel S., McCarthy A., Barnett J. P., Eijlander R. T., Nenninger A., Kuipers O. P., Robinson C. 2008. The Escherichia coli TatABC system and a Bacillus subtilis TatAC-type system recognise three distinct targeting determinants in twin-arginine signal peptides. J. Mol. Biol. 375, 661–672 10.1016/j.jmb.2007.09.087 (doi:10.1016/j.jmb.2007.09.087) [DOI] [PubMed] [Google Scholar]
- 17.Ize B., Gerard F., Wu L. F. 2002. In vivo assessment of the Tat signal peptide specificity in Escherichia coli. Arch. Microbiol. 178, 548–553 10.1007/s00203-002-0488-1 (doi:10.1007/s00203-002-0488-1) [DOI] [PubMed] [Google Scholar]
- 18.Cristobal S., de Gier J. W., Nielsen H., von Heijne G. 1999. Competition between Sec- and Tat-dependent protein translocation in Escherichia coli. EMBO J. 18, 2982–2990 10.1093/emboj/18.11.2982 (doi:10.1093/emboj/18.11.2982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yahr T. L., Wickner W. T. 2001. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 20, 2472–2479 10.1093/emboj/20.10.2472 (doi:10.1093/emboj/20.10.2472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luke I., Handford J. I., Palmer T., Sargent F. 2009. Proteolytic processing of Escherichia coli twin-arginine signal peptides by LepB. Arch. Microbiol. 191, 919–925 10.1007/s00203-009-0516-5 (doi:10.1007/s00203-009-0516-5) [DOI] [PubMed] [Google Scholar]
- 21.Bogsch E., Brink S., Robinson C. 1997. Pathway specificity for a ΔpH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J. 16, 3851–3859 10.1093/emboj/16.13.3851 (doi:10.1093/emboj/16.13.3851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaudeck N., Kreutzenbeck P., Freudl R., Sprenger G. A. 2003. Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J. Bacteriol. 185, 2811–2819 10.1128/JB.185.9.2811-2819.2003 (doi:10.1128/JB.185.9.2811-2819.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tullman-Ercek D., DeLisa M. P., Kawarasaki Y., Iranpour P., Ribnicky B., Palmer T., Georgiou G. 2007. Export pathway selectivity of Escherichia coli twin arginine translocation signal peptides. J. Biol. Chem. 282, 8309–8316 10.1074/jbc.M610507200 (doi:10.1074/jbc.M610507200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLisa M. P., Tullman D., Georgiou G. 2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl Acad. Sci USA 100, 6115–6120 10.1073/pnas.0937838100 (doi:10.1073/pnas.0937838100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panahandeh S., Maurer C., Moser M., DeLisa M. P., Müller M. 2008. Following the path of a twin-arginine precursor along the TatABC translocase of Escherichia coli. J. Biol. Chem. 283, 33 267–33 275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders C., Wethkamp N., Lill H. 2001. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol. Microbiol. 41, 241–246 10.1046/j.1365-2958.2001.02514.x (doi:10.1046/j.1365-2958.2001.02514.x) [DOI] [PubMed] [Google Scholar]
- 27.Santini C. L., Ize B., Chanal A., Muller M., Giordano G., Wu L. F. 1998. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17, 101–112 10.1093/emboj/17.1.101 (doi:10.1093/emboj/17.1.101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigue A., Chanal A., Beck K., Muller M., Wu L. F. 1999. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J. Biol. Chem. 274, 13 223–13 228 [DOI] [PubMed] [Google Scholar]
- 29.Sambasivarao D., Turner R. J., Simala-Grant J. L., Shaw G., Hu J., Weiner J. H. 2000. Multiple roles for the twin arginine leader sequence of dimethyl sulfoxide reductase of Escherichia coli. J. Biol. Chem. 275, 22 526–22 531 10.1074/jbc.M909289199 (doi:10.1074/jbc.M909289199) [DOI] [PubMed] [Google Scholar]
- 30.Hutcheon G. W., Bolhuis A. 2003. The archaeal twin-arginine translocation pathway. Biochem. Soc. Trans. 31, 686–689 10.1042/BST0310686 (doi:10.1042/BST0310686) [DOI] [PubMed] [Google Scholar]
- 31.Rose R. W., Bruser T., Kissinger J. C., Pohlschroder M. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45, 943–950 10.1046/j.1365-2958.2002.03090.x (doi:10.1046/j.1365-2958.2002.03090.x) [DOI] [PubMed] [Google Scholar]
- 32.van der Ploeg R., et al. 2011. Environmental salinity determines the specificity and need for Tat-dependent secretion of the ywbn protein in Bacillus subtilis. PLoS ONE 6, e18140. 10.1371/journal.pone.0018140 (doi:10.1371/journal.pone.0018140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachmann J., Bauer B., Zwicker K., Ludwig B., Anderka O. 2006. The Rieske protein from Paracoccus denitrificans is inserted into the cytoplasmic membrane by the twin-arginine translocase. FEBS J. 273, 4817–4830 10.1111/j.1742-4658.2006.05480.x (doi:10.1111/j.1742-4658.2006.05480.x) [DOI] [PubMed] [Google Scholar]
- 34.De Buck E., Vranckx L., Meyen E., Maes L., Vandersmissen L., Anne J., Lammertyn E. 2007. The twin-arginine translocation pathway is necessary for correct membrane insertion of the Rieske Fe/S protein in Legionella pneumophila. FEBS Lett. 581, 259–264 10.1016/j.febslet.2006.12.022 (doi:10.1016/j.febslet.2006.12.022) [DOI] [PubMed] [Google Scholar]
- 35.Hatzixanthis K., Palmer T., Sargent F. 2003. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol. Microbiol. 49, 1377–1390 10.1046/j.1365-2958.2003.03642.x (doi:10.1046/j.1365-2958.2003.03642.x) [DOI] [PubMed] [Google Scholar]
- 36.Walther T. H., Grage S. L., Roth N., Ulrich A. S. 2010. Membrane alignment of the pore-forming component TatA(d) of the twin-arginine translocase from Bacillus subtilis resolved by solid-state NMR spectroscopy. J. Am. Chem. Soc. 132, 15 945–15 956 [DOI] [PubMed] [Google Scholar]
- 37.Greene N. P., Porcelli I., Buchanan G., Hicks M. G., Schermann S. M., Palmer T., Berks B. C. 2007. Cysteine scanning mutagenesis and disulfide mapping studies of the TatA component of the bacterial twin arginine translocase. J. Biol. Chem. 282, 23 937–23 945 [DOI] [PubMed] [Google Scholar]
- 38.Hicks M. G., de Leeuw E., Porcelli I., Buchanan G., Berks B. C., Palmer T. 2003. The Escherichia coli twin-arginine translocase: conserved residues of TatA and TatB family components involved in protein transport. FEBS Lett. 539, 61–67 10.1016/S0014-5793(03)00198-4 (doi:10.1016/S0014-5793(03)00198-4) [DOI] [PubMed] [Google Scholar]
- 39.Lee P. A., et al. 2006. Cysteine-scanning mutagenesis and disulfide mapping studies of the conserved domain of the twin-arginine translocase TatB component. J. Biol. Chem. 281, 34 072–34 085 [DOI] [PubMed] [Google Scholar]
- 40.Yen M. R., Tseng Y. H., Nguyen E. H., Wu L. F., Saier M. H., Jr 2002. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 177, 441–450 10.1007/s00203-002-0408-4 (doi:10.1007/s00203-002-0408-4) [DOI] [PubMed] [Google Scholar]
- 41.Wexler M., Sargent F., Jack R. L., Stanley N. R., Bogsch E. G., Robinson C., Berks B. C., Palmer T. 2000. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J. Biol. Chem. 275, 16 717–16 722 [DOI] [PubMed] [Google Scholar]
- 42.Jack R. L., Sargent F., Berks B. C., Sawers G., Palmer T. 2001. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J. Bacteriol. 183, 1801–1804 10.1128/JB.183.5.1801-1804.2001 (doi:10.1128/JB.183.5.1801-1804.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jongbloed J. D., Grieger U., Antelmann H., Hecker M., Nijland R., Bron S., van Dijl J. M. 2004. Two minimal Tat translocases in Bacillus. Mol. Microbiol. 54, 1319–1325 10.1111/j.1365-2958.2004.04341.x (doi:10.1111/j.1365-2958.2004.04341.x) [DOI] [PubMed] [Google Scholar]
- 44.Sargent F., Stanley N. R., Berks B. C., Palmer T. 1999. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J. Biol. Chem. 274, 36 073–36 082 [DOI] [PubMed] [Google Scholar]
- 45.Sargent F., Bogsch E. G., Stanley N. R., Wexler M., Robinson C., Berks B. C., Palmer T. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17, 3640–3650 10.1093/emboj/17.13.3640 (doi:10.1093/emboj/17.13.3640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogsch E. G., Sargent F., Stanley N. R., Berks B. C., Robinson C., Palmer T. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273, 18 003–18 006 [DOI] [PubMed] [Google Scholar]
- 47.Hu Y., Zhao E., Li H., Xia B., Jin C. 2010. J Am Chem Soc 132, 15 942–15 944 [DOI] [PubMed] [Google Scholar]
- 48.Chan C. S., Haney E. F., Vogel H. J., Turner R. J. 2011. Towards understanding the Tat translocation mechanism through structural and biophysical studies of the amphipathic region of TatA from Escherichia coli. Biochim. Biophys. Acta 1808, 2289–2296 10.1016/j.bbamem.2011.05.024 (doi:10.1016/j.bbamem.2011.05.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porcelli I., de Leeuw E., Wallis R., van den Brink-van der Laan E., de Kruijff B., Wallace B. A., Palmer T., Berks B. C. 2002. Characterization and membrane assembly of the TatA component of the Escherichia coli twin-arginine protein transport system. Biochemistry 41, 13 690–13 697 10.1021/bi026142i (doi:10.1021/bi026142i) [DOI] [PubMed] [Google Scholar]
- 50.Gouffi K., Gerard F., Santini C. L., Wu L. F. 2004. Dual topology of the Escherichia coli TatA protein. J. Biol. Chem. 279, 11 608–11 615 [DOI] [PubMed] [Google Scholar]
- 51.Chan C. S., Zlomislic M. R., Tieleman D. P., Turner R. J. 2007. The TatA subunit of Escherichia coli twin-arginine translocase has an n-in topology. Biochemistry 46, 7396–7404 10.1021/bi7005288 (doi:10.1021/bi7005288) [DOI] [PubMed] [Google Scholar]
- 52.Pop O. I., Westermann M., Volkmer-Engert R., Schulz D., Lemke C., Schreiber S., Gerlach R., Wetzker R., Muller J. P. 2003. Sequence-specific binding of prePhoD to soluble TatAd indicates protein-mediated targeting of the Tat export in Bacillus subtilis. J. Biol. Chem. 278, 38 428–38 436 [DOI] [PubMed] [Google Scholar]
- 53.De Keersmaeker S., Van Mellaert L., Lammertyn E., Vrancken K., Anne J., Geukens N. 2005. Functional analysis of TatA and TatB in Streptomyces lividans. Biochem. Biophys. Res. Commun. 335, 973–982 [DOI] [PubMed] [Google Scholar]
- 54.Westermann M., Pop O. I., Gerlach R., Appel T. R., Schlormann W., Schreiber S., Muller J. P. 2006. The TatAd component of the Bacillus subtilis twin-arginine protein transport system forms homo-multimeric complexes in its cytosolic and membrane embedded localisation. Biochim. Biophys. Acta 1758, 443–4451 10.1016/j.bbamem.2006.03.018 (doi:10.1016/j.bbamem.2006.03.018) [DOI] [PubMed] [Google Scholar]
- 55.Schreiber S., Stengel R., Westermann M., Volkmer-Engert R., Pop O. I., Muller J. P. 2006. Affinity of TatCd for TatAd elucidates its receptor function in the Bacillus subtilis twin arginine translocation (Tat) translocase system. J. Biol. Chem. 281, 19 977–19 984 [DOI] [PubMed] [Google Scholar]
- 56.Barnett J. P., van der Ploeg R., Eijlander R. T., Nenninger A., Mendel S., Rozeboom R., Kuipers O. P., van Dijl J. M., Robinson C. 2009. The twin-arginine translocation (Tat) systems from Bacillus subtilis display a conserved mode of complex organization and similar substrate recognition requirements. FEBS J. 276, 232–243 10.1111/j.1742-4658.2008.06776.x (doi:10.1111/j.1742-4658.2008.06776.x) [DOI] [PubMed] [Google Scholar]
- 57.Berthelmann F., Mehner D., Richter S., Lindenstrauss U., Lunsdorf H., Hause G., Brüser T. 2008. Recombinant expression of tatABC and tatAC results in the formation of interacting cytoplasmic TatA tubes in Escherichia coli. J. Biol. Chem. 283, 25 281–25 289 10.1074/jbc.M707757200 (doi:10.1074/jbc.M707757200) [DOI] [PubMed] [Google Scholar]
- 58.Frielingsdorf S., Jakob M., Klösgen R. B. 2008. A stromal pool of TatA promotes Tat-dependent protein transport across the thylakoid membrane. J. Biol. Chem. 283, 33 838–33 845 10.1074/jbc.M806334200 (doi:10.1074/jbc.M806334200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warren G., Oates J., Robinson C., Dixon A. M. 2009. Contributions of the transmembrane domain and a key acidic motif to assembly and function of the TatA complex. J. Mol. Biol. 388, 122–132 10.1016/j.jmb.2009.02.060 (doi:10.1016/j.jmb.2009.02.060) [DOI] [PubMed] [Google Scholar]
- 60.Hicks M. G., Lee P. A., Georgiou G., Berks B. C., Palmer T. 2005. Positive selection for loss-of-function tat mutations identifies critical residues required for TatA activity. J. Bacteriol. 187, 2920–2925 10.1128/JB.187.8.2920-2925.2005 (doi:10.1128/JB.187.8.2920-2925.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barrett C. M., Robinson C. 2005. Evidence for interactions between domains of TatA and TatB from mutagenesis of the TatABC subunits of the twin-arginine translocase. FEBS J. 272, 2261–2275 10.1111/j.1742-4658.2005.04654.x (doi:10.1111/j.1742-4658.2005.04654.x) [DOI] [PubMed] [Google Scholar]
- 62.Barrett C. M., Mathers J. E., Robinson C. 2003. Identification of key regions within the Escherichia coli TatAB subunits. FEBS Lett. 537, 42–46 10.1016/S0014-5793(03)00068-1 (doi:10.1016/S0014-5793(03)00068-1) [DOI] [PubMed] [Google Scholar]
- 63.Kreutzenbeck P., Kroger C., Lausberg F., Blaudeck N., Sprenger G. A., Freudl R. 2007. Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 282, 7903–7911 10.1074/jbc.M610126200 (doi:10.1074/jbc.M610126200) [DOI] [PubMed] [Google Scholar]
- 64.Buchanan G., Leeuw E., Stanley N. R., Wexler M., Berks B. C., Sargent F., Palmer T. 2002. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 43, 1457–1470 10.1046/j.1365-2958.2002.02853.x (doi:10.1046/j.1365-2958.2002.02853.x) [DOI] [PubMed] [Google Scholar]
- 65.Allen S. C., Barrett C. M., Ray N., Robinson C. 2002. Essential cytoplasmic domains in the Escherichia coli TatC protein.. J. Biol. Chem. 277, 10 362–10 366 [DOI] [PubMed] [Google Scholar]
- 66.Maldonado B., Buchanan G., Müller M., Berks B. C., Palmer T. 2011. Genetic evidence for a TatC dimer at the core of the Escherichia coli twin arginine (Tat) protein translocase. J. Mol. Microbiol. Biotechnol. 20, 168–175 10.1159/000329076 (doi:10.1159/000329076) [DOI] [PubMed] [Google Scholar]
- 67.Strauch E. M., Georgiou G. 2007. Escherichia coli tatC mutations that suppress defective twin-arginine transporter signal peptides. J. Mol. Biol. 374, 283–291 10.1016/j.jmb.2007.09.050 (doi:10.1016/j.jmb.2007.09.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holzapfel E., et al. 2007. The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46, 2892–2898 10.1021/bi062205b (doi:10.1021/bi062205b) [DOI] [PubMed] [Google Scholar]
- 69.Lee P. A., Buchanan G., Stanley N. R., Berks B. C., Palmer T. 2002. Truncation analysis of TatA and TatB defines the minimal functional units required for protein translocation. J. Bacteriol. 184, 5871–5879 10.1128/JB.184.21.5871-5879.2002 (doi:10.1128/JB.184.21.5871-5879.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barrett C. M., Mangels D., Robinson C. 2005. Mutations in subunits of the Escherichia coli twin-arginine translocase block function via differing effects on translocation activity or Tat complex structure. J. Mol. Biol. 347, 453–463 10.1016/j.jmb.2005.01.026 (doi:10.1016/j.jmb.2005.01.026) [DOI] [PubMed] [Google Scholar]
- 71.Maldonado B., Kneuper H., Buchanan G., Hatzixanthis K., Sargent F., Berks B. C., Palmer T. 2011. Characterisation of the membrane-extrinsic domain of the TatB component of the twin arginine protein translocase. FEBS Lett. 585, 478–484 10.1016/j.febslet.2011.01.016 (doi:10.1016/j.febslet.2011.01.016) [DOI] [PubMed] [Google Scholar]
- 72.Chanal A., Santini C., Wu L. 1998. Potential receptor function of three homologous components, TatA, TatB and TatE, of the twin-arginine signal sequence-dependent metalloenzyme translocation pathway in Escherichia coli. Mol. Microbiol. 30, 674–676 10.1046/j.1365-2958.1998.01095.x (doi:10.1046/j.1365-2958.1998.01095.x) [DOI] [PubMed] [Google Scholar]
- 73.Blaudeck N., Kreutzenbeck P., Müller M., Sprenger G. A., Freudl R. 2005. Isolation and characterization of bifunctional Escherichia coli TatA mutant proteins that allow efficient Tat-dependent protein translocation in the absence of TatB. J. Biol. Chem. 280, 3426–3432 10.1074/jbc.M411210200 (doi:10.1074/jbc.M411210200) [DOI] [PubMed] [Google Scholar]
- 74.Barrett C. M., Freudl R., Robinson C. 2007. Twin arginine translocation (Tat)-dependent export in the apparent absence of TatABC or TatA complexes using modified Escherichia coli TatA subunits that substitute for TatB. J. Biol. Chem. 282, 36 206–36 213 [DOI] [PubMed] [Google Scholar]
- 75.Ki J. J., Kawarasaki Y., Gam J., Harvey B. R., Iverson B. L., Georgiou G. 2004. A periplasmic fluorescent reporter protein and its application in high-throughput membrane protein topology analysis. J. Mol. Biol. 341, 901–909 10.1016/j.jmb.2004.05.078 (doi:10.1016/j.jmb.2004.05.078) [DOI] [PubMed] [Google Scholar]
- 76.Behrendt J., Standar K., Lindenstrauss U., Bruser T. 2004. Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol. Lett. 234, 303–308 10.1111/j.1574-6968.2004.tb09548.x (doi:10.1111/j.1574-6968.2004.tb09548.x) [DOI] [PubMed] [Google Scholar]
- 77.Punginelli C., Maldonado B., Grahl S., Jack R., Alami M., Schroder J., Berks B. C., Palmer T. 2007. Cysteine scanning mutagenesis and topological mapping of the Escherichia Coli twin-arginine translocase TatC component. J. Bacteriol. 189, 5482–5494 10.1128/JB.00647-07 (doi:10.1128/JB.00647-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matos C. F., Di Cola A., Robinson C. 2009. TatD is a central component of a Tat translocon-initiated quality control system for exported FeS proteins in Escherichia coli. EMBO Rep. 10, 474–479 10.1038/embor.2009.34 (doi:10.1038/embor.2009.34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lindenstrauss U., Matos C. F., Graubner W., Robinson C., Bruser T. 2010. Malfolded recombinant Tat substrates are Tat-independently degraded in Escherichia coli. FEBS Lett. 584, 3644–3648 10.1016/j.febslet.2010.07.039 (doi:10.1016/j.febslet.2010.07.039) [DOI] [PubMed] [Google Scholar]
- 80.Qiu J., Yoon J. H., Shen B. 2005. Search for apoptotic nucleases in yeast: role of Tat-D nuclease in apoptotic DNA degradation. J. Biol. Chem. 280, 15 370–15 379 [DOI] [PubMed] [Google Scholar]
- 81.Centore R. C., Lestini R., Sandler S. J. 2008. XthA (Exonuclease III) regulates loading of RecA onto DNA substrates in log phase Escherichia coli cells. Mol. Microbiol. 67, 88–101 [DOI] [PubMed] [Google Scholar]
- 82.Behrendt J., Lindenstrauss U., Brüser T. 2007. The TatBC complex formation suppresses a modular TatB-multimerization in Escherichia coli. FEBS Lett. 581, 4085–4090 10.1016/j.febslet.2007.07.045 (doi:10.1016/j.febslet.2007.07.045) [DOI] [PubMed] [Google Scholar]
- 83.Orriss G. L., Tarry M. J., Ize B., Sargent F., Lea S. M., Palmer T., Berks B. C. 2007. TatBC, TatB, and TatC form structurally autonomous units within the twin arginine protein transport system of Escherichia coli. FEBS Lett. 581, 4091–4097 10.1016/j.febslet.2007.07.044 (doi:10.1016/j.febslet.2007.07.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Leeuw E., Granjon T., Porcelli I., Alami M., Carr S. B., Müller M., Sargent F., Palmer T., Berks B. C. 2002. Oligomeric properties and signal peptide binding by Escherichia coli Tat protein transport complexes. J. Mol. Biol. 322, 1135–1146 10.1016/S0022-2836(02)00820-3 (doi:10.1016/S0022-2836(02)00820-3) [DOI] [PubMed] [Google Scholar]
- 85.Oates J., Barrett C. M., Barnett J. P., Byrne K. G., Bolhuis A., Robinson C. 2005. The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J. Mol. Biol. 346, 295–305 10.1016/j.jmb.2004.11.047 (doi:10.1016/j.jmb.2004.11.047) [DOI] [PubMed] [Google Scholar]
- 86.Cline K., Mori H. 2001. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC–Hcf106 complex before Tha4-dependent transport. J. Cell. Biol. 154, 719–729 10.1083/jcb.200105149 (doi:10.1083/jcb.200105149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barnett J. P., Eijlander R. T., Kuipers O. P., Robinson C. 2008. A minimal Tat system from a Gram-positive organism. A bifunctional TatA subunit participates in discrete TatAC and TatA complexes. J. Biol. Chem. 283, 2534–2542 10.1074/jbc.M708134200 (doi:10.1074/jbc.M708134200) [DOI] [PubMed] [Google Scholar]
- 88.McDevitt C. A., Hicks M. G., Palmer T., Berks B. C. 2005. Characterisation of Tat protein transport complexes carrying inactivating mutations. Biochem. Biophys. Res. Commun. 329, 693–698 10.1016/j.bbrc.2005.02.038 (doi:10.1016/j.bbrc.2005.02.038) [DOI] [PubMed] [Google Scholar]
- 89.Bolhuis A., Mathers J. E., Thomas J. D., Barrett C. M., Robinson C. 2001. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276, 20 213–20 219 [DOI] [PubMed] [Google Scholar]
- 90.Oates J., Mathers J., Mangels D., Kuhlbrandt W., Robinson C., Model K. 2003. Consensus structural features of purified bacterial TatABC complexes. J. Mol. Biol. 330, 277–286 10.1016/S0022-2836(03)00621-1 (doi:10.1016/S0022-2836(03)00621-1) [DOI] [PubMed] [Google Scholar]
- 91.Richter S., Brüser T. 2005. Targeting of unfolded PhoA to the TAT translocon of Escherichia coli. J. Biol. Chem. 280, 42 723–42 730 [DOI] [PubMed] [Google Scholar]
- 92.Mangels D., Mathers J., Bolhuis A., Robinson C. 2005. The core TatABC complex of the twin-arginine translocase in Escherichia coli: TatC drives assembly whereas TatA is essential for stability. J. Mol. Biol. 345, 415–423 10.1016/j.jmb.2004.10.043 (doi:10.1016/j.jmb.2004.10.043) [DOI] [PubMed] [Google Scholar]
- 93.de Leeuw E., Porcelli I., Sargent F., Palmer T., Berks B. C. 2001. Membrane interactions and self-association of the TatA and TatB components of the twin-arginine translocation pathway. FEBS Lett. 506, 143–148 10.1016/S0014-5793(01)02904-0 (doi:10.1016/S0014-5793(01)02904-0) [DOI] [PubMed] [Google Scholar]
- 94.Kostecki J. S., Li H., Turner R. J., DeLisa M. P. 2010. Visualizing interactions along the Escherichia coli twin-arginine translocation pathway using protein fragment complementation. PLoS ONE 5, e9225. 10.1371/journal.pone.0009225 (doi:10.1371/journal.pone.0009225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maurer C., Panahandeh S., Jungkamp A. C., Moser M., Müller M. 2010. TatB functions as an oligomeric binding site for folded tat precursor proteins. Mol. Biol. Cell 21, 4151–4161 10.1091/mbc.E10-07-0585 (doi:10.1091/mbc.E10-07-0585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tarry M. J., Schäfer E., Chen S., Buchanan G., Greene N. P., Lea S. M., Palmer T., Saibil H. R., Berks B. C. 2009. Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc. Natl Acad. Sci. USA 106, 13 284–13 289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma X., Cline K. 2010. Multiple precursor proteins bind individual Tat receptor complexes and are collectively transported. EMBO J. 29, 1477–1488 10.1038/emboj.2010.44 (doi:10.1038/emboj.2010.44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alder N. N., Theg S. M. 2003. Protein transport via the cpTat pathway displays cooperativity and is stimulated by transport-incompetent substrate. FEBS Lett. 540, 96–100 10.1016/S0014-5793(03)00231-X (doi:10.1016/S0014-5793(03)00231-X) [DOI] [PubMed] [Google Scholar]
- 99.Fröbel J., Rose P., Müller M. 2011. Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J. Biol. Chem. 286, 43 679–43 689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McDevitt C. A., Buchanan G., Sargent F., Palmer T., Berks B. C. 2006. Subunit composition and in vivo substrate-binding characteristics of Escherichia coli Tat protein complexes expressed at native levels. FEBS J. 273, 5656–5668 10.1111/j.1742-4658.2006.05554.x (doi:10.1111/j.1742-4658.2006.05554.x) [DOI] [PubMed] [Google Scholar]
- 101.Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., Müller M. 2003. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12, 937–946 10.1016/S1097-2765(03)00398-8 (doi:10.1016/S1097-2765(03)00398-8) [DOI] [PubMed] [Google Scholar]
- 102.Gerard F., Cline K. 2006. Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 281, 6130–6135 10.1074/jbc.M512733200 (doi:10.1074/jbc.M512733200) [DOI] [PubMed] [Google Scholar]
- 103.Gohlke U., Pullan L., McDevitt C. A., Porcelli I., de Leeuw E., Palmer T., Saibil H. R., Berks B. C. 2005. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl Acad. Sci. USA 102, 10 482–10 486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leake M. C., Greene N. P., Godun R. M., Granjon T., Buchanan G., Chen S., Berry R. M., Palmer T., Berks B. C. 2008. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single molecule imaging. Proc. Natl Acad. Sci. USA 105, 15 376–15 381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dabney-Smith C., Mori H., Cline K. 2006. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J. Biol. Chem. 281, 5476–5483 10.1074/jbc.M512453200 (doi:10.1074/jbc.M512453200) [DOI] [PubMed] [Google Scholar]
- 106.Dabney-Smith C., Cline K. 2009. Clustering of C-terminal stromal domains of Tha4 homo-oligomers during translocation by the Tat protein transport system. Mol. Biol. Cell 20, 2060–2069 10.1091/mbc.E08-12-1189 (doi:10.1091/mbc.E08-12-1189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sargent F., Gohlke U., De Leeuw E., Stanley N. R., Palmer T., Saibil H. R., Berks B. C. 2001. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur. J. Biochem. 268, 3361–3367 10.1046/j.1432-1327.2001.02263.x (doi:10.1046/j.1432-1327.2001.02263.x) [DOI] [PubMed] [Google Scholar]
- 108.Jakob M., Kaiser S., Gutensohn M., Hanner P., Klösgen R. B. 2009. Tat subunit stoichiometry in Arabidopsis thaliana challenges the proposed function of TatA as the translocation pore. Biochim. Biophys. Acta 1793, 388–394 10.1016/j.bbamcr.2008.09.006 (doi:10.1016/j.bbamcr.2008.09.006) [DOI] [PubMed] [Google Scholar]
- 109.Berks B. C., Sargent F., Palmer T. 2000. The Tat protein export pathway. Mol. Microbiol. 35, 260–274 10.1046/j.1365-2958.2000.01719.x (doi:10.1046/j.1365-2958.2000.01719.x) [DOI] [PubMed] [Google Scholar]
- 110.White G. F., Schermann S. M., Bradley J., Roberts A., Greene N. P., Berks B. C., Thomson A. J. 2010. Subunit organization in the TatA complex of the twin arginine protein translocase. A site-directed EPR spin labeling study. J. Biol. Chem. 285, 2294–2301 10.1074/jbc.M109.065458 (doi:10.1074/jbc.M109.065458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baglieri J., Beck D., Vasisht N., Smith C., Robinson C. In press Structure of the TatA paralog, TatE, suggests a structurally homogeneous form of Tat protein translocase that transports folded proteins of differing diameter. J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kipping M., Lilie H., Lindenstrauss U., Andreesen J. R., Griesinger C., Carlomagno T., Brüser T. 2003. Structural studies on a twin-arginine signal sequence. FEBS Lett. 550, 18–22 10.1016/S0014-5793(03)00804-4 (doi:10.1016/S0014-5793(03)00804-4) [DOI] [PubMed] [Google Scholar]
- 113.San Miguel M., Marrington R., Rodger P. M., Rodger A., Robinson C. 2003. An Escherichia coli twin-arginine signal peptide switches between helical and unstructured conformations depending on the hydrophobicity of the environment. Eur. J. Biochem. 270, 3345–3352 10.1046/j.1432-1033.2003.03710.x (doi:10.1046/j.1432-1033.2003.03710.x) [DOI] [PubMed] [Google Scholar]
- 114.Holzapfel E., Moser M., Schiltz E., Ueda T., Betton J. M., Müller M. 2009. Twin-arginine-dependent translocation of SufI in the absence of cytosolic helper proteins. Biochemistry 48, 5096–5105 10.1021/bi900520d (doi:10.1021/bi900520d) [DOI] [PubMed] [Google Scholar]
- 115.Graubner W., Schierhorn A., Brüser T. 2007. DnaK plays a pivotal role in Tat targeting of CueO and functions beside SlyD as a general Tat signal binding chaperone. J. Biol. Chem. 282, 7116–7124 10.1074/jbc.M608235200 (doi:10.1074/jbc.M608235200) [DOI] [PubMed] [Google Scholar]
- 116.Jong W. S., ten Hagen-Jongman C. M., Genevaux P., Brunner J., Oudega B., Luirink J. 2004. Trigger factor interacts with the signal peptide of nascent Tat substrates but does not play a critical role in Tat-mediated export. Eur. J. Biochem. 271, 4779–4787 10.1111/j.1432-1033.2004.04442.x (doi:10.1111/j.1432-1033.2004.04442.x) [DOI] [PubMed] [Google Scholar]
- 117.Perez-Rodriguez R., Fisher A. C., Perlmutter J. D., Hicks M. G., Chanal A., Santini C. L., Wu L. F., Palmer T., DeLisa M. P. 2007. An essential role for the DnaK molecular chaperone in stabilizing over-expressed substrate proteins of the bacterial twin-arginine translocation pathway. J. Mol. Biol. 367, 715–730 10.1016/j.jmb.2007.01.027 (doi:10.1016/j.jmb.2007.01.027) [DOI] [PubMed] [Google Scholar]
- 118.Hou B., Frielingsdorf S., Klösgen R. B. 2006. Unassisted membrane insertion as the initial step in ΔpH/Tat-dependent protein transport. J. Mol. Biol. 355, 957–967 10.1016/j.jmb.2005.11.029 (doi:10.1016/j.jmb.2005.11.029) [DOI] [PubMed] [Google Scholar]
- 119.Shanmugham A., Wong Fong Sang H. W., Bollen Y. J., Lill H. 2006. Membrane binding of twin arginine preproteins as an early step in translocation. Biochemistry 45, 2243–2249 10.1021/bi052188a (doi:10.1021/bi052188a) [DOI] [PubMed] [Google Scholar]
- 120.Schlesier R., Klösgen R. B. 2010. Twin arginine translocation (Tat)-dependent protein transport: the passenger protein participates in the initial membrane binding step. Biol. Chem. 391, 1411–1417 10.1515/BC.2010.138 (doi:10.1515/BC.2010.138) [DOI] [PubMed] [Google Scholar]
- 121.Bageshwar U. K., Whitaker N., Liang F. C., Musser S. M. 2009. Interconvertibility of lipid- and translocon-bound forms of the bacterial Tat precursor pre-SufI. Mol. Microbiol. 74, 209–226 10.1111/j.1365-2958.2009.06862.x (doi:10.1111/j.1365-2958.2009.06862.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brüser T., Yano T., Brune D. C., Daldal F. 2003. Membrane targeting of a folded and cofactor-containing protein. Eur. J. Biochem. 270, 1211–1221 10.1046/j.1432-1033.2003.03481.x (doi:10.1046/j.1432-1033.2003.03481.x) [DOI] [PubMed] [Google Scholar]
- 123.Marrichi M., Camacho L., Russell D. G., DeLisa M. P. 2008. Genetic toggling of alkaline phosphatase folding reveals signal peptides for all major modes of transport across the inner membrane of bacteria. J. Biol. Chem. 283, 35 223–35 235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gerard F., Cline K. 2007. The thylakoid proton gradient promotes an advanced stage of signal peptide binding deep within the Tat pathway receptor complex. J. Biol. Chem. 282, 5263–5272 10.1074/jbc.M610337200 (doi:10.1074/jbc.M610337200) [DOI] [PubMed] [Google Scholar]
- 125.Li H., Faury D., Morosoli R. 2006. Impact of amino acid changes in the signal peptide on the secretion of the Tat-dependent xylanase C from Streptomyces lividans. FEMS Microbiol. Lett. 255, 268–274 10.1111/j.1574-6968.2005.00081.x (doi:10.1111/j.1574-6968.2005.00081.x) [DOI] [PubMed] [Google Scholar]
- 126.Zoufaly S., Fröbel J., Rose P., Flecken T., Maurer C., Moser M., Müller M. Mapping the precursor-binding site on the TatC subunit of the twin-arginine-specific protein translocase by site-specific photo cross-linking. J. Biol. Chem. doi: 10.1074/jbc.M112.343798. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fincher V., McCaffery M., Cline K. 1998. Evidence for a loop mechanism of protein transport by the thylakoid Delta pH pathway. FEBS Lett. 423, 66–70 10.1016/S0014-5793(98)00066-0 (doi:10.1016/S0014-5793(98)00066-0) [DOI] [PubMed] [Google Scholar]
- 128.Fröbel J., Rose P., Lausberg F., Moser M., Freudl R., Müller M. In preparation. Transmembrane insertion of twin-arginine signal peptides is driven by TatC and regulated by TatB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Frielingsdorf S., Klosgen R. B. 2007. Prerequisites for terminal processing of thylakoidal Tat substrates. J. Biol. Chem. 282, 24 455–24 462 10.1074/jbc.M702630200 (doi:10.1074/jbc.M702630200) [DOI] [PubMed] [Google Scholar]
- 130.Di Cola A., Robinson C. 2005. Large-scale translocation reversal within the thylakoid Tat system in vivo. J. Cell. Biol. 171, 281–289 10.1083/jcb.200502067 (doi:10.1083/jcb.200502067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ma X., Cline K. 2000. Precursors bind to specific sites on thylakoid membranes prior to transport on the delta pH protein translocation system. J. Biol. Chem. 275, 10 016–10 022 [DOI] [PubMed] [Google Scholar]
- 132.Alami M., Trescher D., Wu L. F., Müller M. 2002. Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 277, 20 499–20 503 [DOI] [PubMed] [Google Scholar]
- 133.Dabney-Smith C., Mori H., Cline K. 2003. Requirement of a Tha4-conserved transmembrane glutamate in thylakoid Tat translocase assembly revealed by biochemical complementation. J. Biol. Chem. 278, 43 027–43 033 10.1074/jbc.M307923200 (doi:10.1074/jbc.M307923200) [DOI] [PubMed] [Google Scholar]
- 134.Mori H., Cline K. 2002. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J. Cell. Biol. 157, 205–210 10.1083/jcb.200202048 (doi:10.1083/jcb.200202048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cline K., McCaffery M. 2007. Evidence for a dynamic and transient pathway through the Tat protein transport machinery. EMBO J. 26, 3039–3049 10.1038/sj.emboj.7601759 (doi:10.1038/sj.emboj.7601759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Brüser T., Sanders C. 2003. An alternative model of the twin arginine translocation system. Microbiol. Res. 158, 7–17 10.1078/0944-5013-00176 (doi:10.1078/0944-5013-00176) [DOI] [PubMed] [Google Scholar]
- 137.Maurer C., Panahandeh S., Moser M., Müller M. 2009. Impairment of twin-arginine-dependent export by seemingly small alterations of substrate conformation. FEBS Lett. 583, 2849–2853 10.1016/j.febslet.2009.07.038 (doi:10.1016/j.febslet.2009.07.038) [DOI] [PubMed] [Google Scholar]
- 138.Mould R. M., Shackleton J. B., Robinson C. 1991. Transport of proteins into chloroplasts: requirements for the efficient import of two lumenal oxygen-evolving complex proteins into isolated thylakoids. J. Biol. Chem. 266, 17 286–17 289 [PubMed] [Google Scholar]
- 139.Kleerebezem M., Crielaard W., Tommassen J. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the proton motive force under stress conditions. EMBO J. 15, 162–171 [PMC free article] [PubMed] [Google Scholar]
- 140.DeLisa M. P., Lee P., Palmer T., Georgiou G. 2004. Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J. Bacteriol. 186, 366–373 10.1128/JB.186.2.366-373.2004 (doi:10.1128/JB.186.2.366-373.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vrancken K., De Keersmaeker S., Geukens N., Lammertyn E., Anne J., Van Mellaert L. 2007. pspA overexpression in Streptomyces lividans improves both Sec- and Tat-dependent protein secretion. Appl. Microbiol. Biotechnol. 73, 1150–1157 10.1007/s00253-006-0571-7 (doi:10.1007/s00253-006-0571-7) [DOI] [PubMed] [Google Scholar]
- 142.Di Cola A., Bailey S., Robinson C. 2005. Bacterial secreted proteins: secretory mechanisms and role in pathogenesis. J. Biol. Chem. 280, 41 165–41 170 [Google Scholar]
- 143.Finazzi G., Chasen C., Wollman F. A., de Vitry C. 2003. Thylakoid targeting of Tat passenger proteins shows no ΔpH dependence in vivo. EMBO J. 22, 807–815 10.1093/emboj/cdg081 (doi:10.1093/emboj/cdg081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Braun N. A., Davis A. W., Theg S. M. 2007. The chloroplast Tat pathway utilizes the transmembrane electric potential as an energy source. Biophys. J. 93, 1993–1998 10.1529/biophysj.106.098731 (doi:10.1529/biophysj.106.098731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Theg S. M., Cline K., Finazzi G., Wollman F. A. 2005. The energetics of the chloroplast Tat protein transport pathway revisited. Trends Plant Sci. 10, 153–154 10.1016/j.tplants.2005.02.001 (doi:10.1016/j.tplants.2005.02.001) [DOI] [PubMed] [Google Scholar]
- 146.Alder N. N., Theg S. M. 2003. Energetics of protein transport across biological membranes. Cell 112, 231–242 10.1016/S0092-8674(03)00032-1 (doi:10.1016/S0092-8674(03)00032-1) [DOI] [PubMed] [Google Scholar]
- 147.Musser S. M., Theg S. M. 2000. Proton transfer limits protein translocation rate by the thylakoid ΔpH/Tat machinery. Biochemistry 39, 8228–8233 10.1021/bi000115f (doi:10.1021/bi000115f) [DOI] [PubMed] [Google Scholar]
- 148.Bageshwar U. K., Musser S. M. 2007. Two electrical potential–dependent steps are required for transport by the Escherichia coli Tat machinery. J. Cell Biol. 179, 87–99 10.1083/jcb.200702082 (doi:10.1083/jcb.200702082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hynds P. J., Robinson D., Robinson C. 1998. Progress in nucleic acid research and molecular biology. J. Biol. Chem. 273, 34 868–34 874 [Google Scholar]
- 150.Roffey R. A., Theg S. M. 1996. Protein trafficking in plant cells. Plant Physiol. 111, 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Musser S. M., Theg S. M. 2000. Characterization of the early steps of OE17 precursor transport by the thylakoid ΔpH/Tat machinery. Eur. J. Biochem. 267, 2588–2598 10.1046/j.1432-1327.2000.01269.x (doi:10.1046/j.1432-1327.2000.01269.x) [DOI] [PubMed] [Google Scholar]
- 152.Matos C. F., Robinson C., Di Cola A. 2008. The Tat system proofreads FeS protein substrates and directly initiates the disposal of rejected molecules. EMBO J. 27, 2055–2063 10.1038/emboj.2008.132 (doi:10.1038/emboj.2008.132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Richter S., Lindenstrauss U., Lucke C., Bayliss R., Brüser T. 2007. Bacterial secreted proteins: secretory mechanisms and role in pathogenesis. J. Biol. Chem. 282, 33 257–33 264 [DOI] [PubMed] [Google Scholar]
- 154.Sargent F. 2007. Constructing the wonders of the bacterial world: biosynthesis of complex enzymes. Microbiology 153, 633–651 10.1099/mic.0.2006/004762-0 (doi:10.1099/mic.0.2006/004762-0) [DOI] [PubMed] [Google Scholar]
- 155.Turner R. J., Papish A. L., Sargent F. 2004. Sequence analysis of bacterial redox enzyme maturation proteins (REMPs). Can. J. Microbiol. 50, 225–238 10.1139/w03-117 (doi:10.1139/w03-117) [DOI] [PubMed] [Google Scholar]
- 156.Tranier S., Mortier-Barriere I., Ilbert M., Birck C., Iobbi-Nivol C., Mejean V., Samama J. P. 2002. Characterization and multiple molecular forms of TorD from Shewanella massilia, the putative chaperone of the molybdoenzyme TorA. Protein Sci. 11, 2148–2157 10.1110/ps.0202902 (doi:10.1110/ps.0202902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jack R. L., Buchanan G., Dubini A., Hatzixanthis K., Palmer T., Sargent F. 2004. Coordinating assembly and export of complex bacterial proteins. EMBO J. 23, 3962–3972 10.1038/sj.emboj.7600409 (doi:10.1038/sj.emboj.7600409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Genest O., Seduk F., Ilbert M., Mejean V., Iobbi-Nivol C. 2006Signal peptide protection by specific chaperone. Biochem. Biophys. Res. Commun. 339, 991–995 10.1016/j.bbrc.2005.11.107 (doi:10.1016/j.bbrc.2005.11.107) [DOI] [PubMed] [Google Scholar]
- 159.Oresnik I. J., Ladner C. L., Turner R. J. 2001. Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 40, 323–331 10.1046/j.1365-2958.2001.02391.x (doi:10.1046/j.1365-2958.2001.02391.x) [DOI] [PubMed] [Google Scholar]
- 160.Dubini A., Sargent F. 2003. Assembly of Tat-dependent [NiFe] hydrogenases: identification of precursor-binding accessory proteins. FEBS Lett. 549, 141–146 10.1016/S0014-5793(03)00802-0 (doi:10.1016/S0014-5793(03)00802-0) [DOI] [PubMed] [Google Scholar]
- 161.Maillard A. P., Lalani S., Silva F., Belin D., Duong F. 2007. Deregulation of the SecYEG translocation channel upon removal of the plug domain. J. Biol. Chem. 282, 1281–1287 10.1074/jbc.M610060200 (doi:10.1074/jbc.M610060200) [DOI] [PubMed] [Google Scholar]
- 162.Sargent F., Berks B. C., Palmer T. 2002. Assembly of membrane-bound respiratory complexes by the Tat protein-transport system. Arch. Microbiol. 178, 77–84 10.1007/s00203-002-0434-2 (doi:10.1007/s00203-002-0434-2) [DOI] [PubMed] [Google Scholar]
- 163.Chan C. S., Chang L., Rommens K. L., Turner R. J. 2009. Their chaperones Tat-specific redox enzyme peptides and differential interactions between. J. Bacteriol. 191, 2091–2101 10.1128/JB.00949-08 (doi:10.1128/JB.00949-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hatzixanthis K., Clarke T. A., Oubrie A., Richardson D. J., Turner R. J., Sargent F. 2005. Signal peptide–chaperone interactions on the twin-arginine protein transport pathway. Proc. Natl Acad. Sci. USA 102, 8460–8465 10.1073/pnas.0500737102 (doi:10.1073/pnas.0500737102) [DOI] [PMC free article] [PubMed] [Google Scholar]