Abstract
Type VI secretion systems (T6SSs) are transenvelope complexes specialized in the transport of proteins or domains directly into target cells. These systems are versatile as they can target either eukaryotic host cells and therefore modulate the bacteria–host interaction and pathogenesis or bacterial cells and therefore facilitate access to a specific niche. These molecular machines comprise at least 13 proteins. Although recent years have witnessed advances in the role and function of these secretion systems, little is known about how these complexes assemble in the cell envelope. Interestingly, the current information converges to the idea that T6SSs are composed of two subassemblies, one resembling the contractile bacteriophage tail, whereas the other subunits are embedded in the inner and outer membranes and anchor the bacteriophage-like structure to the cell envelope. In this review, we summarize recent structural information on individual T6SS components emphasizing the fact that T6SSs are composite systems, adapting subunits from various origins.
Keywords: protein transport, protein trafficking, bacteriophage, secretion, Hcp, VgrG
1. Introduction
Type VI secretion systems (T6SSs) are the most recently described specialized secretion systems. T6SSs are widely distributed in Gram-negative bacteria, especially in proteobacteria, where type VI secretion gene clusters may be found in several copies on the chromosome [1–3]. First thought of as secretion systems dedicated to virulence towards eukaryotic host cells, recent data have shown unambiguously that these systems are regulating bacterial interactions and competition [4–10]. T6SSs are required to kill neighbouring, non-immune bacterial cells by secreting anti-bacterial proteins directly into the periplasm of the target cells upon cell-to-cell contact [4,9]. This intense bacterial warfare indirectly contributes to pathogenesis in plants, fishes, animals and humans as T6SS facilitates the colonization of specific niches where pathogens then develop anti-host defences and toxins. However, a limited number of T6SSs have been shown to be directly responsible for pathogenesis, as they deliver toxin modules interfering with the eukaryotic cytoskeleton [11].
2. Overview of type VI secretion systems
Because T6SSs have only recently been identified, we have only a limited knowledge on their assembly and biogenesis, compared with other secretion systems (see accompanying reviews in this issue). In past years, a number of studies have identified sub-complexes of this secretion apparatus and have reported structural information (table 1 and figure 1). This review will focus on the available structural data and we also refer the reader to recent reviews on the various T6SS aspects [1,2,6,11–16]. Thirteen subunits, called core components, are believed to form the minimal apparatus [1–3]. In several cases, the 13 core components are supplemented with additional proteins [17]. Although the functions of these proteins have not been elucidated yet, it has been proposed that these accessory elements might facilitate or modulate T6SS assembly or might confer additional functions to the secretion machine. Regarding the core components, several of them share structural similarities with bacteriophage tail, spike, sheath, hub or baseplate proteins, whereas a second category groups proteins embedded within the inner or outer membrane. However, a number of core components do not share clear homologies with bacteriophage or membrane-associated proteins; biochemical and structural data are therefore required to better understand their functions. Table 1 summarizes the known and suggested homologies, as well as the structure information currently available. Based on this set of data, it has been proposed that the T6SS core components collectively assemble a structure resembling an upside-down bacteriophage-like structure anchored to the bacterial cell envelope [2,16,18]. Figure 1 summarizes the current model for T6SS assembly, as well as the three-dimensional structures available.
Table 1.
Summary of the structural information available on T6SS core components and homologues.
| T6SS subunit | localization | PDB | homologue/localization | PDB |
|---|---|---|---|---|
| TssA | putative cytosolic protein | — | — | — |
| TssB | soluble protein, OM associated | — | bacteriophage sheath | 1FOH |
| TssC | soluble protein, OM associated | — | bacteriophage sheath | 1FOH |
| TssD/Hcp | soluble, forms hexameric rings, putative pilus | 1Y12, 3HE1, 3EAA | bacteriophage tail gp19 | 2K4Q, 2X8K, 2WZP, 2QWU |
| TssE | putative soluble protein | — | bacteriophage wedge gp25 | 2IA7 |
| TssF | putative soluble protein | — | — | — |
| TssG | putative soluble protein | — | — | — |
| TssH/ClpV | cytosolic protein, AAA+ ATPase | 3ZRI, 3ZRJ | Hsp100/Clp AAA+ ATPase | — |
| TssI/VgrG | cell puncturing device, forms trimers | 2P5Z | bacteriophage tail spike gp27-gp5 | 1K28 |
| TssJ/SciN | outer membrane lipoprotein | 3RX9, 4A1R | transthyretin | 1SN5 |
| TssK | putative cytosolic protein | — | — | — |
| TssL | inner membrane, 1 TM | 3U66 | T4bSS IcmH/DotU protein | — |
| TssM | inner membrane, 3 TMs | — | T4bSS IcmF protein | — |
Figure 1.
Schematic of type VI secretion systems. Centre: schematic of the current general model of T6SS assembly. Left: three-dimensional or model structures of the components of T6SS tube and sheath (top) and of the tube hub and the ATPase (below). Right: three-dimensional or model structures of the components of the T6SS membrane complex. See text and figures 2–5 for details.
3. The bacteriophage-like injection apparatus
(a). The type VI secretion systems’ needle tail and syringe
The Hcp protein is, together with VgrG, the hallmark of T6SS secretion. This component of T6SS is essential to its function and forms a tube of stacked hexamers in vitro. Three structures of Hcp proteins have been reported to date: Hcp1 (PDB 1Y12 [19]) and Hcp3 (PDB 3HE1 [20]) from Pseudomonas aeruginosa as well as EvpC from Edwardsiella tarda (PDB 3EAA [21]). Additionally, the structure of the Francisella tularensis IglC protein, an Hcp-like protein, has also been reported (PDB 2QWU [22,23]). The three former Hcp proteins exhibit an identical fold of two antiparallel β-sheets decorated by a β-hairpin extension (figure 2a), while IglC has a supplemental 30-amino acid N-terminal segment [23]. Six Hcp molecules assemble to form 80–90 Å wide hexameric rings stabilized by an extension acting as an inter-subunit belt. The centre of the hexamer has a diameter of 35–40 Å, which may therefore accommodate a small folded protein or unfolded/partly folded protein (figure 2b). Although the Hcp proteins and IglC share structural similarities, the additional N-terminal segment of IglC interferes with hexamer formation, and therefore the supramolecular assembly of IglC remains to be determined [23]. The Hcp hexamers have been shown to assemble as tubes (red in figure 2c), either by direct observation of Hcp3 tubes by electron microscopy (EM) [26] or by examining the crystal packing of Hcp1, Hcp3 and EvpC (Hcp1 and EvpC hexamers are stacked in a head-to-tail mode [19,21], whereas Hcp3 hexamers are packed in a head-to-head mode [20]). This ability has been used to produce nano-objects, tubes of different determined length, by engineering disulphide bridges at the hexamers’ interfaces [27]. In vivo, Hcp accumulates in the culture supernatant as well as in the periplasm, suggesting that it assembles a tubular structure from the inner membrane which passes through the outer membrane. However, tubular structures of Hcp and its packing mode have not been detected in vivo, leaving open the question on how its assembly is controlled.
Figure 2.
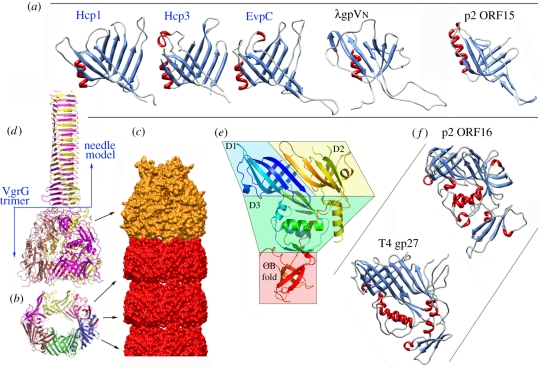
Structure of the tail tube proteins. (a) Ribbon views of the three Hcp proteins of known structures and their bacteriophage homologues (colours according to secondary structures). (b) Ribbon view of the hexameric ring of Hcp. (c) The tube surface model formed of stacked Hcp rings (red) and terminated by a VgrG trimer (orange). (d) Ribbon view of the VgrG trimer (ribbons, colours per monomer) as determined experimentally by X-ray diffraction and the molecular model of the needle domain (modelled from the bacteriophage T4 gp5) attached to it. (e) Ribbon view (rainbow colours from blue (Nt) to red (Ct)) illustrating the VgrG domain topology: the two β-sandwich domains (D1, D2), the α/β domain (D3) and the OB fold. (f) Ribbon view of bacteriophage structural homologues of VgrG (colours according to secondary structures). Figures were drawn with PyMOL [24] or Chimera [25].
The Hcp tertiary structure is very similar to that of gpV, the bacteriophage λ tail tube protein (PDB 2K4Q [28]). Hcp structure resembles the N-terminal domains of Dit proteins from phages SPP1 (PDB 2X8K [29,30]) and p2 (PDB 2WZP [31]), which form hexameric rings at the distal end of the tails (figure 2a). Finally, the Hcp proteins share structural resemblance with phage major tail proteins (MTPs), although the quaternary structure of phage MTPs has not yet been described. These similarities have been reported before [26,28], and we recently proposed that they extend to most bacteriophages tail proteins, a fact that could be a hallmark of a common molecular origin [32].
Current models suggest that the tip of the Hcp tube harbours a trimer of the VgrG protein that functions as a puncturing device towards the targeted cells (figure 2d, and orange in figure 2c). As expected from this model, VgrG proteins are often found in the culture supernatant of bacteria expressing T6SSs [33–36]. Interestingly, surface assemblies of Hcp and VgrG are mutually dependent, as VgrG is not found in the culture supernatant of hcp− cells and Hcp is not found in vgrG− cell supernatant [34–36]. While it is easily conceivable that Hcp assembly pushes out the VgrG trimer located at the tip, the question why Hcp is not found in the culture supernatant of vgrG− cells has not yet been experimentally answered. One may hypothesize that VgrG recruitment to the apparatus triggers Hcp assembly, with polymerization of the Hcp tube then pushing the VgrG protein to the external medium to puncture target cells [2,18]. The N-terminal domain of the VgrG protein from Escherichia coli CFT073 (483 residues out of 824) has been determined by X-ray diffraction (PDB 2P5Z, [26]) (figure 2d,e). VgrG is composed of four domains, two side-to-side β-sandwiches, an α/β domain and the oligonucleotide/oligosaccharide-binding (OB) fold (figure 2e). VgrG architecture is comparable with that of myophage T4 gp27 and the OB-fold domain (PDB 1K28 [37]; figure 2f). By extension, the structural similarity also applies to p2 ORF16, Mu gp44, MuSO2 Q8EDP4 and EGD-e gp18, all of which possess T4 gp27-like topology (figure 2f) [29,31,38]. VgrG forms a trimer in vivo [34,36], as do the similar phage proteins mentioned above. The two β-sandwich domains of VgrG share together a highly similar fold (figure 2e). In the trimer, the three pairs of β-sandwiches form a pseudo sixfold structure that establishes the probable interface with the last Hcp hexamer of the tube (figure 2c). As Hcp resembles the two β-domains of VgrG proteins, this pseudo sixfold symmetry probably eases the transition between the Hcp sixfold and the VgrG threefold symmetries at the interface.
Sequences and structural comparisons suggest that full-length VgrG could be described as a T4 puncturing device in which gp27 and gp5 (the needle) are fused, and the gp5 lysozyme domain removed [26,34] (figure 2d). Although the structure of the gp5-like domain of the E. coli VgrG has not been elucidated, it has been modelled and probably folds as a β-helical prism that will form the puncturing needle (figure 2d). In most bacteria, VgrG follows this scheme, whereas in a few cases, called ‘evolved’ VgrGs, an additional effector domain (most often an enzyme) is carried at the C-terminus [11,34]. The C-terminal additional domains of ‘evolved’ VgrG protein are delivered into the host cell cytosol [39]. Two of these domains have been characterized so far and induce host cell toxicity through a modification of the eukaryotic cytoskeleton [34,39,40]. Based on this observation, it has been proposed that ‘evolved’ VgrGs might target eukaryotic cells, while ‘non-evolved’ VgrGs would target bacterial cells. In this latter case, the lack of homologues of the bacteriophage gp5 lysozyme domain might be detrimental to cell wall perforation. It has been shown recently, however, that the Pseudomonas aeruginosa HSI-1 T6SS (which carries ‘non-evolved’ VgrGs) can inject the Tse1 and Tse3 peptidoglycan hydrolases into the periplasm of targeted Gram-negative bacteria, leading to cell wall destruction and lysis [4,9].
The internal diameter of the Hcp tubes (35–40 Å) is sufficient for the canonical T6SS protein substrates (17 and 43 kDa) to diffuse through it. Importantly, the VgrG proteins do not possess an open internal channel, therefore preventing any toxin protein going through. Obviously, VgrG should disassemble from the Hcp tube after perforation of the recipient outer membrane, or should open by a mechanism analogous to that observed in siphophage p2 [31]. Finally, although the formation of a needle assembling an Hcp tube and a puncturing VgrG trimer seem to be a very likely hypothesis, such assembly has not yet been structurally documented.
(b). The type VI secretion systems’ needle sheath
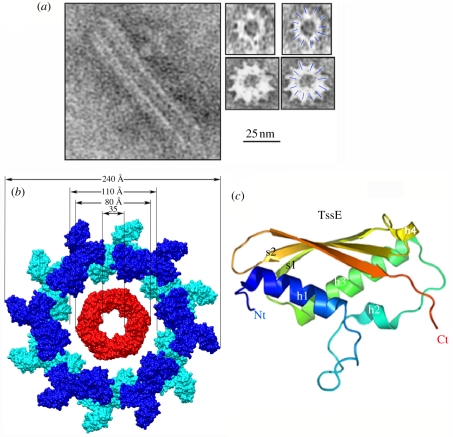
In constrast to siphophages, myophages have a contractile tail sheath enveloping the tail tube through which DNA is injected. The myophage T4 DNA injection process involves a cascade of events triggered by the long fibres attached to the bacterial surface, followed by baseplate conformational change, the attachment of short fibres and contraction of the tail sheath which pushes the tail tube through the cell wall breach formed by the actions of the puncturing device and the lysozyme. We have no structural proof for a similar mechanism in T6SSs. However, it has been observed by electron microscopy (EM) that the Vibrio cholerae TssB and TssC (VipA/VipB) proteins form tubes hundreds of angstroms long, with an approximate external diameter of 300 Å and internal diameter of approximately 100 Å [41] (figure 3a). It seems from the low-resolution pictures that these tubes obey a 12-fold symmetry. The size of these tubes is close to that of T4 tail sheath as observed using single-particle EM maps [26]. After fitting residues 1–510 of the tail sheath gp18 protein (approximately three-quarters of the full length; PDB 1FOA) into the cryo-EM map, Rossmann and colleagues observed that the tube has an external diameter of approximately 250 Å and an internal diameter of approximately 110 Å, allowing a gp19 tail tube to be accommodated (PDB 1FOH [42]). EM thus showed that the bacteriophage tail sheath and the TssB–TssC tubules share a similar organization even though TssB and TssC do not have sequence similarities with gp18. If we consider the T4 tail sheath as a plausible model for the T6SS needle sheath, we can attempt to assemble a molecular model by fitting the Hcp tube structure into that of T4 tail sheath. In this model (figure 3b), the external size of the Hcp tube (80–85 Å) fits well into the internal size of the tail sheath tube (110 Å). Interaction between the TssB and TssC proteins has been demonstrated in several bacteria, including V. cholerae, Burkholderia cenocepacia, F. tularensis, P. aeruginosa and Salmonella enterica [43,44]. The TssB and TssC proteins have been shown to localize both in the cytoplasm as well as at the outer membrane [41,44]. In F. tularensis, the TssB and TssC homologues IglA and IglB have been proposed to assemble structures large enough to be pelleted by ultracentrifugation [23]. The TssB and TssC outer membrane localization might therefore be an artefact due to the size of the TssBC tubules. The data support the hypothesis that a sheath-like cytoplasmic structure assembles from the inner membrane. Interestingly, the Hcp-like IglC protein co-fractionates with IglA and IglB, providing further support to the tail tube and sheath-like hypothesis [23]. Based on these similarities, it is thought that the contraction of the TssBC proteins might provide the force required for pushing the Hcp tube outside the cell [13,26]. This contraction might be provoked by the ClpV AAA+ ATPase that has been shown to depolymerize TssBC tubules ([41]; see §4).
Figure 3.
Structure of tube sheath and hub proteins. (a) Electron microscopy side-view (left) or cross section (right) of the TssB/TssC tubes formed in vitro (with permission from Bönemann et al. [41]). The 25 mm scale bar applies to all the electron microscopy views. (b) Molecular surface model of a cross section of a T6SS tube with Hcp rings in the middle (red) and TssB/TssC sheath around (blue, modelled from phage T4 tail sheath [1FOH] [42]). (c) Ribbon model (rainbow colours) of the hub protein TssE, generated from the structure of Geobacter sulfurreducens gp25 (2IA7; Joint Center for Structural Genomics 2006, unpublished data). Figures were drawn with PyMOL [24] or Chimera [25].
A number of other subunits of the T6SS apparatus, e.g. TssA, TssF and TssG, are not predicted to be anchored to the membrane, but instead are soluble cytoplasmic proteins, suggesting that these subunits might also have structural homologies with bacteriophage components even though no obvious similarities can be inferred from the primary sequence or secondary structure predictions.
(c). The type VI secretion systems’ needle hub
A significant (approx. 40%) sequence similarity was detected between the bacteriophage T4 gp25 component and the T6SS TssE subunit [26,45]. Although T4 gp25 was first suggested to have a lysozyme-like activity [46], it is now clear that this protein is a structural component of the bacteriophage baseplate [47–49]. gp25 contributes to the T4 baseplate structure by interacting with the (gp27–gp5)3 complex (at rest) or the tail tube (in the activated state) [47,50,51]. The structure of a gp25 homologue has been reported (PDB 2IA7) allowing modelling of the TssE subunit (figure 3c); the presence of a homologous protein in the T6SS suggests that it plays a role either in producing a baseplate-like assembly interacting with VgrG and Hcp, or may be a constituent of T6SS hub. Localization studies have shown that TssE is a cytoplasmic protein in P. aeruginosa suggesting that if a baseplate-like structure exists in T6SS, it should be assembled on the cytoplasmic side of the inner membrane.
4. The ClpV AAA+ chaperone
The cytosolic ClpV is a member of the Hsp100 (Clp) family of AAA+ proteins, hexameric ATPases involved in substrate unfolding. The ClpV chaperone, or TssH in the T6SS nomenclature, has been shown to play a crucial role in the T6SS assembly mechanism. When the soluble proteins TssB and TssC are mixed, they assemble spontaneously in long tubes (see §3b and figure 3a) [41] that may prevent the translocation of the monomers into the periplasm to form the putative tube sheath. The ClpV AAA+ chaperone helps solve this problem by using its non-catalytic N-terminal domain (residues 1–159; PDB 3ZRI) to form a complex with an α-helix from TssC (residues 15–28) and thus dissociates it from TssB (PDB 3ZRJ [41,52]) (figure 4). The C-terminal ATP-binding domain of ClpV provides the energy for the dissociation to occur. This chaperone mechanism has been documented in other systems and, interestingly, while the chaperone domains share a structurally conserved α-helical bundle fold, the helices from the substrates bind at quite distinct places [52]. The ClpV ATPase might therefore act at two steps during T6SS assembly and function by (i) depolymerizing the TssBC tubules in the cytosol allowing their transport into the periplasm where TssB and TssC will polymerize to form the sheath-like structure, and then (ii) acting to depolymerize the putative sheath to provide the energy required for its contraction [13,44].
Figure 4.
Structure of the ClpV (TssH) ATPase N-terminal domain in complex with the TssC N-terminal helix. Ribbon view of the N-terminal domain of ClpV (TssH; residues 1–158) from Vibrio cholerae (rainbow colours from blue Nt to red Ct) in complex with the first N-terminal helix of TssC (residues 16–29) (3ZRJ [52]). The critical residues at the ClpV–TssC interface are emphasized in the inset.
5. The membrane complex
Three core genes of T6SS gene clusters are predicted to encode proteins anchored to the cell envelope. Indeed, TssM and TssL localize at the inner membrane [53], whereas TssJ is an outer membrane lipoprotein [54]. Binary interactions have been detected: TssM interacts with TssL [35,53] and TssJ [35,55]. TssM therefore links the inner and outer membrane. Co-immunoprecipitation studies demonstrated that a complex of these three subunits, as well as the accessory TagL protein, assembles in the cell envelope [56].
(a). The inner membrane embedded proteins and their partners
TssL and TssM are prominent features of T6SS. These two proteins share similarities with IcmH/DotU and IcmF, two components associated with type IVB secretion systems [57,58].
Both TssL and TssM proteins are inserted into the inner membrane. Sequence analysis and topological studies have made it possible to determine TssM topology in A. tumefaciens and enteroaggregative E. coli ([53], our unpublished results). The N-terminus of TssM is composed of a few residues located in the cytoplasm, followed by two transmembrane helices separated by a short loop. A cytoplasmic domain of approximately 330 residues, predicted as α-helical, is located between the second and the third TM helices. Except for a few TssM proteins, this cytoplasmic domain carries Walker A and B boxes, suggesting that the TssM proteins have ATP binding and hydrolysing activities. Mutagenesis studies showed that the Walker A motif of A. tumefaciens is indeed required for T6SS assembly [53], while the Walker A mutation has no effect on E. tarda [35]. Several TssM proteins are predicted to possess only a single TM, missing the N-terminal TM hairpin. A large periplasmic domain of approximately 740 residues follows the last helix (figure 1). This domain of the entero-aggregative E. coli TssM protein has been expressed as a soluble fragment [55]. It has been dissected into two subdomains, an N-terminal one of approximately 540 residues, mainly α-helical (as shown by circular dichroism), and a C-terminal domain of approximately 200 residues, formed essentially by β strands [55] (figure 1).
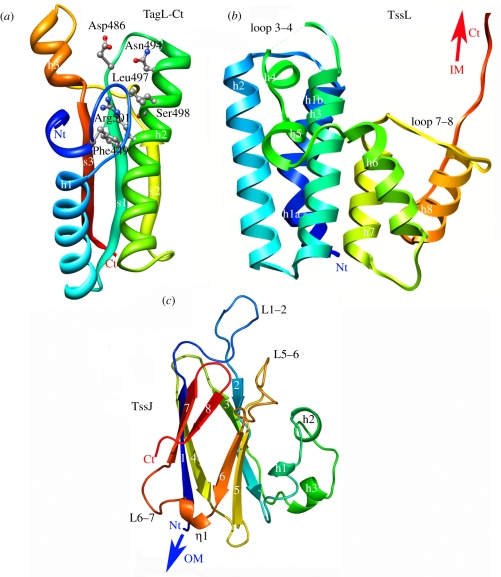
TssL is anchored through a single transmembrane segment [59]. Most TssL proteins share in their C-terminal periplasmic domain a peptidoglycan-binding motif of the OmpA family. When absent, an accessory protein, TagL, compensates by (i) carrying this motif and (ii) interacting with TssL [56,60]. Several structures of peptidoglycan-binding proteins with an OmpA family fold (PDB 1OAP; 3CYQ; 1R1M, 2K1S) have been solved by allowing a modelling of the TssL (or TagL) periplasmic domain (figure 5a). The conserved residues delimitate a groove that accommodates an N-acetylmuramic acid molecule. All the residues involved in groove formation and N-acetylmuramic acid binding are also conserved in the TssL (or TagL) homologues, and indeed the enteroaggregative E. coli TagL protein has been shown to interact with the peptidoglycan in vivo and in vitro [56]. Besides this periplasmic peptidoglycan-binding domain, the TssL subunits share an N-terminal cytosolic domain of approximately 300 residues. We recently solved the crystal structure of the enteroaggregative E. coli TssL cytoplasmic domain (PDB 3U66 [61]; figure 5b). TssL is an eight α-helix protein. The three first long helices form a tight bundle, and are followed by a disordered loop of four polar residues (loop 3–4). Two short helices (h4 and 5) and loops connect the first helix bundle with the second helix bundle, formed of three shorter helices (h6–8). A long stretch of 10 residues protrudes between helices 7 and 8 (loop 7–8), forming two elongated structures separated by a loop. The last helix finishes at residue 157, and is followed by an elongated unstructured segment (residues 158–178). The function of the cytoplasmic domain of TssL is not clear but a hypothesis is that it may form a cytosolic hook to recruit the substrates to be secreted to the T6SS.
Figure 5.
Structures of the membrane complex proteins. (a) Ribbon view (rainbow colours from blue Nt to red Ct) of the C-terminal peptidoglycan-binding domain of the enteroaggregative E. coli TagL protein (residues 414–557) modelled from the X-ray structure of E. coli YiaD (2K1S; T. A. Ramelot & M. A. Kennedy 2008, unpublished data). (b) Ribbon view (rainbow colours from blue Nt to red Ct) of the N-terminal cytoplasmic domain of the TssL protein from enteroaggregative E. coli [61]. (c) Ribbon view (rainbow colours from blue Nt to red Ct) of the C-terminal periplasmic domain of the TssJ lipoprotein from enteroaggregative E. coli [55].
(b). The membrane spanning TssM–TssJ complex
The inner membrane protein TssM also interacts with TssJ [35,55]. Surface plasmon resonance studies demonstrated that the periplasmic domain of TssM interacts with TssJ with a 2–4 μM dissociation constant [55]. TssJ (SciN) is an essential component of T6SS, approximately 180 residues long, and is anchored to the outer membrane by the N-terminal acylated cysteine of the processed form [54]. We have recently determined the structure of the enteroaggregative E. coli TssJ protein (PDB 3RX9 [55]; figure 5c). The first 22 residues of the approximately 150 residues mature protein are not visible in the electron density map, and should therefore form a flexible linker. TssJ has a β-sandwich fold with two four-stranded β-sheets (figure 5c). Sheet 1 is composed of β-strands 4, 1, 7 and 8, and is packed against sheet 2 which contains β-strands 3, 2, 5 and 6. This represents a common fold, namely transthyretin (PDB 1SN5), shared by several dozens of proteins in the protein databank. Compared with similar folds, TssJ owns two additional elements: (i) the external face of β-sheet 2 is covered in part by three short helices (h1–3) occurring between β-strands 2 and 3 and (ii) a loop located between strands 1 and 2 protrudes from the core of the protein [55]. This fold is conserved by the Serratia marcescens TssJ protein (PDB 4A1R [62]). The helical h1–3 domain is required for stability of TssJ, while the L1–2 loop is responsible for efficient interaction with TssM (figure 1) [55]. Interestingly, this fold is also shared by a number of proteins associated with secretion systems: the T3SS-associated ExsB lipoprotein (PDB 2YJL [63]), as well as the type IV plus-associated PilP lipoprotein (PDB 2IVW [64]). This observation suggests that lipoproteins associated with distinct secretion systems or trans-envelope complexes may have evolved from a common ancestor with a β-sandwich fold.
6. Concluding remarks and future direction
We have summarized in this review the current knowledge on the structural characterization of individual type VI secretion components. What emerges from these structures is that T6SSs are patchworks composed of subunits coming from various origins, such as bacteriophage-like components, type IVb-associated proteins, lipoproteins with transthyretin fold or Hsp100/Clp AAA+ ATPases. Although the structures of isolated T6SS subunits provide significant progress in the field, a comprehensive picture of the overall structure is still lacking. The crystal structures of the T6SS-like bacteriophage counterparts provided considerable advances to understanding how T6SSs assemble and raised exciting hypotheses on how T6SSs work. The comparison between T6SS biogenesis and bacteriophage morphogenesis is therefore critical to clearly understand T6SS dynamics during assembly and function. Even though providing new structures will be important, the next step will be to obtain high-resolution images of subassemblies of the T6SS machine, as exemplified for T3SS and T4SS. In vivo and biochemical data aimed at understanding the topology of the proteins and the interaction network will be the foundations for microscopy and structural studies. If it is reasonable to imagine EM or atomic definition of several of the T6SS subassemblies in the future, defining how the different pieces of the jigsaw are connected is also a fascinating challenge.
Acknowledgements
We thank the members of the T6SS groups for discussions and critical reading of the manuscript. We are grateful to Yves-Michel Cully for figure preparation, Axel Mogk and Nature Publishing Group for permissions to reproduce figures, Bernard Hébianca for encouragements, and the two anonymous reviewers for their corrections. Work in E.C.'s laboratory is supported by the CNRS and funded by a grant from the Agence National de la Recherche (ANR-10-JCJC-1303-03). Work in C.C.'s laboratory is supported by the CNRS, the Université Aix-Marseille and by grants from the Marseille-Nice Génopole, IBiSA and the Fondation pour la Recherche Medicale (SPF20101221116 and FRM DEQ2011-0421282).
References
- 1.Bingle L. E., Bailey C. M., Pallen M. J. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8 10.1016/j.mib.2008.01.006 (doi:10.1016/j.mib.2008.01.006) [DOI] [PubMed] [Google Scholar]
- 2.Cascales E. 2008. The Type VI secretion toolkit. EMBO Rep. 9, 735–741 10.1038/embor.2008.131 (doi:10.1038/embor.2008.131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genom. 10, 104. 10.1186/1471-2164-10-104 (doi:10.1186/1471-2164-10-104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hood R. D., et al. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 7, 25–37 10.1016/j.chom.2009.12.007 (doi:10.1016/j.chom.2009.12.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarz S., et al. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathogens 6, e1001068. 10.1371/journal.ppat.1001068 (doi:10.1371/journal.ppat.1001068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarz S., Hood R. D., Mougous J. D. 2010b. What is Type VI secretion doing in all those bugs? Trends Microbiol. 18, 531–537 10.1016/j.tim.2010.09.001 (doi:10.1016/j.tim.2010.09.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jani A. J., Cotter P. A. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 8, 2–6 10.1016/j.chom.2010.06.012 (doi:10.1016/j.chom.2010.06.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacIntyre D. L., Miyata S. T., Kitaoka M., Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl Acad. Sci. USA 107, 19 520–19 524 10.1073/pnas.1012931107 (doi:10.1073/pnas.1012931107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell A. B., Hood R. D., Bui N. K., Leroux M., Vollmer W., Mougous J. D. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 10.1038/nature10244 (doi:10.1038/nature10244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murdoch S. L., Trunk K., English G., Fritsch M. J., Pourkarimi E., Coulthurst S. J. 2011. The opportunistic pathogen Serratia marcescens utilises Type VI secretion to target bacterial competitors. J. Bacteriol. 193, 6057–6069 10.1128/JB.05671-11 (doi:10.1128/JB.05671-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pukatzki S., McAuley S. B., Miyata S. T. 2009. The Type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17 10.1016/j.mib.2008.11.010 (doi:10.1016/j.mib.2008.11.010) [DOI] [PubMed] [Google Scholar]
- 12.Filloux A., Hachani A., Bleves S. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 10.1099/mic.0.2008/016840-0 (doi:10.1099/mic.0.2008/016840-0) [DOI] [PubMed] [Google Scholar]
- 13.Bönemann G., Pietrosiuk A., Mogk A. 2010. Tubules and donuts: a type VI secretion story. Mol. Microbiol. 76, 815–821 10.1111/j.1365-2958.2010.07171.x (doi:10.1111/j.1365-2958.2010.07171.x) [DOI] [PubMed] [Google Scholar]
- 14.Bernard C. S., Brunet Y. R., Gueguen E., Cascales E. 2010. Nooks and crannies in Type VI secretion regulation. J. Bacteriol. 192, 3850–3860 10.1128/JB.00370-10 (doi:10.1128/JB.00370-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung K. Y., Siame B. A., Snowball H., Mok Y. K. 2011. Type VI secretion regulation: crosstalk and intracellular communication. Curr. Opin. Microbiol. 14, 9–15 10.1016/j.mib.2010.09.017 (doi:10.1016/j.mib.2010.09.017) [DOI] [PubMed] [Google Scholar]
- 16.Records A. R. 2011. The Type VI secretion system: a multipurpose delivery system with a phage like machinery. Mol. Plant. Microbe Interact. 24, 751–757 10.1094/MPMI-11-10-0262 (doi:10.1094/MPMI-11-10-0262) [DOI] [PubMed] [Google Scholar]
- 17.Shalom G., Shaw J. G., Thomas M. S. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699 10.1099/mic.0.2007/006585-0 (doi:10.1099/mic.0.2007/006585-0) [DOI] [PubMed] [Google Scholar]
- 18.Kanamaru S. 2009. Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc. Natl Acad. Sci. USA 106, 4067–4068 10.1073/pnas.0901205106 (doi:10.1073/pnas.0901205106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mougous J. D., et al. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 10.1126/science.1128393 (doi:10.1126/science.1128393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osipiuk J., Xu X., Cui H., Savchenko A., Edwards A., Joachimiak A. 2011. Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa. J. Struct. Funct. Genom. 12, 21–26 10.1007/s10969-011-9107-1 (doi:10.1007/s10969-011-9107-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jobichen C., Chakraborty S., Li M., Zheng J., Joseph L., Mok Y. K., Leung K. Y., Sivaraman J. 2010. Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS ONE 5, e12910. 10.1371/journal.pone.0012910 (doi:10.1371/journal.pone.0012910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun P., Austin B. P., Schubot F. D., Waugh D. S. 2007. New protein fold revealed by a 1.65 Å resolution crystal structure of Francisella tularensis pathogenicity island protein IglC. Protein Sci. 16, 2560–2563 10.1110/ps.073177307 (doi:10.1110/ps.073177307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bruin O.,M., et al. 2011. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins, IglA, IglB, IglC, PdpB and DotU, suggest roles in type VI secretion. Microbiology 157, 3470–3478 10.1099/mic.0.052308-0 (doi:10.1099/mic.0.052308-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLano W. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific LLC; See (http://pymol.sourceforge.net/). [Google Scholar]
- 25.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 (doi:10.1002/jcc.20084) [DOI] [PubMed] [Google Scholar]
- 26.Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl Acad. Sci. USA 106, 4154–4159 10.1073/pnas.0813360106 (doi:10.1073/pnas.0813360106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballister E. R., Lai A. H., Zuckermann R. N., Cheng Y., Mougous J. D. 2008. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl Acad. Sci. USA 105, 3733–3738 10.1073/pnas.0712247105 (doi:10.1073/pnas.0712247105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl Acad. Sci. USA 106, 4160–4165 10.1073/pnas.0900044106 (doi:10.1073/pnas.0900044106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veesler D., Robin G., Lichière J., Auzat I., Tavares P., Bron P., Campanacci V., Cambillau C. 2010. Crystal structure of bacteriophage SPP1 distal tail protein (gp19.1): a baseplate hub paradigm in Gram-positive infecting phages. J. Biol. Chem. 285, 36 666–36 673 10.1074/jbc.M110.157529 (doi:10.1074/jbc.M110.157529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulet A., et al. 2011. The opening of the SPP1 bacteriophage tail, a prevalent mechanism in Gram-positive-infecting siphophages. J. Biol. Chem. 286, 25 397–25 405 10.1074/jbc.M111.243360 (doi:10.1074/jbc.M111.243360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciara G., et al. 2010. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl Acad. Sci. USA 107, 6852–6857 10.1073/pnas.1000232107 (doi:10.1073/pnas.1000232107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veesler D., Cambillau C. 2011. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 75, 423–433 10.1128/MMBR.00014-11 (doi:10.1128/MMBR.00014-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., Heidelberg J. F., Mekalanos J. J. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl Acad. Sci. USA 103, 1528–1533 10.1073/pnas.0510322103 (doi:10.1073/pnas.0510322103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl Acad. Sci. USA 104, 15 508–15 513 10.1073/pnas.0706532104 (doi:10.1073/pnas.0706532104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J., Leung K. Y. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206 10.1111/j.1365-2958.2007.05993.x (doi:10.1111/j.1365-2958.2007.05993.x) [DOI] [PubMed] [Google Scholar]
- 36.Hachani A., Lossi N. S., Hamilton A., Jones C., Bleves S., Albesa-Jové D., Filloux A. 2011. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286, 12 317–12 327 10.1074/jbc.M110.193045 (doi:10.1074/jbc.M110.193045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanamaru S., Leiman P. G., Kostyuchenko V. A., Chipman P. R., Mesyanzhinov V. V., Arisaka F., Rossmann M. G. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 10.1038/415553a (doi:10.1038/415553a) [DOI] [PubMed] [Google Scholar]
- 38.Kondou Y., Kitazawa D., Takeda S., Tsuchiya Y., Yamashita E., Mizuguchi M., Kawano K., Tsukihara T. 2005. Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J. Mol. Biol. 352, 976–985 10.1016/j.jmb.2005.07.044 (doi:10.1016/j.jmb.2005.07.044) [DOI] [PubMed] [Google Scholar]
- 39.Ma A. T., McAuley S., Pukatzki S., Mekalanos J. J. 2009b. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 5, 234–243 10.1016/j.chom.2009.02.005 (doi:10.1016/j.chom.2009.02.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suarez G., Sierra J. C., Erova T. E., Sha J., Horneman A. J., Chopra A. K. 2010. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168 10.1128/JB.01260-09 (doi:10.1128/JB.01260-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 10.1038/emboj.2008.269 (doi:10.1038/emboj.2008.269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aksyuk A. A., Leiman P. G., Kurochkina L. P., Shneider M. M., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmannn M. G. 2009. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 28, 821–829 10.1038/emboj.2009.36 (doi:10.1038/emboj.2009.36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bröms J. E., Lavander M., Sjöstedt A. 2009. A conserved α-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191, 2431–2446 10.1128/JB.01759-08 (doi:10.1128/JB.01759-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aubert D., MacDonald D. K., Valvano M. A. 2010. BcsKC is an essential protein for the type VI secretion system activity in Burkholderia cenocepacia that forms an outer membrane complex with BcsLB. J. Biol. Chem. 285, 35 988–35 998 10.1074/jbc.M110.120402 (doi:10.1074/jbc.M110.120402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lossi N. S., Dajani R., Freemont P., Filloux A. 2011. Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system in Pseudomonas aeruginosa. Microbiology 157, 3292–3305 10.1099/mic.0.051987-0 (doi:10.1099/mic.0.051987-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szewczyk B., Bienkowska-Szewczyk K., Kozloff L. M. 1986. Identification of T4 gene 25 product, a component of the tail baseplate, as a 15K lysozyme. Mol. Gen. Genet. 202, 363–367 [DOI] [PubMed] [Google Scholar]
- 47.Kostyuchenko V. A., Leiman P. G., Chipman P. R., Kanamaru S., van Raaij M. J., Arisaka F., Mesyanzhinov V. V., Rossmann M. G. 2003. Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Biol. 10, 688–693 10.1038/nsb970 (doi:10.1038/nsb970) [DOI] [PubMed] [Google Scholar]
- 48.Rossmann M. G., Mesyanzhinov V. V., Arisaka F., Leiman P. G. 2004. The bacteriophage T4 DNA injection machine. Curr. Opin. Struct. Biol. 14, 171–180 10.1016/j.sbi.2004.02.001 (doi:10.1016/j.sbi.2004.02.001) [DOI] [PubMed] [Google Scholar]
- 49.Yap M. L., Mio K., Leiman P. G., Kanamaru S., Arisaka F. 2010. The baseplate wedges of bacteriophage T4 spontaneously assemble into hubless baseplate-like structure in vitro. J. Mol. Biol. 395, 349–360 10.1016/j.jmb.2009.10.071 (doi:10.1016/j.jmb.2009.10.071) [DOI] [PubMed] [Google Scholar]
- 50.Kostyuchenko V. A., Chipman P. R., Leiman P. G., Arisaka F., Mesyanzhinov V. V., Rossmann M. G. 2005. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 12, 810–813 10.1038/nsmb975 (doi:10.1038/nsmb975) [DOI] [PubMed] [Google Scholar]
- 51.Leiman P. G., Arisaka F., van Raaij M. J., Kostyuchenko V. A., Aksyuk A. A., Kanamaru S., Rossmann M. G. 2010. Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 355. 10.1186/1743-422X-7-355 (doi:10.1186/1743-422X-7-355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pietrosiuk A., Lenherr E. D., Falk S., Bönemann G., Kopp J., Zentgraf H., Sinning I., Mogk A. 2011. Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J. Biol. Chem. 286, 30 010–30 021 10.1074/jbc.M111.253377 (doi:10.1074/jbc.M111.253377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma L. S., Lin J. S., Lai E. M. 2009. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its Walker A motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191, 4316–4329 10.1128/JB.00029-09 (doi:10.1128/JB.00029-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aschtgen M. S., Bernard C. S., de Bentzmann S., Lloubes R., Cascales E. 2008. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531 10.1128/JB.00945-08 (doi:10.1128/JB.00945-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Felisberto-Rodrigues C., Durand E., Aschtgen M.-S., Blangy S., Ortiz-Lombardia M., Douzi B., Cambillau C., Cascales E. 2011. Towards a structural comprehension of bacterial Type VI secretion systems: characterization of the TssJ–TssM complex of an Escherichia coli pathovar. PLoS Pathogens. 7, e1002386 10.1371/journal.ppat.1002386 (doi:10.1371/journal.ppat.1002386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aschtgen M. S., Gavioli M., Dessen A., Lloubes R., Cascales E. 2010. The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol. Microbiol. 75, 886–899 10.1111/j.1365-2958.2009.07028.x (doi:10.1111/j.1365-2958.2009.07028.x) [DOI] [PubMed] [Google Scholar]
- 57.Das S., Chaudhuri K. 2003. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 3, 287–300 [PubMed] [Google Scholar]
- 58.Nagai H., Kubori T. 2011. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front. Microbiol. 2, 136. 10.3389/fmicb.2011.00136 (doi:10.3389/fmicb.2011.00136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aschtgen M. S., Zoued A., Lloubés R., Journet L., Cascales E. In press. The C-tail anchored TssL subunit, an essential protein of the enteroaggregative Escherichia coli Sci-1 Type VI secretion system, is inserted by YidC. MicrobiologyOpen. 10.1002/mbo3.8 (doi:10.1002/mbo3.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aschtgen M. S., Thomas M. S., Cascales E. 2010. Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP… what else? Virulence 1, 535–540 10.4161/viru.1.6.13732 (doi:10.4161/viru.1.6.13732) [DOI] [PubMed] [Google Scholar]
- 61.Durand E., Zoued A., Spinelli S., Watson P., Aschtgen M. S., Journet L., Cambillau C., Cascales E. Submitted. Structural characterization of the IcmH/DotU-like TssL protein, a component shared by Type IVb and Type VI secretion systems. J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rao V. A., Shepherd S. M., English G., Coulthurst S. J., Hunter W. N. 2011. The structure of Serratia marcescens Lip, a membrane-bound component of the type VI secretion system. Acta Crystallogr. D 67, 1065–1072 10.1107/S0907444911046300 (doi:10.1107/S0907444911046300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Izoré T., Perdu C., Job V., Attree I., Faudry E., Dessen A. 2011. Structural characterization and membrane localization of ExsB from the Type III secretion system (T3SS) of Pseudomonas aeruginosa. J. Mol. Biol. 413, 236–246 10.1016/j.jmb.2011.07.043 (doi:10.1016/j.jmb.2011.07.043) [DOI] [PubMed] [Google Scholar]
- 64.Golovanov A. P., Balasingham S., Tzitzilonis C., Goult B. T., Lian L. Y., Homberset H., Tønjum T., Derrick J. P. 2006. The solution structure of a domain from the Neisseria meningitidis lipoprotein PilP reveals a new beta-sandwich fold. J. Mol. Biol. 364, 186–195 10.1016/j.jmb.2006.08.078 (doi:10.1016/j.jmb.2006.08.078) [DOI] [PubMed] [Google Scholar]