Abstract
One of the greatest challenges for biodiversity conservation is the poor understanding of species diversity. Molecular methods have dramatically improved our ability to uncover cryptic species, but the magnitude of cryptic diversity remains unknown, particularly in diverse tropical regions such as the Amazon Basin. Uncovering cryptic diversity in amphibians is particularly pressing because amphibians are going extinct globally at an alarming rate. Here, we use an integrative analysis of two independent Amazonian frog clades, Engystomops toadlets and Hypsiboas treefrogs, to test whether species richness is underestimated and, if so, by how much. We sampled intensively in six countries with a focus in Ecuador (Engystomops: 252 individuals from 36 localities; Hypsiboas: 208 individuals from 65 localities) and combined mitochondrial DNA, nuclear DNA, morphological, and bioacoustic data to detect cryptic species. We found that in both clades, species richness was severely underestimated, with more undescribed species than described species. In Engystomops, the two currently recognized species are actually five to seven species (a 150–250% increase in species richness); in Hypsiboas, two recognized species represent six to nine species (a 200–350% increase). Our results suggest that Amazonian frog biodiversity is much more severely underestimated than previously thought.
Keywords: biodiversity, cryptic species, Amazon Basin, amphibians, conservation
1. Introduction
The application of molecular methods to systematics has revolutionized the discovery and description of biodiversity. In particular, DNA sequence data are revealing cryptic diversity—two or more morphologically similar species that are erroneously classified (and hidden) under one species name—in many regions and taxonomic groups [1]. Uncovering cryptic species to more accurately understand diversity patterns is critical from a conservation perspective for two main reasons. First, knowledge of geographical patterns of diversity is essential for identifying regions with high levels of species richness and endemism, sometimes termed biodiversity hotspots, that warrant special conservation status [2,3]. Second, understanding the distribution of individual species is essential for assigning conservation status. For example, species that are considered single, widely distributed species of low conservation concern may actually be multiple morphologically similar cryptic species, each with small ranges that are of high conservation concern [4].
Despite the recognition that cryptic species are widespread, the magnitude of cryptic diversity remains unknown, especially in species rich and relatively poorly explored tropical regions. Part of the reason that cryptic diversity remains poorly characterized in the tropics is insufficient sampling because these regions are often difficult to access. In order to accurately estimate the level of cryptic diversity, intensive sampling is critical. By ‘intensive sampling’, we mean spatially dense sampling (i.e. sampling many localities) and sampling many individuals per locality. Spatially dense sampling is necessary to find species with small ranges, while sampling many individuals is necessary to find locally rare species [5]. By contrast, less intensive broad stroke sampling (few localities and few individuals per locality) will inevitably give estimates of cryptic diversity that are biased low, revealing only the most widespread and common cryptic species.
As the largest and arguably most diverse lowland rainforest on Earth [6], the Amazon Basin probably houses a significant amount of cryptic diversity. Unfortunately, there are few large-scale genetic studies of Amazonian organisms, thus reliable estimates of the level of cryptic diversity are scarce. In particular, the Amazon Basin has one of the most species-rich amphibian faunas in the world [7], yet Amazonian amphibian species richness is still probably underestimated [8]. Understanding amphibian diversity is particularly pressing [9] because amphibians are considered to be in the midst of the sixth great mass extinction event [10], with 41 per cent of amphibian species classified as globally threatened with extinction [11]. By the end of 2004, 427 amphibian species were known from Amazonia [12]. Between 2005 and 2009, another 127 amphibian species were described [13] for a total of 554 species, representing 8.1 per cent of global amphibian species richness according to AmphibiaWeb (downloaded July 2011). Fouquet et al. [14] made the first attempt to estimate the number of cryptic Amazonian amphibians based on an analysis of 420 bp of mitochondrial DNA (mtDNA), and predicted 170–460 undescribed cryptic species (a 22–115% increase in species richness). Fouquet et al.'s study was an important starting point for estimating Amazonian amphibian species richness, but more accurate figures require more exhaustive sampling and phenotypic data because a single mtDNA gene cannot effectively define species boundaries [15]. Accurate species delimitation requires integration of information from diverse datasets (e.g. genetic, morphological and behavioural) and thorough population sampling [16].
Here, we estimated the number of undescribed species in two independent, widely distributed Amazonian frog clades, a terrestrial group (Engystomops) and a treefrog group (Hypsiboas calcaratus species group). We chose these two clades because: (i) they are evolutionarily divergent from each other [17,18] and therefore represent independent tests of the level of cryptic diversity; and (ii) they are relatively well-known taxonomically [14,19–25], thus they should contain fewer cryptic species than other amphibian clades, providing a conservative estimate of cryptic diversity. Our approach was to use intensive sampling and integrative systematics including mtDNA and nuclear DNA (nDNA) sequences and morphological and bioacoustic traits to uncover cryptic diversity. The specific goals of our study were to: (i) test if species richness in these clades was underestimated; and (ii) if so, determine by how much species richness was underestimated.
2. Material and methods
(a). Delimitation of candidate species
We classified frog lineages using the categories defined by Vieites et al. [26]. Confirmed candidate species (CCS) are ‘those differing clearly by morphological and bioacoustic characters and usually showing high genetic differentiation that we hypothesize are distinct, undescribed species’. Vieites et al. found that CCS usually have greater than 3 per cent uncorrected pairwise sequence divergence from other species at 16S mtDNA, but sometimes only 1–2%. Unconfirmed candidate species (UCS) are ‘deep genealogical lineages—bioacoustically and morphologically unstudied and usually derived from geographically distant populations—for which general indications exist that they are distinct, undescribed species’. UCS differ from other species by greater than 3 per cent sequence divergence. We chose to delimit species using the approach of Vieites et al. because it explicitly integrates genetic, morphological and bioacoustic data, which are increasingly recognized as necessary to accurately delimit species [15,16]. First, we identified well-supported clades with a minimum uncorrected sequence divergence of 1–2% from other clades (at 12S and 16S mtDNA). Next, we tested whether these clades differed from other clades morphologically or bioacoustically as detailed below. Formal descriptions of new species are in preparation and will be published elsewhere.
(b). Sampling
We analysed a total of 252 Engystomops from 36 localities and 208 Hypsiboas from 65 localities in six countries in the Amazon Basin (figures 1c and 2c; see the electronic supplementary material, appendix S1 for a list of individuals included). Our sampling was intensive in Amazonian Ecuador and less intensive in the rest of the Amazon Basin. For Engystomops, n = 125, 133 and 51 individuals were included in the DNA, morphological and bioacoustic analyses, respectively. The corresponding sample sizes for Hypsiboas were n = 136, 137 and 26. Tissue samples were either frozen or stored in 95 per cent ethanol or dimethyl sulphoxide buffer. Total genomic DNA was extracted from samples using DNeasy Tissue Kits (Qiagen, Inc., Valencia, CA, USA). Voucher specimens for most samples are available in several museums (electronic supplementary material, appendix S1).
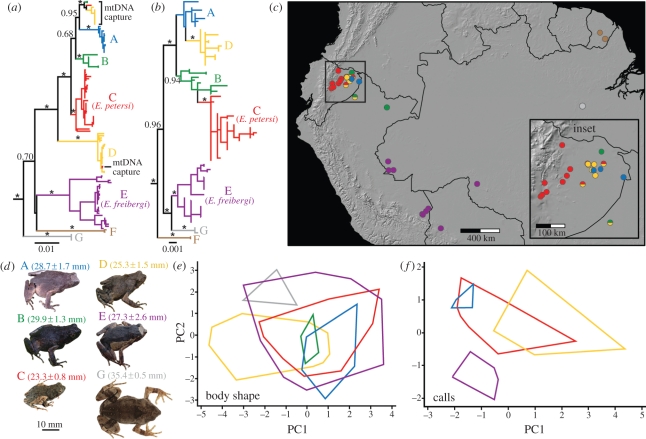
Figure 1.
(a) Engystomops mitochondrial DNA (mtDNA) phylogeny, (b) nuclear DNA (nDNA) phylogeny, (c) map showing the distribution of sampling localities and species, (d) photos of species (in life for clades A–E and of a preserved specimen for clade G) shown to scale, (e) principal component analysis (PCA) using size-corrected morphometric variables showing variation in body shape, and (f) PCA using bioacoustic variables. The mtDNA and nDNA trees are based on maximum-likelihood (ML) analyses of 2358 and 2308 bp, respectively, and asterisks denote major clades with Bayesian posterior probability (bpp) values greater than or equal to 0.99. Detailed mtDNA and nDNA trees with ML bootstrap and bpp values for all nodes and specimen museum numbers are shown in the electronic supplementary material, figures S1 and S2. Photos and morphometric data were unavailable for clade F and bioacoustic data were unavailable for clades B, F and G.
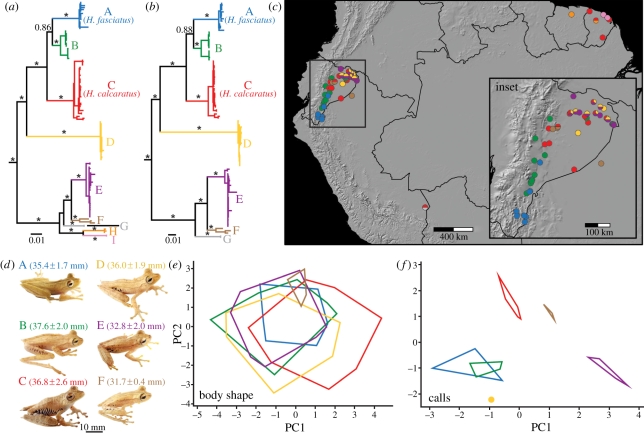
Figure 2.
(a) Hypsiboas 12S + 16S mtDNA phylogeny, (b) 12S + 16S + COI mtDNA phylogeny, (c) map showing the distribution of sampling localities and species, (d) photos of species (all in life) shown to scale, (e) PCA using size-corrected morphometric variables showing variation in body shape and (f) PCA using bioacoustic variables. The 12S + 16S and 12S + 16S + COI trees are based on ML analyses of 1582 and 2221 bp, respectively, and asterisks denote major clades with bpp values greater than or equal to 0.99. Detailed 12S + 16S and12S + 16S + COI trees with ML bootstrap and bpp values for all nodes and specimen museum numbers are shown in the electronic supplementary material, figures S3 and S4. Photos, morphometric and bioacoustic data were unavailable for clades G–I.
(c). DNA sequencing and analysis
We sequenced and analysed mtDNA and nDNA. For Engystomops, we sequenced the mitochondrial 12S, 16S and intervening tRNA genes (2358 kb) and portions of the nuclear a2ab (588 kb), CXCR4 (627 bp), NTF3 (578 bp) and tyrosinase (515 bp) genes. For Hypsiboas, we sequenced portions of the 12S (800 bp), 16S (782 bp) and cytochrome oxidase I (COI, 639 bp) mitochondrial genes and the nuclear proopiomelanocortin (POMC) (564 bp) gene. Primers and their sources are listed in the electronic supplementary material, table S1; and the individuals sequenced and included in the morphological and bioacoustic analyses are shown in the electronic supplementary material, appendix S1. We chose outgroups (Engystomops: Engystomops pustulosus, Engystomops pustulatus, Engystomops puyango, Engystomops montubio and Engystomops coloradorum; Hypsiboas: Hypsiboas multifasciatus, Hypsiboas lanciformis, Hypsiboas pellucens and Hypsiboas crepitans) based on recent phylogenetic analyses [14,18,27].
Phylogenies were inferred separately for mitochondrial and nuclear sequences using maximum-likelihood (ML) and Bayesian approaches. For Engystomops, the four nuclear genes were concatenated. For Hypsiboas, separate analyses were conducted using concatenated 12S and 16S genes (n = 136) and concatenated 12S, 16S and COI genes for a smaller subset of individuals for which COI data were also available (n = 118). ML analyses were conducted using program GARLI v. 0.951 [28]. Model choice was based on Akaike information criterion (AIC) [29] using program MrModeltest v. 2.2 [30]. Analyses were terminated after 10 000 generations without an improvement in tree topology. Support was evaluated using 100 bootstrap replicates with each replicate terminated after 5000 replications without a topology improvement. Bayesian analyses were conducted using MrBayes v. 3.1.2 [31] and using three partitioning strategies for the mitochondrial data (1 partition, partitioning by gene and partitioning by gene and stems and loops) and three partitioning strategies for the nuclear data (1 partition, partitioning by gene and partitioning by gene and codon position). Model choice for each partition was based on AIC in MrModeltest v. 2.2 (see the electronic supplementary material, table S2 for a summary of character variation and MrModeltest results). Bayesian analyses were performed with two replicate searches of 2 × 106 generations each with four Markov chains and trees sampled every 1000 generations. We used a conservative burn-in that was determined by examining stationarity of the likelihood scores and convergence of posterior probabilities between the two runs using the standard deviation of split frequencies. The best partitioning strategy for each dataset was selected by comparing Bayes factors [32].
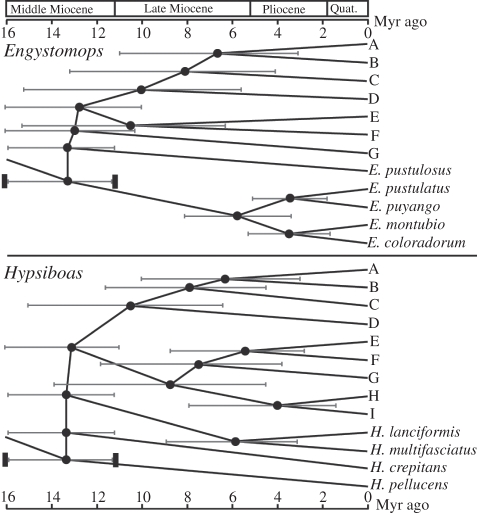
Time to most recent common ancestor (TMRCA) of major nodes was estimated using mitochondrial 12S and 16S genes with the Bayesian Markov Monte Carlo method implemented in program BEAST v. 1.4.8 [33]. Following Weigt et al. [34], we assumed that the rise of the tropical Andes separated the ancestor of the Pacific coast species from the ancestor of the Amazonian species and that this vicariant event occurred at some point in the Mid-Miocene, 16.4–11.2 Myr ago [35,36]. This temporal constraint was incorporated into our analysis by using a uniform prior between 16.4 and 11.2 Myr ago for the root height parameter (shown by heavy black bars in figure 3). For each genus, we ran the analysis for 1 × 107 generations sampling every 1000 generations and removing 10 per cent of the samples as a burn-in. Stationarity of the posterior distributions for all model parameters was determined using program Tracer v. 1.4.1 [33]. We used a GTR + I + G substitution model, an uncorrelated relaxed lognormal molecular clock and a Yule tree prior.
Figure 3.
Divergence time estimates for major nodes using the Bayesian Markov Monte Carlo method implemented in program BEAST, assuming that the rise of the tropical Andes 16.4–11.2 Myr ago precipitated the split of Pacific coast species from Amazonian species. This constraint is represented by heavy black bars on either side of the node connecting Pacific and Amazonian species. Error bars show the lower and upper bounds of the 95% posterior probability distribution.
(d). Analysis of morphological differentiation
We measured nine morphometric variables for male Engystomops and Hypsiboas specimens using digital calipers (accurate to the nearest 0.01 mm). Measurements were taken following Funk et al. [25] for Engystomops, and Duellman [37] for Hypsiboas. Principal component analysis (PCA) and discriminant function analysis (DFA) were performed to assess the degree of morphometric differentiation among candidate species. To visualize variation in shape independent of size, PCA was applied to the residuals of the linear regressions between the other eight variables measured and snout-vent-length (SVL) [25]. DFA was performed using raw morphometric data because the goal of the DFA was to determine the use of all morphometric variables, including SVL, for distinguishing candidate species.
(e). Analysis of bioacoustic differentiation
Advertisement calls in frogs are highly stereotyped within species and are usually species-specific. Because females use them for species discrimination, they are important in pre-mating isolation and thus are particularly useful for defining species boundaries [15,16]. For example in Engystomops, female preference tests demonstrate that variation in calls results in behavioural isolation [38,39]. Call recordings had a sampling rate of 44 kHz and were analysed with program Raven v. 1.2 [40]. Recording temperatures varied little in both Engystomops (mean = 22.1°C, s.d. = 2.2°C) and Hypsiboas (mean = 23.7°C, s.d. = 1.7°C). Fast Fourier transformation size was 2048 and the spectral analysis had a frequency resolution of 21.5 Hz. The measured variables were: (i) dominant and fundamental frequency, (ii) note duration, (iii) number of notes, and (iv) rise time. Engystomops calls have a complex structure consisting of an amplitude-modulated prefix and a whine-like frequency sweep. Thus for Engystomops, we also measured: (i) dominant frequency of the prefix, and (ii) fundamental frequency at the beginning of the whine. Call variables and components are defined in Guerra & Ron [39]. Bioacoustic differentiation among candidate species was also analysed using PCA and DFA.
3. Results
Our integrative analysis of genetic, morphological and bioacoustic data of 252 Engystomops from 36 localities and 208 Hypsiboas from 65 localities uncovered many undescribed, cryptic species (figures 1 and 2). In both Engystomops and Hypsiboas, there were more undescribed than described species.
(a). Cryptic diversity in Engystomops toadlets
In Amazonian Engystomops, our analyses revealed that what were previously considered two species are actually five to seven (depending on whether UCS are included; see below), a 150–250% increase in the number of species (figure 1). Based on type specimen localities, clade C corresponds to Engystomops petersi [41] and clade E to Engystomops freibergi [25,42]. Clades A, C, D, E and G are CCS because they have significant morphological and/or bioacoustic differences from all other clades. Furthermore, clades A, C and D are sympatric, indicating that they are reproductively isolated. Clades B and F are UCS because although they are genetically divergent, well-supported clades, we lack morphological and/or bioacoustic data.
mtDNA and nDNA phylogenies generally agreed and revealed similar distinct genetic groups (figure 1a,b; see the electronic supplementary material, figures S1 and S2 for detailed phylogenies including specimen museum numbers). One difference between mtDNA and nDNA trees, however, was that 12 individuals found north of the Río Napo in northeastern Ecuador formed a clade sister to clade A in the mtDNA tree (figure 1a), but these frogs were strongly supported as belonging to clades B, C or D in the nDNA tree (figure 1b). A possible explanation for this discrepancy is introgression of mtDNA from the ancestor of clade A into localities north of the Río Napo, termed ‘mitochondrial capture’ [43]. Despite this difference, both trees show strong support for the same five clades (C–G). Clades A and B from the mtDNA tree are paraphyletic in the nDNA tree (with respect to clades D and C, respectively), but support for this paraphyly is weak. Moreover, the species status of clades A, C, and D is also supported by morphological and bioacoustic characters (see below) and sympatry. All clades had small geographical ranges with the exception of clade E (E. freibergi; figure 1c). Four clades (A, B, C, and D) were found in sympatry. Sequence divergence among clades at 12S and 16S averaged 3.44 per cent (range: 1.35–4.32%; see the electronic supplementary material, table S3 for pairwise sequence divergence among all clades) and TMRCA among clades was 6.7–13.0 Myr ago, indicating that these are ancient lineages that evolved before the Quaternary (figure 3).
We found significant, albeit subtle, morphometric differences among Engystomops clades. SVL, a measure of body size, varied significantly among clades (ANOVA, F5,127 = 40.66, p < 0.001; figure 1d). Tukey's tests showed that clade C was smaller and clade G was larger than all other clades. PCA of size-corrected morphometric variables also revealed significant differences in shape (figure 1e; see the electronic supplementary material, table S4 for PCA loadings). PC1 differed significantly among clades (ANOVA, F5,127 = 8.31, p < 0.001), as did PC2 (F5,127 = 9.41, p < 0.001). Frogs in clade D had proportionally shorter limbs than those in clades A, B and E. Clade E frogs were wider than those in clades A and D, and clade G frogs were wider than clade A, C and D frogs. In the DFA, frogs from clades A, C, D and G could be assigned fairly accurately (74.2–100% assigned correctly; electronic supplementary material, table S5). However, we were unable to find qualitative morphological characters diagnostic of Engystomops clades.
PCA of bioacoustic variables showed striking differences (figure 1f; see the electronic supplementary material, table S6 for PCA loadings). PC1 varied significantly among clades (ANOVA, F3,47 = 33.24, p < 0.001), with clade D having higher frequency calls than the other three clades included. PC2 also varied significantly (F3,47 = 32.79, p < 0.001), with clade E having shorter calls than the others. In the bioacoustic DFA, frogs from clades A, D and E could be assigned to their true clade with high accuracy (90–100% assigned correctly; electronic supplementary material, table S7). Thus, all clades with morphometric and bioacoustic data could be distinguished and were therefore designated CCS.
(b). Cryptic diversity in Hypsiboas treefrogs
Our analyses also revealed that two previously recognized species in the Hypsiboas calcaratus species group actually represent six to nine species (once again, depending on whether UCS are included; see below), a 200–350% increase in species richness (figure 2). Based on type specimen localities and diagnostic characters, clade A corresponds to Hypsiboas fasciatus [44] and clade C to Hypsiboas calcaratus [45]. Clades A–F are CCS, as they are morphologically and/or bioacoustically differentiated from other clades. Clades G–I are UCS because although they are highly divergent lineages, we lack morphological and/or bioacoustic data.
Both mtDNA trees, one inferred from 12S and 16S genes (figure 2a; see the electronic supplementary material, figure S3 for detailed phylogeny including specimen museum numbers) and the other for a smaller subset of individuals that also had COI sequences (figure 2b and electronic supplementary material, figure S4), were highly concordant and revealed nine distinct, well-supported clades (A–I) nested within two larger clades. These two larger clades were also strongly supported in the nDNA tree based on POMC (see the electronic supplementary material, figure S5), but variation in POMC was insufficient to resolve the nine smaller clades. All clades had small geographical ranges except for clade C, which was found throughout the Amazon Basin (figure 2c). Each clade is sympatric with at least one other clade and up to three clades (C, D and E) occur syntopically along the Río Napo in Ecuador, confirming that each is an independent evolutionary lineage. Sequence divergence among clades at 12S and 16S averaged 6.85 per cent (range: 2.92–9.50%; see the electronic supplementary material, table S3) and TMRCA was 4.0–13.1 Myr ago, again indicating ancient species (figure 3).
Hypsiboas clades also had subtle but significant morphometric differences. SVL varied significantly among clades (F5,131 = 21.90, p < 0.001), with clades A–D being significantly larger than clades E–F (figure 2d). There were also significant differences in shape as revealed by PCA (figure 2e; see the electronic supplementary material, table S4 for PCA loadings). PC1 was significantly different among clades (F5,131 = 10.36, p < 0.001), as was PC2 (F5,131 = 5.19, p < 0.001). Clade C had proportionally longer limbs than clades B, D and E and clade F had proportionally smaller eyes and tympana than clades B–E. All six clades with morphometric data could be assigned fairly accurately using DFA (72.7–100% assigned correctly; electronic supplementary material, table S5). In addition, we found qualitative morphological characters, which in combination are diagnostic of each clade for which data were available (clades A–F; see the electronic supplementary material, table S8).
Bioacoustically, Hypsiboas clades were highly differentiated from each other. PC1 was significantly different among clades (F5,20 = 42.78, p < 0.001), as was PC2 (F5,20 = 38.78, p < 0.001; figure 2f; see the electronic supplementary material, table S6 for PCA loadings). Clade E had significantly longer and higher fundamental frequency calls than all other clades, and clade F had longer and higher frequency calls than clades A–D. Clades C and F had lower dominant frequency calls and fewer notes per call than all other clades. In the bioacoustic DFA, clades were assigned with high accuracy (80–100% assigned correctly; electronic supplementary material, table S7). Moreover, calls varied qualitatively among clades (a quack in clades A–C, a trill in clade E and a whistle in clade F). Thus, all clades with morphometric and bioacoustic data could confidently be distinguished and classified as CCS.
4. Discussion
(a). High levels of cryptic diversity in two Amazonian frog clades
Our integrative analysis of mtDNA, nDNA, morphology and calls uncovered exceptionally high levels of undescribed cryptic diversity, increasing species richness by 150–250% in Engystomops and by 200–350% in Hypsiboas. Moreover, this may be an underestimate for these clades because we were unable to sample some parts of Amazonia, particularly in northwestern Brazil. These two clades are also relatively well-known taxonomically compared with many other Amazonian amphibians, thus cryptic diversity may be even higher in other amphibian clades. Similarly, amphibians and other vertebrates are taxonomically well described compared with other groups such as insects and fungi, suggesting that cryptic diversity will be even higher in many taxa. Indeed, a recent study estimated that 86 per cent of species on Earth await description [46]. Although the Amazon Basin is already recognized as a centre of biodiversity, we provide new evidence that its biodiversity is still vastly underestimated, even among relatively well-studied vertebrates. Nonetheless, it is possible that the level of cryptic diversity found here in Engystomops and Hypsiboas is atypically high. Intensive sampling and integrative systematic analyses of additional clades representing all Amazonian amphibian families are necessary to estimate total amphibian species richness in the region.
Our results also suggest that amphibian biodiversity in Amazonia is partitioned differently than previously thought. Prior to advances in DNA techniques, amphibians were primarily described based on morphology. In the Amazon Basin, these morphologically defined species have large geographical ranges. Although the Amazonian amphibian fauna has high alpha diversity (many species at single localities), it was generally considered to have relatively low beta diversity (similar species composition among localities). However, our analysis and other recent taxonomic studies that include genetic or call data are revealing that apparently widespread species actually consist of several cryptic species with smaller ranges [14,47,48], indicating beta diversity and overall gamma (total) diversity of the Amazon Basin are severely underestimated.
(b). Delimitation of candidate species
Our main conclusion that cryptic species diversity is greatly underestimated in our two focal clades is not changed by the species concept applied. Species delimitation has been termed a ‘Renaissance problem’ in systematics with various approaches proposed [49,50]. Nonetheless, the CCS identified here are all reciprocally monophyletic at mtDNA, genetically divergent and morphologically and/or bioacoustically highly differentiated. Moreover, female choice experiments have shown that Engystomops clades A versus D [38], and C versus D [39] are behaviourally isolated from each other owing to call divergence, which is rarely demonstrated. Thus, the CCS would be considered unambiguous species based on biological, phylogenetic or evolutionary species concepts. Two Engystomops clades and three Hypbsiboas clades were categorized as UCS owing to a lack of morphological and/or bioacoustic data. However, all distinct clades with morphological and bioacoustic data were found to be CCS, suggesting that once these data are obtained for the UCS, they will be elevated to CCS.
Interestingly, we found qualitative, diagnostic morphological characters for Hypbsiboas CCS (electronic supplementary material, table S8), but not for Engystomops CCS, despite similar divergence times for both genera (figure 3). Some authors recommend not recognizing species unless they have diagnostic morphological characters [51]. Nonetheless, we know that Engystomops CCS are bioacoustically differentiated and, as described above, behaviourally isolated based on female preference tests and therefore unambiguous species according to the biological species concept. Thus, strict adherence to requiring qualitatively distinct diagnostic characters would fail to recognize reproductively isolated species in this case, suggesting this is an overly stringent criterion. Fortunately, despite a lack of diagnostic morphological characters, Engystomops CCS can be distinguished in the field based on a combination of dramatic differences in calls, pronounced size variation and geographical location.
A contentious (and unresolved) issue in species delimitation is the minimum sequence divergence necessary to consider a clade a distinct species [14,26]. Although the average uncorrected sequence divergence among our candidate species was fairly high (3.44% for Engystomops and 6.85% for Hypsiboas), some CCS differed by less than 3 per cent sequence divergence. The lowest observed sequence divergence between CCS was 1.68 per cent between Engystomops clades A and C, yet they are well-differentiated species. They have large differences in size (SVLs do not overlap) and are found in sympatry with no evidence of current hybridization. Many other examples of distinct species with low sequence divergence are found in the literature, including other Engystomops species [27], Malagasy frogs [26] and African cichlids [52]. These examples highlight the importance of using criteria in addition to sequence divergence to delimit species.
A new direction in species delimitation is the inference of species trees using coalescent-based approaches [53]. We did not use this approach here because of an insufficient number of nuclear genes for Hypsiboas, but we encourage their use in the future as an additional test of species boundaries.
(c). Comparison with previous estimates of cryptic Amazonian frog diversity
We found a substantially higher percentage increase in the number of cryptic Amazonian frog species (150–350%) than did Fouquet et al. [14] (22–115%). Under the untested assumption that the levels of cryptic diversity found in our study are representative of Amazonian amphibians in general, the estimated number of undescribed amphibians in the Amazon is up to 1385 species (554 known species × 250% increase) based on our Engystomops results and up to 1939 species (554 × 350%) based on our Hypsiboas results. Because these estimates are derived from only two clades, they could be biased and need to be confirmed with integrative systematic analyses of additional clades. Unfortunately, comparable studies with more clades are unlikely to be available in the short term because they are logistically challenging, expensive and time-consuming. Moreover, we recognize that these extrapolations are simplistic and we recommend using more sophisticated and thorough approaches for extrapolating cryptic diversity in the future.
Our results suggest that cryptic frog diversity is even more severely underestimated than previously thought. One possible explanation for this discrepancy is that we were able to sample more intensively than Fouquet et al. For comparison, Fouquet et al. sampled an average of nine individuals per known species, while we were able to sample an average of 115 per known species by focusing on fewer taxa. Because of our intensive sampling, we detected range-restricted and rare species, providing a better approximation of the number of cryptic species. Four out of seven Engystomops species and four out of nine Hypsiboas species were only found at one to four localities (figures 1 and 2), indicating that many have small ranges. This illustrates the importance of intensive sampling for discovering rare and range-restricted cryptic species.
(d). Greater bioacoustic than morphological differences among species
Although there were significant morphological differences among most species examined in size and/or shape, these differences were generally less pronounced than bioacoustic differences. This is consistent with recent studies in frogs [4,54] and suggests that calls evolve faster than morphology, perhaps owing to strong selection for species recognition or sexual selection on calls (or strong stabilizing selection on morphology). These results could explain why there are so many cryptic species: morphological differences among closely related species are generally subtle and are a weak indicator of reproductive isolation. By contrast, calls, which have known importance in causing and maintaining reproductive isolation [15,16,38,39], show pronounced differences among closely related species and seem to be particularly useful for species delimitation.
(e). Conservation implications
Efforts to conserve globally important centres of biodiversity should take into account our finding that Amazonian biodiversity is much greater than previously known, at least for some clades. Unfortunately, despite its unparalleled biodiversity, Amazonia is vulnerable to several ongoing and increasing threats, including timber and petroleum extraction, mining, industrial agriculture and climate change [55,56]. A first step in understanding the potential impacts of these threats would be to accurately characterize the magnitude and spatial distribution of biodiversity in additional clades from other amphibian families and more taxonomic groups.
Identification of cryptic diversity also has important implications for assigning conservation status to individual species. For example, the four recognized species in our study are considered of ‘least concern’ by the IUCN Red List because of their large ranges and abundance (downloaded July 2011). However, our analyses reveal that many species within these two genera have very small ranges, which is one factor that can put them in a higher risk category. Thus, instead of four species of least concern, these two clades may consist of several species of higher conservation concern, depending on additional factors such as evidence of population declines and threats. We predict that as cryptic species continue to be identified, more species of high conservation concern will be identified. Improved species sampling, especially in tropical regions, is almost certain to reveal that the percentage of amphibian species of conservation concern worldwide is even higher than the current estimate of 41 per cent [11].
Acknowledgements
This project was approved by the Institutional Animal Care and Use Committee at Colorado State University.
We thank the many field assistants who helped collect specimens; T. LaDuc, D. C. Cannatella, W. E. Duellman, A. Fouquet, A. Lathrop and R. D. MacCulloch for tissue loans; C. Barnes, S. Bayard de Volo and M. Ordoñez for sequencing; and R. L. Mueller, D. H. Wall, J. M. Robertson, S. W. Fitzpatrick, M. I. Páez, T. D. Price and three anonymous reviewers for helpful comments on the manuscript. This research was funded by a National Geographic Society-Waitt grant, Colorado State University, Secretaría Nacional de Educación Superior, Ciencia, Tecnología e Innovación del Ecuador SENESCYT (grant number PI-C08-0000470), and the Pontificia Universidad Católica del Ecuador. The consortium for the Barcode of Life (CBOL) and the Smithsonian Institution provided funding and laboratory space for DNA sequencing (COI). The Ecuadorian Ministerio de Ambiente provided research and collection permit numbers 004-IC-FAU-DPF, 006-IC-FAU-DBAP/MA, 001-10 IC-FAU-DNB/MA, and 008-09 IC-FAU-DNB/MA. The work was facilitated by ongoing collaborations with the Yanayacu Natural History Research Group.
References
- 1.Bickford D., Lohman D. J., Sodhi N. S., Ng P. K. L., Meier R., Winker K., Ingram K. K., Das I. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155 10.1016/j.tree.2006.11.004 (doi:10.1016/j.tree.2006.11.004) [DOI] [PubMed] [Google Scholar]
- 2.Myers N., Mittermeier R. A., Mittermeier C. G., da Fonseca G. A. B., Kent J. 2000. Biodiversity hotspots for conservation priorities. Nature 403, 853–858 10.1038/35002501 (doi:10.1038/35002501) [DOI] [PubMed] [Google Scholar]
- 3.Orme C. D. L., et al. 2005. Global hotspots of species richness are not congruent with endemism or threat. Nature 436, 1016–1019 10.1038/nature03850 (doi:10.1038/nature03850) [DOI] [PubMed] [Google Scholar]
- 4.Angulo A., Icochea J. 2010. Cryptic species complexes, widespread species and conservation: lessons from Amazonian frogs of the Leptodactylus marmoratus group (Anura: Leptodactylidae). Syst. Biodivers. 8, 357–370 10.1080/14772000.2010.507264 (doi:10.1080/14772000.2010.507264) [DOI] [Google Scholar]
- 5.Crawford A. J., Lips K. R., Bermingham E. 2010. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc. Natl Acad. Sci. USA 107, 13 777–13 782 10.1073/pnas.0914115107 (doi:10.1073/pnas.0914115107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Primack R., Corlett R. 2005. Tropical rain forests: an ecological and biogeographical comparison. Malden, MA: Blackwell Publishing [Google Scholar]
- 7.Duellman W. E. 1999. Distribution patterns of amphibians in South America. In Patterns of distribution of amphibians (ed. Duellman W. E.), pp. 255–328 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 8.Giam X., Scheffers B. R., Sodhi N. S., Wilcove D. S., Ceballos G., Ehrlich P. R. 2012 Reservoirs of richness: least disturbed tropical forests are centres of undescribed species diversity. Proc. R. Soc. B 279, 67–76 10.1098/rspb.2011.0433 (doi:10.1098/rspb.2011.0433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parra G., et al. 2007. Systematics and conservation. In Amphibian conservation action plan (eds Gascon C., Collins J. P., Moore R. D., Church D. R., McKay J. E., Mendelson J. R., III), pp. 45–48 Gland, Switzerland: IUCN [Google Scholar]
- 10.Wake D. B., Vredenburg V. T. 2008. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl Acad. Sci. USA 105, 11 466–11 473 10.1073/pnas.0801921105 (doi:10.1073/pnas.0801921105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart S. N., Chanson J. S., Cox N. A., Young B. E., Rodrigues A. S. L., Fischman D. L., Waller R. W. 2004. Status and trends of amphibian declines and extinctions worldwide. Science 306, 1783–1786 10.1126/science.1103538 (doi:10.1126/science.1103538) [DOI] [PubMed] [Google Scholar]
- 12.Da Silva J. M. C., Rylands A. B., Da Fonseca G. A. B. 2005. The fate of the Amazonian areas of endemism. Conserv. Biol. 19, 689–694 10.1111/j.1523-1739.2005.00705.x (doi:10.1111/j.1523-1739.2005.00705.x) [DOI] [Google Scholar]
- 13.WWF 2010. Amazon alive! A decade of discovery 1999–2009. Godalming, UK: World Wildlife Fund [Google Scholar]
- 14.Fouquet A., Gilles A., Vences M., Marty C., Blanc M., Gemmell N. J. 2007. Underestimation of species richness in neotropical frogs revealed by mtDNA analyses. PLoS ONE 2, e1109. 10.1371/journal.pone.0001109 (doi:10.1371/journal.pone.0001109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vences M., Wake D. B. 2007. Speciation, species boundaries and phylogeography of amphibians. In Amphibian biology: systematics (eds Heatwole H., Tyler M. J.), pp. 2613–2670 Chipping Norton, Australia: Surrey Beatty & Sons Pty Limited [Google Scholar]
- 16.Padial J. M., Miralles A., De la Riva I., Vences M. 2010. The integrative future of taxonomy. Front. Zool. 7, 16. 10.1186/1742-9994-7-16 (doi:10.1186/1742-9994-7-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darst C. R., Cannatella D. C. 2004. Novel relationships among hyloid frogs inferred from 12S and 16S mitochondrial DNA sequences. Mol. Phylogenet. Evol. 31, 462–475 10.1016/j.ympev.2003.09.003 (doi:10.1016/j.ympev.2003.09.003) [DOI] [PubMed] [Google Scholar]
- 18.Frost D. R., et al. 2006. The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 297, 1–370 10.1206/0003-0090(2006)297[0001:TATOL]2.0.CO;2 (doi:10.1206/0003-0090(2006)297[0001:TATOL]2.0.CO;2) [DOI] [Google Scholar]
- 19.Lynch J. D. 1970. Systematic status of the American leptodactylid frog genera Engystomops, Eupemphix, and Physalaemus. Copeia 1970, 488–496 10.2307/1442276 (doi:10.2307/1442276) [DOI] [Google Scholar]
- 20.Duellman W. E. 1973. Frogs of the Hyla geographica group. Copeia 1973, 515–533 10.2307/1443117 (doi:10.2307/1443117) [DOI] [Google Scholar]
- 21.Cannatella D. C., Duellman W. E. 1984. Leptodactylid frogs of the Physalaemus pustulosus group. Copeia 1984, 902–921 10.2307/1445335 (doi:10.2307/1445335) [DOI] [Google Scholar]
- 22.Cannatella D. C., Hillis D. M., Chippindale P. T., Weigt L., Rand A. S., Ryan M. J. 1998. Phylogeny of frogs of the Physalaemus pustulosus species group, with an examination of data incongruence. Syst. Biol. 47, 311–335 10.1080/106351598260932 (doi:10.1080/106351598260932) [DOI] [PubMed] [Google Scholar]
- 23.Faivovich J., Haddad C. F. B., Garcia P. C. A., Frost D. R., Campbell J. A., Wheeler W. C. 2005. Systematic review of the frog family hylidae, with special reference to hylinae: phylogenetic analysis and taxonomic revision. Bull. Am. Mus. Nat. Hist. 294, 1–240 10.1206/0003-0090(2005)294[0001:SROTFF]2.0.CO;2 (doi:10.1206/0003-0090(2005)294[0001:SROTFF]2.0.CO;2) [DOI] [Google Scholar]
- 24.Funk W. C., Caldwell J. P., Peden C. E., Padial J. M., De la Riva I., Cannatella D. C. 2007. Tests of biogeographic hypotheses for diversification in the Amazonian forest frog, Physalaemus petersi. Mol. Phylogenet. Evol. 44, 825–837 10.1016/j.ympev.2007.01.012 (doi:10.1016/j.ympev.2007.01.012) [DOI] [PubMed] [Google Scholar]
- 25.Funk W. C., Angulo A., Caldwell J. P., Ryan M. J., Cannatella D. C. 2008. Comparison of morphology and calls of two cryptic species of Physalaemus (Anura: Leiuperidae). Herpetologica 64, 290–304 10.1655/08-019.1 (doi:10.1655/08-019.1) [DOI] [Google Scholar]
- 26.Vieites D. R., Wollenberg K. C., Andreone F., Kohler J., Glaw F., Vences M. 2009. Vast underestimation of Madagascar's biodiversity evidenced by an integrative amphibian inventory. Proc. Natl Acad. Sci. USA 106, 8267–8272 10.1073/pnas.0810821106 (doi:10.1073/pnas.0810821106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ron S. R., Santos J. C., Cannatella D. C. 2006. Phylogeny of the túngara frog genus Engystomops (=Physalaemus pustulosus species group; Anura: Leptodactylidae). Mol. Phylogenet. Evol. 39, 392–403 10.1016/j.ympev.2005.11.022 (doi:10.1016/j.ympev.2005.11.022) [DOI] [PubMed] [Google Scholar]
- 28.Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation, The University of Texas at Austin, Austin, USA [Google Scholar]
- 29.Posada D., Buckley T. R. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808 10.1080/10635150490522304 (doi:10.1080/10635150490522304) [DOI] [PubMed] [Google Scholar]
- 30.Nylander J. A. A. 2004. MrModeltestv. 2. Uppsala, Sweden: Uppsala University, Evolutionary Biology Centre; Program distributed by the author. [Google Scholar]
- 31.Ronquist F., Huelsenbeck J. P. 2003. MrBayes v. 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 32.Nylander J. A. A., Ronquist F., Huelsenbeck J. P., Nieves-Aldrey J. L. 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53, 47–67 10.1080/10635150490264699 (doi:10.1080/10635150490264699) [DOI] [PubMed] [Google Scholar]
- 33.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weigt L. A., Crawford A. J., Rand A. S., Ryan M. J. 2005. Biogeography of the túngara frog, Physalaemus pustulosus: a molecular perspective. Mol. Ecol. 14, 3857–3876 10.1111/j.1365-294X.2005.02707.x (doi:10.1111/j.1365-294X.2005.02707.x) [DOI] [PubMed] [Google Scholar]
- 35.Hoorn C., Guerrero J., Sarmiento G. A., Lorente M. A. 1995. Andean tectonics as a cause for changing drainage patterns in Miocene northern South America. Geology 23, 237–240 (doi:10.1130/0091-7613(1995)023<0237:ATAACF>2.3.CO;2) [DOI] [Google Scholar]
- 36.Gregory-Wodzicki K. M. 2000. Uplift history of the Central and Northern Andes: a review. Geol. Soc. Am. Bull. 112, 1091–1105 (doi:10.1130/0016-7606(2000)112<1091:UHOTCA>2.3.CO;2) [DOI] [Google Scholar]
- 37.Duellman W. E. 1970. Hylid frogs of Middle America. Monogr. Mus. Nat. Hist. Univ. Kansas 1, 1–753 [Google Scholar]
- 38.Boul K. E., Funk W. C., Darst C. R., Cannatella D. C., Ryan M. J. 2007. Sexual selection drives speciation in an Amazonian frog. Proc. R. Soc. B 274, 399–406 10.1098/rspb.2006.3736 (doi:10.1098/rspb.2006.3736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerra M. A., Ron S. R. 2008. Mate choice and courtship signal differentiation promotes speciation in an Amazonian frog. Behav. Ecol. 19, 1128–1135 10.1093/beheco/arn098 (doi:10.1093/beheco/arn098) [DOI] [Google Scholar]
- 40.Charif R. A., Clark C. W., Fristrup K. M. 2004. Ravenv. 1.2, user's manual. Ithaca, NY: Cornell Laboratory of Ornithology [Google Scholar]
- 41.Jiménez de la Espada M. 1872. Nuevos batrácios Americanos. Anal. Soc. Española Hist. Nat. Madrid 1, 84–88 [Google Scholar]
- 42.Donoso-Barros R. 1969. Un nuevo anuro de Bolivia, Eupemphix freibergi, nov. sp. Bol. Soc. Biol. Concepc. 41, 183–187 [Google Scholar]
- 43.Good J. M., Hird S., Reid N., Demboski J. R., Steppan S. J., Martin-Nims T. R., Sullivan J. 2008. Ancient hybridization and mitochondrial capture between two species of chipmunks. Mol. Ecol. 17, 1313–1327 10.1111/j.1365-294X.2007.03640.x (doi:10.1111/j.1365-294X.2007.03640.x) [DOI] [PubMed] [Google Scholar]
- 44.Günther A. C. L. G. 1858. Neue Batrachier in de Sammlung des britischen museums. Arch. Nat. Berlin 24, 319–328 [Google Scholar]
- 45.Troschel F. H. 1848. Theil 3 Versuch einer Zusammenstellung der Fauna und Flora von Britisch-Guiana. In Reisen in Britisch-Guiana in den Jahren 1840–44. Im Auftrage Sr. Majestät des Königs von Preussen ausgeführt (ed. Schomburgk R.), pp. 645–661 Leipzig, Germany: J. J. Weber [Google Scholar]
- 46.Mora C., Tittensor D. P., Adl S., Simpson A. G. B., Worm B. 2011. How many species are there on Earth and in the ocean? PLoS Biol. 9, e1001127. 10.1371/journal.pbio.1001127 (doi:10.1371/journal.pbio.1001127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elmer K. R., Dávila J. A., Lougheed S. C. 2007. Cryptic diversity and deep divergence in an upper Amazonian leaflitter frog, Eleutherodactylus ockendeni. BMC Evol. Biol. 7, 247. 10.1186/1471-2148-7-247 (doi:10.1186/1471-2148-7-247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fouquet A., Vences M., Salducci M.-D., Meyer A., Marty C., Blanc M., Gilles A. 2007. Revealing cryptic diversity using molecular phylogenetics and phylogeography in frogs of the Scinax ruber and Rhinella margaritifer species groups. Mol. Phylogenet. Evol. 43, 567–582 10.1016/j.ympev.2006.12.006 (doi:10.1016/j.ympev.2006.12.006) [DOI] [PubMed] [Google Scholar]
- 49.Sites J. W., Jr, Marshall J. C. 2003. Delimiting species: a Renaissance issue in systematic biology. Trends Ecol. Evol. 18, 462–470 10.1016/S0169-5347(03)00184-8 (doi:10.1016/S0169-5347(03)00184-8) [DOI] [Google Scholar]
- 50.Sites J. W., Jr, Marshall J. C. 2004. Operational criteria for delimiting species. Annu. Rev. Ecol. Evol. Syst. 35, 199–227 10.1146/annurev.ecolsys.35.112202.130128 (doi:10.1146/annurev.ecolsys.35.112202.130128) [DOI] [Google Scholar]
- 51.Goldstein P. Z., DeSalle R. 2011. Integrating DNA barcode data and taxonomic practice: determination, discovery, and description. BioEssays 33, 135–147 10.1002/bies.201000036 (doi:10.1002/bies.201000036) [DOI] [PubMed] [Google Scholar]
- 52.Meyer A., Kocher T. D., Basasibwaki P., Wilson A. C. 1990. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature 347, 550–553 10.1038/347550a0 (doi:10.1038/347550a0) [DOI] [PubMed] [Google Scholar]
- 53.Ence D. D., Carstens B. C. 2011. SpedeSTEM: a rapid and accurate method for species delimitation. Mol. Ecol. Resour. 11, 473–480 10.1111/j.1755-0998.2010.02947.x (doi:10.1111/j.1755-0998.2010.02947.x) [DOI] [PubMed] [Google Scholar]
- 54.Padial J. M., Köhler J., Muñoz A., de la Riva I. 2008. Assessing the taxonomic status of tropical frogs through bioacoustics: geographical variation in the advertisement calls in the Eleutherodactylus discoidalis species group (Anura). Zool. J. Linn. Soc. 152, 353–365 10.1111/j.1096-3642.2007.00341.x (doi:10.1111/j.1096-3642.2007.00341.x) [DOI] [Google Scholar]
- 55.Laurance W. F., Powell G., Hansen L. 2002. A precarious future for Amazonia. Trends Ecol. Evol. 17, 251–252 10.1016/S0169-5347(02)02484-9 (doi:10.1016/S0169-5347(02)02484-9) [DOI] [Google Scholar]
- 56.Lewis S. L., Brando P. M., Phillips O. L., Van der Heijden G. M. F., Nepstad D. 2011. The 2010 Amazon drought. Science 331, 554. 10.1126/science.1200807 (doi:10.1126/science.1200807) [DOI] [PubMed] [Google Scholar]