Abstract
Harbouring a plasmid often imposes a fitness cost on the bacterial host. Motivated by implications for public health, the majority of studies on plasmid cost are focused on elements that impart antibiotic resistance. Plasmids, however, can provide a wide range of ecologically important phenotypes to their bacterial hosts—such as virulence, specialized catabolism and metal resistance. The Agrobacterium tumefaciens tumour-inducing (Ti) plasmid confers both the ability to infect dicotyledonous plants and to catabolize the metabolites that plants produce as a result of being infected. We demonstrate that this virulence and catabolic plasmid imposes a measurable fitness cost on host cells under resource-limiting, but not resource replete, environmental conditions. Additionally, we show that the expression of Ti-plasmid-borne pathogenesis genes necessary to initiate cooperative pathogenesis is extremely costly to the host cell. The benefits of agrobacterial pathogenesis stem from the catabolism of public goods produced by infected host plants. Thus, the virulence-plasmid-dependent costs we demonstrate constitute costs of cooperation typically associated with the ability to garner the benefits of cooperation. Interestingly, genotypes that harbour derived opine catabolic plasmids minimize this trade-off, and are thus able to freeload upon the pathogenesis initiated by other individuals.
Keywords: cooperation, public good, Ti plasmid, trade-off, Agrobacterium tumefaciens, greenbeard
1. Introduction
Many ecologically important functions—including virulence, antibiotic resistance, bacteriocin production and specialized catabolism—are conferred by genes commonly found on bacterial plasmids. These self-replicating DNA elements tend to carry non-essential genes and can be transmitted vertically, and often horizontally, between host cells [1]. In many cases, the benefits that plasmids confer are only realized under certain environmental conditions [2]. In contrast, plasmids may be costly to their host cell regardless of the environment, because at a minimum their maintenance requires their replication and partitioning during each bacterial generation [3]. Indeed, numerous studies have found that harbouring a plasmid reduces the fitness of host cells [4–6]. The carriage cost of plasmids is centrally important to their population dynamics [2,7]. However, probably motivated by public health implications, most studies have measured carriage costs associated with plasmids that confer antibiotic resistance to host cells. Indeed, relatively few studies have examined the cost of harbouring plasmids conferring other ecologically important functions (see [8,9] for exceptions). Natural plasmids are often relatively large, have low copy number and maintain long-standing associations with bacterial lineages [1]. Each of these factors is likely to lead to the evolution of limited costs associated with these plasmids. Despite this, as well as the fact that these plasmids frequently confer important phenotypes such as the ability to infect plant or animal hosts, few studies have examined how the costs associated with these plasmids shape their population dynamics.
The tumour-inducing (Ti) plasmid of Agrobacterium tumefaciens confers the majority of virulence functions underlying crown gall disease of dicotyledonous plants [10]. Plant-associated cues stimulate agrobacterial cells harbouring this large (approx. 200 kb) plasmid to infect hosts using a Ti plasmid encoded type IV secretion (T4S) system that delivers copies of a subset of Ti plasmid genes (carried on the transferred DNA or T-DNA) into the plant genome [10]. The infection process is predominantly mediated by a large set of more than 30 different Ti plasmid virulence (vir) genes, including those that form the T4S system and others that drive T-DNA replication and processing. Following transformation of the plant host, T-DNA genes are recognized by the plant's expression machinery and expressed strongly within the plant nucleus. Several of these genes increase pools of the plant hormones, auxin and cytokinin, resulting in a rapid proliferation of the infected cells and thereby tumour development [10]. Other T-DNA genes direct the synthesis and release of metabolites called opines—a suite of specialized nutritional resources that cells use via catabolic genes that, importantly, are also encoded on the Ti plasmid [10]. Most opines are unusual ligation products of amino acids with small organic acids or sugar phosphates that can supply the infecting bacteria with carbon, nitrogen and phosphorus.
In this study, we examine the costs associated with the A. tumefaciens Ti plasmid. We find that these costs probably vary over space and time as a consequence of dependence on local environmental conditions. We observed a significant cost associated with carriage of the Ti plasmid when either carbon or nitrogen availability limited population carrying capacity, but not when these resources were abundant. We also demonstrate that the expression of Ti-plasmid-borne pathogenesis genes imposes a dramatic fitness cost on host cells. While it is not unusual for plasmids to have costs, it is striking that in this system the costs associated with the Ti plasmid are costs of cooperation. Agrobacterial pathogenesis is cooperative with the actions of few individuals infecting the host plant, leading to the availability of a public good resource [11,12]. Cells that harbour a Ti plasmid suffer a modest cost of carrying the Ti plasmid, as well as a high cost of infecting plant host cells, but are able to garner the benefits stemming from the catabolism of the public good nutrients produced by infected plant cells. In nature, some agrobacterial genotypes have an opine catabolic plasmid that confers the ability to catabolize opines but lacks the genes required to infect plant hosts [13,14]. Such strains are able to freeload on public goods produced by host plants infected by other individuals, while themselves avoiding the costs associated with A. tumefaciens's virulence plasmid.
2. Material and methods
(a). Strains, plasmids and growth conditions
All strains and plasmids used in this study are described in the electronic supplementary material. We purchased reagents, antibiotics and media components from Fisher Scientific (Pittsburgh, PA), Sigma-Aldrich (St Louis, MO) and New England Biolabs (Ipswich, MA). We obtained oligonucleotide primers from Integrated DNA Technologies (Coralville, IA) and used QIAquick Spin kits (QIAGEN, Valencia, CA) for purification of nucleic acids. DNA manipulations were performed using standard protocols [15]. Plasmids were transferred into Agrobacterium strains via either conjugation or electroporation using standard approaches [16,17]. Unless specified otherwise, A. tumefaciens strains were grown in AT minimal media supplemented with 27.5 mM glucose and 15 mM (NH4)2SO4 (ATGN [18]) and incubated at 28°C in a rotary aerator or on 1.5 per cent agar plates. Unlike the original recipe, we omitted FeSO4•H2O from our AT minimal media [19]. Escherichia coli strains were grown in LB media at 37°C on a shaker platform or 1.5 per cent agar plates. Agrobacterium tumefaciens strains TGP110 and TGP114 are spontaneous nalidixic acid resistant mutants of 15955 and TGP101, respectively, arising on ATGN supplemented with nalidixic acid (100 µg ml–1). Unless specified otherwise, antibiotics were used at the following concentrations: for A. tumefaciens, 50 µg ml–1 nalidixic acid (Nal), 50 µg ml–1 ampicillin and 100 µg ml–1 kanamycin; for E. coli, 100 µg ml–1 ampicillin and 50 µg ml–1 kanamycin.
(b). Curing the tumour-inducing plasmid
In order to isolate the impact of the Ti plasmid on the growth and competitive ability of bacterial cells, we generated a plasmid-free derivative of A. tumefaciens strain 15955 that differs from the parent strain only in that it lacks pTi15955. We cured the Ti plasmid from 15955, using the approach described by Uraji et al. [20]. We introduced the curing vector pTP101 into the 15955 genetic background via conjugation with an E. coli S17-1 λpir donor. Transconjugants of A. tumefaciens were selected for by their ability to grow on ATGN supplemented with kanamycin and ampicillin. The curing vector was constructed by cloning the entire vegetative replication region of pTi15955 (repABC) into pCF117, which contains a counterselectable sacRB gene conferring sucrose sensitivity. The two plasmids, pTP101 and pTi15955, use the same replication and partitioning machinery, and are therefore incompatible [20,21]. As a consequence, transconjugants that contain both plasmids give rise to cells that lack one or the other plasmid. Populations of these cells were subsequently screened for a lack of octopine catabolism, consistent with a loss of pTi15955, by patching colonies onto AT minimal media supplemented with 3.25 mM octopine (Sigma; discontinued) as the sole source of carbon and nitrogen. We then counterselected against the sacRB gene on pTP101 by plating populations of derivatives incapable of octopine catabolism onto AT minimal media supplemented with 0.5 per cent sucrose and 15 mM (NH4)2SO4 to obtain a markerless derivative of 15955 that has been cured of the Ti plasmid and also the curing vector. We confirmed that we successfully cured 15955 of pTi15955 using a modified Eckhardt method [22] to evaluate the large plasmid content of our cured strain relative to that of its parent strain, as well as R10, KYC55 and SA122 as controls.
(c). Characterizing resource-limited growth
We evaluated the population yield of A. tumefaciens strain 15955 in response to increasing amounts of glucose as the sole carbon source or ammonia as the sole nitrogen source. To do this, we prepared A. tumefaciens 15955 inoculum free of nitrogen and carbon sources by growing this strain to mid-log phase, harvesting cells via centrifugation and washing them seven times with 78.6 mM KH2PO4 buffer (pH 7.0). The washed cells were then diluted to an optical density at 600 nm (OD600) of 0.005 in the appropriate media. Glucose was the sole source of carbon in AT minimal media supplemented with 15 mM (NH4)2SO4 and 0, 1, 3, 6, 12, 17 or 27.5 mM glucose. Analogously, ammonium was the sole source of nitrogen in AT minimal media supplemented with 27.5 mM glucose and 0, 0.04, 0.08, 0.16, 0.3, 0.6, 1.2, 2.4, 5, 10 or 20 mM ammonia. We monitored the OD600 of these cultures until their population density, as monitored by optical density, ceased to increase.
(d). Measuring the cost of harbouring the tumour-inducing plasmid in competition
In order to assess the fitness cost associated with bearing the Ti plasmid, we grew pTi+ cells and pTi− cells together under a variety of environmental conditions. pTi+ and pTi− cells were competed in standard ATGN media as well as AT minimal media modified to an acidic pH. This latter media contained standard AT salts, 27.5 mM glucose, 15 mM (NH4)2SO4 and 78.6 mM KH2PO4 (pH 5.6), and was buffered to pH 5.6 by 2-(N-morpholino)ethanesulfonic (MES). Under the stress of these acidic conditions, agrobacterial populations grow markedly slower than they grow under standard, pH neutral ATGN conditions. We also competed pTi+ and pTi− cells under carbon- and nitrogen-limiting conditions. The carbon-limiting media was AT minimal media supplemented with 1 mM glucose and 30 mM ammonia, while the nitrogen-limiting media was AT minimal media supplemented with 27.5 mM glucose and 1.2 mM ammonia.
All competitions were inoculated with approximately 6 × 106 cells, with approximately half pTi+ and half a nearly isogenic pTi-cured derivative. Half of the competitions for each environmental condition competed 15955 (pTi+) against TGP114 (pTi−; NalR), while the other half of the replicates competed TGP110 (pTi+; NalR) and TGP101 (pTi−), thereby controlling for possible effects of the antibiotic resistance label. For all competition experiments, conjugation and loss of the plasmid is very rare under the tested environmental conditions [16,20]. We prepared carbon- and nitrogen-free inocula of each strain as described earlier. These cells were then used to inoculate 2 ml of appropriate fresh media to an approximate OD600 of 0.005. The competition cultures were then incubated at 28°C for 24 h, after which all mixed cultures were sub-cultured 1 : 100 into 2 ml of the appropriate fresh media and incubated as before. We repeated this sub-culturing for a total of five passages. We plated a dilution series of each mixed culture at the start of the experiment, as well as after the initial, first passage and fifth passage populations reached stationary phase, onto ATGN and ATGN supplemented with 50 µg ml–1 Nal so that we could estimate the density and frequency of both strains present. After the fifth passage reached stationary phase, a subset of the mixed cultures were allowed to incubate for an additional 24 h and then plated as described before.
(e). Virulence gene expression
Ti plasmid borne vir genes are expressed in response to acidic conditions, low phosphate levels and, most critically, the presence of plant-produced phenolics, such as acetosyringone (AS) [23]. To confirm that the vir genes are expressed in response to the resource-limiting conditions used in our experiments, we measured the expression levels driven off the virB promoter. We electroporated pSW209Ω into 15955 to obtain a reporter of virulence gene expression. This plasmid contains a copy of lacZ fused to the virB promoter (PvirB::lacZ), such that β-galactosidase levels—as measured by standard assays using cleavage of the colorometric substrate o-nitrophenyl-β-d-galactoside—correspond to levels of expression driven from this promoter [24]. We grew populations of 15955 pSW209Ω to mid-log phase and washed the cells seven times with 20 mM MES pH 5.6, each time collecting the cells by centrifugation. We used these washed cells to inoculate to an OD600 of 0.005 media that contained AT minimal media salts, 20 mM MES pH 5.6 buffer, 500 µM phosphate, 3 mM glucose, 0.05 per cent DMSO and either no AS or 100 µM AS. Under these conditions, 15955 population growth is limited by carbon availability (data not shown). We likewise inoculated washed cells into analogous media that differed only in having 10 mM glucose and 0.6 mM (NH4)2SO4, rather than 3 mM glucose and 15 mM (NH4)2SO4. Under these conditions, agrobacterial population growth is limited by nitrogen availability (data not shown). We allowed these populations to grow to late-log phase before assaying their β-galactosidase activity [24].
We measured the doubling time of pTi+ and pTi− cells under vir-inducing and non-inducing limiting carbon conditions. To do this, we grew clonal populations of each strain to mid-log phase, washed the cells seven times with 20 mM MES pH 5.6 and inoculated low-carbon vir-inducing media to an OD600 of 0.005 with the washed cells (see above for details). We then monitored the population growth of these cultures into stationary phase based on changes in OD600. We calculated the doubling time of each culture based on the population's rate of increase in optical density during mid-log phase.
(f). Measuring the fitness cost of expressing cooperative pathogenesis genes in competition
To confirm that the growth differences we observed between clonal pTi+ and pTi− populations translate into fitness differences between the strains in competition, we measured the cost associated with expressing Ti plasmid borne virulence genes by competing these strains in environments wherein virulence genes are either induced or uninduced. We did this under limiting carbon, limiting nitrogen, carbon- and nitrogen-replete environmental conditions (see §2e). These competition experiments were performed as described above, except that we only passaged each mixed culture one time and inocula were washed with 20 mM MES (pH 5.6). As described earlier, we estimated the frequency and density of each cell type in each mixed culture at the start of the experiment, as well as after the initial and first passage populations reached stationary phase.
(g). Statistical analysis
For both competition experiments, the relative fitness of pTi+ cells (WpTi +) was estimated as the ratio of the number of doublings by pTi+ cells to that of pTi− cells over the course of the experiment [4]. Costs associated with the Ti plasmid (c) were estimated by subtracting one from the relative fitness of pTi+ cells competing with pTi− cells (1 + c = WpTi +). These fitness and plasmid cost data were analysed using Proc GLM based on SAS software. The model included both the environmental condition and the NalR marker orientation as factors. Including the latter factor allowed us to remove variance associated with the marker and thereby isolate the effect of virulence gene expression by comparing least square (LS) means of the different media treatments.
3. Results
(a). Curing the tumour-inducing plasmid
Experimental examination of the growth and fitness consequences of bearing a plasmid requires the comparison of strains that differ only in the presence or absence of the plasmid. Studies that do not carefully make such isogenic derivatives are prone to falsely attributing the effects of other factors to the plasmid. We generated these strains by curing A. tumefaciens 15955 of pTi15955 using plasmid incompatibility. The unmarked derivative we obtained is nearly isogenic with the 15955 parent strain, differing only in that it lacks the Ti plasmid. The absence of the Ti plasmid was confirmed with Eckhardt gel analysis, and the inability to PCR amplify several Ti plasmid specific sequences distributed around the plasmid, in parallel with reactions that did amplify sequences from the 15955 parental strain. We also confirmed that our cured derivative is unable to induce tumourigenesis in a potato-disc virulence assay [25] and cannot activate a bioreporter of acyl-homoserine lactones [26], another Ti plasmid encoded activity. Both of these phenotypes are consistent with the absence of the Ti plasmid. This pTi-cured derivative was designated as TGP101.
(b). Resource-limited growth
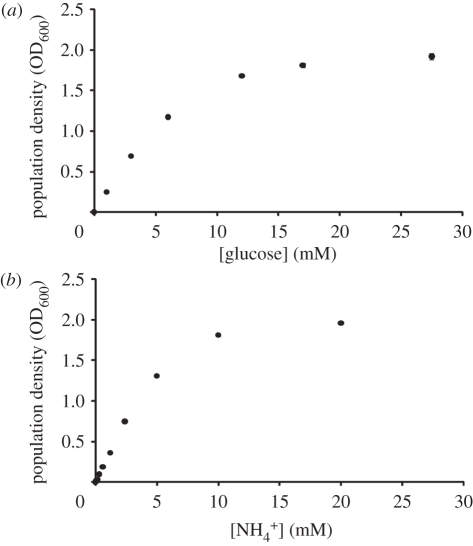
We monitored the population growth of 15955 under a variety of environmental conditions supplied with increasing amounts of glucose as the sole carbon source. We also performed an analogous experiment varying the concentration of ammonia as the sole nitrogen source. The carrying capacity of 15955 populations increased with increasing availability of glucose and ammonia, respectively (figure 1). These effects started to plateau at glucose or ammonia levels greater than approximately 5 mM (figure 1).
Figure 1.
Agrobacterium tumefaciens strain 15955 achieved higher population density in response to higher levels of (a) glucose as the sole source of carbon (quadratic regression, β[glucose] = 0.18, t = 21.61, p < 0.001) and (b) ammonium as the sole source of nitrogen (quadratic regression, β[ammonia] = 0.28, t = 44.3, p < 0.0001). In both cases, this effect eventually levelled off, suggesting that carbon (quadratic regression, 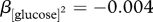 , t = −13.9, p < 0.0001) or nitrogen (quadratic regression,
, t = −13.9, p < 0.0001) or nitrogen (quadratic regression, 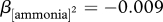 , t = –28.3, p < 0.0001) availability no longer limited population growth. OD600 was measured 45 h after inoculation. Values represent mean ± s.e. of three replicates. All means are statistically different from one another in the glucose dose response (p < 0.05). However, the 0, 0.04, 0.08 and 0.16 mM ammonia treatment means are not significantly different from one another, while all other means in (b) are significantly different (p < 0.05).
, t = –28.3, p < 0.0001) availability no longer limited population growth. OD600 was measured 45 h after inoculation. Values represent mean ± s.e. of three replicates. All means are statistically different from one another in the glucose dose response (p < 0.05). However, the 0, 0.04, 0.08 and 0.16 mM ammonia treatment means are not significantly different from one another, while all other means in (b) are significantly different (p < 0.05).
(c). Fitness cost of harbouring the plasmid
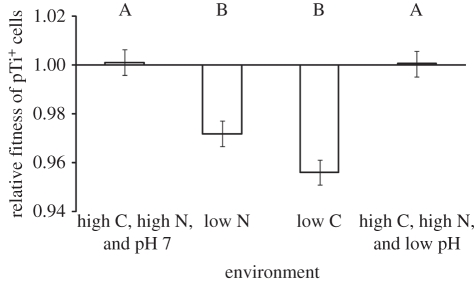
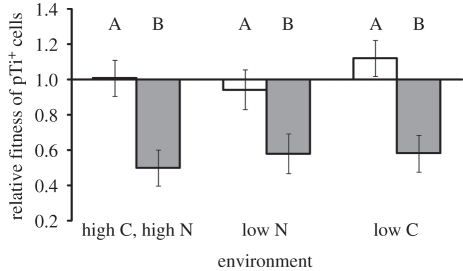
The fitness of cells harbouring the Ti plasmid depended upon the environmental conditions in which they competed with pTi− cells (F3,39 = 18.3, p < 0.0001). Over the course of approximately 35 generations in carbon- or nitrogen-limiting media, pTi− cells began to overtake the pTi+ cells in the population, regardless of which cells were resistant to nalidixic acid. This indicates that cells harbouring the Ti plasmid were at a competitive disadvantage to cells lacking the plasmid under both carbon- and nitrogen-limiting environmental conditions (figure 2). However, we did not observe this when these nutrients were abundant at both neutral and stressful, low pH (figure 2). We also observed that the frequencies of the two strains did not significantly change over the course of 24 h in stationary phase in any of the tested media (F3,23 = 0.65, n.s.). This suggests that the advantage that pTi− cells manifested when resources were limiting stems from differences between the strains during lag and/or log population growth phases.
Figure 2.
Cells harbouring the Ti plasmid have a lower fitness than pTi− cells when competing in low-carbon and low-nitrogen environments for approximately 35 generations. In contrast, neither strain has a competitive advantage in environments with high carbon and nitrogen. ‘High C, high N and pH 7’ media is standard ATGN, and ‘high C, high N and low pH’ media is ATGN modified to be buffered by 20 mM MES to pH 5.6. ‘Low C’ media is AT minimal media supplemented with 1 mM glucose and 15 mM (NH4)2SO4, while ‘low N’ media is AT minimal media supplemented with 27.5 mM glucose and 0.6 mM (NH4)2SO4. Values represent LS means ± s.e. of 10 replicates. Means marked with different letters are significantly different (p < 0.05). The low C and low N treatment means are significantly lower than one (p < 0.0001), whereas the other two treatment means are not significantly different from one (p > 0.05).
(d). Regulation of virulence gene expression
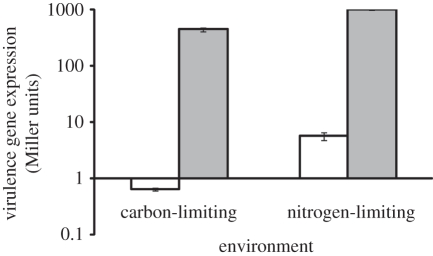
Induction of the vir genes by the plant phenolic acetosyringone (AS) is well established [23]. Our experiments required modifying standard vir induction media in order to achieve either carbon-limiting or nitrogen-limiting conditions. Doing this involved increasing the availability of phosphate and decreasing the availability of glucose and ammonia. We confirmed that the vir genes are strongly induced in our carbon- and nitrogen-limiting modified induction media. When both carbon (F1,9 = 160.2, p < 0.001) and nitrogen (F1,9 = 1282.92, p < 0.001) were limiting, the presence of AS strongly induced the expression of virulence genes as measured by the activity of a PvirB::lacZ fusion (figure 3).
Figure 3.
Virulence gene expression is tightly regulated by environmental stimuli. The presence of the plant phenolic acetosyringone (AS) strongly promoted expression of the virB promoter in pTi+ cells (15955 pSW209Ω) when both carbon (p < 0.05) and nitrogen (p < 0.05) is limiting under acidic, low-phosphate conditions. Values represent mean ± s.e. of five replicates. Filled bars, with AS; open bars, without AS.
(e). Fitness cost of expressing plasmid-borne cooperative virulence genes
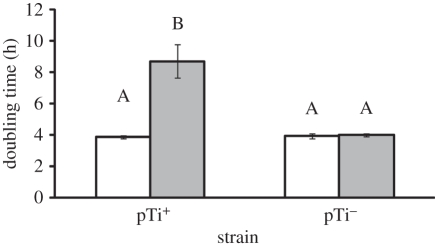
The growth rate of clonal pTi+ 15955 populations slowed dramatically (F1,5 = 20.24, p < 0.05) when the environment contained AS. Under vir-inducing conditions, 15955 populations took nearly twice as long to double in size (figure 4). In contrast, populations of TGP101, which lack the Ti plasmid and therefore are unable to induce expression of the Ti plasmid virulence genes in response to the plant phenolic, were not affected for growth by the presence of AS (F1,5 = 0.11, n.s.). Furthermore, after approximately only 12 generations under vir-inducing conditions, pTi− cells were much more common than pTi+ (15955) cells in populations initially composed of equal numbers of each type of cell. Under vir-inducing conditions, cells harbouring the Ti plasmid were at a striking competitive disadvantage to pTi− cells regardless of nutrient levels (figure 5). In this experiment, we did not observe a significant cost to harbouring the Ti plasmid when AS was absent under carbon-limiting or nitrogen-limiting conditions (figure 5). This cost was observable in similar experiments that ran for approximately 35 generations (figure 2). This suggests that 12 generations is insufficient to observe the modest cost of harbouring the Ti plasmid when expression of the vir genes is not induced or that the differences between the media used for these experiments affect the carriage cost of the Ti plasmid.
Figure 4.
The presence of the plant phenolic acetosyringone (AS) was associated with a large increase in the mid-log phase doubling time of pTi+ populations but not pTi− populations. Environmental conditions are limiting carbon (3 mM glucose), low phosphate (500 µM), acidic pH (pH 5.6) either with (filled bars) or without (open bars) 100 µM AS present. Values represent mean ± s.e. of three replicates. Different letters denote treatment means that are significantly different (p < 0.05).
Figure 5.
The expression of the Ti-plasmid-borne virulence genes was very costly. Cells harbouring the plasmid had dramatically reduced fitness relative to plasmid-free cells when environmental acetosyringone (AS) stimulates the expression of genes involved in pathogenesis. In all cases, media were acidic (pH 5.6) and contained 500 µM phosphate. ‘Low C’ media contained 3 mM glucose, while ‘low N’ media contained 1.2 mM ammonia. All other nutrients were provided in abundance as in the ‘high C, high N’ media. Values represent LS means ± s.e. of at least nine replicates. None of the treatment means were significantly different from one when AS was not present (open bars; high C, high N: t = 0.11, d.f. = 9, n.s.; low N: t = −0.49, d.f. = 9, n.s.; low C: t = 1.19, d.f. = 9, n.s.), whereas all treatment means were less than one when AS was present (filled bars; high C, high N: t = –4.92, d.f. = 9, p < 0.001; low N: t = −3.68, d.f. = 9, p < 0.01; low C: t = −3.97, d.f. = 9, p < 0.01). Different letters denote treatments for which the change in the frequency of pTi+ cells significantly differed (p < 0.05). Note that because the cost of expressing virulence genes is much greater than the cost of harbouring the plasmid, the scale of the y-axis differs from that of figure 2.
4. Discussion
Bacterial plasmids frequently impose a fitness burden on their host cells [4–6]. Most empirical estimates of these costs have focused on plasmids that impart antibiotic resistance on their hosts, while the costs of other important plasmids such as those conferring virulence, bacteriocin and catabolic functions have received much less empirical attention (see [8,9] for exceptions). In this study, we demonstrated that a natural, low-copy-number, ecologically important plasmid—the A. tumefaciens Ti plasmid—is associated with two kinds of fitness costs. We observed a relatively modest cost to harbouring the plasmid when few of its genes are induced under either nitrogen- (LS mean c ± s.e. = −0.028 ± 0.005) or carbon- (−0.044 ± 0.005) limiting conditions. We also demonstrated a more severe burden on host cells associated with the expression of plasmid-borne genes underlying virulence that did not significantly depend on nutrient availability (nutrient replete: LS mean c ± s.e. = –0.500 ± 0.087; nitrogen-limiting: −0.389 ± 0.101; carbon-limiting: −0.410 ± 0.092).
(a). Costs and plasmid population dynamics
Fitness costs associated with plasmids are thought to reflect the consequences of metabolic drain from the expression of plasmid genes and/or the plasmid having adverse effects on host physiology [27]. These costs can vary depending on external environmental conditions [28,29], the genetic background of the cell that harbours them [27] or the population frequency of the plasmid [8]. In this study, we demonstrated that there is a fitness cost associated with the carriage of the A. tumefaciens virulence plasmid under resource-limiting conditions. In contrast, we did not observe this cost when cells harbouring the Ti plasmid competed against plasmidless cells in a resource-abundant environment. The resource dependence of this cost is consistent with the carriage cost of the Ti plasmid stemming from the Ti plasmid imposing a metabolic drain on the host cell. Similar results have also been reported for the antibiotic resistance plasmid pB15 in E. coli, where reduced conjugation rates in high-carbon environments were associated with lower plasmid burden on the host cell [29].
Previous work has found that the long-term association of plasmid and chromosomal genes can diminish the costs associated with plasmids owing to changes in either the plasmid or the chromosomal genetic content [4,27,30]. In this study, we showed that despite the long-term association between pTi15955 and the 15955 chromosomal background, there continue to be fitness costs associated with harbouring this plasmid. However, we also note that these costs were not apparent under resource-replete conditions and thus were not readily apparent in laboratory experiments where saturating nutrients mask the cost of the plasmid. In most natural terrestrial environments, bacteria are resource-limited, and thus the costs of the Ti plasmid are likely to be manifested. Like many natural plasmids, the Ti plasmid exhibits several phenotypes consistent with selection to minimize the burden that it imposes on its host cell, also promoting the fitness of the plasmid itself because it benefits from vertical transmission during host cell division [29,31,32]. Most of the genes on the Ti plasmid are known to be tightly regulated according to biotic or abiotic stimuli such that these gene products are only produced when needed [33]. Natural plasmids are often maintained at a low copy number and typically have stability systems promoting efficient partitioning into both daughter cells, so that they are not lost during cell division [34]. The low copy number of these plasmids is thought to minimize their burden on host cells, thereby promoting the vertical transmission of the plasmid [31,32].
Despite the Ti plasmid generally having a low copy number, expressing genes only when induced by proper environmental stimuli and having a long-standing association with agrobacterial genetic backgrounds, it still imposes a significant fitness burden on host cells when resources are limiting and few plasmid-borne genes are expressed (figure 2). Ti plasmids belong to the repABC plasmid family, whose plasmids are widely distributed among α-proteobacteria, but particularly common in the Rhizobiales, the order that includes Agrobacterium, Rhizobium and Brucella, among others [21]. As with the Ti plasmid, these plasmids tend to be found at low copy number within host cells. Stable vertical transmission of these plasmids depends upon an efficient partitioning system involving the RepA and RepB proteins, which physically links replication of the plasmid to the process of cell division [35]. In addition, recent work has identified a toxin-antidote system on pTiC58 that further enhances the stability of this Ti plasmid [36]. Cells that lose the plasmid after segregation are often killed owing to poor stability of the antidote and unconstrained toxin activity [37]. These replication, partitioning, toxin-antidote and perhaps other stability systems can slow the generation of pTi− cells from pTi+, thereby helping to account for how the Ti plasmid is maintained despite the carriage costs that it imposes on its host cell. Ironically, they may also contribute to the burden that the Ti plasmid imposes on its agrobacterial host because these systems must be expressed each generation in their role of stabilizing the inheritance of the plasmid during cell division [3].
We also show that the expression of the plasmid-borne virulence genes was associated with a dramatic reduction in fitness. Consistent with previous studies on other plasmids [8,38], pTi+ cells were at a much greater competitive disadvantage to pTi− cells when environmental conditions stimulated the expression of plasmid-borne virulence genes than when these genes were not induced (figure 5). Agrobacterial virulence is likely to be a metabolically expensive behaviour because it involves the construction of the elaborate T4S apparatus necessary for agrobacterial pathogenesis. These macromolecular bacterial secretion machines span the inner membrane, periplasm and outer membranes of the bacterial cell and are composed of numerous protein subunits. The T4S system encoded by the vir genes is composed of many copies of 11 distinct proteins [39]. During infection, the T-DNA and several effector proteins, which modify plant cell physiology and promote T-DNA integration, translocate into the plant host cell through the channel formed by this complex structure [10], allowing for subsequent genetic transformation of the plant cell. Additionally, vir-induction has been shown to be associated with an increase in Ti plasmid copy number, potentially adding to the cost associated with harbouring the plasmid [40,41]. This increase in Ti plasmid copy number, in addition to the metabolic drain of producing T4S machinery and associated functions, may contribute to the high plasmid cost that cells experience under vir-inducing conditions. In light of this high fitness cost, it is sensible that the expression of these genes is tightly regulated by external environmental cues associated with the presence of a potential plant host. This regulation minimizes the plasmid's burden on the host cell, thereby benefiting the plasmid in terms of the fitness it gains through vertical transmission during host cell reproduction [31,32].
Expression of the plasmid conjugal machinery used to transfer the plasmid between bacterial cells can also impose a dramatic fitness burden on host cells [29,32,38]. This cost establishes a trade-off between horizontal and vertical transmission of a plasmid because conjugation of the plasmid comes at the expense of lower cell division rates, and thus a lower efficiency of plasmid vertical transmission [32]. The Ti plasmid is able to colonize new agrobacterial genetic backgrounds via conjugative transfer of the plasmid; however, under our experimental conditions, the Ti plasmid conjugal machinery is not expressed—and therefore does not contribute to the plasmid associated costs that we observed. The conjugation of the Ti plasmid is highly regulated such that it only occurs in the disease environment where opines are abundant [16] and in which pTi+ cells have a competitive advantage [42].
The cost associated with carrying a plasmid plays a central role in its population dynamics [2,7,29]. The fitness costs that we have demonstrated in this study probably factor into the observation that the majority of natural agrobacterial isolates lack a virulence-conferring Ti plasmid [43,44], despite the plasmid's high stability [21], ability to conjugate into novel backgrounds [16], and benefits associated with phenolic resistance [45] and opine catabolism [42].
(b). Cooperation and plasmid evolutionary dynamics
While the expression of the genes underlying agrobacterial pathogenesis is very costly, its benefits stem from the Ti plasmid mediated catabolism of a public good—specialized catabolites produced by infected plant tissues [46,47]. Access to these opines depends on a cell expressing opine catabolic genes that are carried on the Ti plasmid. Thus, the Ti plasmid is a greenbeard wherein the benefits of the cooperative act of pathogenesis are intrinsically directed towards genetically similar cells that tend to have both virulence and opine catabolic genes due to their efficient coinheritance during vertical or horizontal transmission of the Ti plasmid [11,12]. Greenbeards were once thought to be rare and improbable; however, several examples have been identified in recent years [12]. The agrobacterial greenbeard provides a clear example with a particularly well-characterized genetic basis, making this an ideal model system for studying the evolutionary dynamics of greenbeard cooperation. Towards this goal, we take advantage of this knowledge and this system's genetic tractability to experimentally dissect the costs of cooperation for this greenbeard system.
The greenbeard-like recognition intrinsic to agrobacterial pathogenesis restricts the availability of opines as a nutritional source. However, once opines become available in the disease environment, there is opportunity for selection favouring cheating agrobacterial genotypes that do not pay the substantial costs of being cooperative but are able to access its benefits (figure 5). These cheaters may either arise de novo within the disease environment or colonize from outside sources. The high cost of expressing vir genes (figure 5) establishes a strong selective pressure favouring mutants that suppress expression of these genes, while maintaining the ability to catabolize opines using other plasmid genes. Many natural isolates of agrobacteria harbour opine catabolic plasmids, which confer the ability to degrade opines but not the ability to infect host plants [13,14]. In addition to these agrobacterial cheaters, several non-agrobacterial soil microbes have also evolved the ability to degrade opines [14,48]. Thus, virulent agrobacteria face both intraspecific and interspecific competition for opine resources from agrobacterial cheaters and non-agrobacterial opine catabolizers, respectively, in the soil.
Genes underlying public good cooperation are overrepresented on mobile genetic elements [49]. One possible explanation for this is that horizontal transmission of these genes into non-cooperative individuals helps stabilize cooperation by forcing potential freeloaders to bear the costs of cooperation [50]. This is akin to the idea that the horizontal transmission of public good cooperation genes effectively increases the relatedness at these loci by causing neighbouring cells to share these genes [49,51,52]. Because chromosomal genes are only vertically transmitted, they do not experience this effect; consequently, chromosomal- and plasmid-borne genes are expected to be in conflict over how cooperatively the cell should behave [49]. Interestingly, the ability of conjugation to force agrobacterial cheaters to cooperate [50] is constrained by the fact that these avirulent, opine catabolic mutants carry an incompatibile plasmid that can prevent the cooperative plasmid from being introduced by conjugation [53]. Thus, the persistence of virulent agrobacteria despite these potential competitors suggests that crown gall communities are sufficiently spatially structured such that they are able to at least transiently access the opine benefits of pathogenesis such that the trait can be favoured by kin selection [11,53].
The ability to infect hosts often comes at a cost for pathogens [50,54]. This cost can manifest itself in many ways, including the expression of virulence factors. Interestingly, many forms of virulence involve cooperative behaviour on the part of infecting pathogens [50,55]. This high cost, coupled with the opportunity of freeloading, establishes a selective pressure favouring cheating strains that avoid the costs of pathogenesis but are able to reap its benefits. Consistent with this, Fortin et al. [56] reported the origin and rapid spread of avirulent mutants in laboratory cultures of A. tumefaciens growing under vir-inducing conditions. In some cases, these avirulent mutants no longer harboured the Ti plasmid present in the parent strain, while others appeared to contain a modified form of the plasmid that had undergone deletion events that abolish or diminish the ability to respond to the plant phenolic cues that trigger wild-type vir gene expression [56]. Similar observations have also been made with some agrobacterial strains growing in the presence of infected apple trees [57].
Several studies indicate that selection favouring cheaters can drive the evolution of less virulent Pseudomonas aeruginosa strains that fail to produce at least some important virulence factors [58–60]. Moreover, several studies have found that avirulent E. coli and Shigella flexneri strains often arise and spread within populations growing under conditions inducing virulence expression [61,62]. These avirulent mutants can result from loss of the entire plasmid [61], deletion of virulence-determining regions of the plasmid [61] or integration of the plasmid into the chromosome, which results in lower expression of virulence genes [62]. Interestingly, the Shigella virulence plasmid counteracts the selective pressure favouring the loss of the plasmid using a toxin-antidote system that leads to the death of daughter cells that do not inherit the plasmid [63]. These results parallel the observation that the Ti plasmid is rapidly lost or undergoes virulence-disabling deletion events under laboratory conditions [56]. Our results suggest that at least one source of the selective pressure favouring the spread of these avirulent mutants stems from the high costs associated with the expression of the genes underlying pathogenesis.
The population dynamics and maintenance of bacterial plasmids depend on the costs they impose and benefits they confer on the cells that host them. The costs (figures 2 and 5) and benefits [42] associated with the A. tumefaciens Ti plasmid, as with other plasmids, are environmentally context-dependent. Consequently, the outcome of competition between pTi+ and pTi− agrobacteria varies with the environmental conditions in which they are competing. This genotype-by-genotype-by-environment interaction suggests that the Ti plasmid may be subject to selective pressures that vary over space and time. Because of the benefits of opine catabolism, disease environments are likely to be sources for pathogenic agrobacteria [42]. In contrast, non-disease environments are predicted to be sinks for virulent agrobacteria owing to Ti-plasmid-associated costs when resources are limiting.
Acknowledgements
We thank Anna Larimer, Elise Morton, Sean Curtis, Peter Merritt and Mike Hibbing for conceptual feedback or technical assistance throughout this study. Helpful conversations with Stephen Farrand, Spencer Hall, Michael Hynes and Jeff Smith also improved this paper. T.G.P. was supported by the NIH Genetics, Cellular and Molecular Sciences Training Grant (GM007757), and this work was funded by the NSF (DEB-0608155) and the NIH (R01 GM092660).
References
- 1.Funnell B. E., Phillips G. J. 2004. Plasmid biology. Washington, DC: ASM Press [Google Scholar]
- 2.Slater F. R., Bailey M. J., Tett A. J., Turner S. L. 2008. Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol. Ecol. 66, 3–13 10.1111/j.1574-6941.2008.00505.x (doi:10.1111/j.1574-6941.2008.00505.x) [DOI] [PubMed] [Google Scholar]
- 3.Ebersbach G., Gerdes K. 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39, 453–479 10.1146/annurev.genet.38.072902.091252 (doi:10.1146/annurev.genet.38.072902.091252) [DOI] [PubMed] [Google Scholar]
- 4.Dahlberg C., Chao L. 2003. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165, 1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouma J. E., Lenski R. E. 1988. Evolution of a bacteria/plasmid association. Nature 335, 351–352 10.1038/335351a0 (doi:10.1038/335351a0) [DOI] [PubMed] [Google Scholar]
- 6.Modi R. I., Adams J. 1991. Coevolution in bacterial plasmid populations. Evolution 45, 656–667 10.2307/2409918 (doi:10.2307/2409918) [DOI] [PubMed] [Google Scholar]
- 7.Bergstrom C. T., Lipsitch M., Levin B. R. 2000. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155, 1505–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams J., Kinney T., Thompson S., Rubin L., Helling R. B. 1979. Frequency-dependent selection for plasmid-containing cells of Escherichia coli. Genetics 91, 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis R. J., Lilley A. K., Lacey S. J., Murrell D., Godfray H. C. J. 2007. Frequency-dependent advantages of plasmid carriage by Pseudomonas in homogeneous and spatially structured environments. ISME J. 1, 92–95 10.1038/ismej.2007.11 (doi:10.1038/ismej.2007.11) [DOI] [PubMed] [Google Scholar]
- 10.Escobar M. A., Dandekar A. M. 2003. Agrobacterium tumefaciens as an agent of disease. Trends Plant Sci. 8, 380–386 10.1016/S1360-1385(03)00162-6 (doi:10.1016/S1360-1385(03)00162-6) [DOI] [PubMed] [Google Scholar]
- 11.Platt T. G., Bever J. D. 2009. Kin competition and the evolution of cooperation. Trends Ecol. Evol. 24, 370–377 10.1016/j.tree.2009.02.009 (doi:10.1016/j.tree.2009.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner A., West S. A. 2010. Greenbeards. Evolution 64, 25–38 10.1111/j.1558-5646.2009.00842.x (doi:10.1111/j.1558-5646.2009.00842.x) [DOI] [PubMed] [Google Scholar]
- 13.Merlo D. J., Nester E. W. 1977. Plasmids in avirulent strains of Agrobacterium. J. Bacteriol. 129, 76–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nautiyal C. S., Dion P. 1990. Characterization of the opine utilizing microflora associated with samples of soil and plants. Appl. Environ. Microbiol. 56, 2576–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- 16.Fuqua W. C., Winans S. C. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176, 2796–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mersereau M., Pazour G. J., Das A. 1990. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90, 149–151 10.1016/0378-1119(90)90452-W (doi:10.1016/0378-1119(90)90452-W) [DOI] [PubMed] [Google Scholar]
- 18.Tempe J., Petit A., Holsters M., Montagu M. V., Schell J. 1977. Thermosensitive step associated with transfer of Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc. Natl Acad. Sci. USA 74, 2848–2849 10.1073/pnas.74.7.2848 (doi:10.1073/pnas.74.7.2848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt P. A., Danhorn T., Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189, 8005–8014 10.1128/JB.00566-07 (doi:10.1128/JB.00566-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uraji M., Suzuki K., Yoshida K. 2002. A novel plasmid curing method using incompatibility of plant pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes Genet. Syst. 77, 1–9 10.1266/ggs.77.1 (doi:10.1266/ggs.77.1) [DOI] [PubMed] [Google Scholar]
- 21.Cevallos M. A., Cervantes-Rivera R., Gutierrez-Rios R. M. 2008. The repABC plasmid family. Plasmid 60, 19–37 10.1016/j.plasmid.2008.03.001 (doi:10.1016/j.plasmid.2008.03.001) [DOI] [PubMed] [Google Scholar]
- 22.Hynes M. F., Simon R., Puhler A. 1985. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid 13, 99–105 10.1016/0147-619X(85)90062-9 (doi:10.1016/0147-619X(85)90062-9) [DOI] [PubMed] [Google Scholar]
- 23.Winans S. C. 1992. Two-way chemical signaling in Agrobacterium–plant interactions. Microbiol. Rev. 56, 12–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- 25.Anand V. K., Heberlein G. T. 1977. Crown gall tumorigenesis in potato tuber tissue. Am. J. Bot. 64, 153–158 10.2307/2442102 (doi:10.2307/2442102) [DOI] [Google Scholar]
- 26.Zhu J., Chai Y. R., Zhong Z. T., Li S. P., Winans S. C. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69, 6949–6953 10.1128/AEM.69.11.6949-6953.2003 (doi:10.1128/AEM.69.11.6949-6953.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Gelder L., Ponciano J. M., Joyce P., Top E. M. 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153, 452–463 10.1099/mic.0.2006/001784-0 (doi:10.1099/mic.0.2006/001784-0) [DOI] [PubMed] [Google Scholar]
- 28.Petersen A., Aarestrup F. M., Olsen J. E. 2009. The in vitro fitness cost of antimicrobial resistance in Escherichia coli varies with the growth conditions. FEMS Microbiol. Lett. 299, 53–59 10.1111/j.1574-6968.2009.01734.x (doi:10.1111/j.1574-6968.2009.01734.x) [DOI] [PubMed] [Google Scholar]
- 29.Turner P. E. 2004. Phenotypic plasticity in bacterial plasmids. Genetics 167, 9–20 10.1534/genetics.167.1.9 (doi:10.1534/genetics.167.1.9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dionisio F., Conceicao I. C., Marques A. C. R., Fernandes L., Gordo I. 2005. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 1, 250–252 10.1098/rsbl.2004.0275 (doi:10.1098/rsbl.2004.0275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas C. M. 2000. Paradigms of plasmid organization. Mol. Microbiol. 37, 485–491 10.1046/j.1365-2958.2000.02006.x (doi:10.1046/j.1365-2958.2000.02006.x) [DOI] [PubMed] [Google Scholar]
- 32.Turner P. E., Cooper V. S., Lenski R. E. 1998. Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52, 315–329 10.2307/2411070 (doi:10.2307/2411070) [DOI] [PubMed] [Google Scholar]
- 33.Christie P. J. 2004. The Agrobacterium Ti plasmids. In Plasmid biology (eds Funnell B. E., Phillips G. J.), pp. 455–472 Washington, DC: ASM Press [Google Scholar]
- 34.Nordstrom K., Austin S. J. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23, 37–69 10.1146/annurev.ge.23.120189.000345 (doi:10.1146/annurev.ge.23.120189.000345) [DOI] [PubMed] [Google Scholar]
- 35.Pappas K. M., Winans S. C. 2003. The RepA and RepB autorepressors and TraR play opposing roles in the regulation of a Ti plasmid repABC operon. Mol. Microbiol. 49, 441–455 10.1046/j.1365-2958.2003.03560.x (doi:10.1046/j.1365-2958.2003.03560.x) [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto S., Kiyokawa K., Tanaka K., Moriguchi K., Suzuki K. 2009. Novel toxin-antitoxin system composed of serine protease and AAA-ATPase homologues determines the high level of stability and incompatibility of the tumor-inducing plasmid pTiC58. J. Bacteriol. 191, 4656–4666 10.1128/jb.00124-09 (doi:10.1128/jb.00124-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayes F. 2003. Toxins–antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301, 1496–1499 10.1126/science.1088157 (doi:10.1126/science.1088157) [DOI] [PubMed] [Google Scholar]
- 38.Haft R. J. F., Mittler J. E., Traxler B. 2009. Competition favours reduced cost of plasmids to host bacteria. ISME J. 3, 761–769 10.1038/ismej.2009.22 (doi:10.1038/ismej.2009.22) [DOI] [PubMed] [Google Scholar]
- 39.Christie P. J., Cascales E. 2005. Structural and dynamic properties of bacterial type IV secretion systems. Mol. Membr. Biol. 22, 51–61 10.1080/09687860500063316 (doi:10.1080/09687860500063316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho H. B., Winans S. C. 2005. VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc. Natl Acad. Sci. USA 102, 14 843–14 848 10.1073/pnas.0503458102 (doi:10.1073/pnas.0503458102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pappas K. M. 2008. Cell–cell signaling and the Agrobacterium tumefaciens Ti plasmid copy number fluctuations. Plasmid 60, 89–107 10.1016/j.plasmid.2008.05.003 (doi:10.1016/j.plasmid.2008.05.003) [DOI] [PubMed] [Google Scholar]
- 42.Platt T. G., Fuqua C., Bever J. D. Submitted Resource and competitive dynamics shape the benefits of public goods cooperation in a plant pathogen. [DOI] [PMC free article] [PubMed]
- 43.Burr T. J., Katz B. H., Bishop A. L. 1987. Populations of Agrobacterium in vineyard and non-vineyard soils and grape roots in vineyards and nurseries. Plant Dis. 71, 617–620 10.1094/PD-71-0617 (doi:10.1094/PD-71-0617) [DOI] [Google Scholar]
- 44.Bouzar H., Moore L. W. 1987. Isolation of different Agrobacterium biovars from a natural oak savanna and tallgrass prairie. Appl. Environ. Microbiol. 53, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brencic A., Eberhard A., Winans S. C. 2004. Signal quenching, detoxification and mineralization of vir gene-inducing phenolics by the VirH2 protein of Agrobacterium tumefaciens. Mol. Microbiol. 51, 1103–1115 10.1046/j.1365-2958.2003.03887.x (doi:10.1046/j.1365-2958.2003.03887.x) [DOI] [PubMed] [Google Scholar]
- 46.Guyon P., Petit A., Tempe J., Dessaux Y. 1993. Transformed plants producing opines specifically promote growth of opine-degrading agrobacteria. Mol. Plant Microbe Interact. 6, 92–98 10.1094/MPMI-6-092 (doi:10.1094/MPMI-6-092) [DOI] [Google Scholar]
- 47.Savka M. A., Farrand S. K. 1997. Modification of rhizobacterial populations by engineering bacterium utilization of a novel plant-produced resource. Nat. Biotechnol. 15, 363–368 10.1038/nbt0497-363 (doi:10.1038/nbt0497-363) [DOI] [PubMed] [Google Scholar]
- 48.Nautiyal C. S., Dion P., Chilton W. S. 1991. Mannopine and mannopinic acid as substrates for Arthrobacter sp. strain MBA209 and Pseudomonas putida NA513. J. Bacteriol. 173, 2833–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nogueira T., Rankin D. J., Touchon M., Taddei F., Brown S. P., Rocha E. P. C. 2009. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr. Biol. 19, 1683–1691 10.1016/j.cub.2009.08.056 (doi:10.1016/j.cub.2009.08.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith J. 2001. The social evolution of bacterial pathogenesis. Proc. R. Soc. Lond. B 268, 61–69 10.1098/rspb.2000.1330 (doi:10.1098/rspb.2000.1330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rankin D. J., Rocha E. P. C., Brown S. P. 2011. What traits are carried on mobile genetic elements, and why? Heredity 106, 1–10 10.1038/hdy.2010.24 (doi:10.1038/hdy.2010.24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rankin D. J., Mc Ginty S. E., Nogueira T., Touchon M., Taddei F., Rocha E. P. C., Brown S. P. 2011. Bacterial cooperation controlled by mobile elements: kin selection and infectivity are part of the same process. Heredity 107, 279–281 10.1038/hdy.2011.59 (doi:10.1038/hdy.2011.59) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mc Ginty S. E., Rankin D. J., Brown S. P. 2011. Horizontal gene transfer and the evolution of bacterial cooperation. Evolution 65, 21–32 10.1111/j.1558-5646.2010.01121.x (doi:10.1111/j.1558-5646.2010.01121.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.West S. A., Diggle S. P., Buckling A., Gardner A., Griffins A. S. 2007. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38, 53–77 10.1146/annurev.ecolsys.38.091206.095740 (doi:10.1146/annurev.ecolsys.38.091206.095740) [DOI] [Google Scholar]
- 55.Brown S. P., Hochberg M. E., Grenfell B. T. 2002. Does multiple infection select for raised virulence? Trends Microbiol. 10, 401–405 10.1016/S0966-842X(02)02413-7 (doi:10.1016/S0966-842X(02)02413-7) [DOI] [PubMed] [Google Scholar]
- 56.Fortin C., Nester E. W., Dion P. 1992. Growth inhibition and loss of virulence in cultures of Agrobacterium tumefaciens treated with acetosyringone. J. Bacteriol. 174, 5676–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Belanger C., Canfield M. L., Moore L. W., Dion P. 1995. Genetic analysis of nonpathogenic Agrobacterium tumefaciens mutants arising in crown gall tumors. J. Bacteriol. 177, 3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandoz K. M., Mitzimberg S. M., Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl Acad. Sci. USA 104, 15 876–15 881 10.1073/pnas.0705653104 (doi:10.1073/pnas.0705653104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diggle S. P., Griffin A. S., Campbell G. S., West S. A. 2007. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411–417 10.1038/nature06279 (doi:10.1038/nature06279) [DOI] [PubMed] [Google Scholar]
- 60.Kohler T., Buckling A., van Delden C. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl Acad. Sci. USA 106, 6339–6344 10.1073/pnas.0811741106 (doi:10.1073/pnas.0811741106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuch R., Maurelli A. T. 1997. Virulence plasmid instability in Shigella flexneri 2a is induced by virulence gene expression. Infect. Immun. 65, 3686–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zagaglia C., Casalino M., Colonna B., Conti C., Calconi A., Nicoletti M. 1991. Virulence plasmids of enteroinvasive Escherichia coli and Shigella flexneri integrate into a specific site on the host chromosome: integration greatly reduces expression of plasmid-carried virulence genes. Infect. Immun. 59, 792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sayeed S., Brendler T., Davis M., Reaves L., Austin S. 2005. Surprising dependence on postsegregational killing of host cells for maintenance of the large virulence plasmid of Shigella flexneri. J. Bacteriol. 187, 2768–2773 10.1128/JB.187.8.2768-2773.2005 (doi:10.1128/JB.187.8.2768-2773.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]