Abstract
Adaptive radiations have helped shape how we view animal speciation, particularly classic examples such as Darwin's finches, Hawaiian fruitflies and African Great Lakes cichlids. These ‘island’ radiations are comparatively recent, making them particularly interesting because the mechanisms that caused diversification are still in motion. Here, we identify a new case of a recent bird radiation within a continentally distributed species group; the capuchino seedeaters comprise 11 Sporophila species originally described on the basis of differences in plumage colour and pattern in adult males. We use molecular data together with analyses of male plumage and vocalizations to understand species limits of the group. We find marked phenotypic variation despite lack of mitochondrial DNA monophyly and few differences in other putatively neutral nuclear markers. This finding is consistent with the group having undergone a recent radiation beginning in the Pleistocene, leaving genetic signatures of incomplete lineage sorting, introgressive hybridization and demographic expansions. We argue that this apparent uncoupling between neutral DNA homogeneity and phenotypic diversity is expected for a recent group within the framework of coalescent theory. Finally, we discuss how the ecology of open habitats in South America during the Pleistocene could have helped promote this unique and ongoing radiation.
Keywords: hybridization, Neotropical birds, phenotypic divergence, recent radiation, speciation
1. Introduction
Bird adaptive radiations have contributed greatly to our understanding of the process of speciation. Morphological innovations under specific ecological conditions have fuelled radiations like those of the drepanidine Hawaiian honeycreepers [1], while sexual selection may have contributed to forming speciose groups such as the Dendroica wood warblers [2]. These, as well as many other celebrated examples (e.g. Phylloscopus warblers [3]; Diglossa flowerpiercers [4]) represent radiations that have accrued species over millions of years. There are comparatively far fewer known monophyletic assemblages of bird species originating recently over a short time span; perhaps, the most notable cases include Darwin's finches [5], the red crossbills of North America [6], African indigobirds [7] and Neotropical cardueline finches [8]. These examples, together with other recent vertebrate radiations such as that of the cichlid fishes from the African Great Lakes [9], do not fully comply with the Biological Species Concept and thus provide key insight as to how permeable mechanisms of reproductive isolation and ultimately introgressive hybridization might contribute to the generation of biodiversity [10–13]. Clearly, additional examples of recent radiations in different ecological and historical contexts will provide further insights into the mechanisms that underpin diversification and speciation.
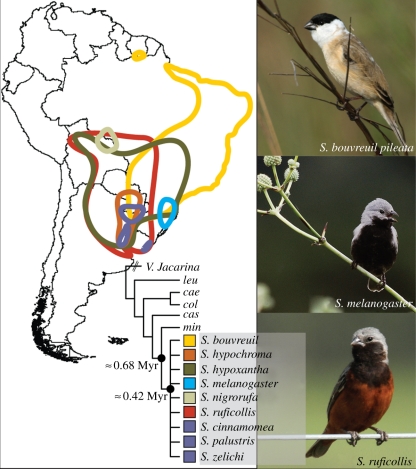
In this study, we identify a novel case of a recent continental bird radiation within the taxonomically complex Neotropical genus Sporophila. The capuchinos, as they are known colloquially, comprise a group of 11 seedeaters that are smaller than their congeners and characterized by cinnamon-based plumage colour patterns in reproductive males [14]. Capuchinos are sexually dimorphic, with divergent plumage patterns among males, while females are mostly brown and olive with little apparent distinction among them. Capuchino species are thus diagnosed based on male nuptial plumage, because they otherwise show little differentiation in size and shape [14]. Typically capuchino species are broadly sympatric (or even syntopic) with several other species of the group [14]. Many are now rare, having limited geographical ranges with populations in decline owing to habitat loss and trapping for the pet trade [15]. Most capuchinos are seasonal migrants, but little is known of the location of their wintering grounds. When not breeding, they are commonly seen in mixed flocks showing similar foraging behaviour and in some cases eclipse plumage [14]. Phylogenetic analyses of mitochondrial DNA (mtDNA) sequence data suggest that the group is monophyletic and indicate that two of the 11 species (Sporophila minuta and Sporophila castaneiventris) are clearly diverged from the other members of the clade [16,17]. However, the remaining nine species (Sporophila bouvreuil, Sporophila cinnamomea, Sporophila hypochroma, Sporophila hypoxantha, Sporophila melanogaster, Sporophila nigrorufa, Sporophila palustris, Sporophila ruficollis and Sporophila zelichi) exhibit extremely low interspecific sequence divergence and apparent lack of reciprocal monophyly. These species are found predominantly south of the Amazon River and hereafter we refer to them as the southern capuchinos (see figure 1 and electronic supplementary material, figure S1 for representative illustrations and range maps).
Figure 1.
Approximate reproductive distribution map for the nine species of southern capuchinos (following the study of Ridgely & Tudor [14] and BirdLife International [15]), examples of adult male plumage for three species, and schematic representation of phylogenetic affinities with other Sporophila species. Distributions for S. cinnamomea, S. palustris and S. zelichi were considered equal for simplicity of rendering the map. Outgroup species names abbreviated as shown in figure 3. Myr, million years.
Funk & Omland [18] and more recently McKay & Zink [19] examined the most commonly cited causes of lack of reciprocal monophyly at mitochondrial loci in animals, identifying imperfect taxonomy as the most frequent, a problem that may be even more severe in groups such as invertebrates that have received less attention. Other major causes are inadequate phylogenetic resolution (caused by too few phylogenetically informative characters or homoplasy), incomplete lineage sorting and interspecific hybridization. We use this conceptual framework to understand the processes responsible for the mitochondrial genetic pattern observed among the southern capuchinos [16,17], augmenting previous genetic analyses to include additional mitochondrial regions and both nuclear DNA microsatellites and sequences. We expand the analyses to include objective quantification of plumage and song traits among males, two key components of bird mate recognition systems [20]. We find marked differences in colour and song among southern capuchino species, suggesting that their current designations are not artefacts of taxonomic practice. This result contrasts with the lack of neutral genetic differentiation among species. Overall, our genetic data imply that the southern capuchinos began to radiate within the Pleistocene but that the process is ongoing, consistent with demographic expansions from small ancestral populations and showing evidence of both incomplete lineage sorting and gene flow between species.
2. Material and methods
(a). Coloration analyses
The distinct differences in reproductive male plumage patterns among southern capuchinos provide the main suite of characters used to diagnose the nine currently recognized species. Because signals can only be understood with reference to the natural receiver, the differences in human and avian visual systems and colour perception can make human description of plumage colour inadequate for the study of many biological questions (see Benites et al. [21] and references therein). Thus, using species designations as a working hypothesis, we objectively quantified relative plumage differences (within the southern capuchinos and with respect to three outgroup species) using an avian visual perspective as modelled by Vorobyev & Osorio [22]. Data were collected from adult male museum study skins: 104 individuals from seven southern capuchino species and 61 specimens from three non-capuchino Sporophila species (see electronic supplementary material, table S1). Plumage coloration reflectances were measured with a spectrometer on seven patches: back, belly, chest, crown, nape, throat and rump. A given colour stimulus can be described by the quantum catches of the light entering the eye by each receptor type present in the avian retina (Q1 = ultraviolet wavelength-sensitive; Q2 = short wavelength-sensitive; Q3 = medium wavelength-sensitive; and Q4 = long wavelength-sensitive). Colour differences were evaluated for each plumage region by estimating a distance in avian perceptual colour space (ΔS), representing the value by which any two plumage colour patches differ based on the quantum catches of each cone receptor and their respective noise-to-signal ratio [22]. Since neither spectral sensitivity data nor cone cell type proportions are available for any of the Sporophila species, the blue tit (Parus caeruleus) was used as a representative passerine visual system (for justifications see the electronic supplementary material, S1 materials and methods). We calculated ΔS between identical patches of each pair of species and values were averaged across the seven measured patches to obtain a global ΔS representing overall plumage differences between taxon pairs. A neighbour-joining (NJ) tree was built from average pairwise ΔS distances, a strategy used throughout this study to clearly display pairwise distance matrices. A ΔS value of 1.0 ‘just noticeable difference’ (jnd) is taken as a general threshold value for the discrimination of two colours [21,22]. We also assessed relative plumage differences among taxa as well as the contribution of different spectral regions through a stepwise discriminant function analysis (DFA). A pairwise distance matrix between species was constructed using Euclidean distances between centroids in the space defined by the first two DFA functions. Statistical differences in DFA1 scores between pairs of taxa were assessed using multiple Mann–Whitney U-tests with sequential Bonferroni corrections [23]. Finally, DFA results were compared with those obtained from a principal components analysis (PCA), which requires no a priori designations of species based on human perception. For details see the electronic supplementary material, S1 materials and methods.
(b). Song analyses
Our song dataset (electronic supplementary material, table S2) included vocalizations belonging to 131 individuals from 10 capuchino species and 30 individuals from three non-capuchino Sporophila species. We measured 14 variables for all songs and differences between species were assessed through a stepwise DFA, predicting group membership for each individual through a jackknifed classification procedure. We also classified syllables into six categories and calculated the overall proportion of each category used in the song of every species. Individual variation was assessed using a modified Jaccard similarity index [24]. We calculated the mean among and within species values for the similarity index as well as 95% confidence intervals (CI). Transforming the index by 1 − x, we generated a species pairwise distance matrix (0 now representing identical syllable use). For details see the electronic supplementary material, S1 materials and methods.
(c). Genetic analyses
All 11 capuchinos were included in our genetic analysis and we used four other sympatric species as outgroups. When available, we included multiple individuals per species and from as many localities of their geographical distribution as possible. Our initial dataset included samples for which we previously obtained cytochrome c oxidase I (COI) sequences [17] to which we added cytochrome b (Cyt b) and mitochondrial control region (CR) data. For analyses based on the frequency of microsatellite DNA alleles, we increased sample sizes by including newly collected specimens and also museum study skins. Several nuclear markers were amplified with the objective of finding fixed single nucleotide polymorphisms between southern capuchino species. Details are provided in the electronic supplementary material, S1 materials and methods and table S3.
We constructed phylogenetic trees with MrBayes v. 3.1.2 [25,26] using concatenated mtDNA sequence data and used haplotype frequencies between southern capuchinos to estimate pairwise FST values for each gene with Arlequin v. 3.5.1.2 [27]. Significance was tested through 1000 random permutations with sequential Bonferroni corrections [23]. Pairwise FST matrices were displayed using NJ trees. Node ages were estimated using time to most recent common ancestor (TMRCA) with the BEAUti/BEAST v. 1.4.8 package [28] and data from Cyt b. For DNA microsatellite loci, genetic differentiation between species was assessed using FST, Nei's standard genetic distance [29] and Structure v. 2.3.3 [30]. We also performed a principal coordinates analysis (PCoA) with GenAlEx v. 6 [31] using distances calculated between all individuals genotyped. The southern capuchinos did not show fixed sites in CHD1Z, Fib5, MUSK, Numt2 or Numt3; however, Numt2 provided good resolution with respect to the outgroups and was used further to build phylogenetic trees. We tested whether southern capuchinos differed in the haplotype frequencies of the nuclear loci sequenced. A combination of the number of segregating sites and our sample sizes made recovery of useful haplotypes from all markers low. To increase the recovery, we arbitrarily divided the sequence of each gene in half and inferred the haplotypes separately for both the 5′ and 3′ portions of each locus using DnaSP v. 5.10 [32]. Differences in haplotype frequencies between species were assessed using FST calculations in both the 5′ and 3′ fraction of each locus separately.
Mitochondrial and nuclear sequence data were used to explore the demographic history of the species in the southern capuchino clade. Our microsatellite markers showed evidence of linkage disequilibrium and deviation from Hardy–Weinberg expectations (see the electronic supplementary material, S1 materials and methods) and were excluded from these analyses. We tested for demographic expansions or contractions by using Fu's F-test [33] and by calculating the exponential growth parameter g [34]. Splitting times between pairs of species, migration (introgression), and ancestral and current effective population sizes were estimated using IMa2 [35]. More information is provided in the electronic supplementary material, S1 material and methods.
3. Results
(a). Male southern capuchinos show significant phenotypic differences in coloration patterns and song
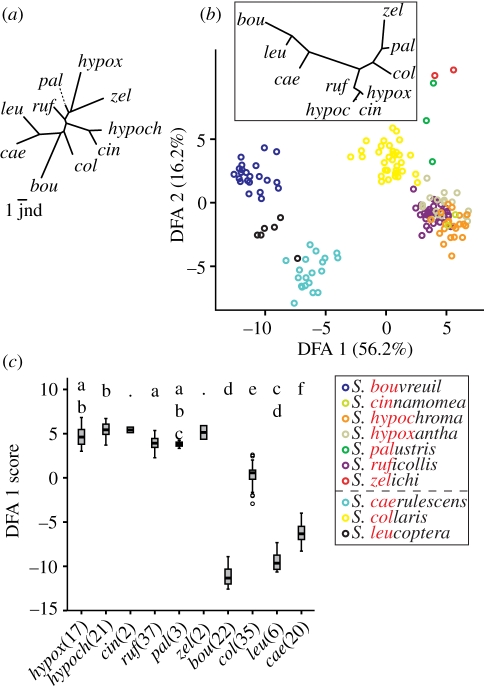
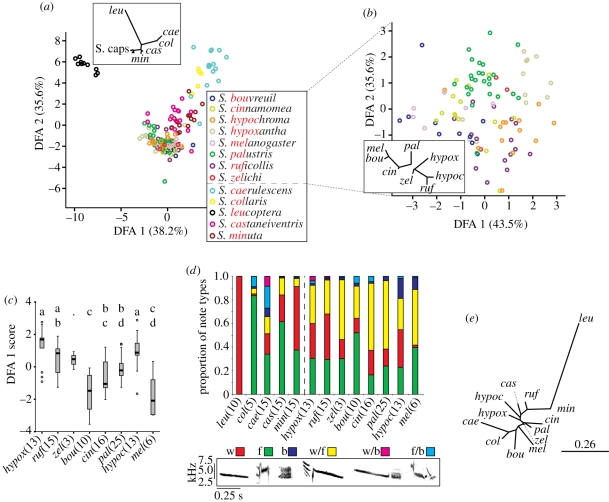
Seven southern capuchino and three other Sporophila species analysed differ markedly in plumage coloration. The average ΔS values (metric of colour differences in avian perceptual colour space) between pairs of species were in all cases above the discernable threshold according to the avian visual model used (figure 2a), suggesting that there are large overall differences in plumage among southern capuchino males. Moreover, males differed significantly in virtually every patch considered (electronic supplementary material, table S4). Interestingly, some southern capuchino males are more similar in plumage attributes to outgroup species than to members of their own clade (figure 2a). Statistically significant differences in plumage coloration among the species surveyed were also found through DFA (Wilk's λ = 1.26 × 10−6, χ1262 = 1888.53, p < 0.0001; figure 2b). Factor loadings for the 14 out of 28 variables retained by the stepwise DFA procedure (electronic supplementary material, table S5) suggest that every portion of the light spectrum and every body part measured (except the rump) accounts for differences among species, with the belly and crown contributing most strongly. Statistically significant differences between southern capuchino species were found in average DFA1 scores (figure 2c). Finally, we obtained similar results when plumage coloration differences were assessed at the individual level through PCA (electronic supplementary material, figure S2).
Figure 2.
Differences in coloration patterns among taxa. (a) NJ tree constructed using average ΔS values between species pairs. (b) DFA with percentage of the total variance explained by each function in parenthesis on its corresponding axis. The inset is a NJ tree constructed from Euclidean distances between species centroids in the space of the first two DFA functions. Species are colour-coded and the three outgroups are separated by the dashed line. (c) Distribution of species DFA1 scores (sample sizes after abbreviated species names); those that share the same letter on the top of the panel do not show statistically significant differences. Dots imply no significant differences with any other taxa. All statistically significant comparisons: p < 0.001; except for S. caerulescens versus S. palustris where p < 0.01.
Songs of southern capuchino and other Sporophila species comprise distinct series of complex syllable types (electronic supplementary material, figure S3). Stepwise DFA (figure 3a) retained 13 out of 14 variables (see the electronic supplementary material, table S6 for factor loadings) and showed statistically significant differences among species (Wilk's λ = 3.36 × 10−4, χ1562 = 1175.82, p < 0.001). Our analysis distinguishes southern capuchino songs from those of S. castaneiventris and S. minuta (figure 3a) and more markedly from non-capuchino species. In comparison, song differences within the southern capuchinos are smaller with 66.5 per cent of cases reclassified to the correct species (41 of 54 misclassifications involving southern capuchinos). Despite this, significant differences are also found when outgroup species are excluded (Wilk's λ = 0.192, χ235 = 154.18, p<0.001; figure 3b). In this analysis performed only with the southern capuchinos, DFA1 represents variation in syllables that span a small range of frequencies, while DFA2 represents longer, more complex syllables that span a wide range of frequencies (see the electronic supplementary material, table S7). The cross-validation procedure had low success in reclassifying songs to the correct species (51.5% with a total of 49 errors). Although there is an overlap of attributes in southern capuchino vocalizations, clustering can be observed between songs from the same species (figure 3b), and significant differences in DFA1 scores exist among various species (figure 3c).
Figure 3.
Differences in song among taxa. (a) DFA for all taxa with a species centroid NJ tree in the inset (S. caps: Southern capuchinos). (b) DFA performed exclusively on the eight southern capuchinos (inset as above). (c) Distribution of species DFA1 scores for southern capuchinos. Statistically significant differences and other details as in figure 2c. Significant comparisons: p < 0.001; except for S. palustris versus S. bouvreuil, S. melanodera versus S, ruficollis and S. palustris versus S. hypoxantha, where p < 0.01. (d) Proportion of six syllable categories used by different species (sample sizes in parenthesis). Syllable examples in the lower panel (w: whistle; f: figure; b: buzz; and combinations of these). (e) NJ tree constructed from a matrix of 1 − x transformed average values of a modified Jaccard similarity index calculated based on syllable usage for species pairs. The scale bar represents the threshold of intraspecific variation (lower 95% CI limit averaged across all intraspecific comparisons).
Sporophila species differ in the use of the six syllable categories in their songs (figure 3d), with larger differences seen between southern capuchinos and outgroup species than within the former group. A modified Jaccard similarity index [24] calculated between every pair of songs allowed us to assess variation in syllable-type usage within and between species. The average intraspecific value was 0.87 (range: 0.74–1), with an average lower 95% CI limit of 0.84. Interspecific values ranged from 0.95 (S. palustris versus S. zelichi) to 0.67 (S. melanogaster versus S. ruficollis). Only the species pair for which our sample sizes were the smallest (S. collaris and S. zelichi) had a 95% CI that included 1, suggesting that all other comparisons differed significantly. Approximately half of the 95% CIs of pairwise inter-southern capuchino comparisons exceeded 0.84, the threshold for intraspecific variation in syllable usage (figure 3e).
(b). Low genetic differentiation between southern capuchino species
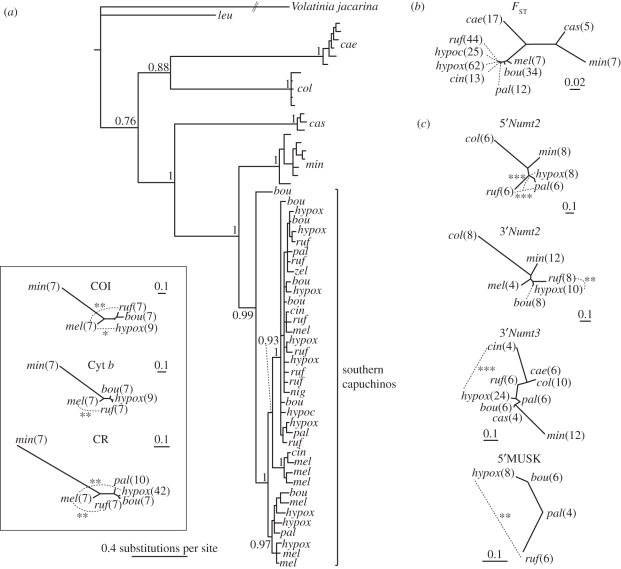
Both the overall capuchinos and southern capuchinos are clearly monophyletic with high support. Sporophila minuta is the sister species to the southern capuchino radiation. By contrast, none of the southern capuchino species is monophyletic for mtDNA, despite including the rapidly evolving CR in our analyses (figure 4a). Indeed, all analyses using a suite of neutral genetic markers reveal extremely low genetic differentiation among southern capuchinos. All southern capuchinos do show statistically significant pairwise FST values with respect to S. minuta (figure 4a; average of 0.89 for COI; 0.76 for Cyt b; and 0.74 for CR; p < 0.01). Sporophila melanogaster has low but significant FST values with two other southern capuchinos consistently across mitochondrial loci: Sporophila hypoxantha (COI: 0.24; and CR: 0.15) and Sporophila ruficollis (COI: 0.47; Cyt b: 0.24; and CR: 0.26).
Figure 4.
Neutral genetic differences among taxa. Species names abbreviated as in figure 3 (nig: S. nigrorufa). (a) Bayesian tree with posterior probabilities indicating node support (some omitted for clarity) based on mtDNA. When members of a species form a monophyletic clade, taxon names are indicated once for simplicity. The inset shows NJ trees built with pairwise FST matrices for each locus separately; sample sizes indicated in parenthesis. Statistically significant comparisons between southern capuchinos are indicated by dashed lines (*p < 0.05; **p < 0.01; ***p < 0.001). (b) NJ trees constructed with FST matrices calculated between Sporophila species; individuals where genotyped for six DNA microsatellite loci and sample sizes are indicated in parenthesis. (c) NJ trees built with FST matrices obtained from nuclear intron haplotype frequencies (only those showing statistically significant differences between southern capuchinos are presented); number of haplotypes used in parenthesis. Statistically significant comparisons as in (a).
For DNA microsatellites, PCoA reveal lack of genetic structure within the southern capuchinos (electronic supplementary material, figure S4). We found evidence of linkage disequilibrium between some loci (see the electronic supplementary material, S1 materials and methods), contra previous studies [36,37]. This observation might result from undetected allele dropout or null alleles. Despite these potential issues, our Structure Bayesian assignment test results corroborate those from the PCoA, implying that the southern capuchino samples most probably comprise a single genetic cluster (K = 1). FST values between southern capuchino species were always below 0.01 and not significantly different from zero (figure 4b). Even though sample sizes for outgroup species were low and homoplasy could have overwritten some genetic signal, we found nearly all FST and Nei's genetic distance values for comparisons involving southern capuchinos and outgroups, or between outgroups, to be statistically significant and higher than intra-southern capuchino comparisons (figure 4b and the electronic supplementary material, figure S4).
Southern capuchino species did not exhibit fixed diagnostic sites in the five nuclear markers used in this study and the Bayesian tree produced using Numt2 DNA sequence data did not resolve below the level of the capuchinos (electronic supplementary material, figure S5). However, frequency-based haplotype differences were evident and most comparisons involving southern capuchinos and outgroups showed high and statistically significant FST values. Some significant FST comparisons were also found between S. hypoxantha and S. ruficollis (5′Numt2: 0.29; 3′Numt2: 0.15; and 5′MUSK: 0.36), S. hypoxantha and S. cinnamomea (3′Numt3: 0.51), and S. palustris and S. ruficollis (5′Numt2: 0.40) (figure 4c).
(c). Demographic history of the southern capuchino clade
Origins of the southern capuchinos date to the Pleistocene. The calculated TMRCA using the coalescent program BEAST (applying a 2.1 per cent per million year divergence time for Cyt b [28,38]) suggests that the mitochondrial ancestor of all southern capuchinos is approximately 422 000 years old (95% high posterior density interval: 211 000–690 000). The age of the ancestor between the southern capuchino clade and its sister species S. minuta was estimated in about 681 000 years before present (95% high posterior density interval: 409 000–989 000). Our analyses using IMa2 [35] suggest that there is considerable gene flow among southern capuchinos (table 1). By contrast, there is practically no gene flow between the southern capuchino clade and its sister species S. minuta. The average splitting time between southern capuchinos calculated with IMa2 is approximately 110 000 years before present. The estimated splitting time between the southern capuchinos and S. minuta is approximately 140 000 years before present. These results must be interpreted with caution as parameters estimated using IMa2 relied on rough molecular clock calibrations and an approach that did not consider all southern capuchinos interacting at the same time in the model. As a consequence of these caveats, we consider that the most conservative interpretation of these results is to say that, overall, our data suggest that the southern capuchino radiation dates approximately to the period stretching from the middle Pleistocene to present. Average current effective population size estimates for southern capuchino species were approximately ninefold larger than the average ancestral population size (table 1), suggesting the possibility of demographic expansions. This is consistent with Fu's F-values, which were negative and statistically significant across markers for the southern capuchino clade itself, and for species for which sample sizes allowed us to perform the test (see the electronic supplementary material, table S8). The exponential growth parameter g was positive for the southern capuchino clade, S. hypoxantha and S. palustris, providing further evidence of demographic expansion during the southern capuchino radiation (see the electronic supplementary material, table S8).
Table 1.
Estimations of ancestral and current effective population sizes (Nancestral, NA and NB; in units of ×1000 individuals), splitting times (t; ×1000 years) and introgressive hybridization (m; number of effective migrants per year). (Parameters were estimated for pairs of species (or southern capuchinos grouped) and mean values as well as estimated 95% highest posterior density interval are shown.)
| A | B | NA | NB | Nancestral | t | m |
|---|---|---|---|---|---|---|
| S. minuta | S. caps | 101 (47–164) | 248 (171–331) | 26 (0–62) | 140 (97–185) | 0.1 (0–0.3) |
| S. hypoxantha | S. caps | 416 (142–769) | 461 (161–858) | 151 (50–267) | 101 (22–182) | 12 (1–27) |
| S. hypoxantha | S. ruficollis | 511 (60–1654) | 489 (41–1763) | 46 (3–108) | 134 (60–213) | 104 (4–201)a |
| S. hypoxantha | S. melanogaster | 267 (56–584) | 342 (33–1043) | 31 (0–85) | 59 (16–105) | 23 (0–70) |
| S. hypoxantha | S. palustris | 364 (114–679) | 947 (104–2333)b | 45 (4–104) | 153 (82–227) | 43 (4–97) |
| S. hypoxantha | S. bouvreuil | 485 (47–1502) | 360 (15–1674) | 59 (0–125) | 153 (51–265) | 56 (10–115) |
| S. ruficollis | S. melanogaster | 578 (11–1808) | 483 (15–1580) | 71 (0–120) | 51 (0–121) | 40 (0–126) |
| S. palustris | S. bouvreuil | 1029 (58–2473)b | 620 (36–2214) | 35 (0–93) | 136 (61–236) | 96 (0–214) |
aMultiple peaks in the posterior density curve.
bPosterior density reaches lower values but not 0 near the upper limit of the prior.
4. Discussion
We find extraordinary variation in phenotype (coloration and song) among southern capuchinos despite extremely low levels of neutral genetic differentiation and no species-level monophyly in any genetic marker assayed among any of the designated capuchino species. Our analyses are also consistent with demographic expansions in the clade as well as gene flow between some of its member species. Taken together, we suggest that this is a compelling example of an extremely rapid, recent and ongoing continental radiation, with species diverging in male plumage coloration patterns and song. Both incomplete lineage-sorting and introgressive hybridization appear to be responsible for the blurry genetic identity of these species, at least at neutral loci. Insufficient data as an explanation for lack of reciprocal monophyly between species [18] are not plausible as we see similar patterns across genetic markers with different modes of inheritance, mutation mechanisms and rates of divergence. Interestingly, the southern capuchinos add another example to the list of known rapid seedeater radiations [5–8], providing further support to the idea that finch-like birds may be particularly inclined to undergo such rapid bursts of speciation [20]. Below, we discuss our findings, arguing that the current practice of recognizing multiple southern capuchino species is correct, and make inferences about the origin of the group, as well as its future in relation to the main conservation threats it faces.
(a). Are the southern capuchinos incipient species?
The southern capuchinos could not be diagnosed using neutral markers, a situation similar to that of Darwin's ground finches [39,40]. This result is expected if speciation events were recent and close together in time and does not necessarily imply panmixia. This genetic pattern has been termed hemiplasy [41] and is the result of rapid bursts of speciation causing ancestral polymorphisms to persist across nodes in a species tree [42]. This imposes limitations on our ability to date speciation events that go beyond the technical difficulties related to the programs and molecular clock calibrations used. In this case, the age of the ancestor between the southern capuchinos and the sister species S. minuta would set the upper bound for speciation times (dating approximately to the middle Pleistocene). The lack of monophyly in neutral markers within species need not imply that these species are not valid biological entities. Differences in vocalizations, plumage or other morphological characters subjected to selection pressures (particularly in isolated populations) may evolve rapidly, creating a lag before the stochastic sorting of neutral molecular markers produces diagnosable lineages [19]. Thus, differences in genes that underpin these diagnostic phenotypic characters would correctly delimit species, while neutral markers show the signature of incomplete lineage sorting or evidence of introgression [43]. Genomic approaches such as amplified fragment length polymorphisms or next-generation sequencing will aid us in finding the crucial genes implicated in the evolution of the phenotypic differences among southern capuchinos.
Contrasting with lack of differentiation in neutral markers, we find striking diversity in male coloration pattern and song, suggesting that the existing species in the group are not a taxonomic artefact but a compelling example of an ongoing but incomplete radiation. This is further substantiated by cryptic plumage differences found between females in the group [21]. Playback experiments are required to test if the observed phenotypic differences result in assortative mating. Thus far, an analysis of aggressive response to playback between S. hypoxantha and S. palustris males suggests that at least some southern capuchinos distinguish between conspecific and heterospecific song, virtually ignoring the latter (P. Benites, L. Campagna & P. L. Tubaro 2010, unpublished data). Future effort should focus on the phenotypic differences among species and their possible role as mechanisms of reproductive isolation.
(b). Inferences from the past and predictions for the future
Our data are consistent with the southern capuchinos originating from small isolated populations, expanding later to occupy their current ranges. Range fragmentation, as a consequence of Late Pleistocene marine ingressions and egressions, accelerated by sexual selection has been proposed as a possible cause for the radiation of the southern capuchinos [16,44]. Alternatively, Fjeldså & Rahbek [45] propose that specific habitat requirements could have kept small local populations isolated in different patches of suitable grassland habitat. Palaeoclimatic reconstructions indicate that open habitats (savannah, Caatinga and Cerrado) dominated South America during the last glacial maximum (approx. 27 000 years before present), accompanied by a marked retraction of the rainforest [46]. There is evidence of south polar air incursions affecting temperatures and precipitation regimes during the various glacial cycles of the Quaternary, causing changes in the length of dry seasons and cycling between overall drier climate and humid conditions. These conditions are thought to have caused expansion and regression of open habitats and rainforest that continued into the Holocene [46–48]. The fluctuation in the prevalence of rainforest over open habitats and vice versa and the inter-digitation of these two biomes could have contributed to isolating small populations in islands of suitable habitat, or grassland refugia, in a scenario analogous to that proposed in the forest refugia hypothesis [49]. Indeed, the contraction of open habitats during the Holocene owing to the changes in forest cover is thought to have affected the composition of communities of savannah specialists, leading to extinctions of several large mammals [50]. Both marine ingressions and the existence of open habitat refugia could have contributed to isolating small ancestral populations of southern capuchinos, making it hard to distinguish these alternatives.
The future of the southern capuchinos is without doubt tied to human use of land. Expanding industrial agriculture is a great threat to grasslands and only small areas remain undisturbed [51], posing a risk to the highly sensitive southern capuchino species [52]. The fact that southern capuchinos have been forced into the remaining patches of suitable habitat carries potential consequences for the viability of small populations with putatively incomplete mechanisms of reproductive isolation [53], and the very real possibility of hybridization. Forced breeding in syntopy could increase opportunities for misimprinting on heterospecifics that would lead to eventual hybridization [54–56]. Moreover, theory predicts that hybridization will increase when species are forced together into small patches of habitat, with habitat disturbance and when potential conspecific mates are rare [57–59], as is now the case for many species of this group. The latter possibility also seems plausible because southern capuchinos differ in their abundance, with taxa like S. hypoxantha and S. ruficollis greatly outnumbering the rare S. palustris, S. melanogaster, S. nigrorufa or S. zelichi. Ultimately, the fate of this unique group of birds may not be as enigmatic as its origin.
Acknowledgements
Field work for this study was authorized by the National Fauna authorities of Argentina, the Argentine National Parks Administration and the Office of Fauna of the Province of Corrientes, Argentina.
For tissue loans, we are indebted to A. T. Peterson and M. B. Robbins (KUNHM); R. T. Brumfield and D. L. Dittmann (LSUMZ); K. Cook and M. Adams (BMNH); C. S. Fontana (MCP); J. Fjeldså and J. B. Kristensen (ZMUC); E. Bermingham (STRI); M. J. Braun and J. Dean (USNM). For authorizing permits in Argentina, we thank the National Fauna authorities, the National Parks Administration and the Office of Fauna of Corrientes. We thank T. Webber (FLMNH), T. Bishop (Macaulay Library, Cornell Laboratory of Ornithology), J. Mazar Barnett and R. Straneck for providing recordings. We thank the Willi Hennig Society for a free edition of TNT 1.1. This project was supported by grants to PLT from ANPCyT, Argentina (PICT 2004-16-25171 and 2010-0805); CONICET, Argentina (PIP 112-200801-00741); UBA, Argentina (UBACyT 2010-2012); IDRC, Canada, and Richard Lounsbery Foundation; and by a grant from NSERC, Canada to SCL. Photographs in figure 1 by R. Güller (S. bouvreuil pileata and S. ruficollis) and P. Fenalti (S. melanogaster). We thank T. Price and two anonymous reviewers for their helpful comments on previous versions of this manuscript.
References
- 1.Lovette I. J., Bermingham E., Ricklefs R. E. 2002. Clade-specific morphological diversification and adaptive radiation in Hawaiian songbirds. Proc. R. Soc. Lond. B 269, 37–42 10.1098/rspb.2001.1789 (doi:10.1098/rspb.2001.1789) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lovette I. J., Bermingham E. 1999. Explosive speciation in the New World Dendroica warblers. Proc. R. Soc. Lond. B 266, 1629–1636 10.1098/rspb.1999.0825 (doi:10.1098/rspb.1999.0825) [DOI] [Google Scholar]
- 3.Price T., Lovette I. J., Bermingham E., Gibbs H. L., Richman A. D. 2000. The imprint of history on communities of North American and Asian warblers. Am. Nat. 156, 354–367 10.1086/303397 (doi:10.1086/303397) [DOI] [PubMed] [Google Scholar]
- 4.Mauck W. M., Burns K. J. 2009. Phylogeny, biogeography, and recurrent evolution of divergent bill types in the nectar-stealing flowerpiercers (Thraupini: Diglossa and Diglossopis). Biol. J. Linn. Soc. Lond. 98, 14–28 10.1111/j.1095-8312.2009.01278.x (doi:10.1111/j.1095-8312.2009.01278.x) [DOI] [Google Scholar]
- 5.Grant P. R. 1999. The ecology and evolution of Darwin's finches. Princeton, NJ: Princeton University Press [Google Scholar]
- 6.Benkman C. W. 2003. Divergent selection drives the adaptive radiation of crossbills. Evolution 57, 1176–1181 10.1111/j.0014-3820.2003.tb00326.x (doi:10.1111/j.0014-3820.2003.tb00326.x) [DOI] [PubMed] [Google Scholar]
- 7.Sorenson M. D., Sefc K. M., Payne R. B. 2003. Speciation by host switch in brood parasitic indigobirds. Nature 424, 928–931 10.1038/nature01863 (doi:10.1038/nature01863) [DOI] [PubMed] [Google Scholar]
- 8.Arnaiz-Villena A., Álvarez-Tejado M., Ruiz-del-Valle V., García-de-la-Torre C., Varela P., Recio M. J., Ferre S., Martínez-Laso J. 1998. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell. Mol. Life Sci. 54, 1031–1041 10.1007/s000180050230 (doi:10.1007/s000180050230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer A. 1993. Phylogenetic relationships and evolutionary processes in East African cichlid fishes. Trends Ecol. Evol. 8, 279–284 10.1016/0169-5347(93)90255-N (doi:10.1016/0169-5347(93)90255-N) [DOI] [PubMed] [Google Scholar]
- 10.Grant B. R., Grant P. R. 1992. Hybridization of bird species. Science 256, 193–197 10.1126/science.256.5054.193 (doi:10.1126/science.256.5054.193) [DOI] [PubMed] [Google Scholar]
- 11.Dowling T. E., Demarais B. D. 1993. Evolutionary significance of introgressive hybridization in cyprinid fishes. Nature 362, 444–446 10.1038/362444a0 (doi:10.1038/362444a0) [DOI] [Google Scholar]
- 12.Dowling T. E., Secor C. L. 1997. The role of hybridization and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 28, 593–619 10.1146/annurev.ecolsys.28.1.593 (doi:10.1146/annurev.ecolsys.28.1.593) [DOI] [Google Scholar]
- 13.Seehausen O. 2004. Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 10.1016/j.tree.2004.01.003 (doi:10.1016/j.tree.2004.01.003) [DOI] [PubMed] [Google Scholar]
- 14.Ridgely R. S., Tudor G. 1989. The birds of South America The oscine passerines, vol. I Austin, TX: University of Texas Press [Google Scholar]
- 15.BirdLife International 2011. Species factsheets. IUCN Red List for birds. See http://www.birdlife.org
- 16.Lijtmaer D. A., Sharpe N. M. M., Tubaro P. L., Lougheed S. C. 2004. Molecular phylogenetics and diversification of the genus Sporophila (Aves: Passeriformes). Mol. Phylogenet. Evol. 33, 562–579 10.1016/j.ympev.2004.07.011 (doi:10.1016/j.ympev.2004.07.011) [DOI] [PubMed] [Google Scholar]
- 17.Campagna L., Lijtmaer D. A., Kerr K. C. R., Barreira A. S., Hebert P. D. N., Lougheed S. C., Tubaro P. L. 2010. DNA barcodes provide new evidence of a recent radiation in the genus Sporophila (Aves: Passeriformes). Mol. Ecol. Resour. 10, 449–458 10.1111/j.1755-0998.2009.02799.x (doi:10.1111/j.1755-0998.2009.02799.x) [DOI] [PubMed] [Google Scholar]
- 18.Funk D. J., Omland K. E. 2003. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 34, 397–423 10.1146/annurev.ecolsys.34.011802.132421 (doi:10.1146/annurev.ecolsys.34.011802.132421) [DOI] [Google Scholar]
- 19.McKay B. D., Zink R. M. 2010. The causes of mitochondrial DNA gene tree paraphyly in birds. Mol. Phylogenet. Evol. 54, 647–650 10.1016/j.ympev.2009.08.024 (doi:10.1016/j.ympev.2009.08.024) [DOI] [PubMed] [Google Scholar]
- 20.Price T. 2007. Speciation in birds. Englewood, CO: Roberts and Company [Google Scholar]
- 21.Benites P., Eaton M. D., Lijtmaer D. A., Lougheed S. C., Tubaro P. L. 2010. Analysis from avian visual perspective reveals plumage colour differences among females of capuchino seedeaters (Sporophila). J. Avian Biol. 41, 597–602 10.1111/j.1600-048X.2010.05205.x (doi:10.1111/j.1600-048X.2010.05205.x) [DOI] [Google Scholar]
- 22.Vorobyev M., Osorio D. 1998. Receptor noise as a determinant of colour thresholds. Proc. R. Soc. Lond. B 265, 351–358 10.1098/rspb.1998.0302 (doi:10.1098/rspb.1998.0302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice W. R. 1989. Analyzing tables of statistical tests. Evolution 43, 223–225 10.2307/2409177 (doi:10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- 24.Chao A., Chazdon R. L., Colwell R. K., Shen J. 2005. A new statistical approach for assessing compositional similarity based on incidence and abundance data. Ecol. Lett. 8, 148–159 10.1111/j.1461-0248.2004.00707.x (doi:10.1111/j.1461-0248.2004.00707.x) [DOI] [Google Scholar]
- 25.Huelsenbeck J. P., Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 10.1093/bioinformatics/17.8.754 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 26.Ronquist F., Huelsenbeck J. P. 2003. MrBayes v. 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 27.Excoffier L., Lischer H. E. L. 2010. Arlequin Suite v. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 10.1111/j.1755-0998.2010.02847.x (doi:10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 28.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei M. 1972. Genetic distance between populations. Am. Nat. 106, 283–292 10.1086/282771 (doi:10.1086/282771) [DOI] [Google Scholar]
- 30.Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peakall R., Smouse P. E. 2006. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295 10.1111/j.1471-8286.2005.01155.x (doi:10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Librado P., Rozas J. 2009. DnaSP v. 5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 10.1093/bioinformatics/btp187 (doi:10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- 33.Fu Y. X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhner M. K. 2006. LAMARC v. 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22, 768–770 10.1093/bioinformatics/btk051 (doi:10.1093/bioinformatics/btk051) [DOI] [PubMed] [Google Scholar]
- 35.Hey J. 2010. Isolation with migration models for more than two populations. Mol. Biol. Evol. 27, 905–920 10.1093/molbev/msp296 (doi:10.1093/molbev/msp296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croteau E. K., Lougheed S. C., Krannitz P. G., Mahony N. A., Walker B. L., Boag P. T. 2007. Genetic population structure of the sagebrush Brewer's sparrow, Spizella breweri breweri, in a fragmented landscape at the northern range periphery. Conserv. Genet. 8, 1453–1463 10.1007/s10592-007-9296-0 (doi:10.1007/s10592-007-9296-0) [DOI] [Google Scholar]
- 37.Campagna L., Geale K., Handford P., Lijtmaer D. A., Tubaro P. L., Lougheed S. C. 2011. A molecular phylogeny of the Sierra-finches (Phrygilus, Passeriformes): extreme polyphyly in a group of Andean specialists. Mol. Phylogenet. Evol. 61, 521–533 10.1016/j.ympev.2011.07.011 (doi:10.1016/j.ympev.2011.07.011) [DOI] [PubMed] [Google Scholar]
- 38.Weir J. T., Schluter D. 2008. Calibrating the avian molecular clock. Mol. Ecol. 17, 2321–2328 10.1111/j.1365-294X.2008.03742.x (doi:10.1111/j.1365-294X.2008.03742.x) [DOI] [PubMed] [Google Scholar]
- 39.Freeland J. R., Boag P. T. 1999. The mitochondrial and nuclear genetic homogeneity of the phenotypically diverse Darwin's ground finches. Evolution 53, 1553–1563 10.2307/2640900 (doi:10.2307/2640900) [DOI] [PubMed] [Google Scholar]
- 40.Sato A., O'hUigin C., Figueroa F., Grant P. R., Grant B. R., Tichy H., Klein J. 1999. Phylogeny of Darwin's finches as revealed by mtDNA sequences. Proc. Natl Acad. Sci. USA 96, 5101–5106 10.1073/pnas.96.9.5101 (doi:10.1073/pnas.96.9.5101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avise J. C., Robinson T. J. 2008. Hemiplasy: a new term in the lexicon of phylogenetics. Syst. Biol. 57, 503–507 10.1080/10635150802164587 (doi:10.1080/10635150802164587) [DOI] [PubMed] [Google Scholar]
- 42.Degnan J. H., Rosenberg N. A. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340 10.1016/j.tree.2009.01.009 (doi:10.1016/j.tree.2009.01.009) [DOI] [PubMed] [Google Scholar]
- 43.Wu C. I. 2001. The genic view of the process of speciation. J. Evol. Biol. 14, 851–865 10.1046/j.1420-9101.2001.00335.x (doi:10.1046/j.1420-9101.2001.00335.x) [DOI] [Google Scholar]
- 44.Nores M. 1989. Patrones de distribución y causas de especiación en aves argentinas. PhD thesis, Universidad Nacional de Córdoba, Córdoba, Argentina [Google Scholar]
- 45.Fjeldså J., Rahbek C. 2006. Diversification of tanagers, a species rich bird group, from lowlands to montane regions of South America. Integr. Comp. Biol. 46, 72–81 10.1093/icb/icj009 (doi:10.1093/icb/icj009) [DOI] [PubMed] [Google Scholar]
- 46.Clapperton C. M. 1993. Nature of environmental changes in South America at the last glacial maximum. Palaeogeogr. Palaeoclimatol. Palaeoecol. 101, 189–208 10.1016/0031-0182(93)90012-8 (doi:10.1016/0031-0182(93)90012-8) [DOI] [Google Scholar]
- 47.Servant M., Maley J., Turcq B., Absy M.-L., Brenac P., Fournier M., Ledru M. P. 1993. Tropical forest changes during the Late Quaternary in African and South American lowlands. Glob. Planet Change 7, 25–40 10.1016/0921-8181(93)90038-P (doi:10.1016/0921-8181(93)90038-P) [DOI] [Google Scholar]
- 48.Ledru M. P., Rousseau D. D., Cruz F. W., Riccomini C., Karmann I., Martin L. 2005. Paleoclimate changes during the last 100,000 yr from a record in the Brazilian Atlantic rainforest region and interhemispheric comparison. Q. Res. 64, 444–450 10.1016/j.yqres.2005.08.006 (doi:10.1016/j.yqres.2005.08.006) [DOI] [Google Scholar]
- 49.Haffer J. 1969. Speciation in Amazonian forest birds. Science 165, 131–137 10.1126/science.165.3889.131 (doi:10.1126/science.165.3889.131) [DOI] [PubMed] [Google Scholar]
- 50.de Vivo M., Carmignotto A. P. 2004. Holocene vegetation change and the mammal faunas of South America and Africa. J. Biogeogr. 31, 943–957 10.1111/j.1365-2699.2004.01068.x (doi:10.1111/j.1365-2699.2004.01068.x) [DOI] [Google Scholar]
- 51.Eva H. D., Belward A. S., De Miranda E. E., Di Bella C. M., Gond V., Huber O., Jones S., Sgrenzaroli M., Fritz S. 2004. A land cover map of South America. Glob. Change Biol. 10, 731–744 10.1111/j.1529-8817.2003.00774.x (doi:10.1111/j.1529-8817.2003.00774.x) [DOI] [Google Scholar]
- 52.Filloy J., Bellocq M. I. 2006. Spatial variations in the abundance of Sporophila seedeaters in the southern neotropics: contrasting the effects of agricultural development and geographical position. Biodivers. Conserv. 15, 3329–3340 10.1007/s10531-005-1341-z (doi:10.1007/s10531-005-1341-z) [DOI] [Google Scholar]
- 53.Rhymer J. M., Simberloff D. 1996. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 27, 83–109 10.1146/annurev.ecolsys.27.1.83 (doi:10.1146/annurev.ecolsys.27.1.83) [DOI] [Google Scholar]
- 54.Grant B. R., Grant P. R. 1996. Cultural inheritance of song and its role in the evolution of Darwin's finches. Evolution 50, 2471–2487 10.2307/2410714 (doi:10.2307/2410714) [DOI] [PubMed] [Google Scholar]
- 55.Grant P. R., Grant B. R. 1997. Hybridization, sexual imprinting, and mate choice. Am. Nat. 149, 1–28 10.1086/285976 (doi:10.1086/285976) [DOI] [Google Scholar]
- 56.Irwin D. E., Price T. 1999. Sexual imprinting, learning and speciation. Heredity 82, 347–354 10.1038/sj.hdy.6885270 (doi:10.1038/sj.hdy.6885270) [DOI] [PubMed] [Google Scholar]
- 57.Hubbs C. L. 1955. Hybridization between fish species in nature. Syst. Zool. 4, 1–20 10.2307/2411933 (doi:10.2307/2411933) [DOI] [Google Scholar]
- 58.Wirtz P. 1999. Mother species-father species: unidirectional hybridization in animals with female choice. Anim. Behav. 58, 1–12 10.1006/anbe.1999.1144 (doi:10.1006/anbe.1999.1144) [DOI] [PubMed] [Google Scholar]
- 59.Randler C. 2002. Avian hybridization, mixed pairing and female choice. Anim. Behav. 63, 103–119 10.1006/anbe.2001.1884 (doi:10.1006/anbe.2001.1884) [DOI] [Google Scholar]