Summary
The objective of this review is to introduce equine clinicians to the rapidly evolving field of clinical genomics with a vision of improving the health and welfare of the domestic horse. For fifteen years a consortium of veterinary geneticists and clinicians has worked together under the umbrella of The Horse Genome Project. This group, encompassing 22 laboratories in 12 countries, has made rapid progress, developing several iterations of linkage, physical and comparative gene maps of the horse with increasing levels of detail. In early 2006, the research was greatly facilitated when the U.S. National Human Genome Research Institute of the National Institutes of Health added the horse to the list of mammalian species scheduled for whole genome sequencing. The genome of the domestic horse has now been sequenced and is available to researchers worldwide in publicly accessible databases. This achievement creates the potential for transformative change within the horse industry, particularly in the fields of internal medicine, sports medicine and reproduction. The genome sequence has enabled the development of new genome-wide tools and resources for studying inherited diseases of the horse. To date, researchers have identified eleven mutations causing ten clinical syndromes in the horse. Testing is commercially available for all but one of these diseases. Future research will probably identify the genetic bases for other equine diseases, produce new diagnostic tests and generate novel therapeutics for some of these conditions. This will enable equine clinicians to play a critical role in ensuring the thoughtful and appropriate application of this knowledge as they assist clients with breeding and clinical decision-making.
Keywords: Medical Genetics, Genomics, Horse, Inherited Disease, Mutation
Introduction
The objective of this review is to assist the equine clinician in navigating the nascent field of equine clinical genomics; definitions of some relevant terminology are found in Table 1. The term “genomics” has supplanted that of “genetics” as research focus has shifted from single genes and their protein products to considerations of how the products of multiple genes interact to produce complex traits and how genes are regulated. The impact of genomic study is far-reaching, encompassing not only the obvious identification of specific disease-causing mutations but also expanded knowledge of normal physiology and insights into the evolution of the horse that contribute to fields as diverse as paleobiology (Orlando et al. 2008) and conservation medicine (Lau et al. 2009; Thirstrup et al. 2008).
Table 1.
Glossary of Terms
| Term | Definition |
|---|---|
| Genetics | The study of genes and heredity; traditionally refers to classic Mendelian principles of inheritance. |
| Genomics | The branch of genetics concerned with the global characterization of all of the genes and DNA sequence of organisms. |
| Polymorphism | Genetic variation; usually refers to genes in which there are two or more alleles (slightly different forms of the same gene) circulating in a population. |
| Microsatellite | Highly repetitive lengths of non-coding DNA involving multiple repeats of shorter segments of nucleotides (e.g. a di- tri-, or tetra-nucleotides). These genetic markers often display length polymorphism based on the number of repeat units. Determination of variation at microsatellite loci is widely used as a measure of polymorphism in a population and to identify near-by coding genes that determine traits of interest. |
| Pleiotropy | The participation of a single gene in two or more unrelated processes; mutation of such genes therefore may produce seemingly unrelated effects in several organs, tissues, or cellular functions. |
| SNP Chip | A single nucleotide polymorphism (SNP, pronounced snip) is a DNA sequence variation occurring when a single nucleotide – A, T, C, or G – in the genome differs between members of the same species. Mammalian genomes contain hundreds of thousands of SNPs dispersed widely throughout the genome. A SNP Chip is a miniaturized molecular device consisting of thousands of single stranded DNA oligonucleotide probes bonded to a silicon support. Each probe can be used to interrogate the DNA sequence at a single SNP locus using DNA hybridization. SNP Chips have become the method of choice for assessing variation across the genome; they are widely used in association and family studies designed to identify genes influencing traits of interest. |
| Expression Array | The expression array is another type of multiplexed, miniaturized hybridization device. Expression arrays contain oligonucleotide probes for the expressed genes of an organism. Expression arrays are used to assess gene activation by providing a semi-quantitative estimate of the amount of mRNA encoding specific genes in a tissue sample, based upon hybridization of fluorescently labeled cDNA. Current equine expression arrays contain probes for nearly all of the estimated 20,000+ horse genes. |
The recently completed horse genome sequence (Wade et al. 2009), in conjunction with ongoing technological advancement, holds the potential to change the way researchers and clinicians investigate, diagnose and treat all diseases, not just those with simple genetic origins. More sophisticated understanding of complex traits and the whole-genome impact on individual fitness could transform the way animals are selected for breeding. Translating this accumulating knowledge into definitive measures that will improve the overall health and welfare of the domestic horse is an immediate and important goal for both researchers and clinicians. These measures include readily accessible diagnostic testing, sound practices for husbandry and medical management of affected animals and breeding strategies that reduce the prevalence of disease-associated alleles while preserving genetic diversity and overall fitness.
The following text provides a brief history of horse genome research, followed by a review of simple, single-gene diseases of the horse and those that are likely to be complex and polygenic. Special focus is given to factors that may not only affect breeding decisions but also have significant impact on genetic diversity and the propagation of deleterious alleles. These include diseases under positive selection due to association with valuable traits like coat color and the emerging use of assisted reproduction techniques. General guidelines are provided for incorporating this knowledge into client discussions. Finally, we try to anticipate how the new tools and methods of genomics may be incorporated into equine medicine.
A Brief History of Horse Genome Research
Knowledge of the horse genome1 has progressed rapidly from a point less than 20 years ago when few genes had been characterised to the present day when the entire genome sequence has been determined. By the mid-twentieth century equine2 genetics was already a dynamic field of study, although the number of researchers in this area was small and information about the horse genome was sparse. Research published during that time investigated the genetic bases of physiology (Braend 1967; Mathai et al. 1966), coat color (Castle 1948) and disease (Dimock 1950; Trommershausen-Smith A. 1977) in the domestic horse. Researchers also took advantage of the unique availability of equid species (e.g. horses and donkeys) and their hybrid offspring (e.g. mules and hinnies) in attempts to understand chromosomal structure and the impact of chromosomal variation on meiosis and fertility (Trujillo et al. 1962). In the 1970s cytogeneticists described gross chromosomal abnormalities often associated with infertility in the horse, including XY and XX sex reversal, X chromosome monosomy, Y chromosome disomy, and sex chromosome mosaicism. Improved chromosome banding techniques later led to the identification of deletions, trisomies and translocations of the autosomes (Lear and Bailey 2008). Moving into the 1980s and early 1990s, efforts were underway to identify genetic causes for several important diseases of the horse including overo lethal white foal syndrome (McCabe et al. 1990; Trommershausen-Smith A. 1977), severe combined immunodeficiency of Arabian foals (Bailey et al. 1997; Wiler et al. 1995) and hyperkalemic periodic paralysis of Quarter Horses (Rudolph et al. 1992a; Spier et al. 1993).
The Horse Genome Project (HGP) was formed in 1995 by a group of equine geneticists and clinicians from 22 laboratories in 12 countries for the purpose of undertaking large-scale studies to characterise several aspects of the genome of the horse. In the United States, research groups from several universities participated, joining academic, private and government-supported laboratories in Australia, New Zealand, France, Sweden, Germany, South Africa, Japan, Poland, Norway, the Netherlands, Ireland and the United Kingdom. This level of international cooperation has been essential to the success of the Horse Genome Project. The Dorothy Russell Havemeyer Foundation convened yearly HGP Workshops and matching research support was provided by a number of local, regional and national funding bodies, including the British Horserace Betting Levy Board, the Morris Animal Foundation and the U.S. Department of Agriculture.
Several important milestones have been achieved by the HGP during the past 15 years, in particular in the development of a variety of genetic maps. Like the various types of maps that are available for geographic information, different experimental approaches yield information that provides complementary views of the genomic landscape of the horse. Furthermore, increasing levels of resolution provide ever more detailed information. The linkage maps of the horse remain useful tools for mapping traits to chromosomes or to sub-chromosomal levels. (Guérin et al. 1999; Penedo et al. 2005; Swinburne et al. 2000). These maps are based upon common, highly polymorphic sites in the genome called microsatellites that are closely linked to genes of interest. Fine mapping and gene identification requires additional or alternative methods. Physical maps represent the linear order of genes on chromosomes. Many such maps have been produced, including detailed maps of the X and Y chromosomes (Chowdhary et al. 2003; Raudsepp et al. 2008; Raudsepp et al. 2004a; Raudsepp et al. 2004b). However, their utility is decreasing now that the entire genome sequence is known. Comparative maps made across genomes provide reference points and identify conserved chromosomal regions. So-called “chromosome painting” has been very informative in these studies. This technique uses fluorescently labeled gene probes from individual chromosomes or chromosome arms that are hybridized to chromosome smears from the same species used to generate the probes, or from different species. In the case of the horse, a high degree of conservation of gene content has been demonstrated between human and horse chromosomes using this technique (Raudsepp et al. 1996; Yang et al. 2004). This has been very useful in predicting gene content and even gene order on individual horse chromosomes.
In 2006 the horse was selected by the U.S. National Human Genome Research Institute of the National Institutes of Health to become one of the mammalian species on a priority list to be sequenced. Sequencing began at the Broad Institute at MIT early in that year and, by January of 2007, a draft sequence was completed.
This achievement represents an important milestone in our attempts to unravel the genomic biology of the horse, but the ultimate contributions of the HGP go beyond the sequence itself. The current sequence reflects a 6.8× coverage of the genome, meaning that on average each section of DNA from the donor was sequenced 6.8 times and approximately 85%–90% of the donor horse’s total DNA has been determined. The less than 100% sequencing outcome reflects the difficulties faced in determining the DNA sequence of certain regions of the genome, such as genes of the immune system that are highly polymorphic, frequently duplicated and have many repetitive sequences. Nonetheless, this is a higher coverage than has been achieved for most sequenced mammalian genomes with the exception of man and mice. The horse sequence consists of 2.6 billion base pairs spread across 31 autosomes and the sex chromosomes. About 20,300 genes have been identified thus far, a number similar to that found in other mammals and the overall polymorphism rate of the equine genome has been estimated at 1/1,000 base pairs (Wade et al. 2009).
The work of the HGP also produced insight into the rapid evolution of the genus Equus. Diploid chromosome numbers within the genus range from 32 to 66 (Piras et al. 2009) in the mountain zebra and Przewalski’s horse respectively; the domestic horse has a diploid number of 64. An “evolutionary new centromere” was identified on equine chromosome 11 (ECA 11) in a region highly conserved across mammalian species, with the horse being the only mammal known to possess a centromere in this location (Wade et al. 2009). Other sites have been identified on ECA 1 and ECA 16 that appear to be fragile and potentially predisposed to rearrangement (Lear et al. 2008). Further investigation revealed repositioned centromeres in the donkey and one species of zebra, further defining the evolutionary relationship between species within this genus (Piras et al. 2009; Piras. et al. 2010).
With the draft sequence in place, the work of gene identification and genome annotation is ongoing and refinements to these complex tasks will continue for years to come. Today’s researchers now have convenient access to genomic data as they continue to elucidate mechanisms of basic physiology, to pursue genetic bases for both single gene and complex diseases and to ascertain whole-genome effects on fitness. Table 2 lists several websites pertaining to the Horse Genome Project, the equine genome sequence, and professional guidelines for using this information. Many of these sites are periodically updated to provide easy access to the most current information on the horse genome.
Table 2.
Websites Relevant to Horse Genetics
| Horse Genome Project (HGP) University of Kentucky | http://www.uky.edu/Ag/Horsemap/ |
| NIH Horse Genome Resources | http://www.ncbi.nlm.nih.gov/genome/guide/horse/ |
| Horse Genome Project (NIH) | http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=11760 |
| Horse Genome Browser Gateway (UC Santa Cruz) | http://genome.ucsc.edu/cgi-bin/hgGateway |
| Ensembl Horse Database | http://www.ensembl.org/Equus_caballus/index.html |
| Horse Genome Project Bacterial Artificial Chromosome (BAC) Resource (Hannover, Germany) |
http://www.tiho-hannover.de/einricht/zucht/hgp/index.htm |
| American Association of Equine Practitioners (AAEP) Statement on Genetic Defects |
http://www.aaep.org/page_editor_page_preview.php?print_friendly=true&page_name=statement_updates |
Simple Genetic Diseases of the Domestic Horse
Simple or monogenetic diseases may be defined as those caused by mutation of a single gene and inherited in a Mendelian pattern (Hardy and Singleton 2009). Virtually all of the equine genetic diseases, fully characterized to date, fall into this category, with eleven identified mutations causing ten clinical syndromes. Although they are relatively few in number, these diseases have had a discernible impact on the fitness of major breeds. Commercial testing is available for all but one of these mutations to facilitate identification of carriers, confirmation of clinical cases and management of the alleles within breeding populations. Most of these disorders have been identified through a candidate gene approach based upon knowledge of similar inherited syndromes in man or mice. The notable exception to this is the most recently characterised equine disorder, Lavender Foal Syndrome, in which the new Single Nucleotide Polymorphism (SNP) Chip was used to localise the causative mutated gene (Brooks et al. 2010)
A review of confirmed genetic diseases of the horse has been published recently (Finno et al. 2009). These are summarised below and in Table 3 with updates and additions as appropriate. Brief mention is made in the text of several additional diseases for which a single gene mutation is suspected or for which a definitive mutation is still uncertain but association with a coat color inherited in an autosomal dominant or recessive pattern has been observed. The genetic classifications of these conditions may be changed as more information becomes available or as advances in genomic study blur the traditional demarcation between “Mendelian” and “non-Mendelian” traits (Plomin et al. 2009). It is likely that identification of causative mutations for these diseases will be facilitated as genomic tools become more widely used.
Table 3.
Characterized Genetic Diseases of the Domestic Horse
| Disease | Major Breed(s) | Chromosome | Gene | Mutation | Inheritance | Testing* |
|---|---|---|---|---|---|---|
| Hyperkalemic Periodic Paralysis (HYPP) (Cannon et al. 1995; Naylor et al. 1999; Rudolph et al. 1992a; Rudolph et al. 1992b) |
Quarter Horse Paint QH-related |
11 | SCN4A | C to G substitution Phe to Leu substitution |
Autosomal Co-dominant | Y |
| Polysaccharide Storage Myopathy (PSSM) (McCue et al. 2009; McCue et al. 2008a; McCue et al. 2008b) |
Quarter Horse QH-related Warmbloods Drafts |
10 | GSY1, +/− others | G to A substitution Arg to His substitution |
Autosomal Dominant | Y |
| Malignant Hyperthermia (Aleman et al. 2005; Aleman et al. 2009) |
Quarter Horse | 10 | RYR1 | C to G substitution Arg to Gly substitution |
Autosomal Dominant | Y |
| Glycogen Branching Enzyme Deficiency (GBED) (Valberg et al. 2001; Ward et al. 2004; Ward et al. 2003) |
Quarter Horse | 26 | GBE1 | C to A substitution Premature stop codon |
Autosomal Recessive | Y |
| Severe Combined Immunodeficiency (SCID) (Bailey et al. 1997; Shin et al. 1997; Wiler et al. 1995) |
Arabian | 9 | DNA-PKcs | 5 bp deletion Unstable protein |
Autosomal Recessive | Y |
| Junctional Epidermolysis Bullosa (JEB) (Graves et al. 2008; Lieto and Cothran 2003) |
American Saddlebred | 8 | LAMA3 | 6589 bp deletion Dysfunctional protein |
Autosomal Recessive | Y |
| Junctional Epidermolysis Bullosa (JEB) (Milenkovic et al. 2003; Spirito et al. 2002) |
Belgian French Drafts |
5 | LAMC2 | C insertion Premature stop codon |
Autosomal Recessive | Y |
| Hereditary Equine Regional Dermal Asthenia (HERDA) (Tryon et al. 2007; Tryon et al. 2005; White et al. 2004; White et al. 2007) |
Quarter Horse | 1 | PPIB | Missense mutation Gly to Arg substitution |
Autosomal Recessive | Y |
| Overo Lethal White Syndrome (OLWS), Ileocolonic Aganglionosis (Metallinos et al. 1998; Santschi et al. 2001; Yang et al. 1998) |
Paint | 17 | EDNRB | TC to AG substitution Ile to Lys substitution |
Autosomal Recessive | Y |
| Gray Horse Melanoma (Pielberg et al. 2008) |
Many | 25 | STX17 | Duplication in intron 6 | Autosomal Dominant | Y |
| Lavender Foal Syndrome (Brooks et al. 2010) |
Arabian | 1 | MYO5A | G138235715del Single bp deletion in exon 30 |
Autosomal Recessive | N |
Y, testing available commercially; N, testing not yet available commercially, but expected soon.
Hyperkalemic Periodic Paralysis (HYPP)
Hyperkalemic periodic paralysis is a disease of skeletal muscle caused by a C to G substitution in the voltage-gated sodium channel (SCN4A) gene on ECA 11. This results in a phenylalanine to leucine substitution in the alpha subunit of the channel, affecting resting membrane potential such that the channel fails to deactivate in response to increasing potassium after initial depolarisation. Continuous depolarisation of myocytes ensues, manifesting clinically as transient paralysis (Cannon et al. 1995; Rudolph et al. 1992a; Rudolph et al. 1992b). The mutation is inherited in an autosomal co-dominant pattern (Naylor et al. 1999).
Clinical signs are episodic, characterised by myotonia, muscle fasciculations, third eyelid prolapse, weakness, respiratory distress and recumbency. Severity of signs may range from asymptomatic to daily episodes to death. Homozygous animals may experience dysphagia and respiratory obstruction and may show more severe signs at an earlier age (Naylor et al. 1999). Transient, infrequent episodes may resolve without treatment but oral corn syrup may be beneficial to initiate insulin release. Severe, acute episodes may require intravenous calcium gluconate, dextrose or bicarbonate. Ongoing management of animals experiencing repeated episodes should include dietary changes to reduce potassium intake and medications such as acetazolamide that promote potassium excretion and insulin release. HYPP status should be determined by genetic testing in suspect horses prior to administering anaesthetics, as this may trigger an episode (Naylor 1994a; Reynolds et al. 1998). Veterinarians should familiarise themselves with the most current drug regulations of relevant equestrian governing bodies prior to prescribing any medications to horses engaging in competition.
The HYPP allele likely has been perpetuated in the Quarter Horse population due to the desirability of the associated muscular phenotype in halter competitions (Naylor 1994b). A recent study reported that while only 1.5% of the Quarter Horse population at large is affected, over one-half of elite halter horses carry the mutation (Tryon et al. 2009). Additionally, 4.5% of the American Paint Horse population also possesses the deleterious allele (Tryon et al. 2009). Efforts intended to reduce the allele frequency in the Quarter Horse breed include exclusion of homozygotes from the American Quarter Horse registry since 2007, but affected animals may still be bred at the discretion of their owners. Laboratories with results acceptable to the American Quarter Horse Association for the purpose of registration are provided on the AQHA web site (http://www.aqha.com).
Polysaccharide Storage Myopathy (PSSM)
Polysaccharide storage myopathy is an autosomal dominant glycogen storage disease of Quarter Horses and related breeds, warmblood, drafts, and several other breeds and crosses. A mutation identified in the glycogen synthase 1 gene (GYS1) on ECA 10 causes a gain-of-function dysregulation of glycogen synthesis resulting in clinical signs of PSSM. This mutation, termed type 1, is identified in a high percentage of PSSM cases in Quarter Horse-related breeds and drafts, but not in others such as Thoroughbreds, Arabians and Standardbreds (Herszberg et al. 2008; McCue et al. 2008a; McCue et al. 2008b). This suggests that a yet undiscovered mutation could cause a clinically similar condition.
Clinical manifestations of PSSM may vary in severity and include stiffness, pain, unwillingness to move, rhabdomyolysis, fasciculations, weakness, gait abnormalities and recumbency. Muscle enzymes may be increased or normal. Acute, severe disease may be managed with rest, fluids and tranquilisers. Proper management of affected horses may alleviate frequency and severity of clinical signs. Diet should be modified to reduce soluble carbohydrates while adding fat as an energy source. A regular exercise regimen is important and stall confinement is not recommended (Firshman et al. 2003; Ribeiro et al. 2004). A more severe form of PSSM has been observed in horses having both the GYS1 mutation and a mutation in RYR1, the gene commonly associated with malignant hyperthermia (McCue et al. 2009).
The prevalence of PSSM type 1 is estimated to be approximately 11% in the Quarter Horse breed, with about 28% prevalence in halter lines (Tryon et al. 2009). Diagnosis traditionally was made by observation of amylase resistant, periodic acid Schiff (PAS) positive polysaccharide inclusions within muscle biopsy tissue. Genetic testing for the type 1 PSSM is now available through the University of Minnesota Veterinary Diagnostic Laboratory (http://www.cvm.umn.edu/vdl/ourservices/equineneuromuscular/home.html).
Malignant Hyperthermia
Malignant hyperthermia is a disease of skeletal muscle characterised by a C to G substitution in exon 46 of the ryanodine receptor gene (RYR1) on ECA 10. The resulting arginine to glycine substitution creates a defect in the calcium release channel of the sarcoplasmic reticulum (Aleman et al. 2009). Clinical disease may be triggered by inhalant anaesthesia and is characterised by severely increased body temperature, acidosis and sometimes death. Treatment of affected animals is often limited to supportive care (Aleman et al. 2005). Typically described in Quarter Horses, the prevalence in one population was 1.3% (Nieto and Aleman 2009). Genetic testing is available through the University of Minnesota Veterinary Diagnostic Laboratory (http://www.cvm.umn.edu/vdl/ourservices/equineneuromuscular/home.html).
Glycogen Branching Enzyme Deficiency (GBED)
A glycogen storage disease consistent with a glycogen branching enzyme deficiency was reported initially in three Quarter Horses, including an aborted fetus, a stillborn foal and a one-month old neonate with a history of weakness since birth (Render et al. 1999). Subsequent research characterised the causative mutation as a C to A substitution at base 102 resulting in a premature stop signal at codon 34 in exon 1 of the gene encoding the glycogen branching enzyme (GBE1) on ECA 26 (Ward et al. 2003). Clinical disease is heritable in an autosomal recessive manner (Ward et al. 2004). The physiologic consequence of the defective glycogen branching enzyme is an inability to form, store and utilise glycogen as required for normal metabolic function. This leads to failure of normal foetal growth and development and, in foals, progressive deterioration of cardiac, muscular and neurologic organ systems (Valberg et al. 2001).
Clinical presentation of GBED can range from abortion and stillbirth to weak foals. Live foals may show flexural limb deformities, seizures, weakness to the point of recumbency, signs of cardiac and respiratory failure and sudden death. Abnormalities on haematology and serum biochemistry panels may include leukopenia, hypoglycaemia and increases in creatine kinase, aspartate transaminase and gamma glutamyl transferase (Render et al. 1999; Valberg et al. 2001). Histopathology of affected organs is characterised by variable amounts of periodic acid Schiff (PAS) positive inclusions, most prominent in cardiac tissue. No appreciable amount of GBE or enzyme activity can be detected in samples from affected individuals (Valberg et al. 2001).
The potential impact of GBED on the fitness of the Quarter Horse and Paint populations may be considerable. In one study of 7 affected foals over 2,500 half siblings that are potential carriers were identified (Valberg et al. 2001). A more recent study estimated the prevalence of heterozygous carriers at 8.3% in Quarter Horses and 7.1% in Paint Horses, with approximately 2.5% of abortions and early neonatal deaths in 2 sample populations attributed to homozygosity. The authors estimated an annual registration of 16,000 heterozygote carriers and 300 deaths of homozygous foals (Wagner et al. 2006). Heterozygous carriers of the GBE1 mutation demonstrate approximately 50% of normal enzyme activity (Valberg et al. 2001), but this has not been shown to have adverse effects on the health of these animals.
Definitive diagnosis of GBED is confirmed by identification of the causative mutation with genetic testing. Testing is available through the University of California at Davis Veterinary Genetics Laboratory (http://www.vgl.ucdavis.edu/services/gbed.php) or vetGen laboratories (http://www.vetgen.com/equine-gbed-service.html) and can be performed on mane and tail hair, oral cheek swabs and blood.
Severe Combined Immunodeficiency (SCID)
Severe combined immunodeficiency (SCID) is an autosomal recessive disorder in Arabians. The causative mutation is a 5 base pair deletion in the gene encoding DNA-protein kinase catalytic subunit (DNA-PKcs) on ECA 9. The effect of this deletion is failure of maturation of both B and T lymphocytes (Bailey et al. 1997; Shin et al. 1997; Wiler et al. 1995). Affected foals are therefore unable to mount pathogen specific cellular or humoral immune responses. These foals may appear clinically normal at birth, but succumb to infection as protection from maternal antibodies wanes. Foals are frequently affected by pathogens such as adenovirus and Pneumocystis carinii that rarely cause disease in normal foals (McGuire and Poppie 1973; McGuire et al. 1974; Studdert 1978).
The SCID gene is reported to be present in a heterozygous carrier state in 8.4% of Arabians in the United States (Bernoco and Bailey 1998). Recently this carrier state has been associated with an increased incidence of sarcoid tumors (Ding et al. 2002). Genetic testing is available through vetGen laboratories (http://www.vetgen.com/equine-scid-service.html).
Junctional Epidermolysis Bullosa (JEB)
Junctional epidermolysis bullosa, sometimes referred to as epitheliogenesis imperfecta or mechanobullous disease, is an autosomal recessive dermatological disorder reported in American Saddlebreds, Belgians and some other draft breeds. Different causative mutations have been identified in American Saddlebreds versus draft breeds; from this perspective 2 genetic diseases exist. Skin samples from affected animals visualised by electron microscopy show separation in the lamina lucida of the basement membrane along with abnormal hemidesmosomes, a lesion characteristic of Herlitz junctional epidermolysis bullosa in humans (Johnson et al. 1988; Lieto et al. 2002).
The molecular defect is a failure to produce normal laminin-5, a heterotrimeric basement membrane protein required for normal adhesion between the dermal and epidermal skin layers (Lieto et al. 2002). In Belgians and the French draft breeds Trait Breton and Trait Comtois, the causative mutation occurs on ECA 5 consisting of a cytosine insertion in the LAMC2 gene that encodes the γ2 chain of the trimer. The mutation creates a premature stop codon and a functional γ2 chain cannot be synthesised (Milenkovic et al. 2003; Spirito et al. 2002). The mutation identified in American Saddlebreds affects a different chain of the trimer (α3), consisting of a 6589 bp deletion across exons 24 through 27 of the LAMA3 gene on ECA 8 (Graves et al. 2008; Lieto and Cothran 2003).
Clinical disease in affected foals is characterised by areas of mucosa and integument that are devoid of epithelium and severe ulcerations occur after minor trauma. Ocular and dental abnormalities are also sometimes present. Foals may suffer from repeated skin infections, failure to thrive due to discomfort associated with nursing and complete sloughing of hooves (Johnson et al. 1988; Lieto et al. 2002; Shapiro and McEwan 1995). There is no curative treatment available for affected foals. Prognosis is poor, with euthanasia typically the most humane option. The recessive LAMA3 vallele is estimated to be carried by 4% of the American Saddlebred breed (Lieto et al. 2002). The heterozygous state has not been associated with disease.
Diagnostic tests have been developed to identify the mutation in both American Saddlebreds (Graves et al. 2008), available through the University of Kentucky (http://www.ca.uky.edu/gluck/AGTRL.asp) and Belgians (Milenkovic et al. 2003) available directly from the University of California at Davis Veterinary Genetics Laboratory (http://www.vgl.ucdavis.edu/services/jeb.php) and through the Belgian Corporation of America (http://www.belgiancorp.com/jeb.html).
Hereditary Equine Regional Dermal Asthenia (HERDA)
Hereditary equine regional dermal asthenia is an autosomal recessive dermatological condition of Quarter Horses and related crosses (Tryon et al. 2005). Though in some reports the condition has been referred to as “hyperelastosis cutis’, this terminology has been discouraged as the defect does not appear to be in the elastic fibers (White et al. 2004). A missense mutation occurs in exon 1 of the cyclophilin B gene (PPIB) on ECA 1 causing a glycine to arginine substitution (Tryon et al. 2007). The mechanism by which this mutation produces the condition is undetermined. Histopathologically, lesions are characterised by thin collagen fibers in the deep dermal tissue that are arranged in clusters instead of having a longitudinal pattern and thicker fibres as seen in normal horses (White et al. 2004).
Clinical signs of disease include excessive elasticity and fragility of the skin, most notably over the dorsal aspect of the trunk, but any area may be affected. Some wounds may heal slowly, though affected colts often recover uneventfully from castration. Hematomas and seromas may be present with eventual chronic secondary infections, abscesses and scarring. Signs typically appear between 1 and 1.5 years of age and may or may not be associated with the use of training tack. No treatment other than supportive care of lesions is available and euthanasia is typically the end result. Mares with clinical HERDA are able to successfully carry a pregnancy to term and give birth without complications involving uterine or perineal tissue (Tryon et al. 2005; White et al. 2007).
The frequency of heterozygous carriers in the Quarter Horse breed is estimated at 3.5%. The mutation is most commonly seen in cutting horse lines within the breed, where a much higher prevalence of 28.3% is reported. This raises speculation of a heterozygote advantage within this discipline. Heterozygotes are not known to experience deleterious effects of the allele (Tryon et al. 2009; Tryon et al. 2007; Tryon et al. 2005). HERDA has also been reported in Quarter Horses in Brazil (Borges et al. 2005).
Skin biopsies are unreliable for diagnosis of HERDA (White et al. 2004), and the genetic tests that have been developed are preferred for diagnosis (Tryon et al. 2007). Genetic testing is available through the University of California at Davis Veterinary Genetics Laboratory (http://www.vgl.ucdavis.edu/services/herda.php).
Ileocolonic Aganglionosis (Overo Lethal White Syndrome)
Overo lethal white syndrome (OLWS) is a congenital disease of neonatal foals characterised by a white hair coat and a functional intestinal obstruction. The causative mutation is a TC to AG dinucleotide substitution at codon 118 of the endothelin receptor B gene (EDNRB) on ECA 17. This results in an isoleucine to lysine substitution in the protein product. The lethal effect of this mutation is developmental failure of the submucosal and myenteric ganglia. The white coat color occurs due to the failure of melanocyte precursors to migrate to their normal position in the skin of the embryo (Metallinos et al. 1998; Santschi et al. 1998; Yang et al. 1998). The prominent clinical sign in addition to the white hair coat is progressive and unrelenting colic due to functional obstruction. There is no treatment for affected foals and euthanasia is the humane option (Lightbody 2002; McCabe et al. 1990).
Prevalence of the deleterious allele is greatest in overo-type color patterns including frame overo, highly white calico overo and frame blend overo; in one study 94% of animals in these categories were heterozygous for the mutation (Santschi et al. 2001). However, carriers have been identified that completely lack white spotting (Santschi et al. 2001). Deafness has been significantly but not absolutely associated with the heterozygous state (Magdesian et al. 2009).
Identification of carriers by genetic testing is critical to avoiding crosses that might produce an affected foal, as relying on the spotting pattern is insufficient due to wide variation in phenotype. Frequent layman’s use of nonspecific terminology to describe equine coat color patterns further complicates the issue. The term “overo” is broadly used to describe any pattern with irregular edges and there are “overo” type patterns that can produce entirely healthy white foals (Brooks and Bailey 2005; Haase et al. 2007). Genetic testing for OLWS is available through the University of Kentucky (http://www.ca.uky.edu/gluck/AGTRL.asp#mutation) and the University of California at Davis Veterinary Genetics Laboratory http://www.vgl.ucdavis.edu/services/coatcolorhorse.php.
Gray Horse Melanoma
Duplication within intron 6 of the syntaxin-17 gene on ECA 25 is the cause of both the gray coat color in horses and the dermal melanomas that develop in most of these animals. The gray color is inherited in an autosomal dominant pattern, with homozygotes showing more rapid and complete graying and a higher incidence of melanoma. The loss of hair pigmentation and development of melanomas over time relates to the regulatory dysfunction of a diverse population of melanocytes. This results in depletion of the terminally differentiated melanocytes of the hair follicle and in contrast, proliferation of dermal melanocytes (Pielberg et al. 2008). While small, dermal gray horse melanomas often do not affect the animals’ quality of life (Seltenhammer et al. 2003), metastasis to other organs including lymph nodes, liver, lung, spleen and muscle can occur (MacGillivray et al. 2002). When treatment is deemed necessary, administration of cimetidine is common (Goetz et al. 1990). Melanomas do occur less frequently in non-gray horses (LeRoy et al. 2005), but a mechanism for this neoplasia is likely different. Genetic testing for the gray genotype is available through the University of Kentucky (http://www.ca.uky.edu/gluck/AGTRL.asp#color) and the University of California at Davis Veterinary Genetics Laboratory http://www.vgl.ucdavis.edu/services/coatcolorhorse.php. While is not possible to have the gray phenotype and therefore probable melanoma without the presence of at least one mutated allele, identification of homozygous breeding stock may facilitate development of a breeding program that minimises production of homozygous offspring who may develop melanoma more rapidly (Pielberg et al. 2008).
Lavender Foal Syndrome (LFS)
Lavender Foal Syndrome, also known as coat color dilution lethal, primarily affects Arabian horses. The disorder is inherited in an autosomal recessive pattern. Clinical signs include a dilute coat color that in some cases appears silver, pink or lavender, seizures, dorsiflexion of the head and neck, hyperesthesia and recumbency. Affected foals show progressive neurologic dysfunction in spite of aggressive treatment and typically succumb or are euthanised. Variable histopathologic signs such as melanin clumping in skin sections have been reported but are not consistent across all cases (Fanelli 2005; Page et al. 2006). The mutation leading to LFS has recently been identified as a single base deletion in the gene encoding myosin Va (MYO5A) on ECA1. Mutations in this gene in humans are associated with Griscelli Syndrome, a disease with phenotypic similarities to LFS (Brooks et al. 2010). Researchers investigating LFS utilised a “SNP Chip” approach rather than the traditional candidate gene approach to identify the causative mutation for this disease. The SNP Chip enables genome wide scanning for single nucleotide polymorphisms that differentiate the affected population from normal animals.
Recurrent Exertional Rhabdomyolysis (RER)
Recurrent exertional rhabdomyolysis, commonly called ‘tying up’ is a myopathy of horses characterised by painful muscle stiffness and contractures. The disease has been most thoroughly described in Thoroughbreds, but also occurs in other breeds including Standardbreds and Arabians. In Thoroughbreds, the disease has been suggested to be heritable in an autosomal dominant pattern, with possible modulation by environmental factors (Dranchak et al. 2005). A dysfunction in myocyte calcium regulation is thought to be involved (Lentz et al. 2002). Several candidate genes have been excluded as potential causative genes for RER (Dranchak et al. 2006) and attempts are ongoing to identify the genetic basis of this important disease.
Fell Pony Immunodeficiency Syndrome
A fatal disorder with multiple hematopoetic abnormalities including anaemia and immune system deficiencies has been identified in Fell pony foals. This disease is presumed to be inherited in an autosomal recessive pattern and it has been estimated that up to 50% of Fell ponies may be carriers (Gardner et al. 2006; Jelinek et al. 2006).
Glanzmann’s Thrombasthenia
Glanzmann’s thrombasthenia is a heritable defect in platelet function that results in platelet-type bleeding (e.g. epistaxis) without a finding of thrombocytopenia. The disorder is well-described in humans and dogs and has been reported rarely in horses (Livesey et al. 2005). Two different mutations involving the platelet glycoprotein complex have been identified in horses showing clinical presentations consistent with Glanzmann’s thrombasthenia (Christopherson et al. 2006; Christopherson et al. 2007). These reported cases include a Thoroughbred cross and 4 Quarter Horses but the prevalence of the deleterious alleles within these breeds has not been investigated. Although rare, Glanzmann’s thrombasthenia should be included in the differential diagnoses for horses that present with platelet-type bleeding and normal thrombocyte counts.
Chronic Progressive Lymphoedema (CPL)
Chronic progressive lymphoedema is a debilitating disease of many draft breeds characterised by thickened lymphatics, oedema, fibrosis of tissue and veins, inflammation, degeneration of elastin, neovascularisation and arteriosclerosis. The mechanism of disease is thought to be a degradation of elastin causing loss of support and subsequent dysfunction of the lymphatics (DeCock et al. 2006; DeCock et al. 2003; DeCock et al. 2009; van Brantegem et al. 2007). Single nucleotide polymorphisms identified within FOXC2, a gene causative for a similar condition in humans, were investigated and showed no association with clinical CPL in the animals studied (Young et al. 2007). Research is ongoing to identify a genetic basis for this disease.
Cerebellar Abiotropy
Cerebellar abiotrophy is a degenerative condition of Arabian foals that typically manifests as progressive neurologic dysfunction beginning at birth or in the immediately following weeks and months. Clinical signs are indicative of cerebellar disease and include ataxia, head tremor and hypermetria. Foals may survive if mildly affected, but are not sound for athletic pursuits. Histopathologically, the cerebellum is characterised by apoptosis of the Purkinje cell layer. A pattern of inheritance consistent with an autosomal recessive trait has been observed, but a causative mutation has not yet been identified (Blanco et al. 2006).
Multiple Congenital Ocular Anomalies (MCOA)
Multiple congenital ocular anomalies, sometimes referred to as anterior segment dysgenesis, is characterised by a variable cluster of defects in the anterior aspect of the globe. Inheritance is co-dominant, with heterozygotes having cysts in the ciliary body, retina or iris and homozygotes additionally suffering from cataracts, hypoplastic iris, prominent corneas, abnormalities of the iridocorneal angles and rarely retinal detachment (Andersson et al. 2008; Grahn et al. 2008).
The condition is primarily found in the recently developed breeds of Rocky Mountain and Kentucky Mountain horses. The locus for MCOA has been mapped to a 4.9 Mb region on ECA 6, and appears to be linked to the silver coat color, itself due to mutations in the gene encoding premelanosomal protein 17 (PMEL17), also on ECA 6 (Brunberg et al. 2006). Further research is required to determine if PMEL17 is the causative gene of MCOA, as ocular anomalies are associated with this gene in other species or if the disease is caused by other genes in this region (Andersson et al. 2008; Brunberg et al. 2006). Because of the popularity of the silver coat color the prevalence of MCOA in affected breeds has been estimated at 50% in both the United States and Canada (Grahn et al. 2008).
Congenital Stationary Night Blindness (CSNB)
Congenital stationary night blindness is a nonprogressive disease of the retina that occurs in Appaloosas and rarely in other breeds such as the Thoroughbred and Paso Fino. Affected animals characteristically show poor vision in dim light and normal vision in bright light, but a wide variation in the severity of visual deficits has been observed (Nunnery et al. 2005; Witzel et al. 1978). More recently clinical diagnosis of CSNB has been reported in the Danish Knabstrupper (de Linde Henriksen et al. 2007).
Studies in Appaloosas suggest an association between inheritance of 2 copies of the leopard complex gene (Lp) involved in the characteristic spotting pattern of this breed and the clinical diagnosis of CSNB. The leopard complex is thought to be inherited as an autosomal dominant trait, but possibly modified by presently unidentified genes that produce the wide variation in Appaloosa coat patterns. It is unclear at this time whether the concurrence of color and disease is due to one gene, a collection of genes, or 2 separate but linked genes. Several candidate genes have been investigated as causative for CSNB, with the most promising thus far being TRPM1. This gene is thought to be involved in calcium channel function and a theory has been proposed that, in CSNB, this creates dysfunction in neural pathways involving the retinal rods (Bellone et al. 2008; Sandmeyer et al. 2007).
Polygenic and Complex Diseases of the Domestic Horse
Polygenic or complex diseases may be defined as those that involve the additive effects of many genes and often the interplay of genetics and environmental factors. In man these types of diseases sometimes are thought to be more common and yet more difficult to characterise, than single gene traits (Hardy and Singleton 2009; Hirschhorn 2005). Type 2 diabetes and rheumatoid arthritis are often cited as examples in man (Hardy and Singleton 2009), whereas in the horse major diseases such as recurrent airway obstruction (RAO) fall into this category. Diseases of the horse suspected to fall into the complex disorder category are summarised briefly below. Although definitive genetic bases have not been established yet for these diseases, investigations are underway. Research likely will continue for an extended period of time due to the complex nature of the target diseases, but the long-term benefits to equine health will be great.
Recurrent Airway Obstruction (RAO)
Recurrent airway obstruction is a respiratory disease of horses characterised clinically by coughing and increased respiratory effort leading to poor performance. Clinical signs tend to worsen upon exposure to moldy hay. Airway hyperreactivity and bronchospasm have been documented and bronchoalveolar fluid cytology is characterised by increased numbers of neutrophils (Couteil et al. 2007). Breed predisposition to RAO has been observed and a genetic basis for this condition has long been proposed, with inheritance patterns variable across families. Environmental factors such as feeding of hay and season may modulate disease (Couteil and Ward 2003; Gerber et al. 2009; Marti et al. 1991). Candidate genes are being investigated (Jost et al. 2007; Swinburne et al. 2009).
Equine Metabolic Syndrome
Equine metabolic syndrome is a multifaceted endocrine disorder characterised by obesity and insulin resistance. It has been theorised that this syndrome predisposes to laminitis (Geor and Frank 2009; Johnson 2002). The underlying genetics of the disorder are unclear at this time, with recent research suggesting either a single dominant gene or multiple genes with reduced penetrance (Treiber et al. 2006). Equine metabolic syndrome has been proposed by some researchers as a model for human metabolic syndrome (Hodavance et al. 2007), although differences are apparent in the 2 conditions (Johnson et al. 2009). This likely will continue to be an active area of research.
Guttural Pouch Tympany of Arabian Foals
Guttural pouch tympany is a disease of foals that results from air becoming trapped within the guttural pouch. The disorder is most commonly seen in Arabian foals, but is also reported in other breeds. Fillies appear to be more frequently affected than colts. Research completed to date suggests that guttural pouch tympany is polygenic in nature and that a different genetic basis may be present in fillies versus colts (Blazyczek et al. 2004; Zeitz et al. 2009).
Genetic Susceptibility to Infectious Diseases
Genetic susceptibility to disease caused by microbial organisms, parasites or insects has been suggested in various species and presents a wide-open area for meaningful study. Furthermore, genetic susceptibility to pathogens has been associated with genome-wide homozygosity in humans and other species (Lyons et al. 2009a; Lyons et al. 2009b; Rijks et al. 2008). Limited work has been conducted in this area in horses. One group of researchers observed an increased susceptibility to respiratory infections in foals possessing a particular transferrin allele (Newton et al. 2007). Susceptibility to papilloma virus-induced sarcoid tumors and hypersensitivity to the biting midge (“sweet itch”) are thought to be associated with the genes of the Major Histocompatibility Complex (MHC) (Lazary et al. 1985; Marti et al. 1992). The causative genes and molecular mechanisms underlying these 2 conditions have not yet been determined.
Orthopaedic Diseases
The horse’s role in society is inextricably linked with athletic performance and as such undesirable variations in conformation as well as diseases of bones, joints, ligaments and tendons are the focus of a significant proportion of current genomic research. The development of orthopaedic disease in horses also is undoubtedly linked to environmental factors such as equestrian discipline, training practices and terrain. Familial tendencies or suggested heritability of both conformational unsoundness and debilitating orthopaedic disease have been observed, including early onset lordosis in American Saddlebreds (Gallagher et al. 2003), equine systemic proteoglycan accumulation in Peruvian Pasos, American Saddlebreds, Arabians and Quarter Horses (Halper et al. 2006) and superficial digital flexor tendon injury in Thoroughbreds (Oki et al. 2008). A whole genome scan using microsatellites has identified a quantitative trait locus on ECA 18 that contributes to fetlock and hock osteochondrosis dissecans in one population of horses (Wittwer et al. 2008). Definitive characterisation of the genetic contribution to orthopedic diseases will likely require sustained research efforts combined with accurate clinical diagnoses.
The Role of Colour in Genetic Disease
With an animal prized for physical beauty, as the horse often is, it is not surprising that the “horse of a different color” would be cherished by many. This is evidenced by the existence of the color breeds; palomino, buckskin and pinto, as well as pedigree-based breeds that include colour as a key defining characteristic such as the American Paint Horse, Appaloosa, Spotted Saddle Horse and Knabstrupper. Coat color inheritance was an early target of equine geneticists (Castle 1948) and recent research suggests that selection for attractive spotting patterns began nearly as early in the relationship between horse and man as domestication itself (Ludwig et al. 2009). Unfortunately human attempts to create a species of superior beauty and functionality have led to unforeseen complications, as genetic disease sometimes manifests in conjunction with specific coat colours. The cultural or symbolic associations that breeders and owners attribute to animals of a particular colour can present a special challenge for veterinarians engaged in breeding or prepurchase counselling. A solid understanding of the science behind this connection may assist veterinarians during these types of discussions.
The association of coat colour with heritable disease has long been observed and may be explained by the concept of genetic “pleiotropy”. Research in the age of genomic sequencing has revealed that an unexpectedly small core set of mammalian genes, only about 20,000, controls the hundreds of thousands of biochemical processes required for life. Many genes participate in 2 or more unrelated processes, so mutation of a single gene may produce seemingly unrelated effects in several organs or tissues. These genes are considered “pleiotropic” (Hodgkin 1998). Selection for any desirable trait therefore carries the potential to produce other unforeseen, undesirable traits by affecting the function of the gene in one of these unrelated processes.
Overo lethal white syndrome is an example of disease caused by mutation of a gene with pleiotropic effects. In the developing horse embryo, melanocytes and myenteric gangliocytes originate in the neural crest and must migrate to their final location in the skin and gastrointestinal tract respectively. A mutation in the gene for endothelin receptor B, which is involved in maturation of both types of cells, results in failure of this migration and the seemingly disparate signs of white coat and functional ileus in homozygotes. A single copy of the allele produces the desirable overo pattern by attenuating the migration of the melanocytes but leaving enough functional signalling to avoid problems in the gut. Melanocytes are also required in cochlear function for reasons that are still uncertain, providing a possible etiology for yet a third related trait, deafness, observed in some horses carrying the endothelin receptor B mutation (Magdesian et al. 2009).
Gray horse melanoma and lavender foal syndrome also exemplify the role of pleiotropy in the association between colour and disease. Two additional traits, multiple congenital ocular anomalies (associated with Silver color) and congenital stationary night blindness (Appaloosa) require additional research to fully elucidate the causative mutations and molecular mechanisms. The genomic locations of these traits were only recently discovered and it is therefore not entirely certain that these diseases involve simple pleiotropic effects of a single gene or different genes linked by very close proximity in the genome. However, the beneficial and detrimental phenotypes of these mutations are nonetheless genetically associated and client education for the purposes of prepurchase or breeding should be approached in a similar manner to known pleiotropic, colour-associated genetic diseases until more information is available. Future work will no doubt better illustrate the true relationship between these positive and negative traits, as well as many more yet to be discovered.
The Role of Assisted Reproduction in Genetic Disease
The intersections between genomics and equine reproduction are numerous. Beyond the obvious goal to select breeding pairs that will produce the most desirable offspring, genetic factors can contribute to reproductive issues such as fertility (Chowdhary et al. 2008). Chromosomal abnormalities can impact fertility as described above (Lear et al. 2008) and there is evidence that a high degree of inbreeding can have a deleterious effect on semen quality (van Eldik et al. 2006).
Assisted reproduction techniques such as artificial insemination, embryo transfer and cloning are available in the horse; a detailed review of these recently has been published (Allen 2005). Contingent upon their manner of use, these techniques can minimise or amplify the propagation of deleterious alleles within a population and can positively or adversely affect genetic diversity. Excessive breeding to a few popular stallions can reduce breed diversity and stallions heterozygous for recessive disease may pass on the undesirable allele to a much greater extent if not restricted to breeding mares that are geographically accessible. Conversely, owners of heterozygote mares may strengthen their breeding programs by having access to geographically remote stallions who are not carriers if none exist in their locale. Embryo transfer enables genetically valuable mares to make a greater contribution to the gene pool by producing more than one offspring per year. Cloning has been successful in the horse (Galli et al. 2003; Hinrichs et al. 2006; Hinrichs et al. 2007) and holds the unique potential to “retrieve” an individual’s genome for breeding purposes, as in the case of a gelding or a mare that becomes unable to breed due to disease.
At this time, there is not a consensus within the horse industry as to the role these technologies should be allowed to play and animals conceived via assisted reproduction are banned from registries in some breeds such as the Thoroughbred (2008). However, genomic research still may improve the health and welfare of animals within these breeds (Gu et al. 2009; Wolc et al. 2006).
Future advances such as accessible pre-implantation screening and sex determination will probably create even more discussion as to the advantages and disadvantages of these technologies. Clinicians are encouraged to familiarise themselves with the regulations of individual breeds and to engage in proactive discussion of assisted reproduction options with breeding clients not only in the context of pedigree or prestige, but as a valuable tool in maintaining the genetic fitness of their breeding stock and progeny.
Guidelines for Management of Genetic Diseases
Discussions of genetic disease between clinicians and their clients often are initiated with the birth of an abnormal foal or the appearance of clinical signs in an animal recently purchased or started in training. Decisions regarding medical treatment or euthanasia of the affected animal and rebreeding of the sire and dam are thus made under conditions of financial and emotional stress. Proactive educational efforts prior to such an event provide clients an opportunity to consider thoroughly the welfare and economic factors associated with a high risk breeding or purchase.
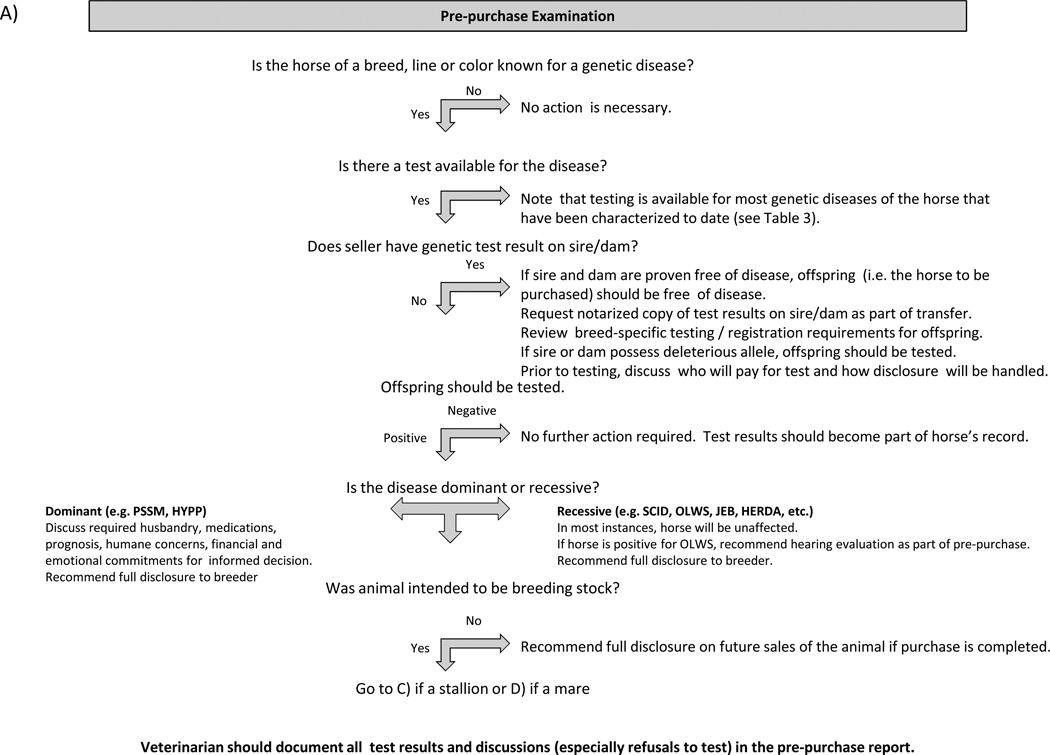
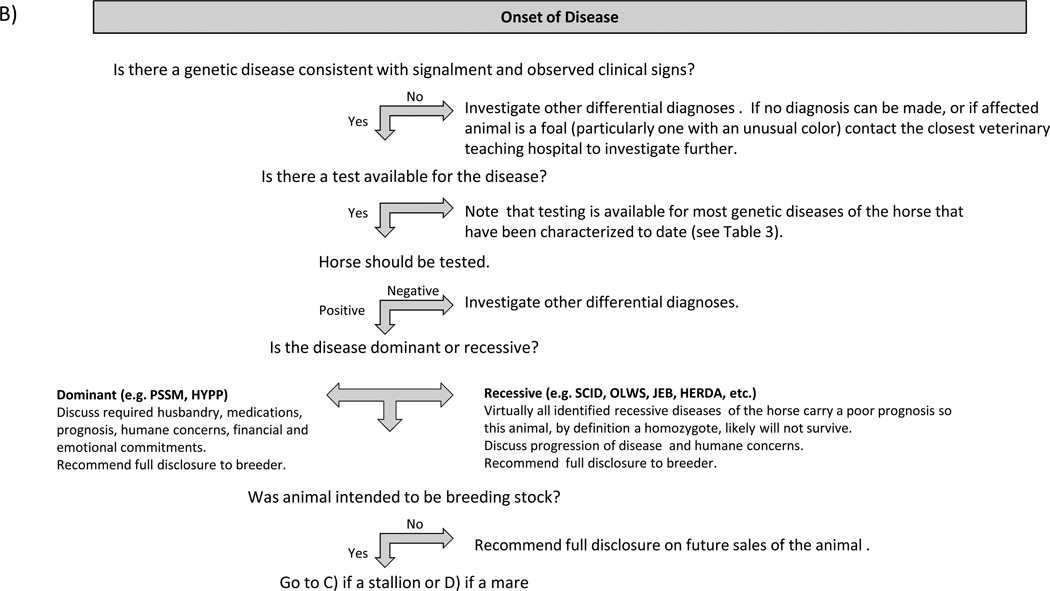
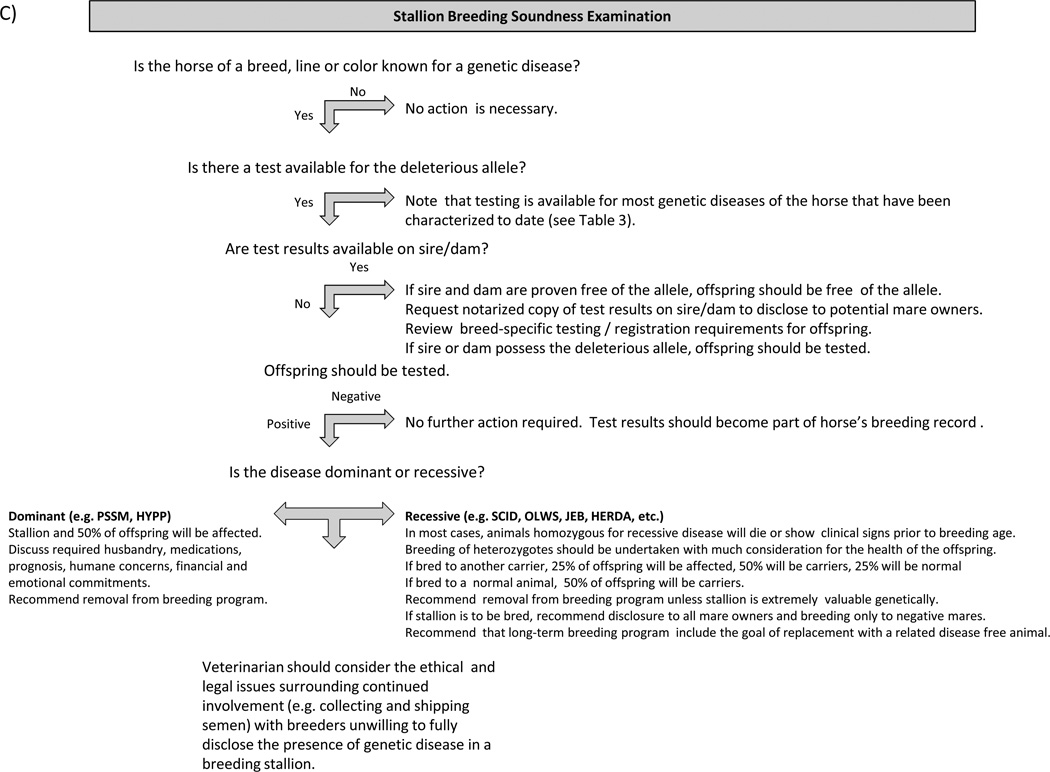
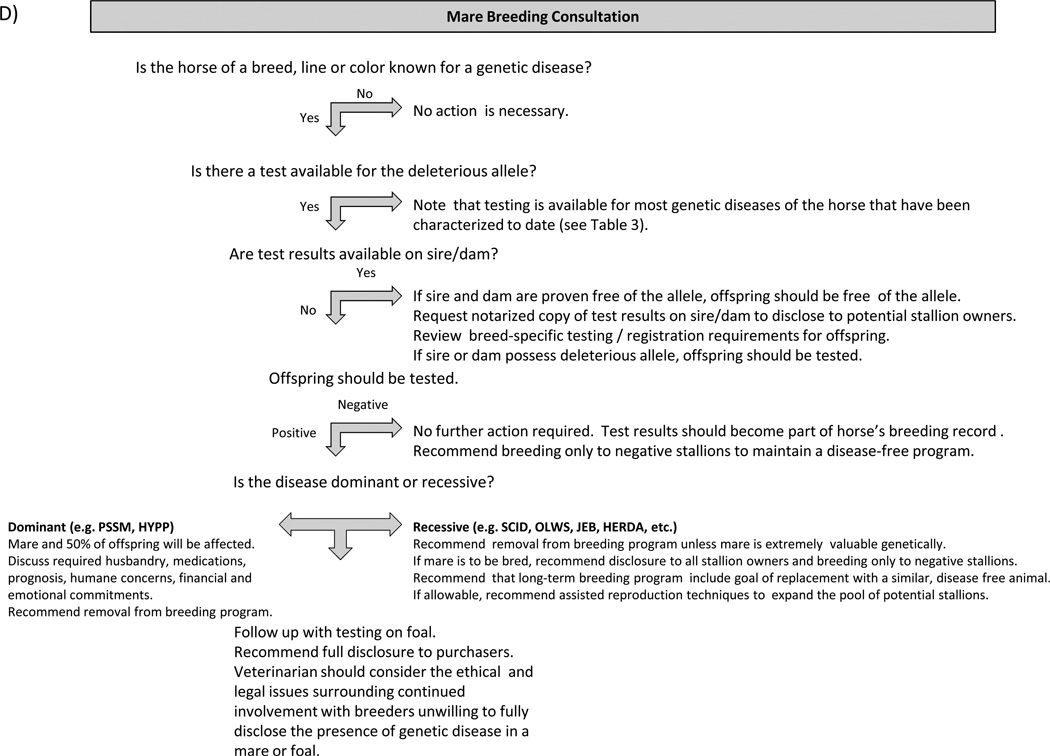
Figure 1 provides guidelines for proactive client discussions regarding the purchase or breeding of animals with a significant risk of possessing or producing offspring with genetic disease. Veterinarians must consider numerous factors when counselling clients in the area of genetic disease, including quality of life, degree of lethality, genetic diversity, deleterious effects on heterozygote carriers, intended use of the animal, the owner’s financial situation, willingness of breeders or sellers to submit animals to testing prior to breeding or sale, ethical and legal considerations pertaining to disclosures and any other circumstances unique to a particular situation. Clinicians must also consider breed registry regulations pertaining to individual diseases where appropriate and overriding laws and regulations that may vary between countries.
Figure 1.
Guidelines for Client Discussions on Genetic Diseases of the Horse Each diagram provides veterinarians with a suggested approach to guide clients through the process of addressing potential genetic diseases a variety of situations.
For genetic diseases characterised to date, situations requiring the most deliberation by breeders, clients and breed registries include:
Management of dominant traits such as HYPP and PSSM that are not invariably lethal, as affected animals routinely survive to breeding age. Propagation of the mutation in a population results in affected animals rather than clinically silent carriers, raising humane concerns.
Management of delayed-onset recessive diseases such as HERDA, where the carrier sire and dam of the affected offspring may have been rebred repeatedly prior to the onset of symptoms in progeny.
Management of heterozygote carriers of recessive alleles. This is perhaps the most challenging situation. Though in most cases the carrier state itself is benign, unchecked breeding of these animals may result in widespread dissemination of an undesirable allele. In smaller breeds or when carrier stallions are popular this could theoretically make carrier-to-carrier matings difficult to avoid. Occasionally new research also uncovers deleterious effects of heterozygosity in addition to the disease condition seen in homozygotes, which can make management of these alleles even more challenging.
Management of polygenic diseases likely will prove to be very difficult, particularly in those cases where onset of disease is delayed or where environmental triggers are a factor in clinical disease. As genes are identified and disease mechanisms elucidated for these diseases, more thorough consideration can be given to the development of relevant guidelines.
Special consideration should be given to the management of genetic diseases associated with particular colour patterns. The prevalence of the colour-associated mutations can be very high within the affected breeds and, therefore, removing animals in possession of the alleles in question potentially would result in a significant loss of genetic diversity. Educational efforts by veterinarians and breed associations emphasising the long-term importance of health relative to colour may be helpful until the conditions are more fully characterised.
Veterinarians and breeders are encouraged in the following ways to become active participants in global efforts to reduce and, when possible eliminate, genetic diseases of the horse:
Make effective use of available genetic testing. This is a key to achieving our common goal of decreasing or eliminating deleterious alleles, while maintaining genetic diversity. This is particularly important in horse breeds with low effective population sizes. Additional considerations for genetic testing recently have been published (Bannasch 2008).
Focus on a common goal of improving the health and welfare of the domestic horse. In recommending genetic testing, veterinarians should work to foster an attitude of breed stewardship rather than stigmatisation in the responsible management of carrier animals. Breeders should show pride in their efforts to reduce genetic disease by fully disclosing genetic test results for breeding stock and foals.
Be proactive in identifying and curtailing the propagation of new genetic diseases. Observations such as unusual colour patterns in aborted, stillborn or weak foals and multiple abnormal offspring from the same breeding pair may be indications of genetic disease. In these cases field clinicians and breeders are encouraged to contact their nearest veterinary school to make arrangements for submission of blood samples for banking or testing and for thorough necropsy of the carcass. Universities have resources including specialists in internal medicine, orthopaedics and genetics as well as expertise in the latest genomics technologies to determine if a genetic basis for the condition exists.
Inevitably, ethical questions will arise as our ability to diagnose and treat genetic diseases improves. Equine clinicians may be drawn into medical and ethical determinations regarding if or when it is appropriate to “cure” individual animals of clinical genetic disease.
Prospects for the Future
In the coming years, the field of medical genomics may contribute much more to equine veterinary medicine than the simple identification of disease causing mutations. The union of well-defined medical questions with cutting-edge genomic technologies will enable deeper investigation into physiology, pathophysiology, pharmacology and many other facets of medicine and disease. Early successes are already being observed, evidenced by recent publications describing the use of ever-improving expression microarray technology (Bright et al. 2009) in the areas of equine medicine (Yuan et al. 2010), orthopaedics (Glaser et al. 2009; Mienaltowski et al. 2010; Mienaltowski et al. 2009; Murray et al. 2010), sports medicine (McGivney et al. 2009) and reproduction (Klein et al. 2101).
Another set of tools that will greatly facilitate future research efforts are the single nucleotide polymorphism (SNP) DNA arrays (commonly referred to as SNP Chips). These permit evaluation of genetic variation across the entire genome in single tests and, furthermore, facilitate dissection and identification of different forms of complex diseases. The SNP Chip supports additional strategies for identifying genes contributing to complex conditions such as family and association studies. In the former affected animals, their parents and other relatives comprise the test population while, in the latter, cohorts of affected and non-affected unrelated individuals are tested. A 56,402 element equine SNP chip was produced (Illumina Inc, San Diego, CA) and evaluated in 2008 and it is becoming widely used in investigations of inherited diseases of the horse (Brooks et al. 2010).
Moving beyond genomics, the newer “-omics” disciplines such as proteomics and metabolomics (Rochfort 2005) will help define a truly integrative biology of the horse. Proteomics approaches to equine disease already have been utilised for investigation of spontaneous equine recurrent uveitis (Deeg 2009) and the technology has been applied to problems in sports medicine such as evaluating responses to exercise conditioning (Bouwman et al. 2010).
Conclusion
Throughout history, man has bred the horse to accentuate both its physical beauty and its incredible athletic ability. We appear to have molded a species with a relatively few but by no means insignificant number of inherited diseases. The Horse Genome Project has provided a genome sequence and powerful tools to help us continue to unravel its mysteries. As stewards of the future health and welfare of the domestic horse, it is imperative that researchers, clinicians, breeders, breed registries and owners work together to achieve maximum benefit from each new discovery. We must ask ourselves whether there are ways to harness this knowledge not simply to eliminate genetic disease but to breed horses that are sounder and healthier than in the past. The possibilities seem limited only by our imagination and our commitment to a species that has contributed immensely to centuries of human work and leisure-time activities. Charles Darwin proposed that species evolve through random mutation and natural selection, with the fittest individuals surviving to reproduce. As we artificially select animals to breed, fitness must be our ultimate goal.
Acknowledgements
We acknowledge financial support from the Harry M. Zweig Memorial Fund for Equine Research in New York State, the US National Institutes of Health (Grants R01- HD049545; T32- RR007059) and the Morris Animal Foundation. DFA is an investigator of the Dorothy Russell Havemeyer Foundation, Inc.
Footnotes
The terms “horse” and “horse genome” used throughout this text refer specifically to the domestic horse, Equus caballus, and the genome thereof.
The terms “equine” and “equid” used throughout this text refer to all animals within the genus Equus, including the domestic horse, Przewalski’s horse, asses, zebras and related hybrids.
References
- The American stud book: principal rules and requirements. Lexington, KY: The Jockey Club; 2008. [Google Scholar]
- Aleman M, Brosnan RJ, Williams DC, LeCouteur RA, Imai A, Tharp BR, Steffey EP. Malignant hyperthermia in a horse anesthetized with halothane. Journal of Veterinary Internal Medicine. 2005;19:263–267. doi: 10.1892/0891-6640(2005)19[363:mhiaha]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Aleman M, Nieto JE, Magdesian KG. Malignant hyperthermia associated with ryanodine receptor 1 (C7360G) mutation in Quarter Horses. Journal of Veterinary Internal Medicine. 2009;23:329–334. doi: 10.1111/j.1939-1676.2009.0274.x. [DOI] [PubMed] [Google Scholar]
- Allen WR. The development and application of the modern reproductive technologies to horse breeding. Reprod Dom Anim. 2005;40:310–329. doi: 10.1111/j.1439-0531.2005.00602.x. [DOI] [PubMed] [Google Scholar]
- Andersson LS, Juras R, Ramsey DT, Eason-Butler J, Ewart S, Cothran G, Lindgren G. Equine multiple congenital ocular anomalies maps to a 4.9 megabase interval on horse chromosome 6. BMC Genetics. 2008;9 doi: 10.1186/1471-2156-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey E, Reid RC, Skow LC, Mathiason K, Lear TL, McGuire TC. Linkage of the gene for equine combined immunodeficiency disease to microsatellite markers HTG8 and HTG4; synteny and FISH mapping to ECA9. Animal Genetics. 1997;28:268–273. doi: 10.1111/j.1365-2052.1997.00152.x. [DOI] [PubMed] [Google Scholar]
- Bannasch D. Genetic testing and the future of equine genomics. Journal of Equine Veterinary Science. 2008;28:645–649. [Google Scholar]
- Bellone RR, Brooks SA, Sandmeyer L, Murphy BA, Forsyth G, Archer S, Bailey E, Grahn B. Differential gene expression of TRPM1, the potential cause of congenital stationary night blindness and coat spotting patterns (LP) in the Appaloosa horse (Equus caballus) Genetics. 2008;179:1861–1870. doi: 10.1534/genetics.108.088807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoco D, Bailey E. Frequency of the SCID gene among Arabian horses in the USA. Animal Genetics. 1998;29:41–42. doi: 10.1046/j.1365-2052.1998.00237.x. [DOI] [PubMed] [Google Scholar]
- Blanco A, Moyano R, Vivo J, Flores-Acuna R, Molina A, Blanco C, Monterde JG. Purkinje cell apoptosis in Arabian horses with cerebellar abiotrophy. Journal of Veterinary Medicine. 2006;53:286–287. doi: 10.1111/j.1439-0442.2006.00836.x. [DOI] [PubMed] [Google Scholar]
- Blazyczek I, Hamann H, Ohnesorge B, Deegen E, Distl O. Inheritance of guttural pouch tympany in the Arabian horse. Journal of Heredity. 2004;95:195–199. doi: 10.1093/jhered/esh041. [DOI] [PubMed] [Google Scholar]
- Borges AS, Conceicao LG, Alves ALG, Fabris VE, Pessoa MA. Hereditary equine regional dermal asthenia in three related Quarter Horses in Brazil. Veterinary Dermatology. 2005;16:125–130. doi: 10.1111/j.1365-3164.2005.00431.x. [DOI] [PubMed] [Google Scholar]
- Bouwman FG, van Ginneken MM, Noben JP, Royackers E, de Graaf-Roelfsema E, Wijnberg ID, van der Kolk JH, Mariman EC, van Breda E. Differential expression of equine muscle biopsy proteins during normal training and intensified training in young standardbred horses using proteomics technology. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:55–64. doi: 10.1016/j.cbd.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Braend M. Genetic variation of horse hemoglobin. Hereditas. 1967;58:385–392. doi: 10.1111/j.1601-5223.1967.tb02163.x. [DOI] [PubMed] [Google Scholar]
- Bright LA, Burgess SC, Chowdhary B, Swiderski CE, McCarthy F. Structural and functional-annotation of an equine whole genome oligoarray. BMC Bioinformatic. 2009;10 doi: 10.1186/1471-2105-10-S11-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA, Bailey E. Exon skipping in the KIT gene causes a Sabino spotting pattern in horses. Mammalian Genome. 2005;16:893–902. doi: 10.1007/s00335-005-2472-y. [DOI] [PubMed] [Google Scholar]
- Brooks SA, Gabreski N, Miller D, Brisbin A, Brown HE, Streeter C, Mezey J, Cook D, Antczak DF. Whole genome SNP association in the horse: identification of a deletion in myosin Va responsible for lavender foal syndrome. PLos Genetics. 2010 doi: 10.1371/journal.pgen.1000909. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunberg E, Andersson L, Cothran G, Sandberg K, Mikko S, Lindgren G. A missense mutation in PMEL17 is associated with the silver coat color in the horse. BMC Genetics. 2006;7 doi: 10.1186/1471-2156-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SC, Hayward LJ, Beech J, Brown RH. Sodium channel inactivation is impaired in equine hyperkalemic periodic paralysis. Journal of Neurophysiology. 1995;73:1892–1899. doi: 10.1152/jn.1995.73.5.1892. [DOI] [PubMed] [Google Scholar]
- Castle WE. The ABC of color inheritance in horses. Genetics. 1948;33:22–35. doi: 10.1093/genetics/33.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary BP, Paria N, Raudsepp T. Potential applications of equine genomics in dissecting diseases and fertility. Animal Reproduction Science. 2008;107:208–218. doi: 10.1016/j.anireprosci.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T, Kata SR, Goh G, Millon LV, Allan V, Piumi F, Guerin G, Swinburne J, Binns M, Lear TL, Mickelson J, Murray J, Antczak DF, Womack JE, Skow LC. The first-generation whole-genome radiation hybrid map in the horse identifies conserved segments in human and mouse genomes. Genome Research. 2003;13:742–751. doi: 10.1101/gr.917503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson PW, Insalaco TA, vanSanten VL, Livesey L, Bourse C, Boudreaux MK. Characterization of the cDNA encoding alphaIIb and beta3 in normal horses and two horses with Glanzmann thrombasthenia. Veterinary Pathology. 2006;43:78–82. doi: 10.1354/vp.43-1-78. [DOI] [PubMed] [Google Scholar]
- Christopherson PW, van Santen VL, Livesey L, Boudreaux MK. A 10-base-pair deletion in the gene encoding platelet glycoprotein IIb associated with Glanzmann's thrombasthenia in a horse. Journal of Veterinary Internal Medicine. 2007;21:196–198. doi: 10.1892/0891-6640(2007)21[196:abditg]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Couteil LL, Hoffman AM, Hodgson J, Buechner-Maxwell V, Viel L, Wood JLN, Lavoie J. Inflammatory airway diseases of the horse. Journal of Veterinary Internal Medicine. 2007:356–361. doi: 10.1892/0891-6640(2007)21[356:iadoh]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Couteil LL, Ward MP. Analysis of risk factors for recurrent airway obstruction in North American horses: 1,444 cases (1990–1999) JAVMA. 2003;223:1645–1650. doi: 10.2460/javma.2003.223.1645. [DOI] [PubMed] [Google Scholar]
- de Linde Henriksen M, Blaabjerg K, Baptiste KE, Flagstad A, Andersen PH. Congenital stationary night blindness (CSNB) in the Danish knabstrubber horse. Veterinary Ophthalmology. 2007;10:326. [Google Scholar]
- DeCock HEV, Affolter VK, Farver TB, Van Brantegem L, Scheuch B, Ferraro GL. Mesurement of skin desmosine as an indicator of altered cutaneous elastin in draft horses with chronic progressive lymphedema. Lymphatic Research and Biology. 2006;4:67–72. doi: 10.1089/lrb.2006.4.67. [DOI] [PubMed] [Google Scholar]
- DeCock HEV, Affolter VK, Wisner ER, Ferraro GL, MacLachlan NJ. Progressive swelling, hyperkeratosis and fibrosis of distal limbs in Clydesdales, Shires and Belgian draft horses, suggestive of primary lymphedema. Lymphatic Research and Biology. 2003;1:191–199. doi: 10.1089/153968503768330238. [DOI] [PubMed] [Google Scholar]
- DeCock HEV, Van Brantegem L, Affolter VK, Oosterlinck M, Ferraro GL, Ducatelle R. Quantitative and qualitative evaluation of dermal elastin of draught horses with chronic progressive lymphoedema. Journal of Comparative Pathology. 2009;140:132–139. doi: 10.1016/j.jcpa.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Deeg CA. A proteomic approach for studying the pathogenesis of spontaneous equine recurrent uveitis. Veterinary Immunology and Immunopathology. 2009;128:132–136. doi: 10.1016/j.vetimm.2008.10.302. [DOI] [PubMed] [Google Scholar]
- Dimock WW. "Wobbles" an hereditary disease in horses. Journal of Heredity. 1950;41:319–323. doi: 10.1093/oxfordjournals.jhered.a106074. [DOI] [PubMed] [Google Scholar]
- Ding Q, Bramble L, Yuzbasiyan-Gurkan V, Bell T, Meek K. DNA-PKcs mutations in dogs and horses: allele frequency and association with neoplasia. Gene. 2002;283:263–269. doi: 10.1016/s0378-1119(01)00880-0. [DOI] [PubMed] [Google Scholar]
- Dranchak PK, Valberg SJ, Onan GW, Gallant EM, Binns MM, Swinburne JE, Mickelson JR. Exclusion of linkage of the RYR1, CACNA1S, and ATP2A1 genes to recurrent exertional rhabdomyolysis in Thoroughbreds. AJVR. 2006;67:1395–1400. doi: 10.2460/ajvr.67.8.1395. [DOI] [PubMed] [Google Scholar]
- Dranchak PK, Valberg SJ, Onan GW, Gallant EM, MacLeay JM, McKenzie EC, De La Corte F, Ekenstedt K, Mickelson JR. Inheritance of recurrent exertional rhabdomyolysis. JAVMA. 2005;227:762–767. doi: 10.2460/javma.2005.227.762. [DOI] [PubMed] [Google Scholar]
- Fanelli HH. Coat color dilution lethal ('lavender foal syndrome'): a tetany syndrome of Arabian foals. Equine Veterinary Education. 2005;17:260–263. [Google Scholar]
- Finno CJ, Spier SJ, Valburg SJ. Equine diseases caused by known genetic mutations. The Veterinary Journal. 2009;179:336–347. doi: 10.1016/j.tvjl.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Firshman AM, Valberg SJ, Bender JB, Finno CJ. Epidemiologic characteristics and management of polysaccharide storage myopathy in Quarter Horses. AJVR. 2003;64:1319–1327. doi: 10.2460/ajvr.2003.64.1319. [DOI] [PubMed] [Google Scholar]
- Gallagher PC, Morrison S, Bernoco D, Bailey E. Measurement of back curvature in American Saddlebred horses: structual and genetic basis for early-onset lordosis. Journal of Equine Veterinary Science. 2003;23:71–76. [Google Scholar]
- Galli C, Lagutina I, Crotti G, Colleoni S, Turini P, Ponderato N, Duchi R, Lazzari G. A cloned horse born to its twin dam. Nature. 2003;424:635. doi: 10.1038/424635a. [DOI] [PubMed] [Google Scholar]
- Gardner RB, Hart KA, Stokol T, Divers TJ, Flaminio MJBF. Fell pony syndrome in a pony in North America. Journal of Veterinary Internal Medicine. 2006;20:198–203. doi: 10.1892/0891-6640(2006)20[198:fpsiap]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Geor R, Frank N. Metabolic syndrome - from human organ disease to laminar failure in equids. Veterinary Immunology and Immunopathology. 2009;129:151–154. doi: 10.1016/j.vetimm.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Gerber V, Baleri D, Klukowska-Rotzler J, Swinburne JE, Dolf G. Mixed inheritance of equine recurrent airway obstruction. Journal of Veterinary Internal Medicine. 2009;23:626–630. doi: 10.1111/j.1939-1676.2009.0292.x. [DOI] [PubMed] [Google Scholar]
- Glaser K, Sun Q, Wells MT, Nixon AJ. Development of a novel equine whole transcript oligonucleotide GeneChip microarray and its use in gene expression profiling of normal articular epiphyseal cartilage. Equine Veterinary Journal. 2009;41:663–670. doi: 10.2746/042516409x412381. [DOI] [PubMed] [Google Scholar]
- Goetz TE, Ogilvie GK, Keegan KG, Johnson PJ. Cimetidine for the treatment of melanoma in three horses. JAVMA. 1990;196:1201–1202. [PubMed] [Google Scholar]
- Grahn BH, Pinard C, Archer S, Bellone R, Forsyth G, Sandmeyer LS. Congenital ocular anomalies in purebred and crossbred Rocky and Kentucky Mountain horses in Canada. Canadian Veterinary Journal. 2008;49:675–681. [PMC free article] [PubMed] [Google Scholar]
- Graves KT, Henney PJ, Ennis RB. Partial deletion of the LAM3 gene is responsible for hereditary junctional epidermolysis bullosa in the American Saddlebred horse. Animal Genetics. 2008;40:35–41. doi: 10.1111/j.1365-2052.2008.01795.x. [DOI] [PubMed] [Google Scholar]
- Gu J, Orr N, Park SD, Katz LM, Sulimova G, MacHugh DE, Hill EW. A genome scan for positive selection in Thoroughbred horses. PLoS One. 2009;4:e5767. doi: 10.1371/journal.pone.0005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérin G, Bailey E, Bernoco D, Anderson I, Antczak DF, Bell K, Binns MM, Bowling AT, Brandon R, Cholewinski G, Cothran EG, Ellegren H, Förster M, Godard S, Horin P, Ketchum M, Lindgren G, McPartlan H, Mériaux JC, Mickelson JR, Millon LV, Murray J, Neau A, Røed K, Ziegle J. Report of the international equine gene mapping workshop: male linkage map. Animal Genetics. 1999;30:341–354. doi: 10.1046/j.1365-2052.1999.00510.x. [DOI] [PubMed] [Google Scholar]
- Haase B, Brooks SA, Schlumbaum A, Azor PJ, Bailey E, Alaeddine F, Mevissen M, Burger D, Poncet P, Rieder S, Leeb T. Allelic heterogeneity at the equine KIT locus in dominant white (W) horses. PLos Genetics. 2007;3:2101–2107. doi: 10.1371/journal.pgen.0030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halper J, Kim B, Khan A, Yoon JH, Mueller POE. Degenerative suspensory ligament desmitis as a systemic disorder characterized by proteoglycan accumulation. BMC Veterinary Research. 2006;2 doi: 10.1186/1746-6148-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Singleton A. Genomewide association studies and human disease. The New England Journal of Medicine. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herszberg B, McCue ME, Larcher T, Mata X, Vaiman A, Chaffaux S, Cherel Y, Valberg SJ, Mickelson JR, Guerin G. A GYS1 mutation is highly associated with polysaccharide storage myopathy in Cob Norman draught horses. Animal Genetics. 2008;40:94–96. doi: 10.1111/j.1365-2052.2008.01778.x. [DOI] [PubMed] [Google Scholar]
- Hinrichs K, Choi YH, Love CC, Chung YG, Varner DD. Production of horse foals via direct injection of roscovitine-treated donor cells and activation by injection of sperm extract. Reproduction. 2006;131:1063–1072. doi: 10.1530/rep.1.01095. [DOI] [PubMed] [Google Scholar]
- Hinrichs K, Choi YH, Varner DD, Hartman DL. Production of cloned horse foals using roscovitine-treated donor cells and activation with sperm extract and/or ionomycin. Reproduction. 2007;134:319–325. doi: 10.1530/REP-07-0069. [DOI] [PubMed] [Google Scholar]
- Hirschhorn J. Genetic approaches to studying common diseases and complex traits. Pediatric Research. 2005;157:74R–77R. doi: 10.1203/01.PDR.0000159574.98964.87. [DOI] [PubMed] [Google Scholar]
- Hodavance MS, Ralston SL, Pelczer I. Beyond blood sugar - the potential of NMR-based metabonomics for type 2 human diabetes, and the horse as a possible model. Analytical and Bioanalytical Chemistry. 2007;387:533–537. doi: 10.1007/s00216-006-0979-z. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Seven types of pleiotropy. International Journal of Developmental Biology. 1998;42:501–505. [PubMed] [Google Scholar]
- Jelinek F, Faldyna M, Jasurkova-Mikutova G. Severe combined immunodeficiency in a Fell pony foal. Journal of Veterinary Medicine. 2006;53:69–73. doi: 10.1111/j.1439-0442.2006.00779.x. [DOI] [PubMed] [Google Scholar]
- Johnson GC, Kohn CW, Johnson CW, Garry F, Scott D, Martin S. Ultrastructure of junctional epidermolysis bullosa in Belgian foals. Journal of Comparative Pathology. 1988;98:329–336. doi: 10.1016/0021-9975(88)90053-9. [DOI] [PubMed] [Google Scholar]
- Johnson PJ. The equine metabolic syndrome peripheral Cushing's syndrome. The Veterinary Clinics Equine Practice. 2002;18:271–293. doi: 10.1016/s0749-0739(02)00006-8. [DOI] [PubMed] [Google Scholar]
- Johnson PJ, Wiedmeyer CE, Messer NT, Ganjam VK. Medical implications of obesity in horses - lessons for human obesity. Journal of Diabetes Science and Technology. 2009;3:163–174. doi: 10.1177/193229680900300119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost U, Klukowska-Rotzler J, Dolf G, Swinburne JE, Ramseyer A, Bugno M, Burger D, Blott S, Gerber V. A region on equine chromosome 13 is linked to recurrent airway obstruction in horses. Equine Veterinary Journal. 2007;39:236–241. doi: 10.2746/042516407x171110. [DOI] [PubMed] [Google Scholar]
- Klein C, Scoggins KE, Ealy AD, Troedsson MH. Transcriptional profiling of equine endometrium during the time of maternal recognition of pregnancy. Biol Reprod. 2101 doi: 10.1095/biolreprod.109.081612. 0.1095/ biolreprod.1109.081612. [DOI] [PubMed] [Google Scholar]
- Lau AN, Peng L, Goto H, Chemnick L, Ryder O, Makova KD. Horse domestication and conservation genetics of Przewalski's horse inferred from sex chromosomal and autosomal sequences. Mol Biol Evol. 2009;26:199–208. doi: 10.1093/molbev/msn239. [DOI] [PubMed] [Google Scholar]
- Lazary S, Gerber H, Glatt PA, Straub R. Equine leucocyte antigens in sarcoid-affected tissue. Equine Veterinary Journal. 1985;17:283–286. doi: 10.1111/j.2042-3306.1985.tb02498.x. [DOI] [PubMed] [Google Scholar]
- Lear TL, Bailey E. Equine clinical cytogenetics: the past and future. Cytogenetic and Genome Research. 2008;120:42–49. doi: 10.1159/000118739. [DOI] [PubMed] [Google Scholar]
- Lear TL, Lundquist J, Zent WW, Fishback WD, Clark A. Three autosomal chromosome translocations associated with repeat early embryonic loss (REEL) in the domestic horse. Cytogenetic and Genome Research. 2008;120:117–122. doi: 10.1159/000118749. [DOI] [PubMed] [Google Scholar]
- Lentz LR, Valberg SJ, Herold LV, Onan GW, Mickelson JR, Gallant EM. Myoplasmic calcium regulation in myotubes from horses with recurrent exertional rhabdomyolysis. AJVR. 2002;63:1724–1731. doi: 10.2460/ajvr.2002.63.1724. [DOI] [PubMed] [Google Scholar]
- LeRoy BE, Knight MC, Eggleston R, Torres-Velez F, Harmon BG. Tail base mass in a horse of a different color. Veterinary Clinical Pathology. 2005;34:69–71. doi: 10.1111/j.1939-165x.2005.tb00014.x. [DOI] [PubMed] [Google Scholar]
- Lieto LD, Cothran EG. The epitheliogenesis imperfecta locus maps to equine chromosome 8 in American Saddlebred horses. Cytogenetic and Genome Research. 2003;102:207–210. doi: 10.1159/000075750. [DOI] [PubMed] [Google Scholar]
- Lieto LD, Swerczek TW, Cothran EG. Equine epitheliogenesis imperfecta in two American Saddlebred foals is a lamina lucida defect. Veterinary Pathology. 2002;39:576–580. doi: 10.1354/vp.39-5-576. [DOI] [PubMed] [Google Scholar]
- Lightbody T. Foal with overo lethal white syndrome born to a registered Quarter Horse mare. Canadian Veterinary Journal. 2002;43:715–717. [PMC free article] [PubMed] [Google Scholar]
- Livesey L, Christopherson P, Hammond A, Perkins J, Toivio-Kinnucan M, Insalaco T, Boudreaux MK. Platelet dysfunction (Glanzmann's thrombasthenia) in horses. Journal of Veterinary Internal Medicine. 2005;19:917–919. doi: 10.1892/0891-6640(2005)19[917:pdgtih]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Pruvost M, Reissmann M, Benecke N, Brockman GA, Castanos P, Cieslak M, Lippold S, Llorent L, Malaspinas A, Slatkin M, Hofreiter M. Coat color variation at the beginning of horse domestication. Science. 2009;324:485. doi: 10.1126/science.1172750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons EJ, Amos W, Berkley JA, Mwangi I, Shafi M, Williams TN, Newton CR, Peshu N, Marsh K, Scott JAG, Hill AVS. Homozygosity and risk of childhood death due to invasive bacterial disease. BMC Medical Genetics. 2009a;10 doi: 10.1186/1471-2350-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons EJ, Frodsham AJ, Zhang L, Hill AVS, Amos W. Consanguinity and susceptibility to infectious diseases in humans. Biology Letters. 2009b;5:574–576. doi: 10.1098/rsbl.2009.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGillivray KC, Sweeney C, Del Piero F. Metastatic melanoma in horses. Journal of Veterinary Internal Medicine. 2002;16:252–256. doi: 10.1892/0891-6640(2002)016<0452:mmih>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- Magdesian KG, Williams DC, Aleman M, LeCouteur RA, Madigan JE. Evaluation of deafness in American Paint Horses by phenotype, brainstem auditory-evoked responses, and endothelin receptor B genotype. JAVMA. 2009;235:1204–1211. doi: 10.2460/javma.235.10.1204. [DOI] [PubMed] [Google Scholar]
- Marti E, Gerber H, Essich G, Oulehla J, Lazary S. The genetic basis of equine allergic diseases: I. chronic hypersensitivity bronchitis. Equine Veterinary Journal. 1991;23:457–460. doi: 10.1111/j.2042-3306.1991.tb03761.x. [DOI] [PubMed] [Google Scholar]
- Marti E, Gerber H, Lazary S. On the genetic basis of equine allergic diseases: II. Insect bite dermal hypersensitivity. Equine Veterinary Journal. 1992;24:113–117. doi: 10.1111/j.2042-3306.1992.tb02794.x. [DOI] [PubMed] [Google Scholar]
- Mathai CK, Ohno S, Beutler E. Sex-linkage of the glucose-6-phosphate dehydrogenase gene in equidae. Nature. 1966;210:115–116. doi: 10.1038/210115a0. [DOI] [PubMed] [Google Scholar]
- McCabe L, Griffin LD, Kinzer A, Chandler M, Beckwith JB, McCabe ER. Overo lethal white foal syndrome: equine model of aganglionic megacolon (Hirschsprung disease) Am J Med Genet. 1990;36:336–340. doi: 10.1002/ajmg.1320360319. [DOI] [PubMed] [Google Scholar]
- McCue ME, Valberg SJ, Jackson M, Borgia L, Lucio M, Mickelson JR. Polysaccharide storage myopathy phenotype in quarter horse-related breeds is modified by the presence of an RYR1 mutation. Neuromuscular Disorders. 2009;19:37–43. doi: 10.1016/j.nmd.2008.10.001. [DOI] [PubMed] [Google Scholar]
- McCue ME, Valberg SJ, Lucio M, Mickelson JR. Glycogen synthase 1 (GYS1) mutation in diverse breeds with polysaccharide storgae myopathy. Journal of Veterinary Internal Medicine. 2008a;22:1228–1233. doi: 10.1111/j.1939-1676.2008.0167.x. [DOI] [PubMed] [Google Scholar]
- McCue ME, Valberg SJ, Miller MB. Glycogen synthase (GYS1) mutation causes a novel skeletal muscle glycogenosis. Genomics. 2008b;91:458–466. doi: 10.1016/j.ygeno.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivney BA, Eivers SS, MacHugh DE, Macleod JN, O'Gorman GM, Park SD, Katz LM, Hill EW. Transcription adaptions following exercise in thoroughbred horse skeletal muscle highlights molecular mechanisms that lead to muscle hypertrophy. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire TC, Poppie MJ. Hypogammaglobulinemia and thymic hypoplasia in horses: a primary combined immunodeficiency disorder. Infection and Immunity. 1973;8:272–277. doi: 10.1128/iai.8.2.272-277.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire TC, Poppie MJ, Banks KL. Combined (B- and T-lymphocyte) immunodeficiency: a fatal genetic disease in Arabian foals. JAVMA. 1974;164:70–76. [PubMed] [Google Scholar]
- Metallinos DL, Bowling AT, RIne J. A missense mutation in the endothelin-B receptor gene is associated with Lethal White Foal Syndrome: an equine version of Hirschsprung disease. Mammalian Genome. 1998;9:426–431. doi: 10.1007/s003359900790. [DOI] [PubMed] [Google Scholar]
- Mienaltowski MJ, Huang L, Bathke AC, Stromberg AJ, MacLeod JN. Transcriptional comparisons between equine articular repair tissue, neonatal cartilage, cultured chondrocytes and mesenchymal stromal cells. Brief Funct Genomic Proteomic. 2010 doi: 10.1093/bfgp/elq007. [DOI] [PubMed] [Google Scholar]
- Mienaltowski MJ, Huang L, Frisbie DD, McIlwraith CW, Stromberg AJ, Bathke AC, MacLeod JN. Transcriptional profiling differences for articular cartilage and repair tissue in equine joint surface lesions. BMC Medical Genomics. 2009;2 doi: 10.1186/1755-8794-2-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D, Chaffaux S, Taourit S, Guerin G. A mutation in the LAMC2 gene causes the Herlitz junctional epidermolysis bullosa (H-jeb) in two French draft horse breeds. Genet Sel Evol. 2003;35:249–256. doi: 10.1186/1297-9686-35-2-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SJ, Santangelo KS, Bertone AL. Evaluation of early cellular influences of bone morphogenic proteins 12 and 2 on equine superficial digital flexor tenocytes and bone marrow-derived mesenchymal stem cells in vitro. American Journal of Veterinary Research. 2010;71:103–114. doi: 10.2460/ajvr.71.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor JM. Equine hyperkalemic periodic paralysis: review and implications. Canadian Veterinary Journal. 1994a;35:279–285. [PMC free article] [PubMed] [Google Scholar]
- Naylor JM. Selection of quarter horses affected with hyperkalemic periodic paralysis by show judges. JAVMA. 1994b;204:926–928. [PubMed] [Google Scholar]
- Naylor JM, Nickel DD, Trimino G, Card C, Lightfoot K, Adams G. Hyperkalemic periodic paralysis in homozygous and heterozygous horses: a co-dominant genetic condition. Equine Veterinary Journal. 1999;31:153–159. doi: 10.1111/j.2042-3306.1999.tb03809.x. [DOI] [PubMed] [Google Scholar]
- Newton JR, Wood JLN, Chanter N. Evidence for transferrin allele as a host-level risk factor in naturally occurring respiratory disease: a preliminary study. Equine Veterinary Journal. 2007;39:164–171. doi: 10.2746/042516407x166954. [DOI] [PubMed] [Google Scholar]
- Nieto JE, Aleman M. A rapid detection method for the ryanodine receptor 1 mutation (C7360G) in Quarter Horses. Journal of Veterinary Internal Medicine. 2009;23:619–622. doi: 10.1111/j.1939-1676.2009.0281.x. [DOI] [PubMed] [Google Scholar]
- Nunnery C, Pickett JP, Zimmerman KL. Congenital stationary night blindness in a Thoroughbred and a Paso Fino. Veterinary Ophthalmology. 2005;8:415–419. doi: 10.1111/j.1463-5224.2005.00416.x. [DOI] [PubMed] [Google Scholar]
- Oki H, Miyake T, Kasashima Y, Sasaki Y. Estimation of heritability for superficial digital flexor tendon injury by Gibbs sampling in the Thoroughbred racehorse. J Anim Breed Genet. 2008;125:413–416. doi: 10.1111/j.1439-0388.2008.00758.x. [DOI] [PubMed] [Google Scholar]
- Orlando L, Male D, Alberdi MT, Prado JL, Prieto A, Cooper A, Hanni C. Ancient DNA clarifies the evolutionary history of American Late Pleistocene equids. J Mol Evol. 2008;66:533–538. doi: 10.1007/s00239-008-9100-x. [DOI] [PubMed] [Google Scholar]
- Page P, Parker R, Harper C, Guthrie A, Neser J. Clinical, clinicopathologic, postmortem examination findings and familial history of 3 Arabians with lavender foal syndrome. Journal of Veterinary Internal Medicine. 2006;20:1491–1494. doi: 10.1892/0891-6640(2006)20[1491:ccpefa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Penedo MC, Millon LV, Bernoco D, Bailey E, Binns M, Cholewinski G, Ellis N, Flynn J, Gralak B, Guthrie A, Hasegawa T, Lindgren G, Lyons LA, Roed KH, Swinburne JE, Tozaki T. International Equine Gene Mapping Workshop Report: a comprehensive linkage map constructed with data from new markers and by merging four mapping resources. Cytogenet Genome Res. 2005;111:5–15. doi: 10.1159/000085664. [DOI] [PubMed] [Google Scholar]
- Pielberg GR, Golovko A, Sundstrom E, Curik I, Lennartsson J, Seltenhammer MH, Druml T, Binns M, Fitzsimmons C, Lindgren G, Sandberg K, Baumung R, Vetterlein M, Stromberg S, Grabherr M, Wade C, Lindblad-Toh K, Ponten F, Heldin C, Solkner J, Andersson L. A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nature Genetics. 2008;40:1004–1009. doi: 10.1038/ng.185. [DOI] [PubMed] [Google Scholar]
- Piras FM, Nergadze SG, Poletto V, Cerutti F, Ryder OA, Leeb T, Raimondi E, Guilotto E. Phylogeny of horse chromosome 5q in the genus Equus and centromere repositioning. Cytogenetic and Genome Research. 2009;126:165–172. doi: 10.1159/000245916. [DOI] [PubMed] [Google Scholar]
- Piras FM, Nergadze SG, Magnani E, Bertoni L, Attolini C, Khoriauli L, Raimondi E, Giulotto E. Uncoupling of satellite DNA and centromeric function in genus Equus. PLos Genetics. 2010;6 doi: 10.1371/journal.pgen.1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nature Reviews. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Fronicke L, Scherthan H, Gustavsson I, Chowdhary BP. Zoo-FISH delineates conserved chromosal segments in horse and man. Chromosome Research. 1996;4:218–225. doi: 10.1007/BF02254963. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Gustafson-Seabury A, Durkin K, Wagner ML, Goh G, Seabury CM, Brinkmeyer-Langford C, Lee EJ, Agarwala R, Stallknecht-Rice E, Schaffer AA, Skow LC, Tozaki T, Yasue H, Penedo MC, Lyon LA, Khazanehdari KA, Binns MM, MacLeod JN, Distl O, Guerin G, Leeb T, Mickelson JR, Chowdhary BP. A 4,103 marker integrated physical and comparative map of the horse genome. Cytogenet Genome Res. 2008;122:28–36. doi: 10.1159/000151313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T, Lee EJ, Kata SR. Exceptional conservation of horse-human gene order on X chromosome revealed by high resolution radiation hybrid mapping. Proc Natl Acad Sci USA. 2004a;101:2386–2391. doi: 10.1073/pnas.0308513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T, Santini A, Wallner B. A detailed physical map of the horse Y chromosome. Proc Natl Acad Sci USA. 2004b;101:9321–9326. doi: 10.1073/pnas.0403011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Render JA, Common RS, Kennedy FA, Jones MZ, Fyfe JC. Amylopectinosis in fetal and neonatal quarter horses. Veterinary Pathology. 1999;36:157–160. doi: 10.1354/vp.36-2-157. [DOI] [PubMed] [Google Scholar]
- Reynolds JA, Potter GD, Greene LW, Wu G, Carter GK, Martin MT, Peterson TV, Murray-Gerzik M, Moss G, Erkert RS. Genetic-diet interactions in the hyperkalemic periodic paralysis syndrome in quarter horses fed varying amounts of potassium: III the relationship between plasma potassium concentration and HYPP symptoms. Journal of Equine Veterinary Science. 1998;18:731–735. [Google Scholar]
- Ribeiro WP, Valberg SJ, Pagan JD, Gustavsson BE. The effect of varying dietary starch and fat content on serum creatine kinase activity and substrate availability in equine polysaccharide storage myopathy. Journal of Veterinary Internal Medicine. 2004;18:887–894. doi: 10.1892/0891-6640(2004)18<887:teovds>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Rijks JM, Hoffman JI, Kuiken T, Osterhaus ADME, Amos W. Heterozygosity and lungworm burden in harbour seals (Phoca vitulina) Heredity. 2008;100:587–593. doi: 10.1038/hdy.2008.18. [DOI] [PubMed] [Google Scholar]
- Rochfort S. Metabolomics reviewed: a new "omics" platform technology for systems biology and implications for natural products research. Journal of Natural Products. 2005;68:1813–1820. doi: 10.1021/np050255w. [DOI] [PubMed] [Google Scholar]
- Rudolph JA, Spier SJ, Byrns G, Hoffman EP. Linkage of hyperkalaemic periodic paralysis in quarter horses to the horse adult skeletal muscle sodium channel gene. Animal Genetics. 1992a;23:241–250. doi: 10.1111/j.1365-2052.1992.tb00136.x. [DOI] [PubMed] [Google Scholar]
- Rudolph JA, Spier SJ, Byrns G, Rojas CV, Bernoco D, Hoffman EP. Periodic paralysis in Quarter Horses: a sodium channel mutation disseminated by selective breeding. Nature Genetics. 1992b;2:144–147. doi: 10.1038/ng1092-144. [DOI] [PubMed] [Google Scholar]
- Sandmeyer LS, Breaux CB, Archert S, Grahn BH. Clinical and electroretinographic characteristics of congenital and stationary night blindness in the Appaloosa and the association with the leopard complex. Veterinary Ophthalmology. 2007;10:368–375. doi: 10.1111/j.1463-5224.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- Santschi EM, Purdy AK, Valberg SJ, Vrotsos PD, Kaese H, Mickelson JR. Endothelin receptor B polymorphism associated with lethal white foal syndrome in horses. Mammalian Genome. 1998;9:306–309. doi: 10.1007/s003359900754. [DOI] [PubMed] [Google Scholar]
- Santschi EM, Vrotsos PD, Purdy AK, Mickelson JR. Incidence of the endothelin receptor B mutation that causes lethal white foal syndrome in white-patterned horses. American Journal of Veterinary Research. 2001;62:97–103. doi: 10.2460/ajvr.2001.62.97. [DOI] [PubMed] [Google Scholar]
- Seltenhammer MH, Simhofer H, Scherzer S, Zechner R, Curik I, Solkner J, Brandt SM, Jansen B, Pehamburger H, Eisenmenger E. Equine melanoma in a population of 296 grey Lippizaner horses. Equine Veterinary Journal. 2003;35:153–157. doi: 10.2746/042516403776114234. [DOI] [PubMed] [Google Scholar]
- Shapiro J, McEwan B. Mechanobullous disease in a Belgian foal in eastern Ontario. Canadian Veterinary Journal. 1995;36:572. [PMC free article] [PubMed] [Google Scholar]
- Shin EK, Perryman L, Meek K. A kinase-negative mutation of DNA-PKcs in equine SCID results in defective coding and signal joint formation. The Journal of Immunology. 1997;158:3565–3569. [PubMed] [Google Scholar]
- Spier SJ, Carlson GP, Harrold D, Bowling A, Byrns G, Bernoco D. Genetic study of hyperkalemic periodic paralysis in horses. JAVMA. 1993;202:933–937. [PubMed] [Google Scholar]
- Spirito F, Charlesworth A, Linder K, Ortonne JP, Baird J, Meneguzzi G. Animal models for skin blistering conditions: absence of laminin-5 causes hereditary junctional mechanobullous disease in the Belgian horse. Journal of Investigative Dermatology. 2002;119:684–691. doi: 10.1046/j.1523-1747.2002.01852.x. [DOI] [PubMed] [Google Scholar]
- Studdert MJ. Primary, severe, combined immunodeficiency disease of Arabian foals. Australian Veterinary Journal. 1978;54:411–417. doi: 10.1111/j.1751-0813.1978.tb05562.x. [DOI] [PubMed] [Google Scholar]
- Swinburne J, Gerstenberg C, Breen M, Aldridge V, Lockhart L, Marti E, Antczak D, Eggleston-Stott M, Bailey E, Mickelson J, Røed K, Lindgren G, von Haeringen W, Guérin G, Bjarnason J, Allen T, Binns M. First comprehensive low-density horse linkage map based on two 3-generation, full-sibling, cross-bred horse reference families. Genomics. 2000;66:123–134. doi: 10.1006/geno.2000.6207. [DOI] [PubMed] [Google Scholar]
- Swinburne JE, Bogle H, Klukowska-Rotzler J, Drogemuller M, Leeb T, Temperton E, Dolf G, Gerber V. A whole-genome scan for recurrent airway obstruction in Warmblood sport horses indicates two positional candidate regions. Mammalian Genome. 2009;20:504–515. doi: 10.1007/s00335-009-9214-5. [DOI] [PubMed] [Google Scholar]
- Thirstrup JP, Pertoldi C, Loeschcke V. Genetic analysis, breed assignment and conservation priorities of three native Danish horse breeds. Animal Genetics. 2008;39:496–505. doi: 10.1111/j.1365-2052.2008.01767.x. [DOI] [PubMed] [Google Scholar]
- Treiber KH, Kronfeld DS, Hess TM, Byrd BM, Splan RK, Staniar WB. Evaluation of genetic and metabolic predispositions and nutritional risk factors for pasture associated laminitis in ponies. JAVMA. 2006;228:1538–1545. doi: 10.2460/javma.228.10.1538. [DOI] [PubMed] [Google Scholar]
- Trommershausen-Smith A. Lethal white foals in matings of overo spotted horses. Theriogenology. 1977;8:303–311. [Google Scholar]
- Trujillo JM, Stenius C, Christian LC. Chromosomes of the horse, donkey and mules. Chromosoma. 1962;13:243–248. doi: 10.1007/BF00577041. [DOI] [PubMed] [Google Scholar]
- Tryon RC, Penedo MCT, McCue ME, Valberg SJ, Mickelson JR, Famula TR, Wagner ML, Jackson M, Hamilton MJ, Nooteboom S, Bannasch DL. Evaluation of allele frequencies of inherited disease genes in subgroups of American Quarter Horses. JAVMA. 2009;234:120–125. doi: 10.2460/javma.234.1.120. [DOI] [PubMed] [Google Scholar]
- Tryon RC, White SD, Bannasch DL. Homozygosity mapping approach identifies a missense mutation in equine cyclophilin B (PPIB) associated with HERDA in the American Quarter Horse. Genomics. 2007;90:93–102. doi: 10.1016/j.ygeno.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Tryon RC, White SD, Famula TR, Schultheiss PC, Hamar DW, Bannasch DL. Inheritence of hereditary equine regional dermal asthenia in Quarter Horses. American Journal of Veterinary Research. 2005;66:437–442. doi: 10.2460/ajvr.2005.66.437. [DOI] [PubMed] [Google Scholar]
- Valberg SJ, Ward TL, Rush B, Kinde H, Hiraragi H, Nahey D, Fyfe J, Mickelson JR. Glycogen branching enzyme deficiency in quarter horse foals. Journal of Veterinary Internal Medicine. 2001;15:572–580. doi: 10.1892/0891-6640(2001)015<0572:gbediq>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- van Brantegem L, Decock HEV, Affolter VK, Duchateau L, Hoogewijs MK, Govaere J, Ferraro GL, Ducatelle R. Antibodies to elastin peptides in sera of Belgian draught horses with chronic progressive lymphoedema. Equine Veterinary Journal. 2007;39:418–421. doi: 10.2746/042516407x205888. [DOI] [PubMed] [Google Scholar]
- van Eldik P, van der Waaij EH, Ducro B, Kooper AW, Stout TAE, Colenbrander B. Possible negative effects of inbreeding on semen quality in Shetland pony stallions. Theriogenology. 2006;65:1159–1170. doi: 10.1016/j.theriogenology.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blocker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MCT, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guerin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Roed KH, Ryder OA, Searle S, Skow L, Swinburne JE, Syvanen AC, Tozaki T, Valberg SJ, Vaudin M, White JR, Zody MC, Lander ES, Lindblad-Toh K Broad Institute Genome Sequencing Platform, Broad Institute Whole Genome Assembly Team. Genomic sequence, comparative analysis and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner ML, Valberg SJ, Ames EG, Bauer MM, Wiseman JA, Penedo MCT, Kinde H, Abbitt B, Mickelson JR. Allele frequency and likely impact of the glycogen branching enzyme deficiency gene in Quarter Horse and Paint Horse populations. Journal of Veterinary Internal Medicine. 2006;20:1207–1211. doi: 10.1892/0891-6640(2006)20[1207:afalio]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ward TL, Valberg SJ, Adelson DL, Abbey CA, Binns MM, Mickelson JR. Glycogen branching enzyme (GBE1) mutation causing equine glycogen storage disease IV. Mammalian Genome. 2004;15:570–577. doi: 10.1007/s00335-004-2369-1. [DOI] [PubMed] [Google Scholar]
- Ward TL, Valberg SJ, Lear TL, Guerin G, Milenkovic D, Swinburne JE, Binns MM, Raudsepp T, Skow L, Chowdhary BP, Mickelson JR. Genetic mapping of GBE1 and its association with glycogen storage disease IV in American Quarter Horses. Cytogenetic and Genome Research. 2003;102:201–206. doi: 10.1159/000075749. [DOI] [PubMed] [Google Scholar]
- White SD, Affolter VK, Bannasch DL, Schultheiss PC, Hamar DW, Chapman PL, Naydan D, Spier SJ, Rosychuk RAW, Rees C, Veneklasen GO, Martin A, Bevier D, Jackson HA, Bettenay S, Matousek J, Campbell KL, Ihrke PJ. Hereditary equine regional dermal asthenia ('hyperelastosis cutis') in 50 horses: clinical, histological, immunohistochemical and ultrastructural findings. Veterinary Dermatology. 2004;15:207–217. doi: 10.1111/j.1365-3164.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- White SD, Affolter VK, Schultheiss PC, Ball BA, Wessel MT, Kass P, Molinaro AM, Bannasch DL, Ihrke PJ. Clinical and pathological findings in a HERDA affected foal for 1.5 years of life. Veterinary Dermatology. 2007;18:36–40. doi: 10.1111/j.1365-3164.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- Wiler R, Leber R, Moore BB, VanDyk LF, Perryman L, Meek K. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc Natl Acad Sci USA. 1995;92:11485–11489. doi: 10.1073/pnas.92.25.11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer C, Dierks C, Hamann H, Distl O. Associations between candidate gene markers at a quantitative trait locus on equine chromosome 4 responsible for osteochondrosis dissecans in fetlock joints of South German coldblood horses. Journal of Heredity. 2008;99:125–129. doi: 10.1093/jhered/esm106. [DOI] [PubMed] [Google Scholar]
- Witzel DA, Smith EL, Wilson RD, Aguirre GD. Congenital stationary night blindness: an animal model. Invest Ophthalmol Visual Sci. 1978;17:788–795. [PubMed] [Google Scholar]
- Wolc A, Bresinka A, Szwaczkowski T. Genetic and permanent environmental variability of twinning in Thoroughbred horses estimated via three threshold models. J Anim Breed Genet. 2006;123:186–190. doi: 10.1111/j.1439-0388.2006.00575.x. [DOI] [PubMed] [Google Scholar]
- Yang F, Fu B, O'Brien PC, Nie W, Ryder OA, Ferguson-Smith MA. Refined genome-wide comparative map of the domestic horse, donkey and human based on cross-species chromosome painting: insight into the occasional fertility of mules. Chromosome Research. 2004;12:65–76. doi: 10.1023/b:chro.0000009298.02689.8a. [DOI] [PubMed] [Google Scholar]
- Yang GC, Croaker D, Zhang AL, Manglick P, Cartmill T, Cass D. A dinucleotide mutation in the endothelin-B receptor gene is associated with lethal white foal syndrome (LWFS); a horse variant of Hirschsprung disease. Hum Mol Genet. 1998;7:1047–1052. doi: 10.1093/hmg/7.6.1047. [DOI] [PubMed] [Google Scholar]
- Young AE, Bower LP, Affolter VK, DeCock HEV, Ferraro GL, Bannasch DL. Evaluation of FOXC2 as a candidate gene for chronic progressive lymphedema in draft horses. The Veterinary Journal. 2007;174:397–399. doi: 10.1016/j.tvjl.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Yuan ZQ, Bennett L, Campos MS, Nasir L. Bovine papillomavirus type E2 and E7 proteins down-regulate Toll Like Receptor 4 (TLR4) expression in equine fibroblasts. Virus Research. 2010;149:124–127. doi: 10.1016/j.virusres.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Zeitz A, Spotter A, Blazyczek I, Diesterbeck U, Ohnesorge B, Deegen E, Distl O. Whole-genome scan for guttural pouch tympany in Arabian and German warmblood. Animal Genetics. 2009;40:917–924. doi: 10.1111/j.1365-2052.2009.01942.x. [DOI] [PubMed] [Google Scholar]