Abstract
This article reviews current advances in the genetics of substance use disorders (SUDs). Both genetic and environmental sources of risk are required to develop a complete picture of SUD etiology. Genetic sources of risk for SUDs are not highly substance specific in their effects. Genetic and environmental risks for SUDs typically do not only add together but also interact with each other over development. Risk gene identification for SUDs has been difficult, with one recent success in identifying nicotinic receptor variants that affect risk for nicotine dependence. The impact of genetic variants on SUD risk will individually be small. Although genetic epidemiologic methods are giving us an increasingly accurate map of broad causal pathways to SUDs, gene discovery will be needed to identify the specific biological systems. Identifying these risk genes and understanding their action will require large clinical samples, and interaction between these studies and work in model organisms.
This article selectively reviews recent advances in the genetic epidemiology and molecular genetics of SUDs. In genetic epidemiology, gene variation is not measured directly. Instead, the action of genetic and environmental factors is inferred from patterns of resemblance in special classes of relatives, particularly twins and adoptees. Molecular genetic studies relate disease risk directly to DNA variation.
SUDs consist of a range of syndromes reflecting harmful and damaging use of psychoactive substances. Current clinical thinking has emphasized two main forms. Substance abuse is defined as “a maladaptive pattern of substance use manifested by recurrent and significant adverse consequences related to the repeated use”1. Substance dependence, the more severe and more frequently studied form of SUDs, is defined as “a cluster of cognitive, behavioral, and physiological symptoms indicating the individual continues use of the substances despite significant substance-related problems. There is a pattern of repeated self-administration that usually results in tolerance, withdrawal and compulsive drug-seeking behavior”1. In this review, we also examine substance use, typically measured by the quantity and frequency of drug consumed. Heavy intermittent or chronic use is necessary but not sufficient for SUDs.
Introduction
Classical twin studies, which compare the similarity of monozygotic and dizygotic twins, are a powerful method for assessing the magnitude of the impact of aggregate genetic risk factors in complex diseases such as SUD. Furthermore, they have been crucial in understanding causes of comorbidity and clarifying the etiologic inter-relationship of genetic and environmental factors. Although they are not without methodological limitations, a careful review of the impact of these limitations suggests that, in most cases, biases are modest2.
Twin studies provide substantial evidence that genetic factors contribute to the etiology of SUDs. Across studies, the weighted mean estimates of SUD heritability—the proportion of variability in risk in a population due to genetic differences between individuals—range from 40% to 70% across different psychoactive substances3–5. The abuse of cocaine may be more heritable than that of other substances. Otherwise, heritability estimates for SUD for most other psychoactive substances in populations of European origin fall largely between 40 and 55%—similar to or greater than that seen for most common chronic biomedical diseases afflicting Western populations.
How specific are the genetic risk factors for SUDs? Studies that address this question have tended to discriminate between illicit and licit psychoactive substances. Illicit substances include drugs that are illegal (for example, cocaine or hallucinogens) and legal prescription drugs (for example, codeine) used in ways other than medically indicated. Western societies have generally deemed three psychoactive drugs to be ‘licit’ and appropriate for use by its adult citizens: caffeine, nicotine and ethanol. Although the specific symptoms of intoxication, tolerance and withdrawal differ substantially across specific psychoactive substances, and many physiological effects differ substantially across drugs, there is no clear physiological division between the effects of licit versus illicit substances.
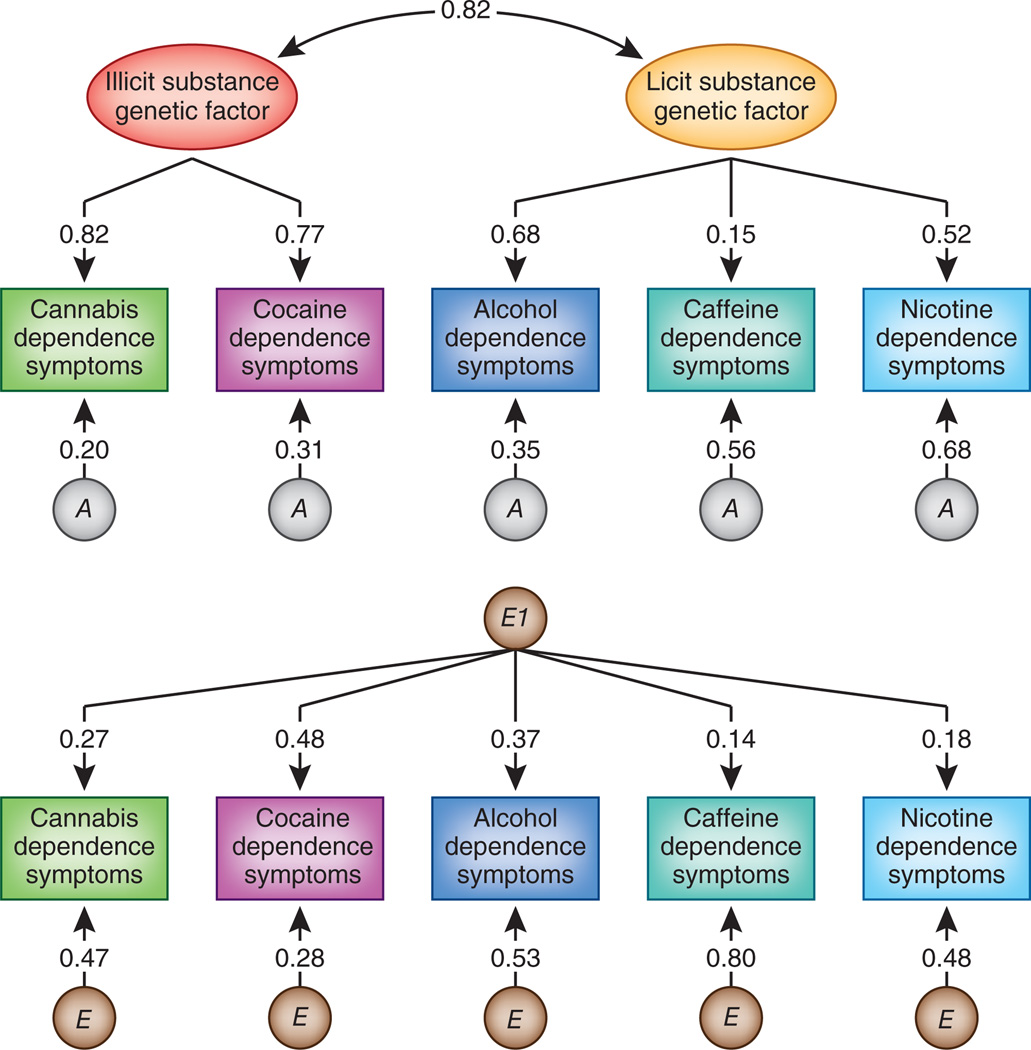
Two large-scale twin studies of illicit drugs showed that most genetic risk is common across the abuse of or dependence on different psychoactive substances6,7. One study examined the specificity of risk factors for both licit and illicit SUDs8 (Fig. 1). Genetic liabilities to licit and illicit SUDs are separable but highly correlated. Cannabis and cocaine dependence are strongly influenced by an illicit substance dependence factor. Loadings for alcohol and nicotine dependence on the licit substance dependence factor—that is, the degree to which the dependence symptoms for these drugs reflect the underlying common factor—are more modest, and the loading for caffeine dependence is low. Genetic influences specific to each substance are substantial for nicotine and caffeine, but less so for the other substances.
Figure 1.
Parameter estimates for the best-fitting confirmatory model for symptoms of cannabis, cocaine, alcohol, caffeine and nicotine dependence. The double-headed arrow connecting the illicit and licit substance genetic factors represents the genetic correlation between these factors. A stands for additive genetic and E for environmental effects unique to each twin. E1 reflects the common factor for the unique environmental effects that affects risk for dependence across all of the psychoactive substances examined. The top part of the figure depicts genetic factors that influence drug dependence and the bottom part reflects environmental factors. The numbers on the paths represent standardized loadings and thus need to be squared to reflect the proportion of variance in the observed SUD accounted for by the factor.
How do the genetic risk factors for SUDs fit into the broader scheme of genetic risk for other psychiatric disorders? Genetic risk factors for SUD are part of a broad genetic liability to “externalizing” disorders, which includes conduct disorder, antisocial personality and probably personality traits related to poor impulse control and sensation seeking9,10. Although the precise mechanism of comorbidity is under debate, the bulk of evidence suggests that it arises because a shared set of risk factors (both genetic and environmental) predispose to both SUD and these externalizing behavioral disorders and traits.
Finally, preliminary studies have examined whether individual SUD diagnoses reflect single or multiple dimensions of genetic risk. Studies of a wide range of alcohol-related responses in rodents indicate that they often reflect independent genetic influences. A recent large-scale twin study found three distinct dimensions of genetic risk underlying the criteria set forth in the Diagnostic and Statistical Manual of Mental Disorders IV (ref. 1) (DSM-IV) for alcohol dependence. These reflect (i) liability to heavy drinking and tolerance, (ii) self-recognition of alcohol-related problems, loss of control, desire to quit, preoccupation and activities given up, and (iii) withdrawal and continued use despite problems. These three dimensions of genetic risk are only modestly correlated in alcohol-dependent subjects and differentially predict risk for other psychiatric and substance use disorders and personality traits.
Genes and environment: how they contribute to SUDs
Genetic epidemiological studies have made progress in clarifying how genetic and environmental factors jointly influence substance use and misuse. Social environmental experiences are critical for SUDs, as psychoactive drugs are almost always provided through social contacts and social environments can powerfully discourage or encourage drug use. Genes and environment jointly affect liability to SUD in three major ways: developmental inter-relationships, gene-environment correlation and gene-environment interaction.
Developmental inter-relationships
Within a developmental framework, how do shared environmental experiences in the home and community compare to genetic factors in the timing of their influence on the use of psychoactive substances? In considering this question, it is critical to move away from the common static concepts of genetic and environmental influences on human behavior. Genes are temporally dynamic, responding both to internal developmental processes (for example, puberty) and to challenges external to the organism. One such prominent challenge is alterations in the social environment that are often particularly dramatic during adolescence.
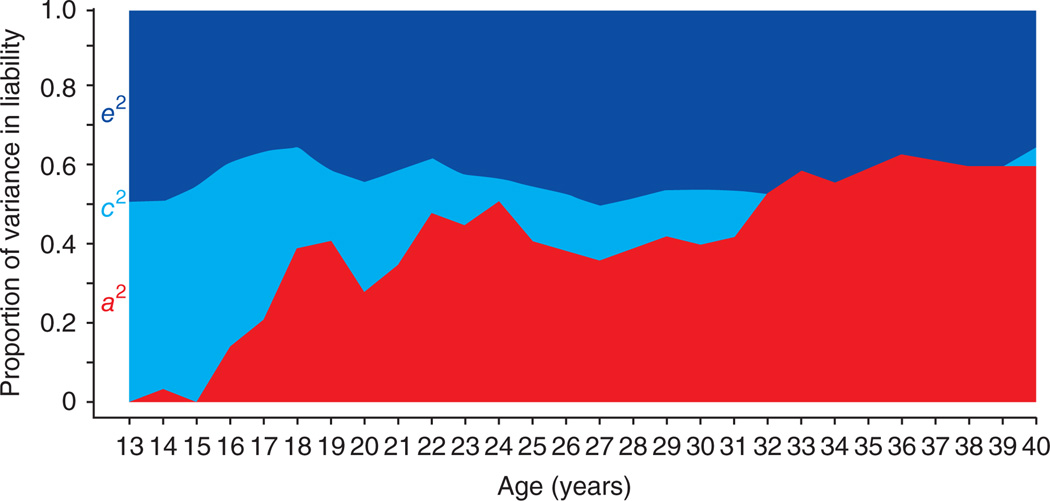
When twins were asked to record their average daily cigarette usage over their lifetime11, analyses of their responses yielded the following results (Fig. 2). In the early years of adolescence, twin resemblance for smoking is entirely the result of environmental experiences shared between the twins; for example, from the home, school and/or from peer groups. As twins age and smoke more, genetic factors begin to emerge (probably because of genetic differences in their responses to nicotine and vulnerability to the development of dependence). From age 16 to 30, genetic factors become progressively more important in their influence on cigarette use, while shared environmental experiences become progressively less important. Similar patterns are seen for other substances of abuse11.
Figure 2.
Parameter estimates for the contributions to variation in liability to psychoactive drug use of additive genetic effects (a2, red), familial environmental factors (c2, light blue) and the individual-specific environment (e2, dark blue) by year for average daily number of cigarettes, ages 13–40. Data from ref. 11.
These results suggest that high levels of SUD typically arise through a two-stage process. First, during the critical developmental period, individuals are exposed to an environment that encourages and supports substance use. Second, once exposed to the psychoactive substance, their genotypes confer vulnerability to its rewarding properties and/or insensitivity to its negative effects so that heavy use can lead to the development of dependence. In Western societies, the critical period for drug experimentation is adolescence to young adulthood, although the particular age for each substance varies as a function of drug availability and the associated societal regulations and sanctions.
Gene-environment correlation
Most individuals think about genes and environment as two separate and independently acting sets of risk factors. For SUD, this is unlikely to be true. Genetic and environmental risk factors for SUD can correlate, interact or both. We provide detailed definitions below, but here is the short version. Gene-environment correlation arises when genetic and environmental risks are positively inter-related within a population; that is, when people who have high genetic risk are on average also exposed to more environmental risk than average. Gene-environment interaction, by contrast, occurs when the impact of genetic risk factors is moderated by the environment. That is, the same set of genetic factors might have a powerful effect on risk for SUD in one environment and a weak effect in another environment.
Gene-environment correlation for SUD (perhaps more accurately termed “genetic controls of exposure to the environment”12) largely arises because genetic risk factors for SUD influence individuals to select themselves into environments that convey a high risk for substance use and abuse. The high-risk environment is here mediational—sitting in the causal pathway between genes and the end-phenotype of SUD. This process is well illustrated by the social construct of ‘peer deviance’, defined as the degree to which close friends engage in antisocial behaviors, ranging from underage smoking, cutting school and cheating on tests to use of hard drugs, stealing and selling drugs. High levels of peer deviance in adolescence strongly predict subsequent SUDs. However, individuals do not associate with their peers at random. Rather, people actively select individuals to socialize with who have similar proclivities. Although an ‘environmental’ measure, peer deviance is substantially influenced by genetic factors, and these genetic factors increase in importance during development13. So, part of the pathway to SUD looks like this: genes → temperament → peer deviance → SUD.
Thus for SUD, which is so clearly influenced by social phenomena, two pathways exist from genes to trait. First is the traditional ‘inside the skin’ pathway typically considered by molecular biology (whereby gene variants influence how the drug is absorbed and interacts with receptors and second messengers, as well as hedonic and motivational pathways). Second is an ‘outside the skin’ pathway, as we illustrated for peer deviance, whereby genetically mediated personality traits affect the degree to which the individual selects themselves into drug-predisposing or protective social environments.
Gene-environment interaction
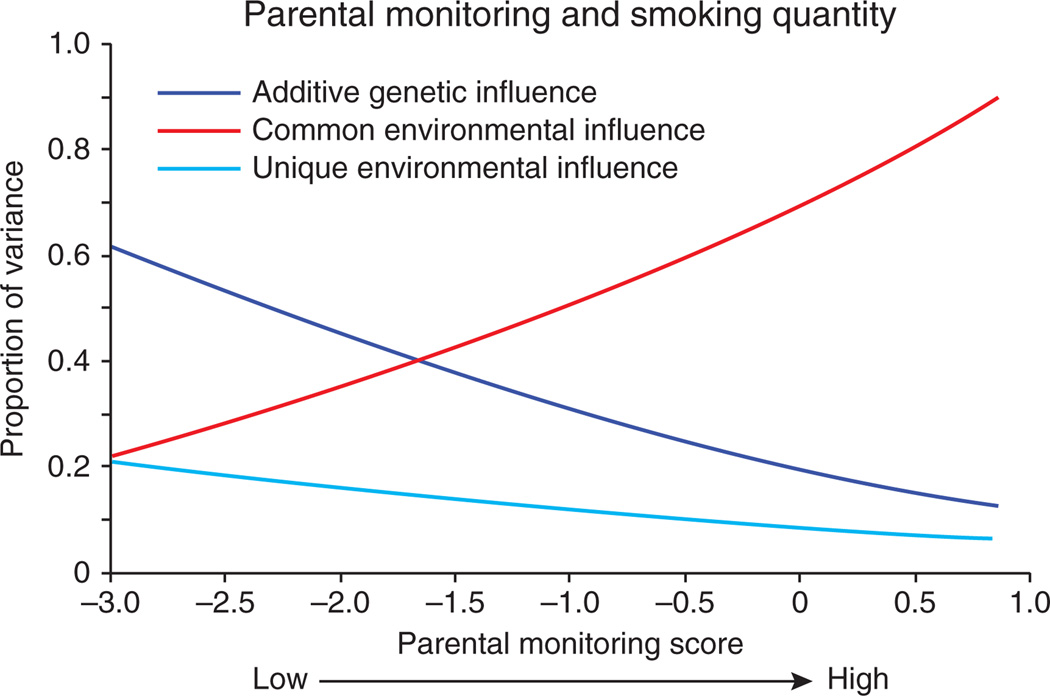
There is accumulating evidence from twin studies that the importance of genetic influences on substance use and SUD varies as a function of environmental exposure (this is logically equivalent to saying that the importance of environmental influences varies as a function of genotype). The development of substance use problems is, in some sense, by definition a gene by environment interaction, as access to the substance is an ‘environmental’ condition that is necessary for the development of problems associated with use of the substance. Accordingly, it is not surprising that environmental conditions that affect access to the substance moderate the degree to which genetic influences act on risk for SUD. For example, genetic influences on adolescent smoking were attenuated after legislation took effect to prohibit smoking in public places14. Similarly, genetic influences on adolescent alcohol use are greater in neighborhoods with higher alcohol sales15, presumably through easier access for those with a predisposition. In fact, many of the environments that reduce genetic influences on substance use seem to operate through social control. Environments that are more controlling and provide greater social structure reduce the impact of genetic risk for SUD. For example, religiosity seems to moderate genetic influences on alcohol use, with genetic factors playing a reduced role among individuals with a religious upbringing16. A similar effect has been reported for smoking, whereby high religiosity attenuates genetic effects on initiation of tobacco use17. The importance of genetic influences on adolescent alcohol use is reduced in rural settings and neighborhoods with less migration and presumably increased stability (which likely leads to enhanced community monitoring of adolescent behavior)15. Genetic influences on adolescent smoking are attenuated under conditions of higher parental monitoring18 (Fig. 3). Greater control and structure, at the levels of legislative restrictions, social institutions such as religion, interpersonal relationships (for adults), parenting (for adolescents) and neighborhoods, can all reduce the importance of individual genetic predispositions toward substance use.
Figure 3.
An example of gene-environment interaction: as parental monitoring increases, the importance of genetic factors significantly decreases, and the influence of common environmental factors becomes more important such that the most important etiological factors affecting adolescent smoking vary markedly as a function of parental monitoring. Data from ref. 18; graph shows results of model-fitting as described in detail in ref. 18.
Conversely, a related mechanism for gene-environment interaction is that of social expression or social triggering. Some environments allow greater opportunity to express genetic predispositions, and in these environments genetic influences become more potent. For example, genetic influences on smoking are highest in schools in which the most popular students smoke19. In Sweden, rates of tobacco use were very low in women born early in the twentieth century, rising over the next 50 years to rates comparable to those in men. The magnitude of genetic influences on tobacco use in women also rose markedly during that time period20. As smoking became more socially acceptable among women, genetic influences had greater opportunity to affect smoking patterns20. Social triggering, the effects of which can be cleanly separated from those of genetic influences, may act by increasing access to the substance or by creating social settings that are more accepting of substance use, both of which would allow enhanced opportunity to express genetic predispositions.
Yet another potential mechanism of relevance for SUDs is the connection to stress. Stress and stress response have long been thought to contribute to SUDs21. One possible mechanism involves the hypothalamic-pituitary-adrenal axis, which is activated by substance use. Circulating glucocorticoids then downregulate hypothalamic-pituitary-adrenal axis activity but also activate extrahypothalamic brain stress systems (for example, the central nucleus of the amygdala and the basolateral amygdala) known to be involved in behavioral responses to stressors.
The impact of stress on genetic risk for SUDs has not been as widely studied in the twin literature as mechanisms related to social control and opportunity; however, one study found that genetic influences on a composite of externalizing disorders (including substance use problems) increase as stressful life events increase22, an effect we have also found in adolescent Finnish twins (D.D. et al., unpublished data). This presumably would fall under a social triggering mechanism; however, the underlying mechanism for stress triggering a genetic predisposition toward alcohol use seems likely to be etiologically different than, for example, peer substance use triggering adolescent substance use. Further delineation of these risk pathways is clearly needed, and more work on the genetic epidemiology of the relationship between stress or adversity and genetic influences on substance use disorders is warranted. This seems especially relevant because most of the human studies testing for gene-environment interaction with specific risk genes in the area of substance use have focused on stress23–25, most having small sample sizes and thus far yielding mixed results. Greater integration of the literature on gene-environment interactions emerging from twin studies and tests of measured gene by environment interaction would benefit the field.
Building contingency in models for substance use disorders
Genetic studies of SUD need to cope with one obvious fact: genes for SUD cannot act in the absence of substance use initiation—that is, the subject starting to actually consume the substance in pharmacologically relevant amounts. Therefore, to understand SUD, we have to study the process of substance use initiation and clarify how genetic and environmental factors influence that critical first stage: the decision whether or not to consume the substance. Then, we can turn our attention to understanding the degree to which risk factors that affect substance use initiation are the same as those that affect SUD given substance use initiation. But how can we do this if we know nothing about the risk of an individual for SUD if they never initiated use of the substance?
Twin studies offer a way around this problem. We can estimate the strength of this relationship in twin data because the co-twin data provide a proxy form of information about the relationship between substance use initiation and SUD, by comparing the rates of SUD in the co-twins of those who do versus those who do not initiate. To model this, each variable has its own genetic and environmental risk factors that are specific to either substance use initiation or progression to SUD. All covariance between these two variables is assumed to arise through a regression path. This contingent causal common pathway (CCC) model has a number of extensions, including the multivariate case and more than two-stage phenomena26. The key is the use of twins to overcome the problem of the systematically missing data of not knowing the risk for SUD of individuals who never initiated.
This CCC model has been most extensively applied to smoking and nicotine dependence27 and more recently to cannabis28,29. The results for nicotine demonstrate a moderate role of genetic factors in smoking initiation, a clear set of genes for nicotine dependence that ‘come on line’ only with smoking initiation (which could reflect changes in gene expression or physiology such that genes serving other functions now affect risk for nicotine dependence) and a moderate to strong relationship between these two stages. The genetic risk for smoking initiation is more highly correlated with the genetic risk for progression to nicotine dependence than many neurobiologists might think. Furthermore, shared environment (for example, rates of smoking in peers) typically affects smoking initiation but not the progression from initiation to dependence. Likewise, genetic risk for symptoms of cannabis abuse also ‘come on line’ conditional on cannabis initiation. Moreover, an extended version of the CCC model that includes ‘upstream’ environmental risk factors has shown that risk of cannabis initiation and subsequent progression to cannabis abuse can be explained, in part, by environmental factors influencing cannabis availability28. One study examined CCC models across different classes of psychoactive substances29. The association between risk factors for initiation and risk factors for abuse or dependence was strongest for cannabis, intermediate for cocaine and weakest for sedatives and non-cocaine stimulants.
Studies on alcohol abuse and dependence have typically not been conditioned on alcohol initiation but instead only include drinkers in the analyses, as most subjects in most populations have been exposed to alcohol. This assumes that the dimensions underlying substance use initiation and SUD are independent. Alternatively, non-users are sometimes included in the analyses but given zero scores on measures of SUD. This can be problematic because non-users really have unknown genetic risk.
This problem of contingency is also important, but sometimes ignored, in molecular genetic studies of SUDs. An appropriate control subject for a genetic study of drug dependence should have used the drug but not developed dependence. Without exposure, their specific genetic risk to dependence is unknown. However, rarely do we know how much exposure is needed to express the genetic potential for SUD. Is one cigarette enough, or one snort of cocaine? Getting a group of subjects exposed to a psychoactive drug without progression to SUD is not difficult for commonly used substances like nicotine and alcohol. It is more difficult for the ‘harder’ drugs, where exposure is rarer. If unexposed subjects are used as controls for such studies, then any differences in marker frequency factors between the SUD and control groups reflects some mixture of genetic effects on both initiation and dependence given initiation.
How to best define an SUD phenotype
Advances in genetic research of SUDs depend heavily on optimal phenotypic measurement. For SUDs, this is a particularly challenging issue. It is not clear that the traditional categorical models used in psychiatry are optimal for SUDs1. It may make little sense to apply—as done in DSM-IV—the same set of criteria for dependence to all substances of abuse. These criteria were selected using expert consensus, but there is no a priori reason to assume that they are optimal or that SUDs are distinct categories in nature rather than continua. Fortunately, however, these issues are open to empirical investigation, and there has been progress, both methodological and substantive, in this area in recent years30,31.
One key question is whether individual differences in SUDs are quantitative (that is, a matter of degree) or qualitative (different in kind), or arise from a combination of both mechanisms. Statistical methods to distinguish between these processes have advanced considerably in the past decade. Space precludes a detailed review of these statistically sophisticated methods32. Tried and true methods such as factor analysis (exploratory and confirmatory forms) are used to see whether SUD symptoms define clear and sensible factors that can be used as quantitative indices for genetic analysis. Latent class analysis attempts to determine whether drug abuse symptoms covary only because the population consists of two or more heterogeneous groups (that is, affected and unaffected) that differ in symptom rates. Historically, latent factor and class approaches have been used independently and for different purposes, when in reality each represents a competing hypothesis regarding why key SUD symptoms correlate with each other. Factor mixture models combine these latent factor and latent class approaches while circumventing the limitations of each31. In principle, this hybrid modeling strategy may fit diagnostic data better and, if so, provide a superior measurement and classification for SUDs. Tests comparing these competing approaches have so far revealed that, for substances like cannabis, a single latent factor remains the optimal means of assessing overall risk when measured by use, abuse, dependence and withdrawal criteria33.
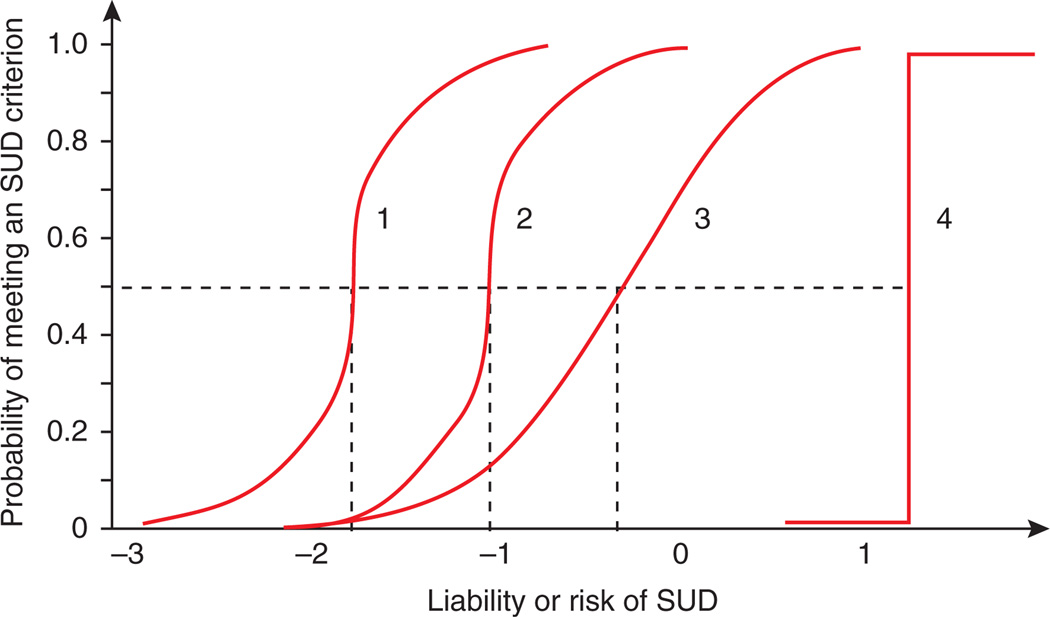
Finally, item-response theory (IRT, where “item” typically refers to a single question or criterion) is being increasingly applied to calibrate and assess the performance of individual drug use symptomatology34. As illustrated using item response curves (Fig. 4), IRT can determine where on the dimension of SUD risk (ranging from low to high) each diagnostic criterion provides information and how well items discriminate between those with high and low liability to SUD. Using this approach, it has been shown that using cocaine is associated with more detrimental and negative consequences, whereas the risk of experiencing such outcomes as a result of cannabis or hallucinogen use is considerably lower34.
Figure 4.
Hypothetical item response curves for SUD diagnostic criteria. The item depicted by curve 1 is a criterion likely to be met by all individuals, except those with very low risk of SUD. It discriminates well between those who meet it and those who do not because the curve is steep, and this is also true curve 2, which has the same steepness but distinguishes between individuals 1 s.d. higher on the liability distribution. Curve 3 is the flattest and is consequently the least discriminating because the probability of showing this symptom changes more slowly as the risk of SUD increases. Curve 4 discriminates perfectly, but it is only useful for measuring individuals close to the 1.5 s.d. point on the distribution.
Application of these methods to large epidemiological databases suggests that substance abuse and dependence are not separate disorders, nor are symptoms of abuse ‘milder’ than those of dependence. Instead, an emerging consensus for most illicit substances is that the diagnostic criteria for abuse and dependence—previously considered to be different manifestations of SUDs—actually measures a single underlying dimension of liability.
IRT modeling can also be used to assess differences in item functioning by sex and age. That is, do the items that are used to measure SUD mean the same thing for men and women and at during adolescence, young adulthood and late adulthood? An application of this type of modeling to nicotine dependence items showed that by allowing for limited measurement differences—due to differential sensitivity of items by sex—inferences about heterogeneity of heritability by sex at the latent factor become more meaningful. In this application, for example, individual items in the scale are somewhat more discriminating of the level of nicotine dependence in men than in women35. An item response framework is to be preferred over a sum score approach in which differential item functioning would be obscured and each item weighted equally regardless of its correlation with the latent construct to be measured. Finally, as was shown previously in using the CCC model to estimate the heritability of nicotine dependence, and repeated in the IRT analysis, it is important to include initiation to obtain unbiased estimates of the factor loadings and thresholds. Factor scores from such an analysis should provide a more accurate quantitative phenotype that may improve the ability to find and validate susceptibility genes for SUD.
A critical related issue in both prevention and treatment of SUDs is that addiction cannot be measured directly before substance use onset, or during remission. Accordingly, it is important to develop measures of addiction propensity36 that could reflect genetic and/or environmental risk and that would predict the likelihood of abuse and dependence should initiation or relapse occur. In both human and animal studies, trait and state measures related to neurobiological systems involved in reward, punishment, motivation, stress response, impulsivity, externalizing and social behavior are useful in this regard37,38.
The molecular genetics of substance use disorders
In this section, we review recent advances in molecular genetic studies of nicotine dependence and alcohol dependence as representative of the kind of results and problems confronted in isolating genetic variants in human populations than contribute to risk for SUDs. Two paradigms dominate the field. Candidate gene association studies examine allelic variation in specific genes, selected on the basis of prior evidence, in SUD cases versus controls. Genome-wide association studies (GWAS) examine up to a million genetic polymorphisms across the genome for association with the phenotype of interest. The latter approach has the advantage of allowing an unbiased assessment of risk variants and the disadvantage of needing very stringent significance levels because of the large number of tests performed: nominal P-values must be less than 5 × 10−8 to remain significant after Bonferroni correction for the very large number of genetic markers tested. To detect genetic effects of modest size that are typical for complex traits like SUD, GWAS studies require very large samples. Efforts are under way to reduce the number of tests performed on GWAS data—for example, by looking at individual genes or networks of genes—and thereby increase statistical power.
Molecular variants, smoking and nicotine dependence
Genetic studies of smoking and nicotine dependence have recently made significant progress with both candidate gene and GWAS approaches. In 2007, the non-synonymous single-nucleotide polymorphism (SNP) rs16969968, which changes the amino acid from aspartate to asparagine in the α5 subunit of the nicotinic acetylcholine receptor (CHRNA5), was identified as a risk variant associated with nicotine dependence39. Several independent studies40–42, including our own43, quickly confirmed the association of this and another highly correlated SNP, rs1051730, with several inter-related measures of nicotine dependence, including number of cigarettes smoked per day (CPD) and nicotine dependence as assessed by the Fagerström test for nicotine dependence44. In more recent investigations, including collaborative studies involving very large samples45–47, the CHRNA5–CHRNA3–CHRNB4 locus has been consistently shown to be associated with CPD with extremely low P-values. Several statistically independent association signals appear across this region46. A priori, it seemed likely that variants in nicotinic receptors would be specific to genetic risk for nicotine dependence. As a reminder of how potentially complex the genetic substrate for SUD may be, however, several studies have now suggested that these genes may contain variants associated with risk for alcohol, cocaine and opiate use and misuse43,48–50.
This same non-synonymous SNP is associated with both lung cancer41,51,52 and chronic obstructive pulmonary disease46,53,54. Many of these associations (but perhaps not all) are mediated through smoking quantity42,46,55,56. This is a powerful illustration of an ‘outside-the-skin gene pathway’ for a cancer risk gene. A genetic variant in a nicotinic receptor increases risk for nicotine dependence, which then causes individuals to go out into the environment, purchase cigarettes and repeatedly inhale the smoke into their lungs, thereby increasing their risk for lung cancer. This is a strange kind of oncogene, one whose effect on cancer is entirely indirect. The gene increases risk for nicotine dependence and it is the drug that is ingested as a result of the dependence that causes all (or at least most) of the oncogenesis.
An ongoing study of these nicotinic receptor variants57 illustrates an important issue concerning phenotypic assessment. Using reports of CPD, the statistical signal detected for SNP rs16969968—although highly significant with very large samples—is small. But when this genetic variant is instead used to predict concentrations of serum cotinine (a long-lasting metabolite of nicotine), the association becomes much stronger. Presumably this difference arises because self-reports of CPDs are a poor measure of the amount of nicotine absorbed. This study may provide important lessons for studies of other SUDs. The power of genetic analysis is closely intertwined with the quality of the phenotypic measures.
In addition to the CHRNA5–CHRNA3–CHRNB4 locus, GWAS have identified, in distinct genomic locations, several SNPs in CHRNAB3–CHRNA6, EGLN2–CYP2A6 and LOC100188947 reaching genome-wide significance for association with CPD47,58. CHRNB3, CHRNA6 and CYP2A6 have been widely studied as candidates for smoking and nicotine dependence, and the results seem convincing. One of these same studies47 also identified brain-derived neurotrophic factor (BDNF) as a candidate gene for smoking initiation.
The establishment of CHRNA5–CHNRA3–CHRNB4 as a risk locus for nicotine dependence has opened a window for functional studies seeking to understand etiologic mechanisms. In HEK-293T human embryonic kidney cells, recombinant receptors for the risk allelic variant of rs16969968 are less responsive to the nicotinic receptor agonist epibatidine, whereas the binding capacity and the amount of protein remain the same as those of the wild-type allele59. Consistent with this result, a study in Xenopus laevis oocytes suggested that the receptor with the asparagine risk allele has lower Ca2+ permeability and desensitizes more quickly than the wild-type aspartate allele in the (α4β2)2α5 receptor, a principal subtype of nicotinic receptor in the central nervous system, but not in (α3β4)2α5 or (α3β2)2α5 subtypes, which are mainly located in the peripheral nervous system60. These results collectively suggest that the α5 subunit modulates the activities of nicotinic receptors and that the asparagine allele makes these receptors less efficient, thereby resulting in an increased consumption of tobacco. Of interest, a human imaging study suggested a possible neural mechanism for the effect of the rs16969968 variant61. It was shown to affect the resting state functional connectivity of the dorsal anterior cingulate–ventral striatum circuit. The risk asparagine allele was associated with decreased signal strength of this circuit, which in turn predicted the severity of nicotine dependence.
Some other lessons can be tentatively learned from the CHRNA5–CHNRA3–CHRNB4 locus62. First, it is now relatively clear that multiple, statistically independent signals coexist in this relatively small genomic region. The associated linkage disequilibrium blocks are substantially larger than conventionally conceived, and they cover several genes. The association signals observed extend to IREB2, covering a genomic distance of more than 200 Kb (refs. 46,53). Second, as noted above, even the same variant affects multiple phenotypes. The CHRNA5–CHNRA3–CHRNB4 locus has been shown to be associated with nicotine dependence, lung cancer, chronic obstructive pulmonary disease, alcohol dependence and cocaine dependence. Third, somewhat troublingly, using different methods for assessing the broad construct of nicotine dependence leads to different results. Traditionally, nicotine dependence is defined either by the American Psychiatric Association’s Diagnostic and Statistical Manuals1 or the Fagerström Test for Nicotine Dependence44. However, these two measures do not agree well. More interestingly, CPD, which was generally considered to be only a rough index of nicotine dependence inferior to more complete assessment tools, has often proven to be the most robust phenotype for this locus.
To make matters more complex, we examined, in a large, population-based twin sample, the association between the key SNPs in this small region and individual items from the Fagerström Test for Nicotine Dependence63. If all the items were reflecting one common underlying genetically influenced trait, they all should have similar associations with these SNPs. Not so. Most of the SNPs had the strongest signals with CPD and time to first cigarette after awakening—typically an item that is quite a sensitive measure of the level of nicotine dependence. But one SNP, rs2869546, had an isolated strong signal from the item about smoking in bed when ill.
Studies of the genetics of SUDs are perforce intertwined with questions of measurement. The hope is that the process might be iterative, whereby genetic findings will help clarify measurement issues, which will in turn increase the significance of genetic findings. However, there may be different spectra of phenotypes associated with different variants, so a ‘one size fits all’ solution to this fundamental measurement problem may not exist.
Molecular variants and alcohol dependence
Gene-discovery studies of human alcohol dependence have evolved considerably from the basic test for differences in the frequency of specific alleles between diagnosed cases and screened controls. Such studies now often include a range of categorical and quantitative phenotypes, reflecting our growing understanding that multiple underlying risk pathways influence alcohol dependence. Some pathways (such as physiological differences in how alcohol affects an individual) are likely specific for alcohol dependence. For example, initial sensitivity to alcohol is strongly and inversely related to risk for alcohol dependence64. However, the only variants that affect sensitivity to alcohol and have known functional impact on alcohol dependence risk are in genes encoding the alcohol dehydrogenase (ADH) 1B and 1C65–68 and aldehyde dehydrogenase 2 (refs. 69,70) enzymes, and they confer substantially reduced, rather than increased, risk for alcohol dependence by causing aversive reactions to ethanol.
In other risk pathways, it remains unclear whether common comorbidities with major depression, conduct disorder and antisocial personality disorder are due to direct etiological overlap between the conditions. Alternatively, those with major depression might have a higher risk for alcohol dependence because they are more likely to seek the anxiolytic effects of alcohol, as those with conduct disorder are more likely to select deviant peer groups who provide them with alcohol. Many different phenotypes are now commonly assessed, and these often provide substantially stronger evidence for the involvement of particular genes in alcohol dependence risk.
We explore one gene that illustrates the complexities and the progress being made in understanding sources of risk for alcohol dependence. GABAA subtype receptors are sensitive to ethanol and mediate several of its behavioral effects (including anxiolysis)71. The region around a cluster of GABAA receptor subunit genes on chromosome 4 was implicated originally by linkage studies in Native American72 and the Collaborative Study on the Genetics of Alcoholism (COGA) family samples73. The GABAA receptor α2 subunit (GABRA2) gene in this cluster became the focus of substantial attention when a three-SNP haplotype was strongly associated with adult alcohol dependence (P = 2.2 × 10−8) and with an electrophysiological endophenotype in family based association in the same sample74. The risk haplotype confers ~1.2 times the risk of developing alcohol dependence, but no high-risk haplotype coding sequence variation was detected. The association with alcohol dependence risk was replicated in United States75 and Russian76 samples, and this risk due to GABRA2 variants is moderated by variables including marital status and anxiety77,78.
GABRA2 has been studied across a range of the phenotypes discussed above. Subsequent studies in the COGA sample gave evidence that GABRA2 variation is associated with other SUDs79,80, antisocial personality disorder77 and conduct disorder80, suggesting that GABRA2 is involved in the predisposition to alcohol dependence through a general externalizing-disorder pathway. The same GABRA2 SNPs originally associated with alcohol dependence in a wide age range of adults also predict early conduct problems (rather than the diagnosis of dependence) in a very young adult sample80. Here too, the effects of GABRA2 variants are moderated by environmental variables, as high levels of parental monitoring diminish the association with conduct disorder81.
GWAS of alcohol dependence have to date yielded modest findings. The combined discovery and replication stage data from a GWAS of 1,511 male German alcohol dependence cases and 2,354 matched controls gave genome-wide significant evidence of association with alcohol dependence for two SNPs, rs7590720 (P = 9.72 × 10−9) and rs1344694 (P = 1.69 × 10−8), in an intergenic region on chromosome 2q35. Both are in linkage disequilibrium with the peroxisomal trans-2-enoyl-coenzymeA reductase (PECR) gene, involved in fatty acid metabolism and most highly expressed in the liver82. Signals in the combined sample were also observed in the prior candidate genes alcohol dehydrogenase 1C (ADH1C, rs1614972, combined P = 1.41 × 10−4) and cadherin 13 (CDH13, rs11640875, combined P = 1.84 × 10−5).
A GWAS of 1,897 cases and 1,932 controls from the COGA sample identified 15 SNPs with evidence of association at P < 10−5, but none were supported at P < 0.05 in two independent replication samples83. Reanalysis of this data set using an index of conduct disorder symptomatology identified four SNPs with genome-wide significant evidence for association, two in the C1q and tumor necrosis factor–related protein 7 (C1QTNF7) gene on chromosome 4p15.3 (ref. 84). In analysis of alcohol dependence symptoms in the 2,357 European American Molecular Genetics of Schizophrenia study control samples, the most significant intragenic SNP was in the potassium large conductance calcium-activated channel, subfamily M, alpha member 1 (KCNMA1, rs717207, 2.17 × 10−5) gene85. KCNMA1 is the human homolog of the slo-1 gene in Caenorhabditis elegans. Mutations in this gene in the worm lead to ethanol resistance and attenuation of the reductions in locomotion and egg-laying that normally follow exposure to ethanol86. The product of the slo-1 gene is known to limit excitatory neurotransmitter release in C. elegans87 and to be potentiated by ethanol88,89, suggesting that ethanol reduces (and slo-1 loss-of-function increases) excitatory neurotransmitter release and ethanol resistance.
Summary
Some lessons can be distilled from our review. SUDs are classic complex traits with strong evidence supporting the etiological roles of both genetic or biological and environmental or social risk factors. Taking a ‘genes only’ perspective on SUDs will miss critical parts of the causal picture. We already know that a large proportion of risk genes for SUDs will not be substance specific in their effects but will rather predispose to addiction generally or even to broader externalizing traits. Genetic and environmental risks for SUD typically do not just add together but act and interact with each other during development. Generally, environmental factors are more important for initiation and less important for progression to dependence, whereas the reverse is seen for genetic factors.
As with other complex human behavioral disorders, risk gene identification has been difficult—with one recent major success in identifying nicotinic receptor variants that affect risk for nicotine dependence. We are still learning how to optimally measure SUDs for genetic studies. It is not clear that our current diagnostic formulations are optimal. Important potential advances can also be made in the assessment of drug consumption, as is well illustrated by the substantial gain in the power to detect risk genes for smoking when measuring the long-lasting nicotine metabolite cotinine. But although genetic epidemiologic methods are giving us an increasingly accurate map of broad causal pathways to SUDs, gene discovery will be needed to identify the specific biological systems involved. Some of these variants will be in systems that might have been predicted a priori (key degradative enzymes and receptors), but most will not be. The impact of an individual genetic variant on SUD risk is likely to be small. Identifying these risk genes and understanding their modes of action will require large and carefully assessed clinical samples, innovations in the statistical analysis of such data and strong interactions between these human studies and work in model organisms.
ACKNOWLEDGMENTS
This work was supported in part by US National Institutes of Health grants DA18673 (M.C.N., H.M.), AA011408, AA017828, DA030005 (K.S.K., B.R.), AA018755, AA15416 (D.D.), DA023549 (N.G.), DA025109, DA022989, DA024413, DA027070, MH084952 (H.M.) and DA019498 (X.C.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th edn. revised. Washington, DC: 2000. [Google Scholar]
- 2.Kendler KS, Prescott CA. Genes, Environment, and Psychopathology: Understanding the Causes of Psychiatric and Substance Use Disorders. Guilford, New York: 2006. [Google Scholar]
- 3.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat. Rev. Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 4.Bienvenu OJ, Davydow DS, Kendler KS. Psychiatric ‘diseases’ versus behavioral disorders and degree of genetic influence. Psychol. Med. 2011;41:33–40. doi: 10.1017/S003329171000084X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 6.Tsuang MT, et al. Co-occurrence of abuse of different drugs in men: the role of drug- specific and shared vulnerabilities. Arch. Gen. Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 7.Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am. J. Psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- 8.Kendler KS, Myers J, Prescott CA. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Arch. Gen. Psychiatry. 2007;64:1313–1320. doi: 10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- 9.Kendler KS, et al. The structure of genetic and environmental risk factors for syndromal and subsyndromal common DSM-IV Axis I and all Axis II disorders. Am. J. Psychiatry. 2011;168:29–39. doi: 10.1176/appi.ajp.2010.10030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks BM, Krueger RF, Iacono WG, McGue M, Patrick CJ. Family transmission and heritability of externalizing disorders: a twin-family study. Arch. Gen. Psychiatry. 2004;61:922–928. doi: 10.1001/archpsyc.61.9.922. [DOI] [PubMed] [Google Scholar]
- 11.Kendler KS, Schmitt E, Aggen SH, Prescott CA. Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch. Gen. Psychiatry. 2008;65:674–682. doi: 10.1001/archpsyc.65.6.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendler KS, Eaves LJ. Models for the joint effect of genotype and environment on liability to psychiatric illness. Am. J. Psychiatry. 1986;143:279–289. doi: 10.1176/ajp.143.3.279. [DOI] [PubMed] [Google Scholar]
- 13.Kendler KS, et al. Creating a social world: a developmental study of peer deviance. Arch. Gen. Psychiatry. 2007;64:958–965. doi: 10.1001/archpsyc.64.8.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boardman JD, Blalock CL, Pampel FC. Trends in the genetic influences on smoking. J. Health Soc. Behav. 2010;51:108–123. doi: 10.1177/0022146509361195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dick DM, Rose RJ, Viken RJ, Kaprio J, Koskenvuo M. Exploring gene-environment interactions: socioregional moderation of alcohol use. J. Abnorm. Psychol. 2001;110:625–632. doi: 10.1037//0021-843x.110.4.625. [DOI] [PubMed] [Google Scholar]
- 16.Koopmans JR, Slutske WS, van Baal GC, Boomsma DI. The influence of religion on alcohol use initiation: evidence for genotype X environment interaction. Behav. Genet. 1999;29:445–453. doi: 10.1023/a:1021679005623. [DOI] [PubMed] [Google Scholar]
- 17.Timberlake DS, et al. The moderating effects of religiosity on the genetic and environmental determinants of smoking initiation. Nicotine Tob. Res. 2006;8:123–133. doi: 10.1080/14622200500432054. [DOI] [PubMed] [Google Scholar]
- 18.Dick DM, et al. Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. J. Abnorm. Psychol. 2007;116:213–218. doi: 10.1037/0021-843X.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boardman JD, Saint Onge JM, Haberstick BC, Timberlake DS, Hewitt JK. Do schools moderate the genetic determinants of smoking? Behav. Genet. 2008;38:234–246. doi: 10.1007/s10519-008-9197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kendler KS, Thornton LM, Pedersen NL. Tobacco consumption in Swedish twins reared apart and reared together. Arch. Gen. Psychiatry. 2000;57:886–892. doi: 10.1001/archpsyc.57.9.886. [DOI] [PubMed] [Google Scholar]
- 21.Koob GF. A role for brain stress systems in addiction. Neuron. 2008;59:11–34. doi: 10.1016/j.neuron.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicks BM, South SC, DiRago AC, Iacono WG, McGue M. Environmental adversity and increasing genetic risk for externalizing disorders. Arch. Gen. Psychiatry. 2009;66:640–648. doi: 10.1001/archgenpsychiatry.2008.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Covault J, et al. Interactive effects of the serotonin transporter 5-HTTLPR polymorphism and stressful life events on college student drinking and drug use. Biol. Psychiatry. 2007;61:609–616. doi: 10.1016/j.biopsych.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Blomeyer D, et al. Interaction between, CRHR1, gene and stressful life events predicts adolescent heavy alcohol use. Biol. Psychiatry. 2008;63:146–151. doi: 10.1016/j.biopsych.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman J, et al. Genetic and environmental predictors of early alcohol use. Biol. Psychiatry. 2007;61:1228–1234. doi: 10.1016/j.biopsych.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 26.Maes HH, et al. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol. Med. 2004;34:1251–1261. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- 27.Kendler KS, et al. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol. Med. 1999;29:299–308. doi: 10.1017/s0033291798008022. [DOI] [PubMed] [Google Scholar]
- 28.Gillespie NA, Neale MC, Kendler KS. Pathways to cannabis abuse: a multistage model from cannabis availability, cannabis initiation and progression to abuse. Addiction. 2009;104:430–438. doi: 10.1111/j.1360-0443.2008.02456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agrawal A, Neale MC, Jacobson KC, Prescott CA, Kendler KS. Illicit drug use and abuse/dependence: modeling of two-stage variables using the CCC approach. Addict. Behav. 2005;30:1043–1048. doi: 10.1016/j.addbeh.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Muthén B, Muthén LK. Integrating person-centered and variable-centered analyses: growth mixture modeling with latent trajectory classes. Alcohol. Clin. Exp. Res. 2000;24:882–891. [PubMed] [Google Scholar]
- 31.Lubke G, Neale M. Distinguishing between latent classes and continuous factors with categorical outcomes: Class invariance of parameters of factor mixture models. Multivariate Behav. Res. 2008;43:592–620. doi: 10.1080/00273170802490673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neale MC, Aggen SH, Maes H, Kubarych TS, Schmitt JE. Methodological issues in the assessment of substance use phenotypes. Addict. Behav. 2006;31:1010–1034. doi: 10.1016/j.addbeh.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 33.Gillespie NA, Kendler KS, Neale MC. Psychometric modeling of cannabis initiation and use and the symptoms of cannabis abuse, dependence and withdrawal in a sample of male and female twins. Drug Alcohol Depend. 2011;118:166–172. doi: 10.1016/j.drugalcdep.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillespie NA, Neale MC, Prescott CA, Aggen SH, Kendler KS. Factor and item-response analysis of DSM-IV criteria for abuse of and dependence on cannabis, cocaine, hallucinogens, sedatives, stimulants and opioids. Addiction. 2007;102:920–930. doi: 10.1111/j.1360-0443.2007.01804.x. [DOI] [PubMed] [Google Scholar]
- 35.Maes HH, Neale MC. Genetic modeling of tobacco use behavior and trajectories. In: Swan GE, et al., editors. NCI Tobacco Control Monograph Series 20: Phenotypes and Endophenotypes: Foundations for Genetic Studies of Nicotine Use and Dependence. US National Institutes of Health; 2009. publication no: 09–6366. [Google Scholar]
- 36.Conway KP, et al. Measuring addiction propensity and severity: the need for a new instrument. Drug Alcohol Depend. 2010;111:4–12. doi: 10.1016/j.drugalcdep.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanyukov MM, et al. Measurement of the risk for substance use disorders: phenotypic and genetic analysis of an index of common liability. Behav. Genet. 2009;39:233–244. doi: 10.1007/s10519-009-9269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berrettini W, et al. α-5/α-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol. Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5–A3 region on chromosome 15q24–25.1 is a risk factor both for nicotine dependence and for lung cancer. J. Natl. Cancer Inst. 2008;100:1552–1556. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, et al. Variants in nicotinic acetylcholine receptors α5 and α3 increase risks to nicotine dependence. Am. J. Med. Genet B. Neuropsychiatr. Genet. 2009;150B:926–933. doi: 10.1002/ajmg.b.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br. J. Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu JZ, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saccone NL, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001053. e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furberg H, et al. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grucza RA, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol. Psychiatry. 2008;64:922–929. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joslyn G, et al. Chromosome 15q25.1 genetic markers associated with level of response to alcohol in humans. Proc. Natl. Acad. Sci. USA. 2008;105:20368–20373. doi: 10.1073/pnas.0810970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erlich PM, et al. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum. Genet. 2010;128:491–499. doi: 10.1007/s00439-010-0876-6. [DOI] [PubMed] [Google Scholar]
- 51.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 53.DeMeo DL, et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am. J. Hum. Genet. 2009;85:493–502. doi: 10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen HM, et al. Fine mapping of chromosome 15q25.1 lung cancer susceptibility in African-Americans. Hum. Mol. Genet. 2010;19:3652–3661. doi: 10.1093/hmg/ddq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, et al. Mediating effects of smoking and chronic obstructive pulmonary disease on the relation between the CHRNA5–A3 genetic locus and lung cancer risk. Cancer. 2010;116:3458–3462. doi: 10.1002/cncr.25085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lips EH, et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals. Int. J. Epidemiol. 2010;39:563–577. doi: 10.1093/ije/dyp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Munafo MR, et al. Chromosome 15 genetic variants are associated with objective measures of tobacco exposure: implications for genetic and epidemiological studies. J. Natl. Cancer Inst. (in the press) [Google Scholar]
- 58.Thorgeirsson TE, et al. Sequence variants at CHRNB3–CHRNA6, and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bierut LJ, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine receptor (AChR) α5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (α4β2)2 α5 AChR function. Mol. Pharmacol. 2011;79:119–125. doi: 10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong LE, et al. A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc. Natl. Acad. Sci. USA. 2010;107:13509–13514. doi: 10.1073/pnas.1004745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Changeux JP. Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nat. Rev. Neurosci. 2010;11:389–401. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- 63.Maes HH, Neale MC, Chen X, Prescott CA, Kendler KS. A twin association study of nicotine dependence with markers in the CHRNA3 and CHRNA5 genes. Behav. Genet. 2011;41:680–690. doi: 10.1007/s10519-011-9476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuckit MA, Smith TL, Tipp JE. The Self-Rating of the Effects of Alcohol (SRE) form as a retrospective measure of the risk for alcoholism. Addiction. 1997;92:979–988. [PubMed] [Google Scholar]
- 65.Shen YC, et al. Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol. Clin. Exp. Res. 1997;21:1272–1277. [PubMed] [Google Scholar]
- 66.Osier M, et al. Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am. J. Hum. Genet. 1999;64:1147–1157. doi: 10.1086/302317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goedde HW, et al. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum. Genet. 1992;88:344–346. doi: 10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- 68.Neale MC, et al. Distinguishing population stratification from genuine allelic effects with Mx: association of ADH2 with alcohol consumption. Behav. Genet. 1999;29:233–243. [Google Scholar]
- 69.Thomasson HR, et al. Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol. Clin. Exp. Res. 1994;18:640–643. doi: 10.1111/j.1530-0277.1994.tb00923.x. [DOI] [PubMed] [Google Scholar]
- 70.Dickson PA, et al. Effects of variation at the ALDH2 locus on alcohol metabolism, sensitivity, consumption, and dependence in Europeans. Alcohol. Clin. Exp. Res. 2006;30:1093–1100. doi: 10.1111/j.1530-0277.2006.00128.x. [DOI] [PubMed] [Google Scholar]
- 71.Grobin AC, Matthews DB, Devaud LL, Morrow AL. The role of GABAA receptors in the acute and chronic effects of ethanol. Psychopharmacology (Berl.) 1998;139:2–19. doi: 10.1007/s002130050685. [DOI] [PubMed] [Google Scholar]
- 72.Long JC, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am. J. Med. Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 73.Reich T, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am. J. Med. Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- 74.Edenberg HJ, et al. Variations in GABRA2, encoding the α2 subunit of the GABAA receptor, are associated with alcohol dependence and with brain oscillations. Am. J. Hum. Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- 76.Lappalainen J, et al. Association between alcoholism and γ-amino butyric acid α2 receptor subtype in a Russian population. Alcohol. Clin. Exp. Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- 77.Dick DM, et al. Marital status, alcohol dependence, and GABRA2: evidence for gene-environment correlation and interaction. J. Stud. Alcohol. 2006;67:185–194. doi: 10.15288/jsa.2006.67.185. [DOI] [PubMed] [Google Scholar]
- 78.Enoch MA, Schwartz L, Albaugh B, Virkkunen M, Goldman D. Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2006;141B:599–607. doi: 10.1002/ajmg.b.30336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Agrawal A, et al. Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behav. Genet. 2006;36:640–650. doi: 10.1007/s10519-006-9069-4. [DOI] [PubMed] [Google Scholar]
- 80.Dick DM, et al. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav. Genet. 2006;36:577–590. doi: 10.1007/s10519-005-9041-8. [DOI] [PubMed] [Google Scholar]
- 81.Dick DM, et al. Role of GABRA2 in trajectories of externalizing behavior across development and evidence of moderation by parental monitoring. Arch. Gen. Psychiatry. 2009;66:649–657. doi: 10.1001/archgenpsychiatry.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Treutlein J, et al. Genome-wide association study of alcohol dependence. Arch. Gen. Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bierut LJ, et al. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dick DM, et al. Genome-wide association study of conduct disorder symptomatology. Mol. Psychiatry. 2010;16:800–808. doi: 10.1038/mp.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kendler KS, et al. Genomewide association analysis of symptoms of alcohol dependence in the Molecular Genetics of Schizophrenia (MGS2) control sample. Alcohol. Clin. Exp. Res. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davies AG, et al. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- 87.Wang ZW, Saifee O, Nonet ML, Salkoff L. SLO-1 potassium channels control quantal content of neurotransmitter release at the C. elegans neuromuscular junction. Neuron. 2001;32:867–881. doi: 10.1016/s0896-6273(01)00522-0. [DOI] [PubMed] [Google Scholar]
- 88.Dopico AM, Widmer H, Wang G, Lemos JR, Treistman SN. Rat supraoptic magnocellular neurones show distinct large conductance, Ca2+ -activated K+ channel subtypes in cell bodies versus nerve endings. J. Physiol. (Lond.) 1999;519:101–114. doi: 10.1111/j.1469-7793.1999.0101o.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gruss M, et al. Ethanol reduces excitability in a subgroup of primary sensory neurons by activation of BKCa channels. Eur. J. Neurosci. 2001;14:1246–1256. doi: 10.1046/j.0953-816x.2001.01754.x. [DOI] [PubMed] [Google Scholar]