Abstract
Background
Overexpression of mitotic kinases has been associated with prognosis, histologic grade and clinical stage in ovarian cancer, but the relationship between inherited variation in these genes and ovarian cancer risk has not been well defined.
Methods
We measured associations between 397 single nucleotide polymorphisms (SNPs) from 67 mitotic kinases and invasive epithelial ovarian cancer risk in two case-control studies (n=671 cases; n=939 controls). Thirty-six candidate SNPs (p< 0.05) were assessed in a replication analysis consisting of three additional studies (n=1094 cases; n=829 controls).
Results
In initial analysis, thirty-six SNPs were suggestive of association with risk of serous ovarian cancer, all subtypes of ovarian cancer, or both (p<0.05). Replication analyses suggested an association between rs2125846 in the Nemo-like kinase gene (NLK) and ovarian cancer (serous odds ratio (OR)=1.36, 95% confidence interval (CI) 1.11 – 1.67, p=1.77 × 10−3; all subtypes OR=1.30, 95% CI 1.08 – 1.56, p=2.97 × 10−3). Furthermore, rs2125846 was associated with risk in the combined discovery and replication sets (serous OR=1.33, 95% CI 1.15 – 1.54; all subtypes OR=1.27, 95% CI 1.12 – 1.45).
Conclusions
Variation in NLK may be associated with risk of invasive epithelial ovarian cancer. Further studies are needed to confirm and understand the biological relationship between this mitotic kinase and ovarian cancer risk.
Impact
An association between SNPs in NLK and ovarian cancer may provide biological insight into the development of this disease.
Keywords: genetic susceptibility, serous, cell cycle, association study, mitotic kinase
Introduction
Ovarian cancer has the highest mortality of gynecologic malignancies. Factors associated with ovarian cancer risk include age, family history, fertility drug use, postmenopausal hormone therapy (1), and inherited factors (2-4). While inherited mutations in BRCA1 and BRCA2 account for 50% of ovarian cancer cases in families with two or more confirmed cases (5), the remaining unexplained familial and sporadic ovarian cancer risk is likely in part attributable to common, low-penetrance alleles (6). Efforts to identify low-penetrance alleles by genome-wide association studies have identified variants in the chromosome 9p22 BNC2 locus (3), a 19p13 locus containing the MERIT40 gene (7), and in 2q31 and 8q24 loci (4).
Mitotic kinases are essential components in the regulation of mitosis and cytokinesis, acting upon various structures involved in mitotic entry, progression, and exit. These kinases phosphorylate proteins involved in centrosome duplication and separation, chromosome condensation, spindle assembly and fidelity, chromosome segregation, and cytokinesis, and have the ability to behave as oncogenes, providing a compelling link between errors in mitosis and oncogenesis (8). Indeed, errors in the choreography of the processes controlled by mitotic kinases disrupt successful division of mammalian cells and can lead to aneuploidy, genetic instability and cancer. More specifically, alterations in these genes and disregulation of protein products have been implicated in cancer development in mouse models (9) and in multiple human tumor types (8).
Mitotic kinases include members of the Aurora, Polo-like, and Nek families as well as individual kinases involved in mitotic checkpoints, mitotic exit and cytokinesis. Within the Aurora kinase family, the overexpression of both AURKA and AURKB has been associated with poor prognosis in epithelial ovarian cancers (10-13). Similarly, overexpression of polo-like kinases such as PLK1 and PLK2 have also been shown to correlate with prognosis, histologic grade and clinical stage in ovarian cancer (14-16). While disregulation of these mitotic kinases has been associated with ovarian cancer prognosis, and one study of polymorphisms found evidence of association with risk and polymorphisms in CDK1, a key mitotic kinase required for entry into mitosis (17), the relationship between alterations in these genes and ovarian cancer risk has not been well-studied.
Here we tested the hypothesis that inherited variation in genes encoding mitotic kinases increases the risk of invasive epithelial ovarian cancer. Single nucleotide polymorphisms (SNPs) from 67 mitotic kinase genes were genotyped in two ovarian cancer case-control studies followed by replication in three additional studies.
Materials and Methods
Study populations
The current hypothesis was tested in a collaborative effort that combined studies of invasive epithelial ovarian cancer from Mayo Clinic (MAY) and the North Carolina Ovarian Cancer Study (NCO). Details of the study protocols have been published previously (18, 19) (Table S1). Questionnaire data obtained from all subjects included established risk factors such as demographics, reproductive history, family history of cancer, medical and surgical history, and lifestyle habits. Candidate SNPs (p< 0.05) identified in this discovery population were assessed in a replication study comprised of three additional ovarian cancer studies: a case-control study from Brigham and Women’s Hospital (BWH), the Tampa Bay Ovarian Cancer Study (TBO), and the Familial Ovarian Tumour Study (TOR) (Table S2). Details of these studies have also been previously described (4). We restricted all analyses to subjects who were self-reported whites.
SNP selection, genotyping and quality control
Mitotic kinase genes were identified using data from a published study in which the authors performed an RNA-interference-based functional screen in Drosophila(20), from the Gene Ontology database and from the current literature for genes encoding proteins with mitotic kinase function. Discovery tagSNPs (n=397) were selected based on position within and 5kb of 67 genes (Table S3), minor allele frequency (MAF) ≥ 0.05 and pairwise linkage disequilibrium (LD) of r2 ≥ 0.8 in unrelated white samples within HapMap Consortium release 22 (Hapmap, 2003), and the predicted likelihood of successful genotyping using Illumina Golden Gate Assay™. These were genotyped as part of a larger investigation of 1,152 SNPs in a variety of pathways using the Illumina GoldenGate™ assay. Genotyping was attempted on 897 genomic DNA samples from MAY participants, 1,279 whole-genome amplified samples from NCO participants, and 129 duplicate samples for a total of 2,047 unique study participants. We excluded 44 samples with call rates <90% and 22 samples due to study ineligibility, leaving 1,981 samples. The sample call rate was 99.74% and the concordance for the 129 duplicate samples was 100%. Eleven SNPs with significant deviations from Hardy-Weinberg equilibrium (HWE) in controls (p<0.001), assessed using Pearson good of fit or Fisher exact tests, were excluded from analyses.
The replication analyses utilized genotype data from a published genome-wide association study of ovarian cancer (4, 21). TBO and TOR samples were genotyped using the Illumina 610K platform, and BWH samples were genotyped using the Illumina 317K platform. Imputation of approximately 2.5 million SNPs using HapMap data as the reference was performed for each replication site using MACH, and imputed genotypes were used where observed genotype data were missing.
Statistical Analysis
SNP-specific analyses were performed to evaluate the association of genotypes at each SNP and serous ovarian cancer risk and risk of all histologic types of ovarian cancer combined. Associations were estimated as odds ratios (OR) with associated 95% confidence intervals (95% CI) using unconditional logistic regression under log-additive genetic models. We also performed haplotype analyses using Haplostat, for SNPs with p< 0.05 when multiple SNPs had p < 0.05 within a gene, estimating haplotype frequencies for each gene using all SNPs within the gene, and then testing the global significance (P<0.05) for haplotype association with risk using a likelihood ratio test. Individual haplotype associations were evaluated using a log-additive model. All discovery single SNP and haplotype models were adjusted for age, geographic location, BMI, oral contraceptive use, hormone replacement therapy, and parity and age at first birth. Single-SNP p<0.05 was used to select SNPs for replication. All replication analyses were adjusted for age and study center (additional covariate data were not available). Assessment of the most promising SNPs in combined analysis by histologic subtype was performed using polytomous logistic regression using control status as the reference outcome. Heterogeneity of SNP associations by histologic subtype was measured by applying polytomous logistic regression to cases only.
As a conservative approach to the large number of statistical tests, we used the following method. First, we sought replication of any SNP with single-SNP p-value <0.05 in discoveray analysis. Second, we used a modified correlation-adjusted Bonferroni adjustment in replication analysis, accounting for multiple testing. Recognizing that some correlation due to linkage disequilibrium exists between the replication SNPs, we first determined the effective number of independent tests using a principal components-based method (22), which indicated that our analysis of the 36 replication SNPs was equal to approximately 35.5 independent tests of hypothesis. Recognizing that any SNP found to be statistically significant in the replication data set but in the opposite direction of the discovery set result would not be considered a replication, our replication analyses are based on one-sided tests of hypothesis. Any SNP in the discovery set with two-sided unadjusted p < 0.05, and statistically significant in the replication set with one-sided adjusted p < 1.41 × 10−3 and with an estimate in the same direction as in the discovery phase, was considered a replication. Finally, for SNPs statistically significant in the replication set using adjusted p-values (1.41 × 10−3), we cautiously interpret the pooled analysis results using odds ratios and confidence intervals but not calculating p-values due to the interpretive complexities of combining such data.
Results
The goal of this analysis was to assess whether common genetic variation in mitotic kinases is associated with risk of invasive epithelial ovarian cancer. To achieve this, we genotyped 397 tagSNPs in 67 genes (Table S3) encoding mitotic kinases in two case-control studies (Table S1). We first restricted our analysis to serous invasive ovarian cancer cases and controls (n=407 cases, n=939 controls): this selection was based on recent findings from the Ovarian Cancer Association Consortium (OCAC) showing that genome wide association study (GWAS) associations with serous ovarian cancer were generally stronger than for all histologic subtypes combined, possibly because of refinement of the phenotype under study (4). Twenty SNPs tested were suggestive of association with risk of serous ovarian cancer in a log-additive model (p<0.05) (Table 1, Table S4). These 20 SNPs were located in 13 different genes: CDC7, CDK6, CSNK2A, SIK3, MAST2, NEK2, NEK4, NEK8, NLK, PRKG2, STK4, TEX14, and TRIB3.
Table 1. Discovery single SNP associations with risk of invasive ovarian cancer in a log-additive model (p<0.01).
| Serous | All subtypes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Chr | MAF | SNP | OR | 95% CI | P-value* | OR | 95% CI | P-value* |
| CDK6 | 7 | 0.07 | rs2282990 | 1.71 | (1.25 - 2.34) | 8.2 × 10−4 | 1.61 | (1.22 - 2.12) | 6.7 × 10−4 |
| NLK | 17 | 0.19 | rs2125846 | 1.35 | (1.09 - 1.66) | 6.0 × 10−3 | 1.28 | (1.07 - 1.54) | 7.2 × 10−3 |
| PLK2 | 5 | 0.11 | rs12513877 | 1.16 | (0.88 - 1.53) | 0.285 | 1.34 | (1.08 - 1.66) | 8.6 × 10−3 |
| SIK3 | 11 | 0.22 | rs7928320 | 0.76 | (0.61 - 0.93) | 8.4 × 10−3 | 0.80 | (0.67 - 0.95) | 0.0117 |
| TEX14 | 17 | 0.06 | rs12944693 | 1.49 | (1.07 - 2.08) | 0.020 | 1.52 | (1.14 - 2.04) | 4.3 × 10−3 |
Results based on 671 cases (407 serous) and 939 controls. Adjusted for age, geographic location, body mass index (BMI), oral contraceptive (OC) use, hormone replacement therapy (HRT), and parity/age at first birth. MAF, minor allele frequency among controls.
Based on a test for trend.
Since four of the 13 genes identified in the single SNP serous analysis contained multiple candidate SNPs, we performed haplotype analyses to better understand the patterns of risk in these genes. CDK6 haplotypes of the four SNPs (rs2282990, rs3731348, rs17690388, and rs2282983) were suggestive of association with risk of serous invasive ovarian cancer (global haplotype association p=0.0034) (Table S5a). The first CDK6 risk haplotype was perfectly tagged by the minor allele (A) of rs17690388, and was associated with a decrease in risk of serous ovarian cancer (OR=0.63, 95% CI 0.40-0.99; p=0.044). The second CDK6 risk haplotype captured the minor alleles of rs2282990 (T) and rs2282983 (C) and the major allele at rs3731348 (G), and was associated with an increase in serous ovarian cancer risk (OR=2.42, 95% CI 1.30 – 4.50; p=0.0054). SIK3 and TEX14 each contained haplotypes associated with risk of serous ovarian cancer that were captured by variation at single SNPs (Tables S5b,c). Thus, associations between variation in SIK3 and TEX14 and serous ovarian cancer were best described by the single SNPs rs7928320 and rs12944693. NLK haplotypes were not associated with risk of serous ovarian cancer (Table S5d).
Having observed possible associations with serous ovarian cancer, we evaluated whether variation in mitotic kinases were also associated with risk of all histologic subtypes of invasive ovarian cancer. Specifically, we tested all 397 SNPs using a larger group of cases (n=671), comprised of 407 serous (60.8%), 28 mucinous (4.2%), 115 endometroid (17.2%), 50 clear cell (7.5%), and 69 other (10.3%) epithelial ovarian cancers. Twenty-three SNPs were suggestive of association with invasive ovarian cancer in 15 different genes (Table 1, Table S4). Only SNPs in six of these genes were also possible candidates in the serous-only analysis: CDK6, SIK3, NEK4, NLK, STK4, and TEX14.
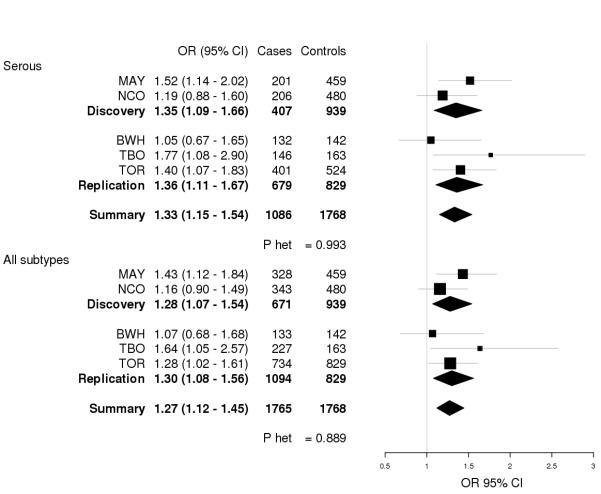
We next evaluated the thirty-six SNPs identified as candidates in the discovery phase (unadjusted p<0.05 in serous or all subtypes) in a replication study of 1,094 invasive ovarian cancer cases and 829 controls (Table S3) utilizing data from a published ovarian cancer GWAS. After adjustment for multiple testing none of these 36 SNPs were statistically significantly associated (p<1.41 × 10−3) with risk of serous or overall ovarian cancer in the replication study (Table 2, Table S6-S7); however, rs2125846 (NLK) showed similar associations with risk of both serous and all subtypes of ovarian cancer and was thus explored further. The G allele of rs2125846 was associated with a 1.36-fold increased risk of serous ovarian cancer (95% CI 1.11 – 1.67, p=1.77 × 10−3) and with a 1.30-fold increased risk of all subtypes (95% CI 1.08 – 1.56, p=2.97 × 10−3). Combining discovery and replication studies, rs2125846 was again associated with risk of serous and all subtypes of ovarian cancer (serous OR=1.33, 95% CI 1.15 – 1.54; all subtype OR=1.27, 95% CI 1.12 – 1.45), with consistent associations across analyses (Figure 1). Risk estimates were also similar across histologic subtypes (p-heterogeneity=0.75) (Figure S1). However, our power to detect associations with subtypes of ovarian cancer was limited due to small sample sizes and this result should be interpreted with caution.
Table 2. NLK and CDK6 SNP associations in discovery, replication, and combined analyses.
| Serous |
All subtypes |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI) | P-value | Cases | Controls | OR (95% CI) | P-value | |
| rs2125846 (NLK) | ||||||||
| Discovery | 407 | 939 | 1.35 (1.10 - 1.66) | 3.87 × 10−3 | 671 | 939 | 1.29 (1.08 - 1.54) | 4.23 × 10−3 |
| Replication* | 679 | 829 | 1.35 (1.11 - 1.67) | 1.77 × 10−3 | 1094 | 829 | 1.29 (1.07 - 1.55) | 2.97 × 10−3 |
| Combined | 1086 | 1768 | 1.33 (1.15 - 1.54) | NA | 1765 | 1768 | 1.27 (1.12 - 1.45) | NA |
One-sided p-values.
Discovery and replication analyses adjusted for age group and study site. Combined analyses adjusted for age group, study site, and replication phase. P-values based on test for trend; p-values were not estimated for combined analyses due to inability to correctly account for xxx
Figure 1. Forest plot of rs2125846 (NLK) association with ovarian cancer.
Forest plots for rs2125846 and risk of (a) serous ovarian cancer and (b) all ovarian cancer subtypes are shown by study. Study-specific odds ratios (95% CIs) are denoted by black boxes (black lines). Combined discovery, replication, and summary OR estimates are represented by black diamonds, where diamond width corresponds to 95% CI bounds. Box and diamond heights are inversely proportional to precision of the OR estimate. P-values for heterogeneity (P het) of odds ratios by study are shown. MAY, Mayo Clinic case-control study of epithelial ovarian cancer; NCO, North Carolina Ovarian Cancer Study; BWH, Brigham and Women’s Hospital case-control study; TBO, Tampa Bay Ovarian Cancer Study; TOR, Familial Ovarian Tumour Study.
Discussion
In an analysis of genes encoding kinases required for normal cell division, we have identified a SNP, rs2125846, in the NLK locus that is associated with risk of ovarian cancer. This SNP showed a very similar influence on risk of ovarian cancer in the discovery and replication studies (OR=1.35) and was also associated with risk of the serous subtype (combined OR=1.33) and all subtypes (combined OR=1.27) of ovarian cancer risk. It is important to interpret this association with caution, since the rs2125846 association did not retain significance after adjustment for multiple testing in the replication phase. However, we have identified a biologically interesting candidate ovarian cancer SNP that warrants replication in larger studies of ovarian cancer.
Nemo-like kinase (NLK) is a MAPK-like kinase belonging to the serine/threonine kinase superfamily. Studies in C. elegans have shown that NLK is involved in the cancer-related Wnt/beta-catenin signalling pathway (23, 24). Further, NLK has been shown to inhibit several transcription factors such as NF-κB, Smads, AP1, and p53 (24, 25). Several functional studies have found a relationship between NLK expression and various cancer types. A prostate cancer study demonstrated in cell lines that NLK expression is decreased in metastases compared to normal prostate epithelium and that over-expression of NLK induces apoptosis particularly among androgen-receptor expressing cells (24). Similarly, over-expression of NLK was shown to induce apoptosis in colon cancer cell lines (26). Finally, NLK is upregulated in hepatocellular carcinomas, and disruption of NLK inhibits hepatocellular carcinoma cell growth (27). However, there are currently no functional data for the intronic NLK SNPs identified in this study. In addition, no associations for NLK variants have been identified in candidate gene studies or GWAS of cancer.
Overall, we report on the evaluation of the contribution of inherited variation in mitotic kinases to ovarian cancer risk. These results warrant further investigation in independent studies of ovarian cancer to understand the biological relationship between NLK and ovarian cancer risk.
Supplementary Material
Acknowledgments
Financial Support – NIH grants R01 CA122340, R01 CA128978, R01 CA88868, R01 CA122443, P50 CA136393, U19 CA148112, U.S. Department of Defense Ovarian Cancer IDEA award W91XWH-10-1-0341, Alberta Cancer Research Institute Projects 23905 and 24258, and the Canadian Institutes of Health Research MSH-87734. The Ovarian Cancer Association Consortium is supported by a grant from the Ovarian Cancer Research Fund
References
- 1.Morch LS, Lokkegaard E, Andreasen AH, Kruger-Kjaer S, Lidegaard O. Hormone therapy and ovarian cancer. JAMA. 2009;302:298–305. doi: 10.1001/jama.2009.1052. [DOI] [PubMed] [Google Scholar]
- 2.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41 doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–9. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramus SJ, Harrington PA, Pye C, DiCioccio RA, Cox MJ, Garlinghouse-Jones K, et al. Contribution of BRCA1 and BRCA2 mutations to inherited ovarian cancer. Hum Mutat. 2007;28:1207–15. doi: 10.1002/humu.20599. [DOI] [PubMed] [Google Scholar]
- 6.Fasching PA, Gayther S, Pearce L, Schildkraut JM, Goode E, Thiel F, et al. Role of genetic polymorphisms and ovarian cancer susceptibility. Mol Oncol. 2009;3:171–81. doi: 10.1016/j.molonc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–4. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 9.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–86. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lassus H, Staff S, Leminen A, Isola J, Butzow R. Aurora-A overexpression and aneuploidy predict poor outcome in serous ovarian carcinoma. Gynecol Oncol. 2011;120:11–7. doi: 10.1016/j.ygyno.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Landen CN, Jr., Lin YG, Immaneni A, Deavers MT, Merritt WM, Spannuth WA, et al. Overexpression of the centrosomal protein Aurora-A kinase is associated with poor prognosis in epithelial ovarian cancer patients. Clin Cancer Res. 2007;13:4098–104. doi: 10.1158/1078-0432.CCR-07-0431. [DOI] [PubMed] [Google Scholar]
- 12.Chen YJ, Chen CM, Twu NF, Yen MS, Lai CR, Wu HH, et al. Overexpression of Aurora B is associated with poor prognosis in epithelial ovarian cancer patients. Virchows Arch. 2009;455:431–40. doi: 10.1007/s00428-009-0838-3. [DOI] [PubMed] [Google Scholar]
- 13.Mendiola M, Barriuso J, Marino-Enriquez A, Redondo A, Dominguez-Caceres A, Hernandez-Cortes G, et al. Aurora kinases as prognostic biomarkers in ovarian carcinoma. Hum Pathol. 2009;40:631–8. doi: 10.1016/j.humpath.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Weichert W, Denkert C, Schmidt M, Gekeler V, Wolf G, Kobel M, et al. Polo-like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br J Cancer. 2004;90:815–21. doi: 10.1038/sj.bjc.6601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24:287–91. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- 16.Takai N, Miyazaki T, Fujisawa K, Nasu K, Hamanaka R, Miyakawa I. Expression of polo-like kinase in ovarian cancer is associated with histological grade and clinical stage. Cancer Lett. 2001;164:41–9. doi: 10.1016/s0304-3835(00)00703-5. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham JM, Vierkant RA, Sellers TA, Phelan C, Rider DN, Liebow M, et al. Cell cycle genes and ovarian cancer susceptibility: a tagSNP analysis. Br J Cancer. 2009;101:1461–8. doi: 10.1038/sj.bjc.6605284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sellers TA, Schildkraut JM, Pankratz VS, Vierkant RA, Fredericksen ZS, Olson JE, et al. Estrogen bioactivation, genetic polymorphisms, and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2536–43. doi: 10.1158/1055-9965.EPI-05-0142. [DOI] [PubMed] [Google Scholar]
- 19.White KL, Sellers TA, Fridley BL, Vierkant RA, Phelan CM, Tsai YY, et al. Variation at 8q24 and 9p24 and risk of epithelial ovarian cancer. Twin Res Hum Genet. 2010;13:43–56. doi: 10.1375/twin.13.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–7. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 21.Permuth-Wey J, Chen Z, Tsai YY, Lin HY, Chen YA, Barnholtz-Sloan J, et al. MicroRNA Processing and Binding Site Polymorphisms Are Not Replicated in the Ovarian Cancer Association Consortium. Cancer Epidemiol Biomarkers Prev. 2011;20:1793–7. doi: 10.1158/1055-9965.EPI-11-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smit L, Baas A, Kuipers J, Korswagen H, van de Wetering M, Clevers H. Wnt activates the Tak1/Nemo-like kinase pathway. J Biol Chem. 2004;279:17232–40. doi: 10.1074/jbc.M307801200. [DOI] [PubMed] [Google Scholar]
- 24.Emami KH, Brown LG, Pitts TE, Sun X, Vessella RL, Corey E. Nemo-like kinase induces apoptosis and inhibits androgen receptor signaling in prostate cancer cells. Prostate. 2009;69:1481–92. doi: 10.1002/pros.20998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasuda J, Yokoo H, Yamada T, Kitabayashi I, Sekiya T, Ichikawa H. Nemo-like kinase suppresses a wide range of transcription factors, including nuclear factor-kappaB. Cancer Sci. 2004;95:52–7. doi: 10.1111/j.1349-7006.2004.tb03170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuda J, Tsuchiya A, Yamada T, Sakamoto M, Sekiya T, Hirohashi S. Nemo-like kinase induces apoptosis in DLD-1 human colon cancer cells. Biochem Biophys Res Commun. 2003;308:227–33. doi: 10.1016/s0006-291x(03)01343-3. [DOI] [PubMed] [Google Scholar]
- 27.Jung KH, Kim JK, Noh JH, Eun JW, Bae HJ, Xie HJ, et al. Targeted disruption of Nemo-like kinase inhibits tumor cell growth by simultaneous suppression of cyclin D1 and CDK2 in human hepatocellular carcinoma. J Cell Biochem. 2010;110:687–96. doi: 10.1002/jcb.22579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.