Abstract
Background
Genome-wide association studies (GWAS) for epithelial ovarian cancer (EOC), the most lethal gynecologic malignancy, have identified novel susceptibility loci. GWAS for survival after EOC have had more limited success. The association of each single nucleotide polymorphism (SNP) individually may not be well-suited to detect small effects of multiple SNPs, such as those operating within the same biological pathway. Gene set analysis (GSA) overcomes this limitation by assessing overall evidence for association of a phenotype with all measured variation in a set of genes.
Methods
To determine gene sets associated with EOC overall survival, we conducted GSA using data from two large GWASes (N cases = 2,813, N deaths = 1,116), with a novel Principal Component – Gamma GSA method. Analysis was completed for all cases and then separately for high grade serous (HGS) histological subtype.
Results
Analysis of the HGS subjects resulted in 43 gene sets with p<0.005 (1.7%); of these, 21 gene sets had p < 0.10 in both GWASes, including intracellular signaling pathway (p = 7.3 × 10−5) and macrolide binding (p = 6.2 ×10−4) gene sets. The top gene sets in analysis of all cases were meiotic mismatch repair (p=6.3 ×10−4) and macrolide binding (p=1.0×10−3). Of 18 gene sets with p<0.005 (0.7%), eight had p < 0.10 in both GWASes.
Conclusion
This research detected novel gene sets associated with EOC survival.
Impact
Novel gene sets associated with EOC survival might lead to new insights and avenues for development of novel therapies for EOC and pharmacogenomic studies.
Keywords: pathway analysis, genetic association, GWAS, SNPs, gynecologic neoplasm
Introduction
Epithelial ovarian cancer (EOC) is the fifth leading cause of cancer mortality among women in the United States, accounting for five percent of cancer deaths (1). Most patients are diagnosed with advanced disease, and for, the three-quarters of women diagnosed with stage III or IV disease, the likelihood of long-term disease-free survival is less than 20 percent (2–4). Stage, grade, and other clinical features of disease, such as degree of debulking and presence of ascites, are key to predicting prognosis; however, much variation in outcome is unexplained. As women may inherently vary in their ability to eradicate disease or tolerate treatment, genetic association studies have sought to identify inherited variants related to outcome. Candidate gene studies of angiogenesis, inflammation, or chemoresistance pathways show promising results, although not always consistently across populations (5–7). Similarly, genome-wide association studies (GWAS) of ovarian cancer have not yet found outcome-associated loci despite large sample sizes and comprehensive coverage of common genomic variation (8).
One explanation for the lack of findings from GWAS is that the analysis strategy commonly used, testing for association of the phenotype with each SNP individually, is not well-suited for detecting multiple variants with small effects (9–12). The application of novel methods which incorporate biological knowledge into analyses of GWAS data has proven useful to many studies (13, 14). One approach is gene set analysis (GSA) which assesses the overall evidence of association of a phenotype with all measured variation in a pre-defined set of genes, such as a biological pathway (15–17). A gene set is simply any user-defined group of genes; for example, with GWAS data, GSA allows for the use of standardized biological classifications, such as those from the Kyoto Encyclopedia of Genes and Genomes (KEGG). Because numerous genes can be combined into a limited number of gene sets for analysis, the multiple-testing burden is greatly reduced. We have recently shown that the PC-GM approach to modeling SNPs within genes using principal component (PC) analysis (18) and then combining gene-level p-values using the Gamma method (GM) (19) is a powerful GSA method (20). Therefore, in order to identify novel avenues for investigation of this lethal condition, we conducted a GSA of EOC overall survival using the PC-GM approach and data from two large ovarian cancer GWASes.
Methods and Materials
Study Participants
GSAs were conducted using data from two independent multi-site ovarian cancer GWASes. The North American GWAS data was derived from three case-control studies of EOC, as described previously (21), including: the Mayo Clinic Ovarian Cancer Study (MAY) (Rochester, MN), the North Carolina Ovarian Cancer Study (NCO) (Durham, NC), and the Tampa Bay Ovarian Cancer Study (TBO) (Tampa, FL). NCO and TBO used population-based ascertainment with linkage to state cancer registries and the National Death Index; MAY was clinic-based and linked to medical records and the National Death Index.
The UK GWAS has also been described in detail (8, 22) and included invasive epithelial ovarian cancer cases from four studies: SEARCH Ovarian Cancer Study (SEA) (Cambridge, UK), United Kingdom Ovarian Cancer Population Study (UKO) (London, UK), Cancer Research UK Familial Ovarian Cancer Register (UKR) (London, UK), and Royal Marsden Hospital (RMH) (London, UK). These studies recruited cases via regional and nationwide registries with follow-up via linkages to national vital statistics. Study protocols were approved by an institutional review board or ethics committee at each center, and all study participants provided written informed consent.
Genotyping, Quality Control, Definition of Gene Sets
Although both GWASes used the Illumina Infinium 610K array, genotyping and quality control was performed separately. Samples were removed with call rate <95%, ambiguous gender, unresolved identical genotypes, self reported as non-Caucasian, or predicted by STRUCTURE (23) analysis to have less than 80% European ancestry. SNPs were excluded from each GWAS with call rate < 95%, Hardy-Weinberg Equilibrium (HWE) p-value < 10−4, or no variation. Genotypes were coded as 0, 1 or 2 in terms of the number of minor alleles present.
SNPs were mapped to genes based on physical location; in particular, if they resided within 20 kb of the 5′ or 3′ end of a gene based on RefSeq Build 29 (NCBI genome build 36.3). This allowed for a SNP to be mapped to multiple genes and did not consider LD blocks or gene lengths. Genes were then mapped to gene sets using the following standarized sources: the Gene Ontology (GO) project (24) which categorizes genes by function using biological, cellular, and molecular schemes (“GO:BIO, “GO:CELL”, “GO:MOLE”), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (25), and PharmGKB (26) which group genes into biological pathways. GO uses a hierarchical ontology that classifies genes and therefore a level of the hierarchy (or specificity) must be specified for defining gene sets. We relied on Level 4 to determine GO gene sets, as a compromise between specificity and sensitivity. Across these sources, we identified a total of 2,566 gene sets containing approximately 16,500 unique genes. Supplementary Table 1 summarizes the numbers of SNPs, genes, and gene sets by source for the GSA.
Gene Set Analysis using PC-GM method
To assess association between predefined gene sets with overall survival from EOC, we performed GSA of each GWAS and combined results using Fisher’s method, a meta-analytic technique. We used a self-contained method which tests the null hypothesis Ho: SNPs/genes in the gene set of interest are NOT associated with the phenotype versus the alternative hypothesis Ha: SNPs/genes in the gene set are associated with the phenotype. GSA was completed in two steps, first using a principal component analysis (PCA) (18) in combination with a model of the phenotype, then a summarization step using the Gamma method (19), a generalization of Fisher’s method (27), which we refer to as the PC-GM method. Due to computational issues, redundant markers within genes (r2 = 1.0) were removed prior to PCA (28). For the first step in a two-step GSA (assessing gene-level associations), the PCs that explained 80% of the genetic variation within each gene were used to assess the significance of the gene with overall survival. For the gene-level analysis, the average number of PCs included in the analysis was 3.91 for North American analysis (4.75 for North American HGS cases) and 3.74 for UK analysis (3.82 for UK HGS cases). Gene-level association testing was completed using Cox proportional hazards regression (29) considering time from date of diagnosis to death with censoring at last follow-up. We accounted for the existence of left truncated data due to delayed enrollment of some cases (average 94.4 days between diagnosis and enrollment) using the Cox regression start-stop follow-up approach (30). Covariates included age at diagnosis, study site, and the first eigenvector adjusting for possible population stratification analysis from a PCA of the genome-wide SNPs using EigenSTRAT (31). Exploratory analysis of gene set adjusting for stage and grade were also completed. More extensive clinical data, such as treatment detail and degree of debulking, was missing on a large proportion of cases and thus not included in any model.
Following determination of the gene-level association p-values for genes within each gene set, p-values were summarized to the gene set using a meta-analytical method which can be applied to p-values. We chose to use the Gamma method with soft truncation threshold value (STT) of 0.15, a generalization of Fisher’s method. The Gamma method is based on summing transformed p-values, using an inverse Gamma(ω, 1) transformation. For a particular shape parameter ω, the test statistic is defined as , where G−1 is the inverse of a Gamma(ω, 1) cumulative distribution function (19). For the Gamma method the shape parameter, ω, controls the STT. When ω is 1, the transformed p-values follow a Chi-Square distribution, and therefore the Gamma method becomes equivalent to Fisher’s Method with a STT value of 1/e. Simulation studies have shown this approach with STT = 0.15 to be powerful for testing a self-contained gene set hypothesis under a variety of genetic models (20).
To account for the correlation between genes within a gene set and the size of the gene sets, empirical gene set association p-values were determine from ten-thousand permutations. First, the response variable was randomly permuted 10,000 times keeping the genotypic data fixed (and thus keeping the correlation structure between SNPs and genes fixed). The association test for each gene within the gene set was then computed based on the data set with the permuted phenotypes and the non-permuted phenotype, followed by computation of the gene set analysis statistics. The gene set statistics based on the permuted phenotypes then represent the empirical distribution of the gene-set test statistic. The proportion of permutations in which the empirical GSA test-statistic was greater than the observed GS statistic provided the empirical estimate of the p-value for the GS test for association. It should be noted that this GSA approach does not allow for estimation of the size or direction of gene set effect.
Following GSA of each GWAS, a meta-analysis was completed using Fisher’s method to combine the gene set p-values between the North American and UK GSA. Finally, to assist in the interpretation of results, as multiple SNPs may map to multiple gene sets, we completed hierarchical clustering of the gene sets with p < 0.005. The clustering was based on a distance measure of 1 − μ (μ = average proportion of SNPs shared between gene sets).
Results
Characteristics of invasive EOC cases and those with HGS subtype are described in Table 1. To elucidate gene sets related to overall survival, we completed a combined analysis in which the gene set p-values from each survival GWAS were combined using Fisher’s method for meta-analysis. The combined HGS analysis resulted in 43 (1.7%) gene sets with p < 0.005 (Table 2), with many of the top GSs from GO. However, this apparent “enrichment” of significant GO GSs is largely due to the fact that the set of GO GSs is much larger than the set of GSs in KEGG or PharmGKB. Assuming independence in GSs, we would have expected only 12.83 GSs to have p < 0.005 out of the 2,566 GSs tested by chance alone. The top gene sets were the intracellular signaling pathway (p = 7.3 × 10−5), regulation of cell-substrate junction assembly (p = 4.0 × 10−4), anatomical structure formation involved in morphogenesis (p = 5.3 × 10−4), and organelle outer membrane (p = 5.8 × 10−4). Of the 43 gene sets with p < 0.005, 21 had p < 0.10 in both the North American and the UK analyses, including the top gene sets of intracellular signaling pathway (North American p = 1.7 × 10−3, UK p = 3.3 × 10−3, combined p = 7.3 × 10−5) and macrolide binding (North American p = 9.0 × 10−4, UK p = 6.4 × 10−2, combined p = 6.2 × 10−4). Using a conservative Bonferroni adjustment for multiple testing (α = 2.0 × 10−5), the intracellular signaling pathway was very close to being statistically significant. The top results were similar when adjusting for stage and grade (Supplemental Table 2).
Table 1.
Clinical characteristics of epithelial ovarian cancer cases.
| Variables | All Cases | HGS Cases |
|---|---|---|
| N Subjects | 2,813 | 899 |
| N Deaths (%) | 1,116 (40%) | 473 (53%) |
| Stage (FIGO) | ||
| I | 800 (34%) | 73 (9%) |
| II | 234 (10%) | 60 (7%) |
| III | 1,147 (49%) | 611 (73%) |
| IV | 170 (7%) | 96 (11%) |
| missing | 462 | 59 |
| Grade (3/4 combined) | ||
| 1 | 330 (14%) | 0 |
| 2 | 637 (27%) | 0 |
| 3 | 1,351 (58%) | 899 (100%) |
| Missing/unknown | 18 | 0 |
| Histology | ||
| Serous | 1,491 (53%) | 899 (100%) |
| Mucinous | 241 (9%) | 0 |
| Endometriod | 497 (18%) | 0 |
| Clear cell | 265 (9%) | 0 |
| Other | 319 (11%) | 0 |
| Age at diagnosis | ||
| Mean (SE) | 57.8 (10.8) | 60.6 (9.8) |
| Days from diagnosis to enrollment | ||
| Mean (SE) | 663.1 (807.3) | 376.1 (519.4) |
| Median (range) | 443 (0–7,598) | 92 (0–3,885) |
| Years from diagnosis to last follow-up | ||
| Mean (SE) | 5.7 (4.1) | 4.0 (2.7) |
| Median (range) | 4.6 (<0.1–30.6) | 3.3 (<0.1–16.8) |
| North American GWAS study sites | ||
| MAY | 352 (13%) | 204 (23%) |
| NCO | 492 (18%) | 162 (18%) |
| TBO | 213 (8%) | 118 (13%) |
| UK GWAS study sites | ||
| RMH | 143 (5%) | 20 (2%) |
| SEA | 1,087 (39%) | 251 (28%) |
| UKR | 32 (1%) | 0 (0%) |
| UKO | 494 (18%) | 144 (16%) |
Values reported as number (percent) unless otherwise indicated. HGS = high grade serous.
Table 2.
Association between gene sets and ovarian cancer survival in cases with high-grade serous (HGS) histological subtype (p < 0.005).
| Source | Gene Set | No. SNPs | No. Genes | GS P-value |
|---|---|---|---|---|
| GO:BIO | intracellular signaling pathway | 22,715 | 857 | 7.3 ×10−5 |
| GO:BIO | regulation of cell-substrate junction assembly | 195 | 8 | 4.0 ×10−4 |
| GO:BIO | anatomical structure formation involved in morphogenesis | 9,701 | 375 | 5.3 ×10−4 |
| GO:CELL | organelle outer membrane | 2,023 | 109 | 5.8 ×10−4 |
| GO:BIO | negative regulation of focal adhesion assembly | 114 | 5 | 5.8 ×10−4 |
| GO:MOLE | macrolide binding | 126 | 8 | 6.2 ×10−4 |
| GO:MOLE | cis-trans isomerase activity | 318 | 34 | 7.5 ×10−4 |
| GO:BIO | negative regulation of odontogenesis | 41 | 2 | 8.2 ×10−4 |
| GO:BIO | osteoblast differentiation | 1,976 | 70 | 8.4 ×10−4 |
| GO:BIO | axis specification | 420 | 29 | 1.0 ×10−3 |
| GO:CELL | photoreceptor inner segment | 211 | 9 | 1.1 ×10−3 |
| GO:CELL | outer membrane | 2,131 | 113 | 1.1 ×10−3 |
| GO:BIO | negative regulation of molecular function | 5,927 | 331 | 1.6 ×10−3 |
| GO:BIO | regulation of response to cytokine stimulus | 24 | 3 | 1.7 ×10−3 |
| GO:MOLE | SH3/SH2 adaptor activity | 1,283 | 51 | 1.7 ×10−3 |
| GO:BIO | homeostatic process | 17,434 | 769 | 1.8 ×10−3 |
| GO:BIO | embryo implantation | 507 | 26 | 1.9 ×10−3 |
| GO:BIO | tissue homeostasis | 1,818 | 70 | 1.9 ×10−3 |
| GO:BIO | chromatin disassembly | 30 | 5 | 1.9 ×10−3 |
| GO:BIO | ossification | 3,808 | 161 | 2.0 ×10−3 |
| GO:MOLE | protein dimerization activity | 13,527 | 540 | 2.1 ×10−3 |
| GO:BIO | bone development | 3,913 | 169 | 2.2 ×10−3 |
| GO:BIO | regulation of odontogenesis | 194 | 8 | 2.3 ×10−3 |
| GO:BIO | regulation of cell adhesion | 3,760 | 123 | 2.5 ×10−3 |
| GO:BIO | anatomical structure morphogenesis | 33,110 | 1244 | 2.5 ×10−3 |
| GO:BIO | signal transmission | 78,767 | 3523 | 2.5 ×10−3 |
| GO:BIO | somatic stem cell maintenance | 244 | 12 | 2.7 ×10−3 |
| GO:BIO | multicellular organismal homeostasis | 2,259 | 92 | 2.8 ×10−3 |
| GO:MOLE | protein binding, bridging | 2,037 | 96 | 2.9 ×10−3 |
| GO:BIO | organ morphogenesis | 12,461 | 554 | 2.9 ×10−3 |
| GO:BIO | peptide transport | 2,067 | 87 | 3.0 ×10−3 |
| PharmGKB | Glucocorticoid & Inflammatory genes (PD) | 146 | 9 | 3.1 ×10−3 |
| GO:MOLE | protein transmembrane transporter activity | 97 | 14 | 3.2 ×10−3 |
| GO:BIO | cell migration | 11,535 | 386 | 3.4 ×10−3 |
| GO:BIO | negative regulation of biological process | 38,075 | 1805 | 3.4 ×10−3 |
| GO:BIO | intracellular transport | 12,735 | 700 | 3.5 ×10−3 |
| GO:BIO | multicellular organismal macromolecule metabolic process | 954 | 41 | 3.5 ×10−3 |
| GO:BIO | signal transduction | 66,542 | 3130 | 3.7 ×10−3 |
| GO:BIO | muscle homeostasis | 989 | 13 | 3.8 ×10−3 |
| GO:CELL | beta-catenin destruction complex | 87 | 6 | 4.0 ×10−3 |
| GO:BIO | negative regulation of response to cytokine stimulus | 8 | 1 | 4.7 ×10−3 |
| KEGG | TGF-beta signaling pathway | 1,359 | 86 | 4.7 ×10−3 |
| GO:BIO | stem cell maintenance | 414 | 25 | 4.7 ×10−3 |
Adjusted for age, study site, and population structure.
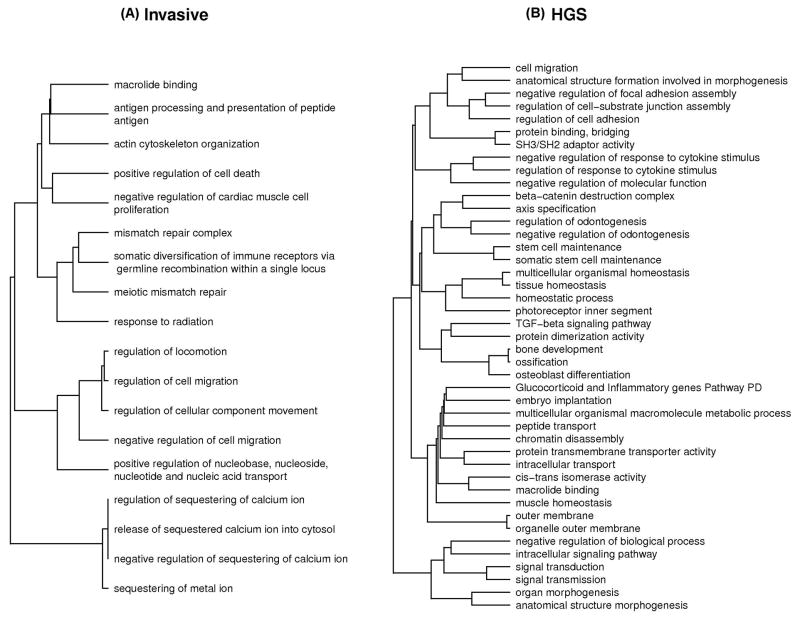
The top gene sets in combined analysis of all cases, regardless of histological subtype, were (Table 3) meiotic mismatch repair (p = 6.3 × 10−4), macrolide binding (p = 1.0 × 10−3), antigen processing and presentation of peptide antigen (p = 1.1 × 10−3); mismatch repair complex (p = 1.3 × 10−3); and regulation of cell migration (p = 1.7 × 10−3). Of the 18 gene sets with combined p < 0.005 (0.7%), eight had p < 0.10 in both the North American and the UK analyses. Similar results were observed in the analysis adjusting for stage and grade (Supplemental Table 2). After adjusting for multiple testing, none of the gene sets were statistically significant in the combined analysis. To aid in the interpretation of the gene set results, Figure 1 presents the results from hierarchical clustering of gene sets with p < 0.005 (based on proportion of SNPs in common) for both the analysis of all cases and HGS cases.
Table 3.
Association between gene sets and ovarian cancer survival (p < 0.005).
| Source | Gene Set | No. SNPs | No. Genes | GS P-value |
|---|---|---|---|---|
| GO:BIO | meiotic mismatch repair | 13 | 1 | 6.3 ×10−4 |
| GO:MOLE | macrolide binding | 126 | 8 | 1.0 ×10−3 |
| GO:BIO | antigen processing and presentation of peptide antigen | 610 | 25 | 1.1 ×10−3 |
| GO:CELL | mismatch repair complex | 135 | 7 | 1.3 ×10−3 |
| GO:BIO | regulation of cell migration | 5,266 | 171 | 1.7 ×10−3 |
| GO:BIO | negative regulation of cardiac muscle cell proliferation | 78 | 4 | 1.7 ×10−3 |
| GO:BIO | positive regulation of cell death | 10,363 | 432 | 2.2 ×10−3 |
| GO:BIO | release of sequestered calcium ion into cytosol | 680 | 24 | 2.4 ×10−3 |
| GO:BIO | regulation of sequestering of calcium ion | 680 | 24 | 2.4 ×10−3 |
| GO:BIO | negative regulation of sequestering of calcium ion | 680 | 24 | 2.4 ×10−3 |
| GO:BIO | sequestering of metal ion | 781 | 31 | 2.5 ×10−3 |
| GO:BIO | regulation of locomotion | 5,869 | 192 | 4.0 ×10−3 |
| GO:BIO | regulation of cellular component movement | 5,986 | 193 | 4.1 ×10−3 |
| GO:BIO | response to radiation | 3,676 | 190 | 4.2 ×10−3 |
| GO:BIO | somatic diversification of immune receptors via germline recombination within a single locus | 480 | 35 | 4.2 ×10−3 |
| GO:BIO | negative regulation of cell migration | 1,365 | 57 | 4.2 ×10−3 |
| GO:BIO | actin cytoskeleton organization | 7,554 | 265 | 4.8 ×10−3 |
| GO:BIO | positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid transport | 56 | 2 | 5.0 ×10−3 |
Adjusted for age, study site, and population structure.
Figure 1.
Hierarchical clustering dendrogram of gene sets with p < 0.005 (distance measure based on proportion of SNPs in common between gene sets) for the analysis of (A) all cases and (B) HGS cases.
Next, for the top gene sets (p < 0.01), we examined which particular gene(s) in these gene set may be most associated with overall survival. Table 4 presents the genes with p < 0.0005 in top gene sets. For the combined analysis the top most significant genes were: HLA-C (p = 1.3 × 10−4), MYH3 (p = 1.7 × 10−4), WNT5A (p = 3.7 × 10−4) and ZSCAN23 (p = 3.8 × 10−4).
Table 4.
Genes with p < 0.0005 in gene sets with p < 0.01 from the combined GSA.
| Analysis | Gene | Combined P | US P | UK P |
|---|---|---|---|---|
| All | HLA-C | 1.3 ×10−4 | 1.9 ×10−4 | 5.3 ×10−2 |
| MYH3 | 1.7 ×10−4 | 1.5 ×10−5 | 9.6 ×10−1 | |
| WNT5A | 3.7 ×10−4 | 5.5 ×10−2 | 6.0 ×10−4 | |
| ZSCAN23 | 3.8 ×10−4 | 4.6 ×10−4 | 7.2 ×10−2 | |
|
| ||||
| HGS | COL28A1 | 3.2 ×10−5 | 6.2 ×10−2 | 3.7 ×10−5 |
| ZNF331 | 1.1 ×10−4 | 2.0 ×10−2 | 4.6 ×10−4 | |
| GNAT3 | 1.3 ×10−4 | 1.6 ×10−4 | 6.2 ×10−2 | |
| NMNAT3 | 2.5 ×10−4 | 1.8 ×10−3 | 1.2 ×10−2 | |
| ARMS2 | 3.6 ×10−4 | 1.5 ×10−1 | 2.1 ×10−4 | |
| PPIH | 3.8 ×10−4 | 8.1 ×10−3 | 4.2 ×10−3 | |
| WWOX | 4.4 ×10−4 | 6.2 ×10−4 | 6.4 ×10−2 | |
Adjusted for age, study site, and population structure. HGS = high grade serous.
To better interpret the combined GSA results, we also examined the GSA results for the individual GWASes. In the North American GWAS, five gene sets with p-values of association with survival < 0.005 were identified among cases with HGS histological subtype, with the top gene set for analysis of HGS being inflammation related (p = 0.0007), cell migration (p =0.0009) and macrolide binding (p = 0.0009). Similarly, four gene sets were found to be associated with survival (gene set p-value < 0.001; Supplemental Table 3) in the overall group. Genetic variation in meiotic mismatch repair, as defined in the biological class of GO, and in mismatch repair complex, as defined in the cellular class of GO, were the most significantly associated gene sets (p-values = 2.0 × 10−4). Of note, the meiotic mismatch repair gene set included only MSH6 and was contained within the mismatch repair complex gene set. Other gene sets with p < 0.001 were positive regulation of cell death (p = 0.0004) and multicellular organismal aging (p = 0.0009). There was no overlap of the top gene sets (p < 0.001) from the analyses of all versus HGS cases. It should be noted that none of these results are statistically significant at the Bonferroni significance level of 2×10−5.
GSA of the UK GWAS revealed 11 gene sets with p < 0.001 from the analysis of the HGS cases and three gene sets with p < 0.001 from the analysis of all cases (Supplemental Table 3). Similar to the North American GSA, there was limited overlap between the top gene sets between the analyses of all individuals and the HGS subgroup. For the analysis of the HGS, the top gene sets were photoreceptor inner segment (p = 0.0002), organelle outer membrane (p = 0.0002), and somatic stem cell maintenance (p = 0.0004). The top gene set in the overall analysis involved regulation of the calcium ion (p = 0.001). However, none of these gene sets were significant at a Bonferroni level of 2 ×10−5. As Supplemental Table 3 illustrates, there was little agreement in top gene sets between the North American and UK analyses.
Discussion
In this manuscript, we present results from the first application of GSA to ovarian cancer. As ovarian cancer has high mortality, we assessed gene set associations with overall survival, hypothesizing that use of standardized groupings of genes based on known biology may identify novel inherited determinants of outcome and identify avenues for mechanistic study. GSA is an increasingly applied approach for secondary analysis of GWAS data, as this analysis approach reduces the number of tests and thus the impact of multiple testing on inferences; in addition, it incorporates prior biological knowledge into the analysis (15). To complete the GSA, we used a novel approach that combines the use of principal components analysis and the Gamma Method, referred to as the PC-GM approach. Simulation studies have found this approach to out perform other self-contained gene set approaches, such as Fisher’s method (32, 33), for a variety of genetic models and scenarios. Although these methods are not designed to identify specific genes or genetic variants that are associated with the trait of interest, results from a GSA can be used to plan further, in-depth, investigation focused on specific gene sets of interest and may uncover additional genetic causes of complex traits.
When attention was confined to cases with the HGS subtype, the top gene set associated with overall survival was the intracellular signaling pathway (p = 7.3 ×10−5) achieving borderline Bonferroni-corrected statistical significance. This is a large gene set containing 22,715 SNPs mapped to 857 genes. The definition of this gene set from GO’s biological classification states that it contains genes involved in “the process in which a signal is passed to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell”(34). A “child” in the hierarchical structure of GO is the “signal transduction by p53 class mediator” gene set containing genes involved in the signaling process induced by the cell cycle regulator phosphoprotein p53. In addition to genes WWOX and APC which have been implicated in response to therapy (35), additional genes with modest gene-level p-values within the intracellular signaling pathway gene set included SMAD4 (p = 0.009), IL6 (p = 0.0145), ERBB4 (p = 0.018), and JAK2 (p = 0.023). Thus, as the most statistically significant gene set in our report, even based on a smaller sample size of HGS cases alone, this particular collection of genes merits additional follow-up.
One of the most significant gene sets in analysis of all cases was macrolide binding (all cases p =1.0 × 10−3; HGS cases p =6.2 ×10−4) which contained 126 SNPs mapped to eight genes (including FKBP1B [p=0.018], FKBP3 [p = 0.086], NFATC1 [p = 0.021] and FKBP6 [p = 0.088]). This gene set originated from the GO molecular classification which indicates that the gene set is a “child” of the drug binding gene set and a “parent” to the FK506 binding gene set which contains genes that interact “selectively and non-covalently with the immunosuppressant FK506”(36). Henriksen et al (37) found that the FK506 binding protein 65 (FKBP65) was highly expressed in ovarian epithelium and in benign ovarian tumor cells, while the expression levels were lower in invasive tumor cells. FKBP65 was also found to be inversely associated with expression of p53 (37).
While this GSA of GWAS data provides novel insight and findings into the association of genome-wide germline variation with overall survival, this type of analysis has limitations. One limitation is that our definitions of gene sets and pathways are limited to our knowledge about the genome, and pathways are continually evolving. Another limitation is the fact that GSA assumes that SNPs can be assigned to relevant genes, particularly in light of the fact that many phenotype-associated SNPs identified to date do not lie in genes. Lastly, this GSA does not allow one to determine the direction of the gene set effect on the outcome. However, as this study illustrates, novel and biologically plausible association can be detected using GSA and thus contributes to our understanding of the relationship between genetic variation and mortality from EOC. Moreover, these results have led to possible gene sets and novel genes that can be followed up in future studies, in particular: replication studies, pharmacogenomic studies, and studies investigating the development of novel EOC therapies.
Supplementary Material
Acknowledgments
We thank all the individuals who took part in this study and all the researchers, clinicians and administrative staff who have made possible the many studies contributing to this work. In particular we thank A. Ryan and J. Ford (UKOPS); P. Harrington and the Studies of Epidemiology and Risk Factors in Cancer Heredity team (SEARCH); the Minnesota Partnership for Biotechnology and Medical Genomics; the Mayo Foundation; the National Institute for Health Research; Cambridge Biomedical Research Centre and the Royal Marsden Hospital. We would also like to thank Joanna Biernacka for her contributions to the development of the PC-GM method for assessing the association of gene sets with a phenotype.
Grant Support
Funding provided by the Cambridge Biomedical Research Centre, Cancer Research UK (C490/A10119), the US National Cancer Institute (CA148112, CA122443, CA114343, CA140879, GM86689), and a pilot project award from the Mayo Clinic SPORE in Ovarian Cancer (CA136393).
Footnotes
Disclosure of Potential Conflicts of Interest
The authors have no conflict of interest to declare.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Barnholtz-Sloan JS, Schwartz AG, Qureshi F, Jacques S, Malone J, Munkarah AR. Ovarian cancer: changes in patterns at diagnosis and relative survival over the last three decades. Am J Obstet Gynecol. 2003;189:1120–7. doi: 10.1067/s0002-9378(03)00579-9. [DOI] [PubMed] [Google Scholar]
- 3.McGuire V, Jesser CA, Whittemore AS. Survival among U. S. women with invasive epithelial ovarian cancer. Gynecol Oncol. 2002;84:399–403. doi: 10.1006/gyno.2001.6536. [DOI] [PubMed] [Google Scholar]
- 4.Hoskins PJ, O’Reilly SE, Swenerton KD, Spinelli JJ, Fairey RN, Benedet JL. Ten-year outcome of patients with advanced epithelial ovarian carcinoma treated with cisplatin-based multimodality therapy. J Clin Oncol. 1992;10:1561–8. doi: 10.1200/JCO.1992.10.10.1561. [DOI] [PubMed] [Google Scholar]
- 5.Goode EL, Maurer MJ, Sellers TA, Phelan CM, Kalli KR, Fridley BL, et al. Inherited determinants of ovarian cancer survival. Clin Cancer Res. 2010;16:995–1007. doi: 10.1158/1078-0432.CCR-09-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnatty SE, Beesley J, Paul J, Fereday S, Spurdle AB, Webb PM, et al. ABCB1 (MDR 1) polymorphisms and progression-free survival among women with ovarian cancer following paclitaxel/carboplatin chemotherapy. Clin Cancer Res. 2008;14:5594–601. doi: 10.1158/1078-0432.CCR-08-0606. [DOI] [PubMed] [Google Scholar]
- 7.Krivak TC, Darcy KM, Tian C, Armstrong D, Baysal BE, Gallion H, et al. Relationship between ERCC1 polymorphisms, disease progression, and survival in the Gynecologic Oncology Group Phase III Trial of intraperitoneal versus intravenous cisplatin and paclitaxel for stage III epithelial ovarian cancer. J Clin Oncol. 2008;26:3598–606. doi: 10.1200/JCO.2008.16.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–4. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardon LR, Bell JI. Association study designs for complex diseases. Nature reviews. 2001;2:91–9. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 10.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature reviews. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 11.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nature reviews. 2005;6:109–18. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 12.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menashe I, Maeder D, Garcia-Closas M, Figueroa JD, Bhattacharjee S, Rotunno M, et al. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 70:4453–9. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–72. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridley BL, Biernacka JM. Gene set analysis of SNP data: benefits, challenges, and future directions. Eur J Hum Genet. 2011;19:837–43. doi: 10.1038/ejhg.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nature reviews. 11:843–54. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 17.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gauderman WJ, Murcray C, Gilliland F, Conti DV. Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol. 2007;31:383–95. doi: 10.1002/gepi.20219. [DOI] [PubMed] [Google Scholar]
- 19.Zaykin DV, Zhivotovsky LA, Czika W, Shao S, Wolfinger RD. Combining p-values in large-scale genomics experiments. Pharm Stat. 2007;6:217–26. doi: 10.1002/pst.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biernacka JM, Jenkins GD, Wang Y, Moyer AM, Fridley BL. Use of the Gamma Method for self-contained gene set analysis of SNP data. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.236. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Permuth-Wey J, Chen YA, Tsai YY, Chen Z, Qu X, Lancaster JM, et al. Inherited variants in mitochondrial biogenesis genes may influence epithelial ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2011;20:1131–45. doi: 10.1158/1055-9965.EPI-10-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein TE, Chang JT, Cho MK, Easton KL, Fergerson R, Hewett M, et al. Integrating genotype and phenotype information: an overview of the PharmGKB project. Pharmacogenetics Research Network and Knowledge Base. Pharmacogenomics J. 2001;1:167–70. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 27.Fisher RA. Statistical Methods for Research Workers. London: Oliver and Boyd; 1932. [Google Scholar]
- 28.Jolliffe IT. Principal components analysis. 2. New York: Springer–Verlag; 2002. [Google Scholar]
- 29.Cox DR. Regression models and life tables (with discussion) Journal of the Royal Statistical Society, Series B (Methodological) 1972;34:187–220. [Google Scholar]
- 30.Therneau TM, Grambsch PM. Modeling survival data: extending the Cox model. New Yor, NY: Springer; 2000. [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 32.Chai HS, Sicotte H, Bailey KR, Turner ST, Asmann YW, Kocher JP. GLOSSI: a method to assess the association of genetic loci-sets with complex diseases. BMC Bioinformatics. 2009;10:102. doi: 10.1186/1471-2105-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De la Cruz O, Wen X, Ke B, Song M, Nicolae DL. Gene, region and pathway level analyses in whole-genome studies. Genet Epidemiol. 2009 doi: 10.1002/gepi.20452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gene Ontology Project. Gene set intracellular signaling pathway. [cited; Available from: http://www.ebi.ac.uk/QuickGO/GTerm?id=GO:0023034.
- 35.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–28. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gene Ontology Project. Gene set FK506 binding. [cited; Available from: http://gowiki.tamu.edu/wiki/index.php/Category:GO:0005528_!_FK506_binding.
- 37.Henriksen R, Sorensen FB, Orntoft TF. Birkenkamp-Demtroder K. Expression of FK506 binding protein 65 (FKBP65) is decreased in epithelial ovarian cancer cells compared to benign tumor cells and to ovarian epithelium. Tumour Biol. 2011 doi: 10.1007/s13277-011-0167-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.