Abstract
The synthesis of substituted 3-(5-imidazo[2,1-b]thiazolylmethylene)-2-indolinones and analogues is reported. Their cytotoxic activity was evaluated according to protocols available at the National Cancer Institute (NCI), Bethesda, MD. The action of selected compounds was examined for potential inhibition of tubulin assembly in comparison with the potent colchicine site agent combretastatin A-4. The most potent compounds also strongly and selectively inhibited the phosphorylation of the oncoprotein kinase Akt in cancer cells. The effect of the most interesting compounds was examined on the growth of HT-29 colon cancer cells. These compounds caused the cells to arrest in the G2/M phase of the cell cycle, as would be expected for inhibitors of tubulin assembly.
Introduction
In the last paper of this series2 we described the synthesis of substituted 3-(5-imidazo[2,1-b]thiazolylmethylene)-2-indolinones and 3-(5-imidazo[2,1-b]thiadiazolylmethylene)-2-indolinones. The best results in terms of inhibition of cell proliferation were obtained with the derivatives bearing methyl groups at positions 2 and 6 of the imidazothiazole or imidazothiadiazole system. Therefore, we planned the synthesis of new derivatives (Scheme 1) bearing this scaffold condensed with indolinones, benzoindolinone or coumaranone (3–10).
Scheme 1.
CBR = condensed benzene ring.
We also demonstrated2 that the methyl group at the 6 position of the imidazothiazole or imidazothiadiazole system could be replaced without loss of activity by p-methylphenyl and p-chlorophenyl groups. To confirm the effect of these substituents on biological activity, a new series of derivatives modified only in the indolinone portion was prepared (11–20).
From a further analysis of the previous results, we decided that it would be interesting to study the effects of other substituents at the 6 position of the imidazothiazole system, such as methoxy, pyridinyl, thienyl and 4-nitrophenyl groups (26–42). Moreover the activity of 2,5-dimethoxyphenyl derivatives3 prompted us to prepare analogues bearing a trimethoxyphenyl group (21–25), since it is present, free or hindered, in well known antitumor agents such as combretastatin A-4, colchicine and podophyllotoxin.
Chemistry
Compounds 3–42 were prepared by means of a single step Knoevenagel reaction between 1 and 2 in methanol/piperidine (triethylamine was used in place of piperidine for compounds 6, 10, 15, and 20 to improve the yield). The structures of the final compounds were confirmed by means of IR and 1H-NMR spectra. Most compounds were obtained as almost pure geometrical isomers, which, according to the usual NOE experiments described previously,3,4 were assigned the E configuration. Compounds 10, 15, 20, 29, 30 and 37–39 were obtained as E/Z mixtures and used as such in the biological assays. The E/Z ratio in solution is time dependent and tends to equilibrate at 50/50. In a previous paper, we described the separation of the two isomers by fractional crystallization,2 but no significant difference in pharmacological behavior was noted. The indolinones 2b, 2k, 2l and 2-coumaranone (2d) are commercially available, whereas the other starting compounds were prepared according to the literature,5–23 except indolinone 2o and aldehydes 1g, 1h, 1i, whose synthesis is reported in the Supporting Information. The aforementioned aldehydes were obtained by means of the Vilsmeier reaction on the corresponding imidazo[2,1-b]thiazole, prepared in turn from 2-amino-thiazole or 2-amino-5-methylthiazole and 2-bromo-1-(3,4,5-trimethoxyphenyl)ethanone or 2-bromo-1-(2-bromo-3,4,5-trimethoxyphenyl)ethanone.
Biology
a. Cell-Based Assays
Compounds 3–42 were initially tested at a single high concentration (10−5 M) in the full NCI 60 cell panel (NCI 60 Cell One-Dose Screen). This panel is organized into subpanels representing leukemia, melanoma and cancers of lung, colon, kidney, ovary, breast, prostate and central nervous system. Only compounds which satisfy predetermined threshold inhibition criteria in a minimum number of cell lines will progress to the full 5-log concentration assay. The threshold inhibition criteria for progression to the five-concentration screen were selected to efficiently capture compounds with antiproliferative activity based on the analysis of historical DTP screening data. The result is expressed as the percent growth of treated cells relative to the control following a 48 h incubation. Twenty-two of forty evaluated compounds were active in the preliminary test and progressed to the five-concentration assay, ranging from 10−9 to 10−4 M. TABLE 2 reports the results obtained (vincristine is reported for comparison purposes), at three assay endpoints: 50% growth inhibition (GI50), total cytostatic effect (TGI=Total Growth Inhibition) and cytotoxic effect (LC50, loss of 50% of the initial cell protein). For some compounds, the 5-concentration test was repeated, and no significant differences were found. For these compounds, the data reported in TABLE 2 are the mean values of the two experiments. The tested compounds showed a mean GI50 range between 0.04 and 93 μM, and compounds 5, 29–31, 33–36, 38 and 39 were submitted to the Biological Evaluation Committee of the NCI for possible future development.
TABLE 2.
Nine subpanels at five concentrations: growth inhibition, cytostatic and cytotoxic activity (μM) of selected compounds.
| Comp a | Modes | Leukemia | NSCLC | Colon | CNS | Melanoma | Ovarian | Renal | Prostate | Breast | MG-MIDb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | GI50 | 1.07 | 2.29 | 0.98 | 2.40 | 1.58 | 7.41 | 4.79 | 2.29 | 1.20 | 2.00 |
| TGI | 37.15 | - | 66.07 | 25.70 | 57.54 | - | 70.79 | - | 15.85 | 51.29 | |
| 5 c | GI50 | 0.21 | 0.31 | 0.11 | 0.17 | 0.14 | 0.85 | 0.23 | 0.21 | 0.18 | 0.21 |
| TGI | 53.70 | 28.18 | 58.88 | 5.37 | 50.12 | 14.13 | 12.02 | 16.22 | 5.62 | 19.95 | |
| LC50 | - | 93.33 | - | 74.13 | - | 74.13 | 70.79 | - | 50.12 | 81.28 | |
| 7 | GI50 | 2.34 | 19.50 | 4.17 | 20.89 | 26.92 | 33.88 | 26.92 | - | 6.31 | 15.14 |
| TGI | - | - | - | 87.10 | - | - | 91.20 | - | 57.54 | 91.20 | |
| 9 c | GI50 | 0.17 | 0.87 | 0.35 | 0.39 | 0.68 | 0.91 | 0.83 | 0.38 | 0.30 | 0.50 |
| TGI | 3.31 | 15.49 | 6.61 | 4.90 | 16.98 | 11.75 | 10.47 | 5.13 | 6.31 | 8.51 | |
| LC50 | 66.07 | 74.13 | 48.98 | 46.77 | 70.79 | 56.23 | 56.23 | 15.14 | 75.86 | 58.88 | |
| 11 | GI50 | 52.48 | 18.62 | 60.26 | 11.75 | 33.88 | 10.47 | 18.20 | 21.38 | 12.59 | 22.91 |
| TGI | 83.18 | 72.44 | - | 91.20 | - | 63.10 | - | - | 63.10 | 83.18 | |
| 12 | GI50 | 7.08 | 23.99 | 20.42 | 19.05 | 17.38 | 32.36 | 15.14 | 6.46 | 13.49 | 16.60 |
| TGI | 63.10 | 89.13 | - | 75.86 | 87.10 | 89.13 | 85.11 | - | 75.86 | 83.18 | |
| 13 | GI50 | 4.37 | 14.79 | 6.03 | 39.81 | 52.48 | 56.23 | 4.79 | 5.37 | 12.88 | 13.80 |
| GI50 | 56.23 | 85.11 | 33.11 | - | - | - | 70,79 | - | 72.44 | 74.13 | |
| 16 | GI50 | - | - | - | 69.18 | - | - | 81.28 | - | 95.50 | 93.33 |
| 17 | GI50 | 25.12 | 60.26 | 38.02 | 20.42 | 66.07 | 21.88 | 26.30 | - | 36.31 | 37.15 |
| TGI | - | - | - | 79.43 | - | - | 72.44 | - | 95.50 | 93.33 | |
| 24 | GI50 | 36.31 | 32.36 | 26.30 | 23.44 | 29.51 | 39.81 | 30.90 | 30.20 | 19.05 | 28.84 |
| TGI | - | 89.13 | 75.86 | 54.95 | 85.11 | 85.11 | 75.86 | 85.11 | 63.10 | 77.62 | |
| 28 | GI50 | 0.35 | 1.58 | 0.49 | 0.81 | 2.45 | 2.88 | 2.34 | 1.02 | 0.59 | 1.15 |
| TGI | 14.79 | 70.79 | 52.48 | 20.89 | 91.20 | 37.15 | 95.50 | - | 36.31 | 50.12 | |
| LC50 | - | - | - | 89.13 | - | 91.20 | - | - | - | 97.72 | |
| 29 c | GI50 | 0.28 | 1.78 | 0.52 | 0.71 | 0.68 | 1.62 | 1.15 | 1.26 | 0.45 | 0.83 |
| TGI | 7.08 | 18.20 | 10.47 | 22.39 | 11.75 | 17.38 | 10.47 | 19.05 | 7.76 | 12.88 | |
| LC50 | 69.18 | 58.88 | 33.88 | 66.07 | 51.29 | 50.12 | 43.65 | 47.86 | 42.66 | 50.12 | |
| 30 c | GI50 | 0.06 | 0.33 | 0.11 | 0.17 | 0.12 | 0.27 | 0.25 | 0.15 | 0.21 | 0.18 |
| TGI | 4.57 | 28.84 | 21.88 | 6.03 | 15.14 | 11.48 | 15.85 | 31.62 | 12.59 | 14.13 | |
| LC50 | - | 48.98 | 46.77 | 38.02 | 38.90 | 36.31 | 42.66 | - | 46.77 | 43.65 | |
| 31 c | GI50 | 0.02 | 0.11 | 0.02 | 0.03 | 0.02 | 0.04 | 0.07 | 0.03 | 0.04 | 0.04 |
| TGI | 0.58 | 6.46 | 1.35 | 2.57 | 6.76 | 2.19 | 4.68 | 3.98 | 8.32 | 3.39 | |
| LC50 | - | 39.81 | 15.14 | 34.67 | 38.90 | 16.98 | 28.84 | - | 46.77 | 31.62 | |
| 33 c | GI50 | 0.50 | 0.43 | 0.29 | 0.25 | 0.47 | 0.44 | 0.54 | 0.55 | 0.28 | 0.39 |
| TGI | 21.38 | 51.29 | 13.80 | 7.94 | 40.74 | 20.42 | 38.90 | 29.51 | 13.18 | 22.91 | |
| LC50 | 60.26 | 93.33 | 89.13 | 50.12 | 95.50 | - | - | - | 91.20 | 85.11 | |
| 34 c | GI50 | 2.19 | 7.08 | 3.72 | 6.31 | 4.90 | 4.68 | 7.08 | 13.18 | 3.24 | 4.90 |
| TGI | 64.57 | 70.79 | 91.20 | 32.36 | 60.26 | 43.65 | 48.98 | - | 50.12 | 57.54 | |
| LC50 | - | - | - | 87.10 | - | 97.72 | 89.13 | - | 93.33 | 95.50 | |
| 35 c | GI50 | 1.32 | 4.27 | 1.41 | 3.31 | 3.72 | 3.47 | 3.31 | 3.98 | 2.45 | 2.82 |
| TGI | 26.30 | 89.13 | 64.57 | 40.74 | 40.74 | 46.77 | 58.88 | 81.28 | 74.13 | 54.95 | |
| 36 c | GI50 | 0.14 | 1.29 | 0.33 | 0.35 | 0.21 | 0.65 | 0.62 | 0.39 | 0.54 | 0.44 |
| TGI | 5.37 | 79.43 | 22.39 | 16.22 | 34.67 | 21.38 | 48.98 | 69.18 | 58.88 | 31.62 | |
| LC50 | 89.13 | - | 79.43 | 75.86 | 93.33 | 89.13 | 85.11 | - | - | 89.13 | |
| 37 c | GI50 | 3.39 | 6.03 | 3.16 | 6.46 | 6.03 | 5.62 | 6.76 | 12.02 | 2.88 | 5.01 |
| TGI | 52.48 | 66.07 | 61.66 | 42.66 | 47.86 | 44.67 | 47.86 | 93.33 | 51.29 | 52.48 | |
| 38 c | GI50 | 0.02 | 0.16 | 0.04 | 0.04 | 0.04 | 0.10 | 0.14 | 0.05 | 0.17 | 0.07 |
| TGI | 4.57 | 44.67 | 7.59 | 15.49 | 20.89 | 12.02 | 31.62 | 40.74 | 34.67 | 19.05 | |
| LC50 | 91.20 | - | 66.07 | - | 81.28 | - | - | - | - | 93.33 | |
| 39 c | GI50 | 0.16 | 1.02 | 0.35 | 0.38 | 0.79 | 0.63 | 0.71 | 0.55 | 0.49 | 0.54 |
| TGI | 4.90 | 58.88 | 23.99 | 18.62 | 72.44 | 31.62 | 53.70 | - | 48.98 | 34.67 | |
| LC50 | 79.43 | - | 87.10 | - | 91.20 | - | - | - | - | 95.50 | |
| 40 c | GI50 | 3.89 | 7.94 | 2.24 | 10.96 | 8.71 | 6.46 | 15.85 | 21.38 | 2.19 | 6.31 |
| TGI | 43.65 | 95.50 | 60.26 | 77.62 | 77.62 | 72.44 | 85.11 | - | 60.26 | 72.44 | |
| Vincristine sulfate d | GI50 | 0.10 | 0.25 | 0.10 | 0.13 | 0.16 | 0.32 | 0.32 | 0.13 | 0.32 | 0.20 |
| TGI | 15.85 | 15.85 | 3.98 | 6.31 | 7.94 | 19.95 | 19.95 | 6.31 | 7.94 | 10.00 |
Highest conc. tested = 10−4 M. The GI50 and TGI values are reported only when <100 μM. The compound exposure time was 48 h.
Mean Graph MIDpoint: average value for all cell lines tested; i.e., mean GI50.
Mean of two separate experiments.
Highest conc. tested = 10−3 M.
The first group of compounds prepared for further study of the biological activity of 2,6-dimethyl substituted imidazothiazoles and imidazothiadiazoles confirmed that these moieties can generate active derivatives. However, substitution of indolinone with coumaranone (6, 10) and the introduction of a dimethylamino group in the indolinone system (3, 8) led to compounds that had weak activity. The most active compounds in this group bear a benzoindole system (5 and 9, with mean GI50 values of 0.21 and 0.50 μM, respectively).
Considering the 4-chlorophenyl and 4-methylphenyl substituents at the 6 position of the imidazole ring (11–20), it is clear that the latter leads to increased activity (11–13). From the data obtained with compounds 11–20, it is also clear that replacement of indolinone with coumaranone (15, 20) led to inactive compounds.
Considering other substituents on the imidazole ring (21–42), the 4-nitrophenyl and the 2-thienyl groups were detrimental for antiproliferative activity, while the most active compounds had either a 2-pyridyl (28–32) or a methoxy (33–42) group, with mean GI50 values of 0.55 μM for the former and 2.56 μM for the latter. In particular compound 31 was the most active agent in the entire series (GI50, 0.04 μM), although it had a relatively high LC50 (32 μM).
b. Tubulin
A COMPARE analysis24 of a number of the compounds indicated that tubulin was the likely target of the most active agents. We therefore examined most of the compounds with mean GI50 values less than 1 μM for potential inhibition of tubulin assembly (TABLE 3) in comparison with the potent colchicine site agent combretastatin A-4.25 In the assay we used,26 the IC50 is defined as the concentration of compound that inhibits by 50% the extent of assembly of 10 μM tubulin after a 20 min incubation at 30 °C. All compounds inhibited the reaction, although an IC50 could not be obtained with compound 29. None of the compounds was as active as combretastatin A-4, but most of them had substoichiometric (i.e., < 10 μM) IC50 values.
Table 3.
Inhibitory effects of selected compounds on tubulin assembly and colchicine binding to tubulin.
| Compound | Inhibition of tubulin assembly IC50 (μM) ± SD |
Inhibition of colchicine binding % Inhibition ± SD |
|---|---|---|
| 5 μM inhibitor | ||
| Combretastatin A-4 | 1.1 ± 0.1 | 99 ± 0.06 |
| 5 (NSC 743420) | 5.3 ± 0.3 | 49± 2 |
| 9 (NSC 742500) | 16 ± 1 | 45 ± 4 |
| 29 (NSC 748117) | > 20 | 13 ± 3 |
| 30 (NSC 748119) | 9.3 ± 0.1 | 48 ± 4 |
| 31 (NSC 748118) | 9.1 ± 1 | 36 ± 5 |
| 33 (NSC 744489) | 3.7 ± 0.02 | 48 ± 3 |
| 36 (NSC 748115) | 13 ± 2 | 53 ± 4 |
| 38 (NSC 748113) | 2.9 ± 0.3 | 62 ± 2 |
| 39 (NSC 748114) | 3.8 ± 0.01 | 45 ± 5 |
The compounds were also evaluated for their effects on the binding of [3H]colchicine to tubulin26 (Table 3). As in the assembly assay, none of the compounds was as active as combretastatin A-4, but, except for the relatively inactive 29, the other compounds had similar inhibitory effects on colchicine binding, with inhibition ranging from 36% to 62%. With these agents, there was relatively little correlation between the two assays, except that compound 38 was the most active as both an inhibitor of assembly and an inhibitor of colchicine binding.
Structural basis for tubulin activity
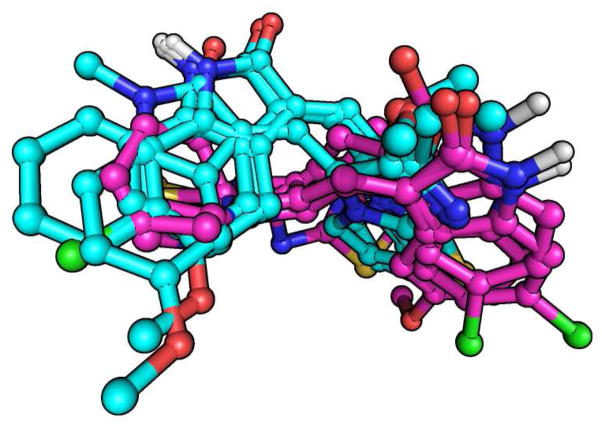
The binding modes of the compounds in Table 3 were modeled using the 3G2N crystal structure of αβ-tubulin in complex with NSC 613863 as a template. These models revealed structural insights into the in vitro activities of these compounds. The models show that compounds where R is methyl or methoxy assume a common binding mode relative to compounds where R is a 2-pyridyl group (Figure 1). Compounds that possess methyl and methoxy substituents include 5, 9, 33, 36, 38, and 39, and, for these compounds, their binding poses exhibit a significant overlap in their indolinone systems. The one exception is compound 39, which is characterized by a N-methyl substitution of its indolinone and accordingly, has a binding mode that is slightly offset from the other compounds. The larger 2-pyridine substituent at the R position of compounds 30, 31, and 29 results in alternative binding conformations for these compounds. This may explain their weak activity against tubulin relative to the other compounds, since they do not assume the optimal binding poses represented by methyl- and methoxy-substituted compounds.
Figure 1.
Two distinct orientations for the binding modes of the compounds in β-tubulin. Carbon atoms of 5, 9, 33, 36, 38, and 39 are colored cyan, and those of 30, 31, and 29 are purple. Sulfur, oxygen, nitrogen, chlorine and hydrogen atoms are colored yellow, red, blue, green and white, respectively.
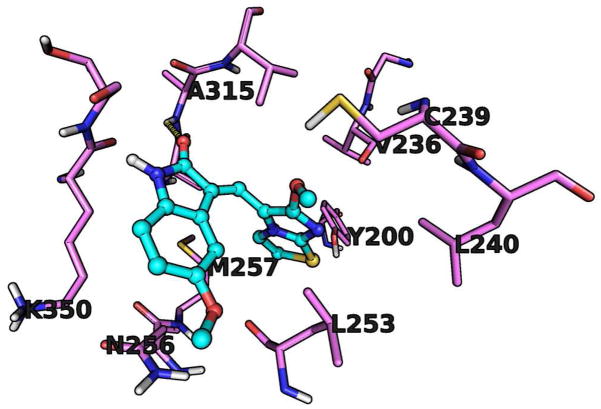
As shown for compound 38 in Figure 2, the other active compounds form a number of common stabilizing interactions with β-tubulin. The first is a hydrogen bond between the indolone oxygen atom and the backbone –NH of Ala315. Additionally, the aryl portion of the indolone ring is packed against the alipathic side chain of Lys350. At the interior of the binding pocket, the imidazothiazole system is wedged against Tyr200, with the sulfur atom potentially forming a weak hydrogen bond to the hydroxyl group of Tyr200.
Figure 2.
Binding model of the most active compound 38, with its carbon atoms rendered in cyan. The carbon atoms of the binding site amino acids are rendered in purple. Other colors as in Figure 1.
Compounds 5 and 9 are distinguishable from the others in that they are characterized by a condensed benzene ring at the R3–R4 positions and a methyl substituent at the R position. The binding models showed that the condensed benzene ring of 5 and 9 can be accommodated at the binding site and that the condensed benzene ring forms stabilizing interactions with the alipathic side chain of Lys350.
In contrast to compounds 5 and 9, the two most active compounds, these being compounds 36 and 38, are characterized by methoxy substituents at the R position. In the binding models, these methoxy groups are favorably wedged against the side chains of Leu240 and Leu253, and this provides more favorable binding contacts than for the methyl groups of compounds 5 and 9. The binding models also indicate favorable binding interactions of the R2-methoxy of 38 and the R3-Cl of 36 against the Lys350 side chain, and this is reflected in the similar % inhibition of colchicine binding for 36 and 38 of 53% and 62%, respectively. In contrast, compound 33, which is unsubstituted at the R1–R4 positions and, accordingly, does not gain favorable interactions with Lys350, is characterized by a weaker inhibition of 48%. Additionally, the weaker inhibitory activity of compound 39, which differs by only a N-methyl group from the most active compound 38, may be explained by the loss of a hydrogen bond to Ala315 due its offset binding mode.
c. Effect on growth inhibition of cancer cells
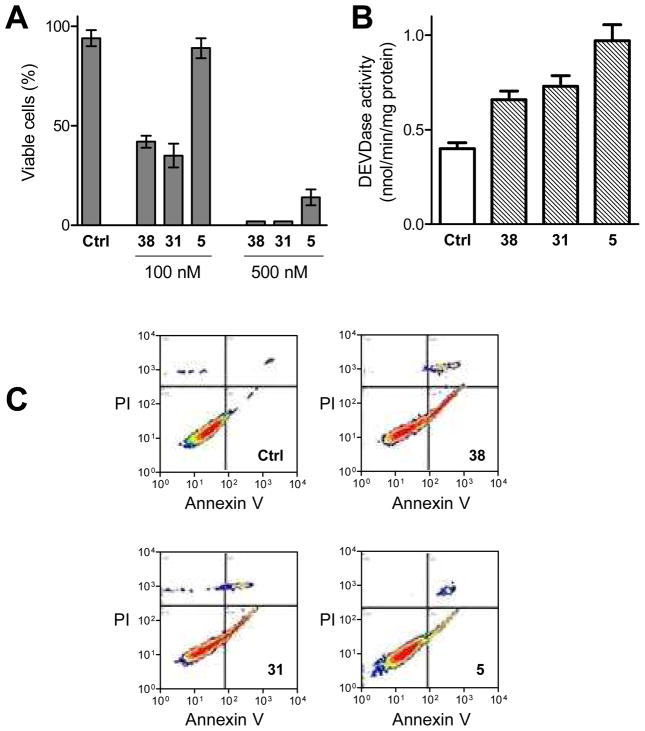
We examined the effect of the very active compounds 38, 31 and 5 on the growth of HT-29 colon cancer cells. First, in order to assess whether these compounds interfered with cell cycle progression, DNA profiles of cultured cells were examined by flow cytometry. Figure 3A shows that compounds 38 and 31 induced accumulation of HT-29 cells in the G2/M phase at 100 nM; compound 5 produced a similar effect only at a higher concentration, 500 nM.
Figure 3.
Effects of 38, 31 and 5 on cell cycle distribution in HT-29 cells. (A) Cell cycle distribution was determined in cells treated with the indicated concentration of compounds for 24 h. The graph depicts the results obtained in one experiment representative of three. (B) Effects of compounds on p21 cellular levels detected by immunofluorescence confocal microscopy. Nuclei were stained with propidium iodide (red fluorescence). p21 was stained with FITC antibody and results in green fluorescence. The image is representative of three experiments.
Next, the expression levels of the p21 protein were evaluated by immunofluorescence analysis, since this protein can induce cell cycle arrest.27 Upon p21 staining with FITC-conjugated antibody, fluorescence was detected in entire cells by confocal microscopy. Figure 3B shows a significant increase in p21 expression after treatment with all examined compounds.
The cell cycle block was associated with a significant reduction in cell viability. Figure 4A shows that 38 and 31 triggered cell death at 100 nM, while compound 5 elicited a cytotoxic effect at 500 nM. This was similar to the effects observed with G2/M accumulation. Moreover, propidium iodide (PI) staining (Figure 3B) revealed chromatin condensation and nuclear fragmentation, suggesting the activation of apoptosis. However, identification of the mode of cell death requires multiple assays.28 Therefore, to determine whether treatment with 38, 31, or 5 caused cell death by activation of the apoptotic program, the activity of caspase proteases, a marker of apoptosis, was assayed. In cells treated for 48 h with these compounds, the activity of effector caspases was significantly increased (Figure 4B). The exposure of phosphatidylserine from the inner to the outer side of the plasma membrane is an early marker of apoptosis.28 To further confirm the onset of apoptosis, we utilized the Annexin V-FLUOS staining method. This technique measures the extent of phosphatidylserine externalization. Annexin is a protein with high affinity for phospholipids, and cells undergoing apoptosis are marked from early stages with Annexin V-FLUOS, which binds to externalized phosphatidylserine. On the other hand, PI enters cells and stains DNA only if there is a permeabilized plasma membrane, a later event in cell death. As shown in Figure 4C, after 24 h of incubation, only 3.9% of control HT-29 cells were Annexin V positive and PI negative (early apoptotic cells). In contrast, this percentage rose to 27%, 23% and 12.5% in cells incubated in the presence of compounds 38, 31 or 5, respectively.
Figure 4.
Induction of apoptosis in HT-29 cells after treatment with compounds 38, 31 or 5. (A) The cells were treated for 24 h with 100 or 500 nM compound, then the percentage of viable cells was determined by the trypan blue exclusion assay. Data are means ± s.e.m. of three determinations. (B) The activity of caspase proteases acting on the peptide sequence Asp-Glu-Val-Asp (DEVD), indicated as DEVDase activity, was measured in extracts obtained from cells treated for 48 h with the indicated compound (500 nM). Results are means ± s.e.m. of four determinations. (C) Cells treated for 24 h, as indicated (derivative concentration was 500 nM), were labeled with Annexin V-FLUOS and PI and analyzed by flow cytometry. The exposure of phosphatidylserine on the outer side of the plasma membrane in PI-negative cells, marking early apoptotic cells, is demonstrated by the cell population with high AnnexinV and low PI fluorescence (lower right section of each panel).
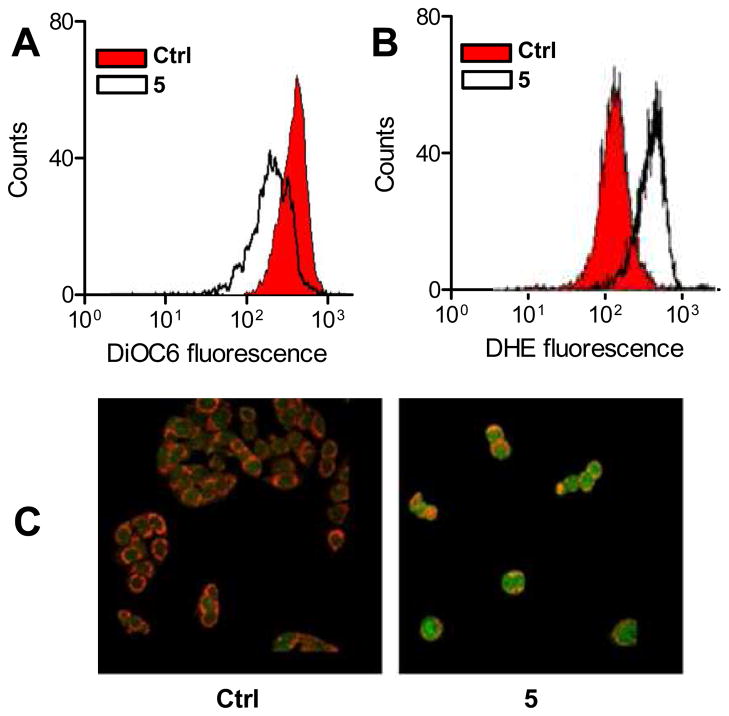
Next, the mechanisms involved in activation of apoptosis were examined. In order to clarify whether the apoptotic pathway triggered by the new derivatives was mediated by mitochondrial events,29 we investigated the effects of compound 5. The mitochondrial membrane potential (ΔΨm) can be measured in intact cells by using the fluorescent probe 3,3′-dihexyloxacarbocyanine iodide (DiOC6). This compound accumulates and aggregates in mitochondria, producing a green fluorescence. Following ΔΨm collapse in apoptotic cells, DiOC6 no longer enters mitochondria, resulting in a decrease in green fluorescence. Figure 5A shows that a 24 h treatment with 5 caused a marked disruption of ΔΨm. Since depolarization of the mitochondrial membrane is associated with mitochondrial production of reactive oxygen species (ROS), intracellular ROS content was evaluated by using dihydroethidium (DHE). DHE is a specific probe for superoxide species, since it binds with superoxide to form the highly fluorescent derivative 2-hydroxyethidium. The treatment of colon cancer cells with compound 5 caused a marked increase in superoxide formation, as shown in Figure 5B. Finally, we studied the involvement of the proapoptotic Bax protein by measuring its expression levels and its translocation from cytosol to mitochondria in intact cells. Bax was detected with a fluorescent FITC-conjugated antibody, and the resulting fluorescence was observed in the cells by confocal microscopy. Figure 5C shows that compound 5 caused a significant increase in the cellular content of Bax, which was largely localized to mitochondria.
Figure 5.
Induction of apoptosis by compound 5 in HT-29 colon cancer cells is accompanied by mitochondrial events. The cells were treated for 24 h with 500 nM 5 before each evaluation. All panels depict the result obtained in one experiment that was representative of three to five. (A) Change in mitochondrial membrane potential as estimated from DiOC6 fluorescence histograms. ΔΨm collapse is evidenced by a decrease in DiOC6 fluorescence in whole cells. (B) Analysis of intracellular ROS content measured by flow cytometry. Reaction of DHE with superoxide radicals causes the formation of a derivative with increased fluorescence. (C) Confocal laser scanning micrographs showing staining for Bax (green fluorescence) and mitochondria (red fluorescence) in control (left) and treated cells (right). Localization of Bax with mitochondria is demonstrated by orange pixels.
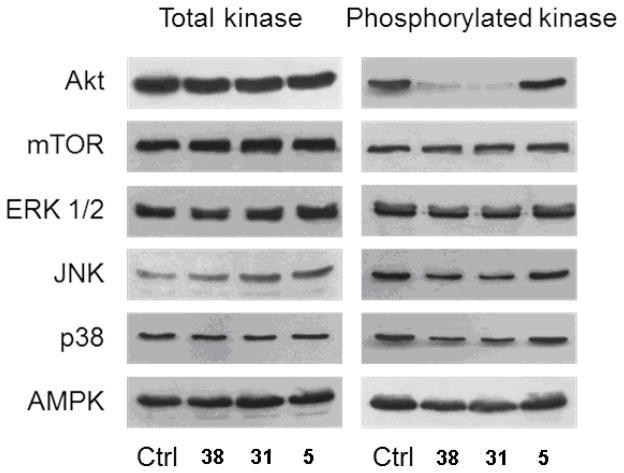
The activity of compounds 38, 31 and 5 on signal transduction pathways was studied by examining effects on the activation of a panel of kinases involved in cell growth and survival. In these studies, cervical carcinoma HeLa cells were treated with 1 μM of the different agents for 3 h, afterward kinase activation was determined by detecting their specific phosphorylation by Western blotting (Figure 6). At this time point, all cells were viable but committed to death. The most potent compounds 38 and 31 strongly reduced Akt(Ser473) phosphorylation, whereas no significant effect was detectable on the phosphorylation of the other oncoprotein mTOR(Ser2448), master regulator of protein synthesis.30
Figure 6.
Effect of 38, 31, and 5 on the activation of protein kinases associated with cell growth and/or cell survival. HeLa cells were incubated for 3 h in the presence of a 1 μM concentration of the indicated compounds. The content and phosphorylation of the indicated protein kinases in cell extracts were determined by Western blotting.
Compounds 38 and 31 also slightly decreased phosphorylation/activation of all three members of the mitogen-activated protein kinase family ERK1/2(Thr202/Tyr204), JNK(Thr183/Tyr185), and p38MAPK(Thr180/Tyr182) that, depending of the circumstances, can be associated either with cell growth or with activation of apoptosis in cancer cells.31
Similarly, the phosphorylation of AMPK(Thr172), a metabolic stress sensitive kinase often associated with cell survival,32 was only slightly inhibited by 38 and 31. In conclusion, at the concentration used, compound 5 did not significantly influence the signaling pathways examined, whereas 38 and 31 selectively and strongly inhibited Akt activation.
Conclusion
In conclusion, we have described the synthesis of a new class of colchicine site agents that inhibit tubulin polymerization. In the colon cancer cell line HT-29, we studied compounds 5, 31 and 38 in detail. These compounds caused the cells to arrest in the G2/M phase of the cell cycle, as would be expected for inhibitors of tubulin assembly. This cell cycle arrest was associated with activation of apoptosis, as occurs with most inhibitors of tubulin assembly. The apoptosis in the HT-29 cells was accompanied by caspase activation and phosphatidylserine externalization. In these cells there was disruption of the mitochondrial membrane potential, increased ROS production, and Bax translocation into mitochondria. Interestingly, the most potent compounds 31 and 38 strongly inhibited the activation of the kinase Akt, an oncoprotein associated with cell survival and proliferation. Akt is upregulated in several cancers and responsible for resistance to cell death.30 Considering that the tubulin effects were relatively modest, compared with combretastatin A-4, the cytotoxic effects of compounds 38 and 31 may be substantially mediated by effects on the AKT pathway.
Experimental section
1. Chemistry
All the compounds prepared had a purity of at least 95%, as determined by combustion analysis. The melting points are uncorrected. TLC was performed on Fluka plates (art. 99577) and column chromatography on Silica gel 70–200 μ 60A (Fluorochem): the eluent was a mixture of petroleum ether/acetone in various proportions. The IR spectra were recorded in nujol on a Nicolet Avatar 320 E.S.P.; νmax is expressed in cm−1. The 1H-NMR spectra were recorded on a Varian Gemini (300 MHz); the chemical shift (referenced to solvent signal) is expressed in δ (ppm) and J in Hz (abbreviations: im=imidazole, th=thiazole, ind=indole, bzind=benzoindole, coum=coumaranone, ph=phenyl, thio=thiophene, py=pyridine). For spectra and elemental analyses that are not reported here, see Supporting Information. The oxindoles 2b, 2k, 2l, 2-coumaranone (2d) and 1-(3,4,5-trimethoxyphenyl)ethanone are commercially available. The following compounds were prepared according to the literature: 1a–f,5,8,10,14,15,18 1j–m,19,20,10,22 2a,6 2c,7 2e,9 2f–h,11–13 2i,16 2j,17 2m21 and 2n.23
General procedure for the synthesis of compounds 3–42
The appropriate compound 2 (10 mmol) was dissolved in methanol (100 mL) and treated with the equivalent of the appropriate aldehyde 1 and piperidine (1 mL). The reaction mixture was refluxed for 3–5 h (progress of the reaction followed by TLC), and the precipitate that formed on cooling was collected by filtration with a yield of 20% for compound 40 and 42, 40–50% for compounds 3, 7–9, 11–14, 16–19, 21–23, 27, 33–39, and 41 and 70–80% for compounds 4, 5, 24–26, 28 and 29–32.
For compounds 6, 10, 15, and 20 the yield was much lower (5%), and an improvement (15–25%) was obtained by replacing piperidine with triethylamine.
Compounds 6, 10, 15 and 37 were purified by column chromatography with petroleum ether/acetone as the eluent. All the crude products were crystallized from methanol except 6, 8, and 12 (acetone/petroleum ether), 15, 20, and 38 (CHCl3/petroleum ether) and 42 (ethanol).
Data for 4. I.R.: 2707, 1706, 1607, 1156, 725. 1H-NMR: 2.20 (3H, s, CH3im), 2.39 (3H, d, CH3th, J=1.2), 6.73 (1H, d, ind-4, J=8.4), 6.89 (1H, d, ind-7, J=2.1), 6.96 (1H, dd, ind-5, J=8.4, J=2.1), 7.52 (1H, q, th, J=1.2), 7.61 (1H, s, CH), 10.75 (1H, s, NH). Anal. Calcd for C16H12ClN3OS (MW 329.80): C, 58.27; H, 3.67; N, 12.74. Found: C, 57.98; H, 3.72; N, 13.03.
Data for 5. I.R.: 2727, 1695, 1618, 818, 725. 1H-NMR: 2.25 (3H, s, CH3), 2.37 (3H, s, CH3), 6.99 (1H, d, ind-4/5, J=8.7), 7.45 (1H, d, ind-4/5, J=8.7), 7.51 (2H, m, bzind), 7.55 (1H, s, th), 7.61 (1H, s, CH), 7.86 (1H, m, bzind), 8.14 (1H, m, bzind), 11.38 (1H, s, NH). Anal. Calcd for C20H15N3OS (MW 345.41): C, 69.54; H, 4.38; N, 12.16. Found: C, 69.85; H, 4.47; N, 11.92.
Data for 7. I.R.: 2714, 1706, 1633, 1233, 712. 1H-NMR: 2.26 (3H, s, CH3), 2.72 (3H, s, CH3), 6.74 (1H, dd, ind-4, J=9.0, J=2.4), 6.85 (1H, dd, ind-7, J=9.0, J=4.4), 7.07 (1H, td, ind-6, J=9.0, J=2.4), 7.57 (1H, s, CH), 10.66 (1H, s, NH). Anal. Calcd for C15H11FN4OS (MW 314.34): C, 57.31; H, 3.53; N 17.82. Found: C, 56.98; H, 3.87; N, 18.00.
Data for 9. I.R.: 3375-2715, 1699, 815, 723. 1H-NMR: 2.27 (3H, s, CH3), 2.70 (3H, s, CH3), 7.16 (1H, d, ind-4/5, J=8.6), 7.44 (1H, d, ind-4/5, J=8.6), 7.51 (2H, m, bzind), 7.56 (1H, s, CH), 7.88 (1H, m, bzind), 8.13 (1H, m, bzind), 11.39 (1H, s, NH). Anal. Calcd for C19H14N4OS (MW 346.40): C, 65.88; H, 4.07; N, 16.17. Found: C, 66.02; H, 4.67; N, 15.96.
Data for 11. I.R.: 3151, 1696, 1614, 1189, 824. 1H-NMR: 2.09 (3H, s, CH3), 2.32 (3H, s, CH3), 2.39 (3H, d, CH3th, J=1.4), 6.08 (1H, s, ind), 6.60 (1H, s, ind), 7.14 (1H, q, th, J=1.4), 7.25 (2H, d, ph, J=8.0), 7.39 (1H, s, CH), 7.60 (2H, d, ph, J=8.0), 8.77 (1H, s, OH), 10.30 (1H, s, NH). Anal. Calcd for C23H19N3O2S (MW 401.48): C, 68.81; H, 4.77; N, 10.47. Found: C, 69.01; H, 5.01; N, 10.73.
Data for 12. I.R.: 1695, 1242, 1149, 824, 765. 1H-NMR: 2.30 (3H, s, CH3ph), 2.38 (3H, d, CH3th, J=1.2), 3.24 (3H, s, NCH3), 6.52 (1H, d, ind-4, J=7.5), 6.86 (1H, t, ind-5/6, J=7.5), 7.06 (2H, d, ph, J=7.6), 7.25 (1H, q, th, J=1.2), 7.27 (3H, m, 2Hph+1Hind), 7.59 (2H, m, CH+1Hind). Anal. Calcd for C23H19N3OS (MW 385.48): C, 71.66; H, 4.97; N, 10.90. Found: C, 71.35; H, 5.06; N, 11.13.
Data for 13. I.R.: 1690, 1593, 1117. 1H-NMR: 2.30 (3H, s, CH3ph), 2.40 (3H, d, CH3th, J=1.4), 3.21 (3H, s, NCH3), 3.46 (3H, s, OCH3), 6.10 (1H, d, ind-4, J=2.2), 6.86 (1H, dd, ind-6, J=8.4, J=2.2), 6.96 (1H, d, ind-7, J=8.4), 7.23 (2H, d, ph, J=7.6), 7.35 (1H, q, th, J=1.4), 7.59 (2H, d, ph, J=7.6), 7.66 (1H, s, CH). Anal. Calcd for C24H21N3O2S (MW 415.50): C, 69.38; H, 5.09; N, 10.11. Found: C, 69.76; H, 4.89; N, 10.34.
Data for 16. I.R.: 3140-2663, 1696, 1199, 717. 1H-NMR: 2.73 (3H, s, CH3), 3.38 (3H, s, OCH3), 6.20 (1H, s, ind-4), 6.74 (2H, s, ind-6+7), 7.44 (2H, d, ph, J=8.4), 7.62 (1H, s, CH), 7.71 (2H, d, ph, J=8.4), 10.47 (1H, s, NH). Anal. Calcd for C21H15ClN4O2S (MW 422.88): C, 59.64; H, 3.58; N, 13.25. Found: C, 59.83; H, 3.87; N, 13.67.
Data for 17. I.R.: 3124-2725, 1704, 1183, 721. 1H-NMR: 2.74 (3H, s, CH3), 6.42 (1H, dd, ind-4, J=9.0, J=2.4), 6.80 (1H, dd, ind-7 J=9.0, J=4.6), 7.00 (1H, dt, ind-6, J=9.0, J=2.4), 7.46 (2H, d, ph, J=8.4), 7.65 (1H, s, CH), 7.69 (2H, d, ph, J=8.4), 10.67 (1H, s, NH). Anal. Calcd for C20H12ClFN4OS (MW 410.86): C, 58.61; H, 2.71; N, 13.67. Found: C, 58.38; H, 2.98; N, 13.98.
Data for 24. I.R.: 3300-2800, 1706, 1110, 999. 1H-NMR: 2.44 (3H, d, CH3, J=1.3), 3.34 (3H, s, OCH3), 3.71 (3H, s, OCH3), 3.72 (3H, s, OCH3), 6.54 (1H, d, ind-4, J=7.5), 6.70 (1H, t, ind-5/6, J=7.5), 6.79 (1H, d, ind-7, J=7.5), 6.92 (1H, s, ph), 7.13 (1H, t, ind-5/6, J=7.5), 7.43 (1H, s, CH), 7.57 (1H, q, th, J=1.3), 10.56 (1H, s, NH). Anal. Calcd for C24H20BrN3O4S (MW 526.40): C, 54.76; H, 3.83; N. 7.98. Found: C, 54.91; H, 4.00; N, 8.04.
Data for 28. I.R.: 3250-2800, 1696, 1583, 717. 1H-NMR: 6.28 (1H, d, ind-4, J=7.6), 6.80 (1H, t, ind-5/6, J=7.6), 6.89 (1H, d, ind-7, J=7.6), 7.20 (1H, t, ind-5/6, J=7.6), 7.32 (1H, d, th, J=4.5), 7.31 (1H, m, py), 7.45 (1H, d, th, J=4.5), 7.90 (1H, td, py, J=7.7, J=1.8), 8.16 (1H, d, py, J=7.7), 8.18 (1H, s, CH), 8.63 (1H, m, py), 10.68 (1H, s, NH). Anal. Calcd for C19H12N4OS (MW 344.39): C, 66.26; H, 3.51; N, 16.27. Found: C, 66.72; H, 3.22; N, 15.99.
Data for 29. I.R.: 3200-2720, 1688, 1601, 721. 1H-NMR: 6.16 (1H, d, ind-4, J=2.2), 6.88 (2H, m, ind-6+7), 7.25 (1H, m, py), 7.43 (1H, d, th, J=4.4), 7.52 (1H, d, th, J=4.4), 7.90 (1H, t, py, J=7.7), 8.16 (1H, d, py, J=7.7), 8.26 (1H, s, CH), 8.61 (1H, m, py), 10.80 (1H, s, NH). Anal. Calcd for C19H11ClN4OS (MW 378.83): C, 60.24; H, 2.93; N, 14.79. Found: C, 60.56; H, 3.03; N, 14.98.
Data for 30. I.R.: 3200-2720, 1692, 1603, 722. 1H-NMR: 6.27 (1H, d, ind, J=8), 6.87 (2H, m, ind), 7.33 (1H, m, py), 7.34 (1H, d, th, J=4.4), 7.47 (1H, d, th, J=4.4), 7.91 (1H, t, py, J=7.7), 8.16 (1H, d, py, J=7.7), 8.23 (1H, s, CH), 8.63 (1H, m, py), 10.72 (1H, s, NH). Anal. Calcd for C19H11ClN4OS (MW 378.83): C, 60.24; H, 2.93; N, 14.79. Found: C, 60.57; H, 3.01; N, 14.42.
Data for 31. I.R.: 3180-2660, 1689, 1034, 722. 1H-NMR: 3.76 (3H, s, OCH3), 6.81 (2H, m, ind), 7.33 (2H, m, py+ind), 7.34 (1H, d, th, J=4.4), 7.56 (1H, d, th, J=4.4), 7.90 (1H, t, py, J=7.7), 8.16 (1H, d, py, J=7.7), 8.33 (1H, s, CH), 8.65 (1H, m, py), 10.37 (1H, s, NH). Anal. Calcd for C20H14N4O2S (MW 374.41): C, 64.16; H, 3.77; N, 14.96. Found: C, 64.59; H, 3.47; N, 15.02.
Data for 33. I.R.: 3500-2800, 1692, 1598, 1081. 1H-NMR: 4.05 (3H, s, OCH3), 6.83 (1H, d, ind-4/7, J=7.7), 6.93 (1H, t, ind-5/6, J=7.7), 7.13 (1H, t, ind-5/6, J=7.7), 7.33 (1H, d, th, J=4.4), 7.52 (1H, s, CH), 7.58 (1H, d, ind-4/7, J=7.7), 7.77 (1H, d, th, J=4.4), 10.47 (1H, s, NH). Anal. Calcd for C15H11N3O2S (MW 297.33): C, 64.16; H, 3.77; N, 14.96. Found: C, 63.99; H, 3.23; N, 15.03.
Data for 34. I.R.: 3400-2700, 1688, 1523, 722. 1H-NMR: 4.06 (3H, s, OCH3), 6.84 (1H, d, ind-5/7, J=7.8), 6.98 (1H, d, ind-5/7, J=7.8), 7.13 (1H, t, ind-6, J=7.8), 7.34 (1H, d, th, J=4.4), 7.65 (1H, d, th, J=4.4), 8.20 (1H, s, CH), 10.74 (1H, s, NH). Anal. Calcd for C15H10ClN3O2S (MW 331.77): C, 54.30; H, 3.04; N, 12.67. Found: C, 54.02; H, 2.89; N, 12.86.
Data for 35. I.R.: 3300-2800, 1688, 1076, 723. 1H-NMR: 4.07 (3H, s, OCH3), 6.83 (1H, d, ind-7, J=8.2), 6.96 (1H, d, ind-4, J=2.1), 7.18 (1H, dd, ind-6, J=8.2, J=2.1), 7.43 (1H, d, th, J=4.3), 7.67 (1H, s, CH), 8.18 (1H, d, th, J=4.3), 10.57 (1H, s, NH). Anal. Calcd for C15H10ClN3O2S (MW 331.77): C, 54.30; H, 3.04; N 12.67. Found: C, 54.67; H, 2.97; N, 12.39.
Data for 36. I.R.: 3300-2700, 1712, 1004, 722. 1H-NMR: 4.04 (3H, s, OCH3), 6.85 (1H, s, ind), 6.96 (2H, m, ind), 7.41 (1H, d, th, J=4.4), 7.62 (1H, s, CH), 8.10 (1H, d, th, J=4.4), 10.58 (1H, broad, NH). Anal. Calcd for C15H10ClN3O2S (MW 331.77): C, 54.30; H, 3.04; N 12.67. Found: C, 54.62; H, 2.96; N, 12.82.
Data for 37. I.R.: 3300-2700, 1693, 1599, 722. 1H-NMR: 4.06 (3H, s, OCH3), 6.79 (2H, m, ind), 6.95 (1H, m, ind), 7.43 (1H, d, th, J=4.5), 7.66 (1H, s, CH), 8.15 (1H, d, th, J=4.5), 10.45 (1H, s, NH). Anal. Calcd for C15H10FN3O2S (MW 315.33): C, 57.14; H, 2.71; N, 13.67. Found: C, 56.98; H, 2.97; N, 13.45.
Data for 38. I.R.: 3300-2700, 1707, 1687, 722. 1H-NMR: 3.65 (3H, s, OCH3ind), 4.04 (3H, s, OCH3th), 6.71 (1H, s, ind), 6.73 (2H, s, ind), 7.55 (1H, s, CH), 7.72 (1H, d, th, J=4.2), 8.04 (1H, d, th, J=4.2), 10.25 (1H, s, NH). Anal. Calcd for C16H13N3O3S (MW 327.35): C, 58.71; H, 4.00; N, 12.84. Found: C, 58.98; H, 4.37; N, 12.99.
Data for 39. I.R.: 1681, 1523, 1157, 1070, 722. 1H-NMR: 3.19 (3H, s, NCH3), 3.68 (3H, s, OCH3ind), 4.04 (3H, s, OCH3th), 6.64 (1H, d, ind-4, J=2.4), 6.82 (1H, dd, ind-6, J=8.4, J=2.4), 6.90 (1H, d, ind-7, J=8.4), 7.42 (1H, d, th, J=4.4), 7.64 (1H, s, CH), 8.06 (1H, d, th, J=4.4). Anal. Calcd for C17H15N3O3S (MW 341.38): C, 59.81; H, 4.43; N, 12.31. Found: C, 60.01; H, 4.68; N, 12.57.
Data for 40. I.R.: 1701, 1603, 1153, 999, 717. 1H-NMR: 3.21 (3H, s, NCH3), 4.05 (3H, s, OCH3), 7.00 (2H, m, ind), 7.22 (1H, t, ind, J=7.4), 7.33 (1H, d, th, J=4.4), 7.56 (1H, s, CH), 7.65 (1H, d, ind, J=7.4), 7.80 (1H, d, th, J=4.4). Anal. Calcd for C16H13N3O2S (MW 311.35): C, 61.72; H, 4.21; N, 13.50. Found: C, 62.01; H, 4.43; N, 13.68.
2. Biology
2a. Cell-Based Screening Assay
The NCI screening process occurs in two stages,33 beginning with the evaluation of all compounds against the 60 cell lines at 10−5 M. Compounds exhibiting significant growth inhibition are subsequently evaluated against the 60 cell lines at five concentration levels according to standard procedures (http://dtp.nci.nih.gov/branches/btb/ivclsp.html). In both cases, the exposure time is 48 h.
2b. Tubulin
Combretastatin A-4 was a generous gift of Dr. G. R. Pettit, Arizona State University. Bovine brain tubulin was purified as described previously.34 The tubulin assembly assay was performed with 10 μM tubulin and varying compound concentrations as described previously.26 The IC50 is the compound concentration that inhibits extent of assembly after 20 min at 30 °C. The colchicine binding assay was performed with 1.0 μM, 5.0 μM [3H]colchicine and 5.0 μM inhibitor. Incubation was for 10 min at 37 °C. At this time point, about 40–60% maximum colchicine binding occurs in the control reactions. Details of the method were described previously.35
Molecular Modeling
The Maestro 9 (Schrödinger, LLC, New York, NY) modeling software running on a Dell (Round Rock, TX) Precision 690 with Red Hat (Raleigh, NC) Enterprise Linux 4 was used to perform the modeling studies. Simulations were performed in vacuo using a distance-dependent dielectric with a nonbonded interaction limited to within 13 Å in an OPLS 2005 force field. Minimizations involved up to 500 steps of Polak-Ribière conjugate gradient. The 3G2N crystal structure of αβ-tubulin in complex with NSC 613863 and a stathmin fragment was selected as the template for docking studies. More precisely, the B-subunit (β-tubulin) of the 3G2N structure was extracted and utilized in the docking studies. The β-tubulin structure contains the full binding site for NSC 613863. The missing segments in the β-tubulin crystal structure were modeled, and the offset amino acid sequence of the crystal structure relative to the uniprot code D0VWY9 was corrected to give a model of β-tubulin in complex with NSC 613863. The D0VWY9-NSC 613863 complex was prepared for modeling by addition of bond orders and hydrogen atom and then energy minimized with limited atom movements of less than 0.3 Å.
Compound 38, which was the most active in the series, was selected for the initial docking. The Glide program was used to flexible dock compound 38. The top 5 binding poses were individually inspected and unfavorable intermolecular contacts were identified. Manual changes to the position or the torsional bond angles of the ligands were used to relieve unfavorable protein-ligand contacts, if possible. The binding poses were subsequently refined using the Glide program using the highest precision setting. A high scoring 38 pose was selected and subjected to further stepwise refinement. First, with tubulin fixed in Cartesian space, the conformation of 38 was energy minimized. Secondly, with 38 fixed, β-tubulin was minimized. This was followed by energy minimization of the complex in which all atoms were unconstrained. The resulting binding model was evaluated for unfavorable intra- and intermolecular contacts. If unfavorable contacts were evident, the refinement cycle was iterated. Upon determination of satisfactory intra- and intermolecular contacts in the model, compound 38 was extracted from the pocket and a docking grid was created based on the refined β-tubulin model. Using the Glide program, the compound was re-docked into the colchicine site. The final binding pose scored a favorable binding Gscore of −6.0.
The docking grid based on the refined β-tubulin structure was used as a common template for generating docking poses for the other compounds. The binding scores (Gscores) for these compounds were as follows: compound 5 = −4.3; 9 = −4.1; 29 = −3.9; 30 = −4.3; 31 = −2.9; 33 = −5.4; 36 = −6.0; 38 = −6.0; 39 = −4.3.
The binding models indicate that the two most active compounds in terms of % inhibition of colchicine binding (Table 3), 36 and 38, have the most favorable binding scores of −6.0, while the least active compounds, 29 and 31, have the least favorable binding scores of −3.9 and −2.9, respectively.
2c. Cell culture and treatment
Human colon adenocarcinoma HT-29 and cervical carcinoma HeLa cells were cultured in RPMI 1640 medium (Labtek Eurobio, Milan, Italy), supplemented with 10% fetal bovine serum (PAA Laboratories GmbH, Pashing, Austria) and 2 mM Lglutamine (Sigma-Aldrich, St Louis MO), in humidified air at 37 °C with 5% CO2.
Compounds were dissolved in DMSO at 10 mmol/L and diluted with medium to obtain the desired concentration. DMSO concentration in the medium was kept constant at 0.01%. The cells were plated at 2×104 cells/cm2 in a plastic well (60 cm2) and treated for 24 h in triplicate. In control cells, only DMSO was added to the culture medium. After incubation for the indicated times, viable cells were detected and counted by trypan blue exclusion.
Confocal Microscopy
Cells were seeded at 1×104 cells/cm2 on glass cover-slips and treated with test compounds for 24 h. They were washed three times with phosphate buffered saline (PBS), fixed with 3% paraformaldehyde, washed with 0.1 M glycine in PBS, and permeabilized with 70% ice-cold ethanol. Cells were then washed three times with 1% bovine serum albumin in PBS and incubated with the respective primary antibody (anti-Bax, anti-p21, anti-tubulin and anti-actin) for 1 h at room temperature. The cells were washed again, and incubated with FITC-conjugated antibody (secondary antibody) for 1 h at room temperature in the dark. In experiments in which Bax intracellular localization was investigated, cells were first incubated with 150 nM Mito-Tracker Red CM-H2 XRos in fresh medium at 37 °C for 10 min and then fixed. Finally, we used PI to stain the nuclei. All preparations were embedded in Mowiol containing antibleaching DABCO and analyzed by using a laser scanning confocal microscope NIKON C1s, equipped with a NIKON Eclipse TE300.
Cell cycle and apoptosis
Cells were fixed with 70% ethanol at −20 °C for at least 12 h. After two washes with PBS, the cells were incubated in RNaseA/PBS (10 μg/mL) at 37 °C for 30 min. Intracellular DNA was labeled with PI (50 μg/mL). Cell cycle analysis was performed using Modfit 5.0 software. Surface exposure of phosphatidylserine in apoptotic cells was measured with an Annexin V-FLUOS staining kit (Roche Diagnostics Deutschland GmbH, Mannheim, Germany), following the manufacturer’s instructions. Briefly, cells were washed twice with cold PBS and resuspended in binding buffer at a concentration of 1×105 cells in a total volume of 100 μL. Next, 5 μL of Annexin V-FLUOS and 5 μL of PI (50 μg/mL) solutions were added. The cells and reagents were gently mixed and incubated for 20 min at room temperature. PI and Annexin V-FLUOS were quantitated by flow cytometry, with the data acquired with logarithmic amplification. The enzymatic activity of caspases hydrolyzing the peptide sequence Asp-Glu-Val-Asp (DEVD) is indicated as DEVDase activity, which was measured in cell extracts by a fluorometric assay.36
Mitochondrial Membrane Potential (ΔΨm)
To measure ΔΨm, mitochondria were selectively probed with the potential-sensitive dye DiOC6. After treatment, cells were incubated with medium containing 4 nM DiOC6 for 40 min at a cell concentration of 1×106 cell/mL at 37 °C in the dark. Cells were counterstained by PI at 5 μg/mL to permit elimination of dead cells. Fluorescence was analyzed by flow cytometry with logarithmic amplification. Flow cytometric analysis was performed with a Beckmann Coulter Epics XL MCL cytometer (USA) equipped with a 15 mW argon ion laser.
ROS measurement
In order to detect intracellular superoxide levels,37 the cells were incubated with 5 μM DHE (Molecular Probes, Leiden, The Netherlands), made as a 10 mM stock in DMSO, for 30 min at 37 °C. Cells were analyzed by measuring red fluorescence with logarithmic amplification.
Western Blotting
The cells were lysed and the homogenates were centrifuged at 15,000 × g for 15 min. The supernatant was diluted in loading buffer and then denatured by boiling for 4 min. Aliquots corresponding to 80 μg protein were analyzed by SDS-PAGE. Proteins were transferred onto a nitrocellulose membrane and probed with the specific primary antibodies (Cell Signaling). After further washing, the membrane was incubated for 1 h with peroxidase-conjugated goat anti-rabbit IgG (Santa Cruz). Immunoreactive bands were visualized by chemiluminescence with the ECL reagent (Amersham).
Supplementary Material
TABLE 1.
Compounds 3–42.
| Comp | Formula | MW | Mp, °C |
|---|---|---|---|
| 3 | C18H18N4OS | 338.42 | 223–226 |
| 4 | C16H12ClN3OS | 329.80 | >310 |
| 5 | C20H15N3OS | 345.41 | 267–271 |
| 6 | C16H12N2O2S | 296.34 | 175–177 |
| 7 | C15H11FN4OS | 314.34 | >310 |
| 8 | C17H17N5OS | 339.41 | 264–265 |
| 9 | C19H14N4OS | 346.40 | >300 |
| 10 | C15H11N3O2S | 297.33 | 176–178 |
| 11 | C23H19N3O2S | 401.48 | >310 |
| 12 | C23H19N3OS | 385.48 | 227–230 |
| 13 | C24H21N3O2S | 415.50 | 200–204 |
| 14 | C25H16ClN3OS | 441.93 | >310 |
| 15 | C21H13ClN2O2S | 392.85 | 248–250 |
| 16 | C21H15ClN4O2S | 422.88 | 301–303 |
| 17 | C20H12ClFN4OS | 410.86 | >310 |
| 18 | C22H18ClN5OS | 435.93 | >310 |
| 19 | C24H17ClN4O4S | 492.93 | 283–285 |
| 20 | C20H12ClN3O2S | 393.84 | 220–223 |
| 21 | C24H21N3O5S | 463.50 | 250–255 |
| 22 | C24H20BrN3O5S | 542.40 | 193–195 |
| 23 | C25H23N3O5S | 477.53 | 220–223 |
| 24 | C24H20BrN3O4S | 526.40 | 270–273 dec. |
| 25 | C25H22BrN3O5S | 556.43 | 196–200 |
| 26 | C21H14N4O3S | 402.42 | >310 |
| 27 | C19H13N3OS2 | 363.45 | >310 |
| 28 | C19H12N4OS | 344.39 | >310 |
| 29 | C19H11ClN4OS | 378.83 | >310 |
| 30 | C19H11ClN4OS | 378.83 | >310 |
| 31 | C20H14N4O2S | 374.41 | 286–288 |
| 32 | C21H16N4O2S | 388.44 | 301–303 |
| 33 | C15H11N3O2S | 297.33 | 214–216 |
| 34 | C15H10ClN3O2S | 331.77 | 267–269 |
| 35 | C15H10ClN3O2S | 331.77 | 263–265 |
| 36 | C15H10ClN3O2S | 331.77 | 267–269 |
| 37 | C15H10FN3O2S | 315.33 | 260–262 |
| 38 | C16H13N3O3S | 327.35 | 216–218 |
| 39 | C17H15N3O3S | 341.38 | 188–190 |
| 40 | C16H13N3O2S | 311.35 | 187–189 |
| 41 | C24H21N3O4S | 447.50 | 172–173 |
| 42 | C15H10N2O3S | 298.31 | 180–182 |
Acknowledgments
This work was supported in part by a grant from the University of Bologna, Italy (RFO) and from MIUR (PRIN 2009). We are grateful to the National Cancer Institute (Bethesda, MD) for the anticancer tests. It has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute.
Abbreviations
- NCI
National Cancer Institute
- DTP
Developmental Therapeutics Program
- DMSO
dimethylsulfoxide
- GI
growth inhibition
- TGI
total growth inhibition
- LC
lethal concentration
- PI
propidium iodide
- DiOC6
3,3′-dihexyloxacarbocyanine iodide
- ROS
reactive oxygen species
- DHE
dihydroethidium
- FITC
fluorescein isothiocyanate
- PBS
phosphate-buffered saline
Footnotes
Supporting Information Available. Synthesis of precursors, additional IR and 1H NMR spectra (Table S1), additional elemental analysis results (Table S2), NSC numbers (Table S3) and an example of a mean graph (compound 31) where all the cell lines employed are reported (Figure S1). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Potential Antitumor Agents. 48. For part 47 see the following: Andreani A, Granaiola M, Leoni A, Locatelli A, Morigi R, Rambaldi M, Varoli L, Lannigan D, Smith J, Scudiero D, Kondapaka S, Shoemaker RH. Imidazo[2, 1-b]thiazole Guanylhydrazones as RSK2 Inhibitors. Eur J Med Chem. 2011;46:4311–4323. doi: 10.1016/j.ejmech.2011.07.001.
- 2.Andreani A, Burnelli S, Granaiola M, Leoni A, Locatelli A, Morigi R, Rambaldi M, Varoli L, Calonghi N, Cappadone C, Voltattorni M, Zini M, Stefanelli C, Masotti L, Shoemaker RH. Antitumor Activity of New Substituted 3-(5-Imidazo[2, 1b]thiazolylmethylene)-2-indolinones and 3-(5-Imidazo[2, 1-b]thiadiazolylmethylene)-2-indolinones: Selectivity Against Colon Tumor Cells and Effect on Cell Cycle-Related Events. J Med Chem. 2008;51:7508–7513. doi: 10.1021/jm800827q. [DOI] [PubMed] [Google Scholar]
- 3.Andreani A, Locatelli A, Leoni A, Rambaldi M, Morigi R, Bossa R, Chiericozzi M, Fraccari A, Galatulas I. Synthesis and Potential Coanthracyclinic Activity of Substituted 3-(5-Imidazo[2, 1-b]thiazolylmethylene)-2-indolinones. Eur J Med Chem. 1997;32:919–924. [Google Scholar]
- 4.Andreani A, Granaiola M, Leoni A, Locatelli A, Morigi R, Rambaldi M, Giorgi G, Salvini L, Garaliene V. Synthesis and Antitumor Activity of Substituted 3-(5-Imidazo[2,1-b]thiazolylmethylene)-2-indolinones. Anticancer Drug Design. 2001;16:167–174. [PubMed] [Google Scholar]
- 5.Andreani A, Rambaldi M, Mascellani G, Rugarli P. Synthesis and Diuretic Activity of Imidazo[2,1-b]thiazole Acetohydrazones. Eur J Med Chem. 1987;22:19–22. [Google Scholar]
- 6.Minisci F, Galli R, Cecere M. Amminazione Radicalica di Composti Aromatici Attivati: Acetammidi. Nuovo Processo per la Sintesi di para-Ammino-N,N-dialchilaniline. La Chimica e l’Industria. 1966;48:1324–1326. [Google Scholar]
- 7.Mayer F, Oppenheimer T. Über Naphthyl-essigsäuren. 3. Abhandlung: 1-Nitronaphthyl-2-brenztraubensäure und 1-Nitronaphthyl-2-essigsäure. Chem Ber. 1918;51:1239–1245. [Google Scholar]
- 8.Andreani A, Leoni A, Locatelli A, Morigi R, Rambaldi M, Simon WA, Senn-Bilfinger J. Synthesis and Antisecretory Activity of 6-Substituted 5-Cyanomethylimidazo[2, 1-b]thiazoles and 2, 6-Dimethyl-5-hydroxymethylimidazo[2, 1-b][1, 3, 4]thiadiazole. Arzneim Forsch. 2000;50:550–553. doi: 10.1055/s-0031-1300247. [DOI] [PubMed] [Google Scholar]
- 9.Zakrzewska A, Kolehmainen E, Osmialowski B, Gawinecki R. 4-Fluoroanilines: Synthesis and Decomposition. J Fluorine Chem. 2001;111:1–10. [Google Scholar]
- 10.Andreani A, Rambaldi M, Locatelli A, Andreani F. 5-Formylimidazo[2,1-b]thiazoles and Derivatives with Herbicidal Activity. Collect Czech Chem Comm. 1991;56:2436–2447. [Google Scholar]
- 11.Andreani A, Granaiola M, Leoni A, Locatelli A, Morigi R, Rambaldi M, Garaliene V. Synthesis and Antitumor Activity of 1,5,6-Substituted 3-(2-Chloro-3-indolylmethylene)1,3-dihydroindol-2-ones. J Med Chem. 2002;45:2666–2669. doi: 10.1021/jm011123c. [DOI] [PubMed] [Google Scholar]
- 12.Hodges R, Shannon JS, Jamieson WD, Taylor A. Chemical and Biological Properties of Some Oxindol-3-ylidene methines. Can J Chem. 1968;46:2189–2194. [Google Scholar]
- 13.Porter JC, Robinson R, Wyler M. Monothiophthalimide and Some Derivatives of Oxindole. J Chem Soc. 1941:620–624. [Google Scholar]
- 14.Andreani A, Rambaldi M, Carloni P, Greci L, Stipa P. Imidazo[2,1-b]thiazole Carbamates and Acylureas as Potential Insect Control Agents. J Heterocyclic Chem. 1989;26:525–529. [Google Scholar]
- 15.Meyer H, Horstmann H, Moeller E, Garthoff B. Imidazoazolealkenoic Acid Amides, Their Intermediate Products and Their Use in Drugs. CODEN: GWZZBX DE 3020421 A1 19811210 CAN 96:85561 AN 1982:85561. Ger Offen. 1981:64.
- 16.Koelsch CF. A Synthesis of Ethyl Quininate from m-Cresol. J Amer Chem Soc. 1944;66:2019–2020. [Google Scholar]
- 17.Nakagawa K, Sato T, Nishi T, Oshiro Y, Yamamoto K. Jpn. Kokai Tokkyo Koho: CODEN: JKXXAF JP 52073866 19770621. Carbostyril and Oxindole Derivatives. 1977:5. Showa. Patent written in Japanese. Application: JP 75-150935 19751216. CAN 87:167899 AN 1977:567899 CAPLUS.
- 18.Budriesi R, Ioan P, Locatelli A, Cosconati S, Leoni A, Ugenti MP, Andreani A, Di Toro R, Bedini A, Spampinato S, Marinelli L, Novellino E, Chiarini A. Imidazo[2,1-b]thiazole System: A Scaffold Endowing Dihydropyridines with Selective Cardiodepressant Activity. J Med Chem. 2008;51:1592–1600. doi: 10.1021/jm070681+. [DOI] [PubMed] [Google Scholar]
- 19.Andreani A, Granaiola M, Leoni A, Locatelli A, Morigi R, Rambaldi M, Giorgi G, Garaliene V. Potential Antitumor Agents. 34. Synthesis and Antitumor Activity of Guanylhydrazones From Imidazo[2,1-b]thiazoles and From Diimidazo[1,2-a:1,2-c]pyrimidine. Anticancer Res. 2004;24:203–212. [PubMed] [Google Scholar]
- 20.Andreani A, Burnelli S, Granaiola M, Leoni A, Locatelli A, Morigi R, Rambaldi M, Varoli L, Calonghi N, Cappadone C, Farruggia G, Zini M, Stefanelli C, Masotti L, Radin NS, Shoemaker RH. New Antitumor Imidazo[2,1-b]thiazole Guanylhydrazones and Analogues. J Med Chem. 2008;51:809–816. doi: 10.1021/jm701246g. [DOI] [PubMed] [Google Scholar]
- 21.Andreani A, Rambaldi M, Bonazzi D, Greci L, Andreani F. Potential Antitumor Agents. III. Hydrazone Derivatives of 5-Substituted 2-Chloro-3-formyl-6-methylindole. Farmaco. 1979;34:132–138. doi: 10.1002/chin.197924211. [DOI] [PubMed] [Google Scholar]
- 22.Andreani A, Rambaldi M, Leoni A, Locatelli A, Bossa R, Chiericozzi M, Galatulas I, Salvatore G. Synthesis and Cardiotonic Activity of Imidazo[2,1-b]thiazoles Bearing a Lactam Ring. Eur J Med Chem. 1996;31:383–387. [Google Scholar]
- 23.Romeo A, Corrodi H, Hardegger E. Umsetzungen des o-Nitrophenylessigesters und des 2-Chlor-6-nitro-phenyl-brenztraubensäureesters. Helv Chim Acta. 1955;38:463–467. [Google Scholar]
- 24.Paull KD, Lin CM, Malspeis L, Hamel E. Identification of Novel Antimitotic Agents Acting at the Tubulin Level by Computer-assisted Evaluation of Differential Cytotoxicity Data. Cancer Res. 1992;52:3892–3900. [PubMed] [Google Scholar]
- 25.Lin CM, Ho HH, Pettit GR, Hamel E. The Antimitotic Natural Products Combretastatin A-4 and Combretastatin A-2: Studies on the Mechanism of Their Inhibition of the Binding of Colchicine to Tubulin. Biochemistry. 1989;28:6984–6991. doi: 10.1021/bi00443a031. [DOI] [PubMed] [Google Scholar]
- 26.Hamel E. Evaluation of Antimitotic Agents by Quantitative Comparisons of Their Effects on the Polymerization of Purified Tubulin. Cell Biochem Biophys. 2003;38:1–21. doi: 10.1385/CBB:38:1:1. [DOI] [PubMed] [Google Scholar]
- 27.Abbas T, Dutta A. p21 in Cancer: Intricate Networks and Multiple Activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kepp O, Galluzzi L, Lipinski M, Yuan J, Kroemer G. Cell Death Assays for Drug Discovery. Nat Rev Drug Discov. 2011;10:221–237. doi: 10.1038/nrd3373. [DOI] [PubMed] [Google Scholar]
- 29.Indran IR, Tufo G, Pervaiz S, Brenner C. Recent Advances in Apoptosis, Mitochondria and Drug Resistance in Cancer Cells. Biochim Biophys Acta. 2011;1807:735–745. doi: 10.1016/j.bbabio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Wu P, Hu YZ. PI3K/Akt/mTOR Pathway Inhibitors in Cancer: A Perspective on Clinical Progress. Curr Med Chem. 2010;17:4326–4341. doi: 10.2174/092986710793361234. [DOI] [PubMed] [Google Scholar]
- 31.Kim EK, Choi EJ. Pathological Roles of MAPK Signaling Pathways in Human Diseases. Biochim Biophys Acta. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Luo Z, Zang M, Guo W. AMPK as a Metabolic Tumor Suppressor: Control of Metabolism and Cell Growth. Future Oncol. 2010;6:457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A, Gray-Goodrich M, Campbell H, Mayo J, Boyd M. Feasibility of a High-Flux Anticancer Drug Screen Using a Diverse Panel of Cultured Human Tumor Cell Lines. J Natl Cancer Inst. 1991;83:757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 34.Hamel E, Lin CM. Separation of Active Tubulin and Microtubule-associated Proteins by Ultracentrifugation, and Isolation of a Component Causing the Formation of Microtubule Bundles. Biochemistry. 1984;23:4173–4184. doi: 10.1021/bi00313a026. [DOI] [PubMed] [Google Scholar]
- 35.Verdier-Pinard P, Lai J-Y, Yoo H-D, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, Day BW, Hamel E. Structure-activity Analysis of the Interaction of Curacin A the Potent Colchicine Site Antimitotic Agent with Tubulin and Effects of Analogs on the Growth of MCF-7 Breast Cancer Cells. Mol Pharmacol. 1998;53:62–67. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 36.Stefanelli C, Bonavita F, Stanic I, Pignatti C, Farruggia G, Masotti L, Guarnieri C, Caldarera CM. Inhibition of Etoposide-induced Apoptosis with Peptide Aldehyde Inhibitors of Proteasome. Biochem J. 1998;332:661–665. doi: 10.1042/bj3320661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, Kalyanaraman B. Detection and Characterization of the Product of Hydroethidine and Intracellular Superoxide by HPLC and Limitations of Fluorescence. Proc Natl Acad Sci USA. 2005;102:5727–5732. doi: 10.1073/pnas.0501719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.