Abstract
In pre-clinical studies, targeted radioimmunotherapy using 212Pb-TCMC-trastuzumab as an in vivo generator of the high energy α-particle emitting radionuclide 212Bi is proving an efficacious modality for the treatment of disseminated peritoneal cancers. To elucidate mechanisms associated with this therapy, mice bearing human colon cancer LS-174T i.p. xenografts were treated with 212Pb-TCMC-trastuzumab and compared to the non-specific control 212Pb-TCMC-HuIgG, unlabeled trastuzumab, and HuIgG, as well as untreated controls. 212Pb-TCMC-trastuzumab treatment induced significantly more apoptosis and DNA double stranded breaks (DSBs) at 24 h. Rad51 protein expression was down-regulated, indicating delayed DNA double strand damage repair compared to 212Pb-TCMC-HuIgG, the non-specific control. 212Pb-TCMC-trastuzumab treatment also caused G2/M arrest, depression of the S phase fraction and depressed DNA synthesis that persisted beyond 120 h. In contrast, the effects produced by 212Pb-TCMC-HuIgG appeared to rebound by 120 h. In addition, 212Pb-TCMC-trastuzumab treatment delayed open chromatin structure and expression of p21 until 72 h, suggesting a correlation between induction of p21 protein and modification in chromatin structure of p21 in response to 212Pb-TCMC-trastuzumab treatment. Taken together, increased DNA DSBs, impaired DNA damage repair, persistent G2/M arrest, and chromatin remodeling were associated with 212Pb-TCMC-trastuzumab treatment and may explain its increased cell killing efficacy in the LS-174T i.p. xenograft model for disseminated intraperitoneal disease.
Keywords: 212Pb-Radioimmunotherapy, DNA DSBs, DNA repair, Apoptosis, Micrometastatic disease
INTRODUCTION
Targeted radiation therapy with monoclonal antibodies (mAbs) using β−-emitting radionuclides has been demonstrated to be an efficacious strategy for the treatment and management of cancer patients (1, 2). Due to the combination of shorter path length (50–80 μm) and higher linear energy transfer (100 KeV μm−1), targeted α-particle RIT offers the potential for more specific tumor cell killing with less damage to surrounding normal tissue than β−-emitters While this strategy has been successfully applied towards the eradication of leukemia (3, 4), the same physical properties suggest that targeted α-therapy would be suitable for the elimination of minimal residual or micrometastatic disease in other types of human malignancies (5–7).
Targeted and pre-targeted RIT using α-emitters such as 212Bi (T1/2 = 1.01 h) and 212Pb (T1/2 =10.6 h) have demonstrated significant therapeutic efficacy in both in vitro and in vivo model systems (8–10). 212Pb is the longer-lived parental radionuclide of 212Bi and, as such, it serves as an in vivo generator of 212Bi. The 212Pb/212Bi system, therefore, is a promising α-particle emitting source that provides an alternative option for the treatment and management of cancer (8, 11).
Trastuzumab (Herceptin®) is a humanized mAb that targets HER2 and has been well demonstrated to have antitumor activity for the management of breast cancer (12, 13). Previously, this laboratory demonstrated the efficacy of α-particle RIT using the CHX-A″ DTPA linker with 213Bi in intraperitoneal models for pancreatic and ovarian cancer using trastuzumab as the targeting moiety (6). Complementary to those results, suitable chelation chemistry for the retention of 212Pb with the protein was also designed and synthesized, i.e., 2-(4-isothiocyanatobenzyl)-1,4,7,10-tetraaza-1,4,7,10-tetra-(2-carbamoylmethyl)-cyclododecane (4-NCS-Bz-TCMC) (TCMC) to overcome the limitations associated with the direct use of the shorter half-life bismuth radioisotopes, 213Bi or 212Bi, and to obviate a source of toxicity originating from intracellular dissociation of 212Pb from 1, 4, 7, 10-tetraazacyclododecane-N,N′N″,N‴-tetraacetic acid (DOTA) (8, 14). Studies utilizing 212Pb showed the feasibility of this isotope in RIT for the treatment of disseminated intraperitoneal disease; 212Pb-TCMC-trastuzumab given as a single injection demonstrated therapeutic efficacy which increased with multiple injections given at approximately monthly intervals (8).
The mechanism by which α-particle RIT induces cells death is not completely understood (15–18). A recent report indicated that upon exposure to α-particles, cell survival was not impacted by traversals of the cytoplasm, but by traversal of nuclei (19). Seidl et al reported that RIT with an α-emitting radionuclide induced non-apoptotic cell death at 72 h post-treatment (20). It is noteworthy that there have been few studies related to the determination of the actual mechanisms involved in the α-particle RIT cytotoxicity. The literature to date has been effectively restricted to in vitro studies, which, by their very nature, are self-limiting and not reflective of RIT treatment of tumors in a complex environment (21, 22).
The studies reported herein were designed to gain an understanding of the underlying mechanism(s) of action of 212Pb-TCMC-trastuzumab therapy in a systematic fashion using the murine model currently under investigation in our laboratory. The ultimate objective will be to incorporate the knowledge gained into the design of future therapy studies and to improve the therapeutic benefit of targeting HER2 with α-particle emitting radionuclides. The studies reported herein describe the apoptotic response, cell cycle distribution, DNA repair, and changes in chromatin remodeling in LS-174T i.p. xenograft tumors following RIT with 212Pb. The studies suggest that 212Pb-TCMC-trastuzumab therapy-induced cell killing in the LS-174T i.p. xenograft model occurred principally by G2/M arrest, accompanied by a delay in DNA damage repair.
MATERIALS AND METHODS
Cell line
The human colon carcinoma cell line (LS-174T) was used for all in vivo studies. LS-174T was grown in a supplemented DMEM as previously described (23). All media and supplements were obtained from Lonza. The cell line has been screened for mycoplasma and other pathogens before in vivo use according to NCI Laboratory Animal Sciences Program policy. No authentication of the cell line was conducted by the authors.
Chelate synthesis, mAb conjugation, and radiolabeling
The synthesis, characterization, and purification of the bifunctional ligand TCMC have been previously described (8, 14). Trastuzumab (Herceptin®; Genentech) was conjugated with TCMC by established methods using a 10-fold molar excess of ligand to mAb as previously reported (8). The final concentration of trastuzumab was quantified by the method of Lowry (24). The number of TCMC molecules linked to the mAb was determined using a spectrophotometric-based assay (25). A 10 mCi 224Ra/212Pb generator was supplied by AlphaMed, Inc. The preparation of the generator and radiolabeling procedure has been previously described (8).
Tumor model, treatment, and tumor harvesting
Studies were performed with 19–21 g female athymic mice (NCI-Frederick) bearing 3 d i.p. LS-174T xenografts as previously reported (8). The viability of the LS-174T cells (> 95 %) was determined using trypan-blue. Mice were injected intraperitoneally (i.p.) with 1x 108 LS-174T cells in 1 mL of DMEM. The inoculum size for this cell line represented the minimum amount of cells required for tumor growth in 100 % of the mice (6). 212Pb-TCMC-trastuzumab (10 μCi in 0.5 mL PBS) was administered to the mice 3 days post-implantation of tumor (n = 10–15). This treatment group was compared with sets of mice that received 212Pb-TCMC-HuIgG, unlabeled trastuzumab or HuIgG, or no treatment. Mice receiving trastuzumab or HuIgG were injected 3 d after tumor implantation with 10 μg of the respective material. Mice utilized for the cell cycle and proliferation studies were injected i.p. with 5-bromo-2′-deoxyuridine (BrdU; 1.5 mg in 0.5 ml PBS; Sigma) 4 h prior to euthanasia. Tumors were harvested from mice bearing i.p. LS-174T xenografts at 6, 24, 48, 72, 96 and 120 h. The amount of tumor collected was not measured; however, based on previous studies, the tumor burden at 7 d is typically 128.5 ± 205.6 mg (26). The tumors at each time point were pooled together, macroscopically inspected and adherent tissues were removed. The tumor tissues were then thoroughly rinsed in ice cold PBS three times, divided and processed accordingly for each assay. The tumor tissues were either stored at −80 °C until use or paraffin-embedded after fixing in 1 % formalin. All animal protocols were approved by the National Cancer Institute Animal Care and Use Committee.
Flow cytometry
Cell cycle distribution and DNA synthesis was determined by flow cytometry as previously described with some modifications (27). The tumors were fixed in cold 70 % ethanol at 4 °C, washed in PBS twice, minced finely, and incubated in 1 mL 0.04 % pepsin (Sigma) w/v in 0.1 N HCl (Mallinckrodt, Inc.) for 1 h at 37 °C with shaking. The digest was filtered through a 70 μm nylon mesh after passage through a 25 gauge needle. Following centrifugation at 10,000 x g for 10 min, the resulting pellet was re-suspended in 1 mL of 2 N HCl (Mallinckrodt, Inc.) and incubated at 37 °C for 20 min with shaking. The nuclear suspension was neutralized with 0.1 M sodium tetraborate (Sigma), washed twice in PBS containing 0.5 % bovine serum albumin and 0.5 % Tween-20 (PBTB) and re-suspended in PBTB. The nuclei (100 μL) were incubated with 20 μL of FITC-labeled anti-BrdU monoclonal antibody (BD Biosciences-Pharmingen) for 1 h at 4 °C, followed by two washes in cold PBS. The samples were then re-suspended in 2 mL of propidium iodide (50 μg/mL in PBTB; Sigma) containing RNAse A (50 μg; Sigma) and incubated for 30 min at 4 °C. Flow cytometry was performed using a FACSCalibur (BD Biosciences), collecting 15,000 events with cell debris excluded from data collection. DNA content (propidium iodide) and DNA synthesis (BrdU content) were analyzed using two parameter data collection with CellQuest (BD Biosciences) software while single parameter DNA distribution was performed and analyzed using Modfit LT ver. 3.0 (Verity Software House, Inc).
Determination of apoptosis
Apoptotic bodies were scored using hematoxylin and eosin (H&E) staining as described previously (28). Five fields were analyzed per tumor section, and the number of apoptotic bodies per 100 nuclei scored expressed as a percentage. The following criteria were used to distinguish apoptotic bodies: (a) isolated distribution of apoptotic bodies; (b) shrunken cells usually with empty space between neighboring cells; (c) eosinophilic cytoplasm; (d) condensation of nuclei into dense particles; (e) fragmentation of the nuclei into several bodies; and (f) the absence of inflammatory reaction surrounding the apoptotic cells (non-necrotic cells).
The presence of apoptotic bodies on tumor sections was also determined using the Dead End Fluorometric TUNEL System (Promega). The sections were incubated in 100 μL of a 20 μg/mL proteinase solution at room temperature for 10 min, washed twice in PBS and fixed in 4% formaldehyde. The sections were labeled by incubation in 100 μL of 200 mM potassium cacodylate, 25 mM Tris-HCl, 0.2 mM DTT, 2.5 mM cobalt chloride and 0.25 mg/mL BSA for 10 min, and then incubated with 50 μL of TdT reaction mixture for 60 min at 37 °C in a humidified chamber; immersion in 2x SSC halted the reaction. Tissue sections were mounted using Vectashield with DAPI (Vector Laboratories) to counterstain DNA.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as described in the manufacturer’s instructions (Cell Signaling) with some modifications. After formalin fixation, antigen unmasking was performed by subjecting the sections to 80 °C for 10 min in 10 mM sodium citrate, pH 6.0. After washing in distilled water three times, the slides were treated with 3 % hydrogen peroxide for 10 min followed by 1 h incubation with 100 μL of PBS with Tween 20 (PBST) containing 5 % normal goat serum at room temperature. Thereafter, 200 μL of γH2AX antibody (Cell Signaling) in PBST with 5 % normal goat serum was added to the sections and incubated overnight at 4 °C. After three washes, the sections were developed with 100 μL DAB (3,3′-diaminobenzidine) substrate, washed twice, dehydrated in 95 % and 100 % ethanol, and dipped in xylene two times. Coverslips were mounted using Cytoseal XYL (Thermo Scientific).
Comet assay
Sample preparation was performed according to the tissue preparation protocol in the manufacturer’s instructions (Comet Assay®, Trevigen). For the best test of whether cells are in a satisfactory condition for comet assay, control (untreated cells) should give comets with a background level of breaks (mostly class 0, or around 10 % DNA in the tail). The neutral comet assay was performed as described in the manufacturer’s instructions (Comet Assay®, Trevisen). In brief, the incubation times (4 °C) were as follows: 30 min lysis, 15 min in 1x TBE and 40 min electrophoresis in 1x TBE at 1 V/cm. Sixty comets per individual preparation were scored and all measurement parameters were calculated on Comet IV automatic scoring system (Perceptive Instruments).
Chromatin immunoprecipitation
The chromatin immunoprecipitation assay (ChIP) was utilized to determine whether histone methylation was altered by 212Pb-TCMC-trastuzumab therapy at the promoter region of the p21 gene. The ChIP assay was performed with the ChIP Assay Kit (Upstate Biotechnology) according to the manufacturer’s instructions with minor adjustments. Formaldehyde was added directly to the tissue immediately after harvesting, incubated for 15 minutes at 37 °C and washed three times in cold PBS. The lysates in 10 mM Tris-HCl, pH 8.0, 1 % SDS containing phosphatase and protease inhibitors were sonicated (Branson Sonifier 450; Branson) and the supernatant split into several aliquots. Ten μL (1:100) of each antibody for H3 methyl K4 and H3 methyl K9 (Upstate Biotechnology) was added; the resulting DNA-protein complexes were isolated on protein G agarose beads and eluted with 1 % SDS in 0.1 M NaHCO3. Crosslinking was reversed by incubation at 65 °C for 5 h. The samples were treated with proteinase K, the DNA extracted with phenol/chloroform, and dissolved in elution reagent. Immunoprecipitated DNA was analyzed by quantitative real time PCR (qPCR) using p21 promoter specific primers (Applied Biosystems).
Western blotting
Immunoblot analysis following standard procedures was performed with total protein isolates using T-PER tissue protein extraction reagent (Thermo Scientific) with protease inhibitors (Roche). Fifty μg of total protein per lane was separated on a 4–20 % tris-glycine gel and transferred to a nitrocellulose membrane. Antibodies against cleaved Caspase-3 (Cell Signaling), Rad51 and DNA-PKcs (Abcam) were used at a dilution of 1:1000 in PBS containing 5 % BSA and 0.05 % Tween-20. Horseradish peroxidase conjugated rabbit secondary antibodies were used at 1:5000 in 3 % non-fat dry milk. The blots were developed using the ECL Plus chemoluminescent detection kit (GE Healthcare) and the images acquired by Fuji LAS 4000.
Statistics
At least three independent experiments were done for each point described. All values were expressed as mean ± S.D. Student’s test was used for paired data, and multiple comparisons were performed with the ANOVA. A p-value < 0.05 was considered statistically significant.
RESULTS
212Pb-TCMC-trastuzumab induces apoptosis in i.p. human colon carcinoma treated xenografts
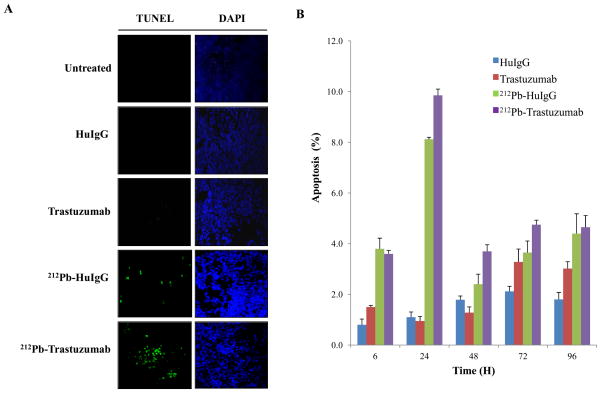
Anticancer drugs and radiation have been shown to activate apoptosis pathways in solid tumors (29, 30). To determine whether 212Pb-TCMC-trastuzumab induces apoptosis in the human colon carcinoma LS-174T xenografts, the TUNEL assay was performed using paraffin-embedded tumor sections. The TUNEL assay results obtained at 24 h after exposure to 212Pb-TCMC-trastuzumab indicated a clear induction of apoptosis (Fig. 1A) that was greater than the other treatments. Next, the apoptotic bodies were quantitated using IHC and H&E staining. Apoptosis was noted based on the morphological criteria described in the methods. Quantitation of the apoptotic bodies showed that 212Pb-TCMC-trastuzumab treatment significantly increased in apoptotic rates (Fig. 1B) compared to the 212Pb-TCMC-HuIgG non-specific control treatment at 24–72 h (p < 0.05), suggesting greater effective targeted cell killing by 212Pb-TCMC-trastuzumab in the tumor tissues.
Figure 1. Induction of apoptosis in LS-174T i.p. xenografts following 212Pb-TCMC-trastuzumab.
A. Mice bearing i.p. LS-174T xenografts were treated with 212Pb-TCMC-trastuzumab and the tumors collected over a 120 h period. Additional groups included untreated, HuIgG, and trastuzumab, and 212Pb-TCMC-HuIgG as a non-specific control with the representative fluorescence microscopy images at 24 h. Left, TUNEL staining; Right, DAPI counter staining (40x)
B. Apoptosis induced by 212Pb-TCMC-trastuzumab. Mice bearing i.p. LS-174T xenografts were treated with 212Pb-TCMC-trastuzumab at the indicated times. Paraffin-embedded sections were stained with H&E and the apoptotic nuclei were counted under light microscopy; 500 nuclei were scored per tumor. Results represent the average of a minimum of three replications (± SD).
212Pb-TCMC-trastuzumab interferes with DNA repair
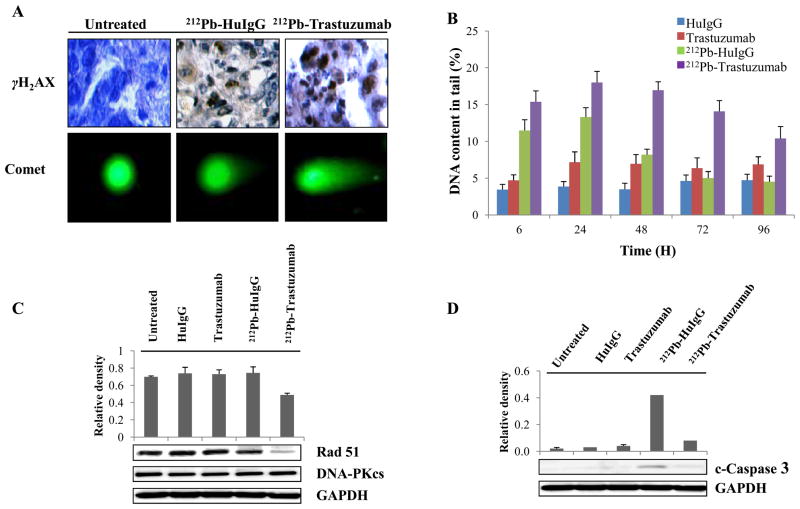
Radiation causes DNA damage, the most harmful in terms of cellular cytotoxicity being double strand breaks (DSBs). An increase in DNA DSBs and impaired DNA damage repair has been invoked to explain the synergy between drugs and ionizing radiation (31). To investigate DNA damage by 212Pb-TCMC-trastuzumab, immunohistochemistry was performed using paraffin-embedded tumor sections. Induction of DNA DSB damage was evident as measured by phosphorylated H2AX, a marker for DNA double strand damage (Figure 2A). The neutral comet assay was also used to demonstrate physical DNA strand damage. Enhancement of the fluorescence intensity of tail was observed in both 212Pb-TCMC-trastuzumab and 212Pb-TCMC-HuIgG treated tumors. Next, to determine the effect of 212Pb-TCMC-trastuzumab on DNA damage repair, DNA content in the tails were compared at different time points up to 96 h after the various treatments. After 24 h, 212Pb-TCMC-trastuzumab showed significantly higher percent DNA in the tail than any other treatment including 212Pb-TCMC-HuIgG. At 24 h, 18 % of the tumor sample presented with DNA strand breaks. There was a steady decrease in the percentage of DNA damage and by 96 h the percentage was 10. Thus, by 96 h, inhibition of DNA damage repair elicited by 212Pb-TCMC-trastuzumab has not yet been completely reversed. In comparison, the 212Pb-TCMC-HuIgG resulted in 13 % DNA strand breaks at 24 h. By the 96 h time point, ?? % DNA in the tail for 212Pb-TCMC-HuIgG was similar to that of unlabeled HuIgG indicating that DNA damage repair had returned to normal levels following treatment. This demonstrates that DNA damage repair was compromised to a greater extent by the treatment with 212Pb-TCMC-trastuzumab (Fig. 2B). These results suggest that there was a general effect attributable to α-radiation; however, there was a greater specific effect attributable to the 212Pb-TCMC-trastuzumab (p < 0.001).
Figure 2. 212Pb-TCMC-trastuzumab-induced DNA damage and repair delay in LS-174T i.p. xenografts.
A. Light and fluorescence microscopy images of untreated, 212Pb-TCMC-HuIgG and 212Pb-TCMC-trastuzumab at 24 h. Top, IHC staining (γH2AX); bottom, neutral comet assay (40x)
B. DNA content in the tail. DNA damage was quantified by % DNA content in the tail using comet assay at the indicated times. Results represent the average of a minimum of three replications (± SD).
C. Down-regulation of Rad51 induced by 212Pb-TCMC-trastuzumab. Immunoblot analysis for Rad51 and DNA-PKcs were performed at the 24 h time point. Rad51 was detected at 37 kDa and DNA-PKcs was detected at 450 kDa. The equivalent protein loading control was GAPDH.
D. Down-regulation of cleaved caspase-3 induced by 212Pb-TCMC-trastuzumab. Immunoblot analysis for cleaved caspase-3 was performed at the 24 h time point. Cleaved caspase-3 was detected at 17 kDa. The equivalent protein loading control was GAPDH.
DSBs can be repaired by different pathways, the most important of which are homologus repair (HR) and non-homologous end-joining (NHEJ) (32). To identify the DNA repair pathways involved in the study presented herein, Rad51 and DNA-PKcs, which play important roles in HR and NHEJ, respectively, were investigated. The densitometric analysis of the western blots illustrated in Figure 2C shows that at the protein expression level, Rad51 was significantly down-regulated by 212Pb-TCMC-trastuzumab (p < 0.05) treatment. However, Rad51 was not affected in the presence of the unlabeled trastuzumab, HuIgG, or 212Pb-TCMC-HuIgG at 24 h. DNA-PKcs was not affected following any of the treatments, suggesting that differences in induction of the HR pathway protein Rad51 may at least in part be responsible for the delayed DNA damage repair following the 212Pb-TCMC-trastuzumab treatment.
To examine the involvement of caspase-3, which plays an important role in the condensation and degradation of chromatin of apoptotic cells in 212Pb-TCMC-trastuzumab induced apoptosis, cleaved caspase-3 was analyzed using immunoblot techniques. Cleaved caspase-3 was observed in the tumor tissues 24 h after treatment with 212Pb-TCMC-HuIgG (Fig. 2D). In contrast to this result, it appeared that 212Pb-TCMC-trastuzumab treatment significantly reduced the level of cleaved caspase-3 (p < 0.001), suggesting that 212Pb-TCMC-trastuzumab-induced apoptosis occurred via a caspase-3 independent mechanism.
212Pb-TCMC-trastuzumabattenuates proliferation in S phase and induces G2 cell cycle arrest
In response to radiation, cells typically require cell cycle arrest at G1 and a slow S phase to allow DNA repair. As a consequence of reduced intracellular signaling, trastuzumab as a sole agent induces early escape from this cycle arrest and thereby promotes an accumulation of DNA damage (33). The effect of 212Pb-TCMC-trastuzumab treatment on cell cycle distribution was examined. In the same study, mice were injected with BrdU 4 h prior to tumor collection to pulse-label the tumor xenografts to evaluate DNA synthesis. DNA content and BrdU incorporation was then determined using a two parameter analysis by flow cytometry. As shown in Table 1, tumor cells harvested from untreated mice were found to have the expected uptake of BrdU (23.8 ± 1.3 %); the resulting cell cycle distribution was also in the expected range (Table 2). On the other hand, 6 h after 212Pb-TCMC-trastuzumab treatment, there was a noticeable decrease in BrdU incorporation (13.7 ± 1.2 %). BrdU incorporation decreased further to 2.6 ± 0.7 % at 48 h and remained at this low level throughout the remainder of the 120 h study period. In contrast to 212Pb-TCMC-trastuzumab treatment, however, DNA synthesis after treatment with 212Pb-TCMC-HuIgG did re-initiate by 120 h after decreasing at the early time points. The continuation of DNA synthesis by the tumors that were treated with unlabeled trastuzumab, or HuIgG alone, provided evidence that the cessation of DNA synthesis is specific to the α-radiation and that continued depression of DNA synthesis after 120 h was specific to the targeted 212Pb-TCMC-trastuzumab treatment. In addition, there was a decrease in the S phase fraction with a corresponding increase in the G2/M phase fraction beginning at 24 h compared to the unlabeled analogs. The lower S phase and the elevated G2/M phase fractions were maintained throughout the 120 h study period for the tumors upon 212Pb-TCMC-trastuzumab treatment. In contrast, the cell cycle distribution rebounded in those tumors that were collected from mice treated with 212Pb-TCMC-HuIgG. The initial depression of the S phase fraction and elevation of G2/M phase fraction appeared to be α-radiation related since tumors collected from mice treated with either antibody alone (not labeled with 212Pb) did not appear to experience any alteration in cell cycle distribution.
Table 1.
Analysis of DNA synthesis in LS-174T tumor xenografts following treatment with 212Pb-TCMC-trastuzumab.
| Time point (h)
|
|||||||
|---|---|---|---|---|---|---|---|
| 0 | 6 | 24 | 48 | 72 | 96 | 120 | |
| None | 23.8±1.3 | ||||||
| 212Pb-Trastuzumab | 13.7±.2 | 7.3±1.4 | 2.6±0.7 | 7.5±0.5 | 1.1±0.2 | 1.6±0.3 | |
| 212Pb-HuIgG | 16.4±1.2 | 8.1±0.9 | 2.4±0.1 | 2.8±0.5 | 4.2±0.4 | 13.2±0.4 | |
| Trastuzumab | 15.1±1.0 | 21.4±1.1 | 14.8±12.9 | 27.5±1.5 | 20.6±1.8 | 18.2±2.3 | |
| HuIgG | 15.9±0.8 | 19.0±0.4 | 20.7±1.3 | 23.4±1.1 | 22.1±0.6 | 16.7±1.3 | |
Results represent the average of a minimum of three replications (± SD).
Table 2.
Cell cycle distribution analysis in i.p. tumor xenografts of LS-174T following treatment with 212Pb-TCMC-trastuzumab.
| Treatment | Phase | Time point (h)
|
||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 24 | 48 | 72 | 96 | 120 | ||
| G1 | 67.5±2.7 | |||||||
| S | 17.7±2.0 | |||||||
| G2-M | 14.8±0.7 | |||||||
| 212Pb-Trastuzumab | G1 | 68.6±3.8 | 66.9±1.3 | 67.7±4.0 | 68.1±1.3 | 66.6±0.7 | 73.2±0.1 | |
| S | 14.3±5.4 | 5.9±0.1 | 6.3±1.8 | 7.9±0.7 | 3.6±0 | 4.6±1.9 | ||
| G2-M | 17.1±1.6 | 27.3±1.2 | 26.1±2.1 | 23.9±0.6 | 29.7±0.7 | 22.2±2.0 | ||
| 212Pb-HuIgG | G1 | 63.9±5.2 | 65.1±2.3 | 64.6± | 67.1±0.5 | 63.1±0.3 | 65.5±2.6 | |
| S | 20.0±5.3 | 7.4±1.3 | 5.8± | 5.5±0.1 | 7.8±0.7 | 17.1±3.6 | ||
| G2-M | 16.0±0.1 | 27.5±1.0 | 29.6± | 27.4±0.6 | 29.1±1.0 | 17.5±0.9 | ||
| Trastuzumab | G1 | 69.5±2.6 | 63.3±1.6 | 68.0±0.1 | 61.3±1.8 | 65.1±3.3 | 73.3±4.3 | |
| S | 21.7±3.4 | 26.0±3.2 | 22.0±1.8 | 28.1±1.6 | 23.5±4.3 | 14.8±2.9 | ||
| G2-M | 8.9±0.9 | 10.7±1.6 | 10.1±1.7 | 10.7±3.4 | 11.5±1.0 | 11.9±1.4 | ||
| HuIgG | G1 | 72.3±3.4 | 70.3±3.4 | 70.6±2.3 | 60.6±1.6 | 73.9±1.0 | 74.6±2.2 | |
| S | 22.0±3.4 | 19.8±2.1 | 20.9±1.9 | 26.6±1.8 | 19.6±2.2 | 18.2±2.3 | ||
| G2-M | 5.7±0 | 9.8±1.4 | 8.4±0.4 | 12.8±0.2 | 6.6±1.2 | 7.2±4.5 | ||
Results represent the average of a minimum of three replications (± SD).
212Pb-TCMC-trastuzumab induces modification in chromatin structure of p21
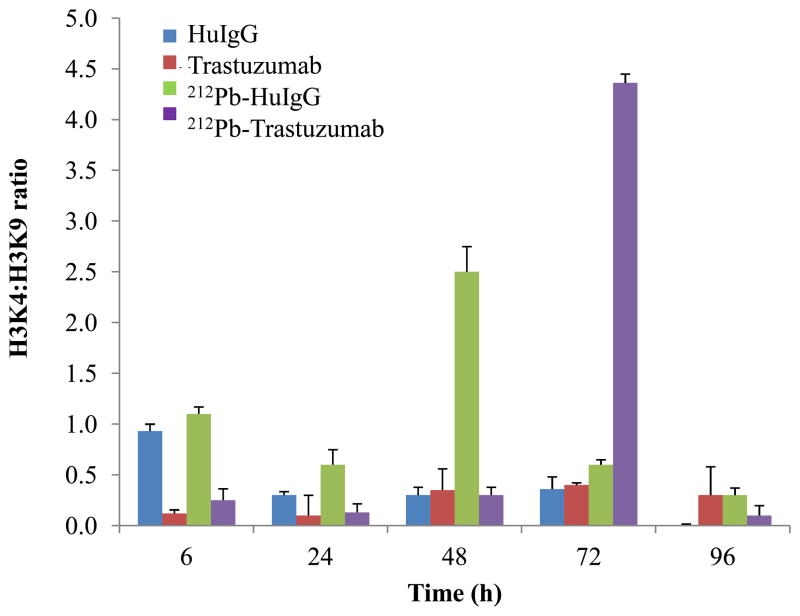
Chromatin remodeling is significantly altered in tumors, suggesting a direct role for methylation in cellular transformation. γH2AX facilitates the recruitment of damage responsive proteins and chromatin remodeling complexes to the sites of DNA damage and influences both the efficiency and fidelity of DNA repair (34). To determine whether changes occur in chromatin remodeling following 212Pb-TCMC-trastuzumab treatment, the ChIP assay was employed using p21 promoter specific primers, one of the known radiation response genes, and immunoprecipitated DNA was analyzed by a quantitative real time PCR (qPCR). The ratio between H3K4 methylation and H3K9 methylation was used as a measurement of change (open/close) in chromatin structure (35–37). The abundance of histone modifications identified with transcriptionally activated chromatin states, such as H3 methylated at lysine 4 was observed at 48 h after treatment with 212Pb-TCMC-HuIgG, and at 72 h after 212Pb-TCMC-trastuzumab treatment (Figure 3). It appeared that at earlier time points, histone modifications associated with transcriptionally repressed chromatin states, such as H3 methylated at lysine 9, prevailed on treatment with 212Pb-TCMC-trastuzumab, indicating that 212Pb-TCMC-trastuzumab induced the delayed open chromatin structure until 72 h.
Figure 3. Effect of 212Pb-TCMC-trastuzumabon chromatin remodeling.
ChIP assay was performed using p21 promoter specific primers and quantified using a quantitative real time PCR (qPCR) at the indicated times. Results represent the average of a minimum of three replications (± SD).
212Pb-TCMC-trastuzumab induces reduction of p21at protein level
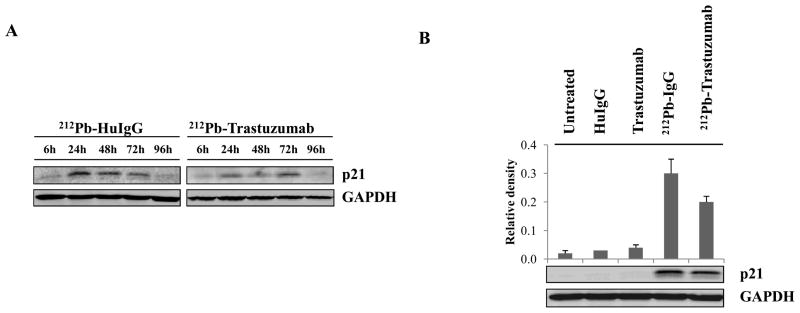
To assess the induction of p21 in response to DNA damage, western blot analysis of p21 was performed. Enhanced protein expression of p21 was observed at 24–48 h after 212Pb-TCMC-HuIgG treatment whereas 212Pb-TCMC-trastuzumab treatment resulted in enhanced p21 expression at 72 h (Figure 4A). These results suggested there were correlations between induction of p21 protein and modification in chromatin structure in response to 212Pb-TCMC-trastuzumaband 212Pb-TCMC-HuIgG treatment. Selecting the 24 h time point, further analysis was performed to compare the effect of all the treatments on the LS-174T xenograft. There was also a significant reduction of p21 protein at 24 h (p < 0.05) compared to a non-specific control, indicating that reduction of p21 protein expression was specific to 212Pb-TCMC-trastuzumab treatment (Figure 4B).
Figure 4. Effect of 212Pb-TCMC-trastuzumabon p21 protein expression.
A. Effect of 212Pb-TCMC-trastuzumab on p21 protein expression. Immunoblot analysis for p21 was performed at the indicated time point. p21 was detected at 21 kDa. The equivalent protein loading control was GAPDH.
B. Down-regulation of p21 by 212Pb-TCMC-trastuzumab treatment. Immunoblot analysis for p21 was performed at 24 h. Results represent the average of a minimum of three replications (± SD).
DISCUSSION
Unlike β−-particle or photon irradiation, α-particle radiation is cytotoxic at dose rates as low as 1 Gy/hour. The shorter path of α particles may also have the advantage of limiting toxicity to normal tissue adjacent to tumor. Studies from this laboratory have shown the exquisite effectiveness of both 213Bi- and 212Pb-labeled trastuzumab for the treatment of low HER2 expressing intraperitoneal disease (6–8). A unique advantage of RIT versus monotherapy with trastuzumab is that neither high expression nor homogenous expression of HER-2 throughout the tumor is required to affect therapy. One strategy to overcome the limitations of the shorter half-lives of the bismuth radioisotopes is to treat with 212Pb (t1/2= 10.6 hr), which essentially serves as an in vivo generator. The application of 212Pb-labeled trastuzumab in the appropriate setting has demonstrated advantages over 213Bi-labeled trastuzumab (6–8). These studies provided sufficient impetus for the in vivo assessment of the mechanisms of therapeutic efficacy of 212Pb-TCMC-trastuzumab therapy.
The effect of 212Pb-TCMC-trastuzumab treatment on apoptosis appeared to be more pronounced in the targeted cells at 24 h (p < 0.05) compared to the non-specific control, 212Pb-TCMC-HuIgG. In prior observations, trastuzumab was reported to enhance radiation-induced apoptosis of cells in an HER2 level dependent manner (38). While the results obtained in this study with 212Pb-TCMC-trastuzumab treatment appears to be similar to the prior report with respect to the enhancement of apoptosis, the two situations are inherently different. In the experiments presented here, trastuzumab was used as a vector to specifically direct α-particle radiation payload to the tumor. In this instance, trastuzumab was not expected to exert any pharmacologic effects. Irradiated cells release signals and induce responses in cells whose nuclei were not hit by radiation, resulting in genetic damage, genomic instability or cell death. A high apoptotic rate was also observed for the non-specific control 212Pb-TCMC-HuIgG treated group. Consideration must be given to this indirect effect of radiation because transmissible biological effects resulting from the radiation insult are pronounced following high LET radiation, such as α-particle irradiation.
Direct injury to DNA is generally attributed radiation therapy. Increased DNA repair has been shown in tumor cells resistant to radiation and anticancer drugs in comparison with tumor cells sensitive to these modalities. Synergy between drugs and ionizing radiation is attributed to impaired repair of residual DNA DSBs (39). As shown from the percent DNA in the comet tail comparison, DNA damage following 212Pb-HuIgG radiation demonstrated gradual recovery beginning at 24 h post-treatment. On the other hand, DNA repair inhibition was more pronounced in the mice treated with 212Pb-TCMC-trastuzumab (p < 0.001) as indicated by the persistently higher % DNA in the comet tail at all time points studied. There appeared to be recovery of DNA repair for the 212Pb-TCMC-HuIgG treated tumors, while recovery was not evident after the 212Pb-TCMC-trastuzumab treatment. These results were further investigated to understand which repair systems were involved. NHEJ and HR are predominant mechanisms in DNA DSB repair. High-level Rad51 expression has been reported in chemoresistant or radioresistant carcinomas (40). Up-regulation of Rad51 after irradiation has been previously observed in eukaryotic cells (41). In this study, Rad51 was not affected in the presence of 212Pb-TCMC-HuIgG at 24 h. In contrast to this result, densitometric analysis of the western blots clearly indicated that the 212Pb-TCMC-trastuzumab treatment significantly reduced Rad51 at the protein level (p < 0.05). Therefore, inhibition of DNA damage repair induced by 212Pb-TCMC-trastuzumab treatment, evidenced by the reduction of Rad51 protein expression and by the persisted inhibition of DNA damage repair, may be an explanation for the increased cell killing efficacy of 212Pb-TCMC-trastuzumab treatment.
After irradiation with 212Pb, cleaved caspase-3 was observed albeit at a lower level after 212Pb-TCMC-trastuzumab treatment (p < 0.001) compared to 212Pb-TCMC-HuIgG treatment, suggesting that apoptosis induced by 212Pb-TCMC-trastuzumab treatment was not dependent on caspase-3. Various studies also reported that high LET α-particle immunoconjugates induced apoptosis, and that the mode of cell death triggered by α-particle emitters seemed to be dependent on the type of cells irradiated (42–46).
The lower levels of DNA synthesis that were observed following 212Pb-TCMC-trastuzumab treatment at the early time points persisted beyond 120 h, but appeared to rebound for the 212Pb-TCMC-HuIgG treated tumors by the same time point. A similar temporal progression was observed for the phase distribution of cells where cells were arrested in the G2 phase with a severely depressed S phase at the early time points on 212Pb treatment which appeared to rebound for 212Pb-TCMC-HuIgG treated mice by 120 h, but not for the group treated with 212Pb-TCMC-trastuzumab. Notably, tumors from mice given 212Pb-TCMC-trastuzumab remained arrested at the G2-M phase with depressed S phase beyond 120 h. Synchronization in the G2 phase after 120 h treatment together with persistent reduced DNA synthesis seemed the most prominent difference between the targeted 212Pb-TCMC-trastuzumab and the control 212Pb-TCMC-HuIgG treatment. Such synchronization has been described as the major course of synergy between chemotherapy and external beam irradiation, although radiosensitization may not be achieved in all cell lines or tumors (47). The arrest of cells in the radiosensitive G2-M phase of the cell cycle induced by 212Pb-TCMC-trastuzumab treatment may be another explanation for the increased cell killing efficacy of α-particle RIT. These results are bolstered by the persistently delayed DNA repair on 212Pb-TCMC-trastuzumab treatment indicated by the comet assay compared to the recovery of DNA repair at later time points on 212Pb-TCMC-HuIgG treatment.
Epigenetic markers for open and closed chromatin status using H3K4/H3K9 ratio revealed correlations between the induction of p21 protein and modification in chromatin structure of p21 in response to 212Pb-TCMC-trastuzumab and 212Pb-TCMC-HuIgG treatment. In contrast to the results of 212Pb-TCMC-HuIgG control, induction of p21 protein and open chromatin structure was delayed until 72 h by 212Pb-TCMC-trastuzumab, indicating that inhibition of p21 protein at early time points was associated with histone modifications that correlated with repressed transcription. In fact, there was a significant reduction of p21 at the protein level at 24h by 212Pb-TCMC-trastuzumab (p < 0.05). These results indicate that the increased killing efficacy of 212Pb-TCMC-trastuzumab treatment was, in part, associated with the delay in open chromatin structure of p21 that correlated to less active transcription at earlier time points. Wendt et al reported that radiation induced p21 expression and G2 arrest results in resistance to apoptosis and inhibition of p21 displayed enhanced radiation-induced apoptosis (48).
Understanding the mechanisms of cell death and DNA repair is critical to the design of novel strategies that combine chemotherapy with targeted α-particle radiation. Studies of this nature help refine and optimize all of the components to improve efficacy and minimize toxicity. Carefully planned pre-clinical investigation and improved targeting strategies will facilitate translation into clinical evaluation to move the field forward. Studies are currently underway to evaluate the potential mechanism(s) of chemotherapeutics in combination with α-particle RIT.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Park SI, Press OW. Radioimmunotherapy for treatment of B-cell lymphomas and other hematologic malignancies. Curr Opin Hematol. 2007;14:632–38. doi: 10.1097/MOH.0b013e3282efb17c. [DOI] [PubMed] [Google Scholar]
- 2.DeNardo GL, DeNardo SJ, Balhorn R. Systemic radiotherapy can cure lymphoma: A paradigm for other malignancies? Cancer Biother Radiopharm. 2008;23:383–97. doi: 10.1089/cbr.2007.0523-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang M, Yao Z, Garmestani K, Axworthy DB, Zhang Z, Mallett RW, et al. Pretargeting radioimmunotherapy of a murine model of adult T-cell leukemia with the α-emitting radionuclide, bismuth 213. Blood. 2002;100:208–16. doi: 10.1182/blood-2002-01-0107. [DOI] [PubMed] [Google Scholar]
- 4.Zhang M, Zhang Z, Garmestani K, Schultz J, Axworthy DB, Goldman CK, et al. Pre-target radiotherapy with an anti-CD25 antibody-streptavidin fusion protein was effective in therapy of leukemia/lymphoma xenografts. Proc Natl Acad Sci USA. 2003;100:1891–95. doi: 10.1073/pnas.0437788100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurtzman SH, Russo A, Mitchell JB, DeGraff W, Sindelar WF, Brechbiel MW, et al. 212Bi linked to an anti-pancreatic carcinoma antibody: Model for α-particle-emitter radioimmunotherapy. J Natl Cancer Inst. 1988;80:449–52. doi: 10.1093/jnci/80.6.449. [DOI] [PubMed] [Google Scholar]
- 6.Milenic DE, Garmestani K, Brady ED, Albert PS, Ma D, Abdulla A, et al. Targeting of HER2 antigen for the treatment of disseminated peritoneal disease. Clin Cancer Res. 2004;10:7834–41. doi: 10.1158/1078-0432.CCR-04-1226. [DOI] [PubMed] [Google Scholar]
- 7.Milenic DE, Brechbiel MW. Targeting of radioisotopes for cancer therapy. Cancer Biol Ther. 2004;3:361–70. doi: 10.4161/cbt.3.4.790. [DOI] [PubMed] [Google Scholar]
- 8.Milenic DE, Garmestani K, Brady ED, Albert PS, Ma D, Abdulla A, et al. α-particle radioimmunotherapy of disseminated peritoneal disease using a 212Pb-labeled radioimmunoconjugate targeting HER2. Cancer Biother Radiopharm. 2005;20:557–68. doi: 10.1089/cbr.2005.20.557. [DOI] [PubMed] [Google Scholar]
- 9.Su FM, Beaumier P, Axworthy D, Atcher R, Fritzberg A. Pre-targeted radioimmunotherapy in tumored mice using an in vivo212Pb/212Bi generator. Nucl Med Biol. 2005;32:741–47. doi: 10.1016/j.nucmedbio.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Roger M, Macklis MD, William D, Kaplan MD, James LM, Ferrara MD, et al. Resident’s essay award: alpha particle radio-immunotherapy: animal models and clinical prospects. Int J Radiation oncology Biol Phys. 1988;16:1337–1387. doi: 10.1016/0360-3016(89)90938-3. [DOI] [PubMed] [Google Scholar]
- 11.Yong K, Brechbiel MW. Towards translation of 212Pb as a clinical therapeutic; getting the lead in! Dalton Trans. 2011;40:6068–76. doi: 10.1039/c0dt01387k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baselga J. Phase I and II clinical trials of trastuzumab. Ann Oncol. 2001;12:49–55. doi: 10.1093/annonc/12.suppl_1.s49. [DOI] [PubMed] [Google Scholar]
- 13.Aqus DB, Bunn PA, Jr, Franklin W, Garcia M, Ozols RF. Her-2/neu as a therapeutic target in non-small cell lung cancer, prostate cancer, and ovarian cancer. Semin Oncol. 2000;27:53–63. [PubMed] [Google Scholar]
- 14.Chappell LL, Dadachova E, Milenic DE, Garmestani K, Wu C, Brechbiel MW. Synthesis, characterization, and evaluation of a novel bifunctional chelating agent for the lead isotopes 203Pb and 212Pb. Nucl Med Biol. 2000;27:93–100. doi: 10.1016/s0969-8051(99)00086-4. [DOI] [PubMed] [Google Scholar]
- 15.Nunez MI, Villalobos M, Olea N, et al. Radiation-induced DNA double-strand breaks rejoining in human tumor cells. Br J Cancer. 1995;71:311–16. doi: 10.1038/bjc.1995.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolesnick RN, Haimovitz-Friedman A, Fuks Z. The sphingomyelin signal transduction pathway mediates apoptosis for tumor necrosis factor, Fas, and ionizing radiation. Biochem Cell Biol. 1994;72:471–74. doi: 10.1139/o94-063. [DOI] [PubMed] [Google Scholar]
- 17.Pandita TK, Lieberman HB, Lim DS, Dhar S, Zheng W, Taya Y, et al. Ionizing radiation activates the ATM kinase throughout the cell cycle. Oncogene. 2000;19:1386–91. doi: 10.1038/sj.onc.1203444. [DOI] [PubMed] [Google Scholar]
- 18.Munro TR. The relative radiosensitivity of the nucleus and cytoplasm of Chinese hamster fibroblasts. Radiat Res. 1970;42:451–70. [PubMed] [Google Scholar]
- 19.Søyland C, Hassfjell SP. Survival of human lung epithelial cells following in vitro α-particle irradiation with absolute determination of the number of alpha-particle traversals of individual cells. Int J Radiat Biol. 2000;76:1315–22. doi: 10.1080/09553000050151583. [DOI] [PubMed] [Google Scholar]
- 20.Seidl C, Schrock H, Seidenschwang S, Beck R, Schmid E, Abend M, et al. Cell death triggered by α-emitting 213Bi-immunoconjugates in HSC-M2 gastric cancer cells is different from apoptotic cell death. Eur J Nucl Med Mol Imaging. 2005;32:274–85. doi: 10.1007/s00259-004-1653-3. [DOI] [PubMed] [Google Scholar]
- 21.Friesen C, Glatting G, Koop B, et al. Breaking chemoresistance and radioresistance with 213Bi anti-CD45 antibodies in Leukemia cells. Cancer Res. 2007;67:1950–58. doi: 10.1158/0008-5472.CAN-06-3569. [DOI] [PubMed] [Google Scholar]
- 22.Suoio S, Gouard S, Charrier J, Apostolidis C, Chatal JF, Barbet J, et al. Mechanisms of cell sensitization to α-radioimmunotherapy by doxorubicin or paclitaxel in multiple myeloma cell lines. Clin Cancer Res. 2005;11:7047–52. doi: 10.1158/1078-0432.CCR-1004-0021. [DOI] [PubMed] [Google Scholar]
- 23.Tom BH, Rutzky LP, Jakstys MM, Oyasu R, Kaye CI, Kahan BD. Human colonic adenocarcinoma cells. I. Establishment and description of a new cell line. In Vitro. 1976;12:180–91. doi: 10.1007/BF02796440. [DOI] [PubMed] [Google Scholar]
- 24.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 25.Dadachova E, Chappell LL, Brechbiel MW. Spectrophotometric method for determination of bifunctional macrocyclic ligands in macrocyclic ligand-protein conjugates. Nucl Med Biol. 1999;26:977–82. doi: 10.1016/s0969-8051(99)00054-2. [DOI] [PubMed] [Google Scholar]
- 26.Milenic DE, Wong KJ, Baidoo KE, Nayak TK, Regino CA, Garmestani K, et al. Mabs. 2010;2:550–64. doi: 10.4161/mabs.2.5.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grégoire V, Van NT, Stephens LC, Brock WA, Milas L, Plunkett W. The role of fludarabine-induced apoptosis and cell cycle synchronization in enhanced murine tumor radiation response in vivo. Cancer Res. 1994;54:6201–09. [PubMed] [Google Scholar]
- 28.Stephens LC, Ang KK, Schultheiss TE, Milas L, Meyn RE. Apoptosis in irradiated murine tumors. Radiat Res. 1991;127:308–16. [PubMed] [Google Scholar]
- 29.Kaufmann SH, Earnshaw WC. Introduction of apoptosis by cancer therapy. Exp Cell Res. 2000;256:42–49. doi: 10.1006/excr.2000.4838. [DOI] [PubMed] [Google Scholar]
- 30.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–76. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 31.Leonard CE, Chan DC, Chou TC, Kumar R, Bunn PA. Paclitaxel enhances in vitro radiosensitivity of squamous carcinoma cell lines of the head and neck. Cancer Res. 1996;56:5198–04. [PubMed] [Google Scholar]
- 32.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair. 2007;6:923–35. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Spector NN, Blackwell KL. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor-2 positive breast cancer. J Clin Oncol. 2009;27:5838–47. doi: 10.1200/JCO.2009.22.1507. [DOI] [PubMed] [Google Scholar]
- 34.Podhorecka M, Skladanowski A, Bozko P. H2AX Phosphorylation: its Role in DNA Damage Response and Cancer Therapy. J Nucleic Acids. 2010;2010:1–9. doi: 10.4061/2010/920161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 36.Kondo Y, Issa JP. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J Biol Chem. 2003;278:27658–62. doi: 10.1074/jbc.M304072200. [DOI] [PubMed] [Google Scholar]
- 37.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol Cell Biol. 2003;23:206–15. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang K, Lu Y, Jin W, Ang Kk, Milas L, Fan Z. Sensitization of breast cancer cells to radiation by trastuzumab. Mol Cancer Ther. 2003;2:1113–20. [PubMed] [Google Scholar]
- 39.Pietras RJ, Fendly BM, Chazin VR, Pegram MD, Howell SB, Slamon DJ. Antibody to Her2/neu receptor blocks DNA repair after cisplatin in human breast and ovarian cancer cells. Oncogene. 1994;9:1829–38. [PubMed] [Google Scholar]
- 40.Ko JC, Hong JH, Wang LH, Cheng CM, Ciou SC, Lin ST, et al. Role of repair protein Rad51 in regulating the response to gefitinib in human non-small cell lung cancer cells. Mol Cancer Ther. 2008;7:3632–41. doi: 10.1158/1535-7163.MCT-08-0578. [DOI] [PubMed] [Google Scholar]
- 41.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 42.Kelly MP, Lee FT, Tahtis K, Smyth FE, Brechbiel MW, Scott AM. Radioimmunotherapy with α-particle emitting 213Bi-C-functionized trans-cyclohexyl-diethylenetriaminepentaacetic acid-humanized 3S193 is enhanced by combination with paclitaxel chemotherapy. Clin Cancer Res. 2007;13:5604s–12s. doi: 10.1158/1078-0432.CCR-07-1071. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Cozzi PJ, Qu CF, Zhang DY, Abbas Rizvi SM, Raja C, et al. Cytotoxicity of human prostate cancer cell lines in vitro and induction of apoptosis using 213Bi-Herceptin alpha conjugate. Cancer Lett. 2004;205:161–71. doi: 10.1016/j.canlet.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 44.Macklis RM, Lin JY, Beresford B, Atcher RW, Hines JJ, Humm JL. Cellular kinetics, dosimetry, and radiobiology of α-particle radioimmunotherapy: Induction of apoptosis. Radiat Res. 1992;130:220–26. [PubMed] [Google Scholar]
- 45.Pouget JP, Mather SJ. General aspects of the cellular response to low- and high-LET radiation. Eur J Nucl Med. 2001;28:541–61. doi: 10.1007/s002590100484. [DOI] [PubMed] [Google Scholar]
- 46.Abend M. Reasons to reconsider the significance of apoptosis for cancer therapy. Int J Radiat Biol. 2003;79:927–41. doi: 10.1080/09553000310001632958. [DOI] [PubMed] [Google Scholar]
- 47.Sinclair WK, Morton RA. X-ray sensitivity during the cell generation cycle of cultured Chinese hamster cells. Radiat Res. 1966;29:450–74. [PubMed] [Google Scholar]
- 48.Wendt J, Radetzki S, von Haefen C, Hemmati PG, Guner D, Schulze-Osthoff K, et al. Induction of p21CIP/WAF-1 and G2 arrest by ionizing irradiation impedes caspase-3-mediated apoptosis in human carcinoma cells. Oncogene. 2006;25:972–80. doi: 10.1038/sj.onc.1209031. [DOI] [PubMed] [Google Scholar]