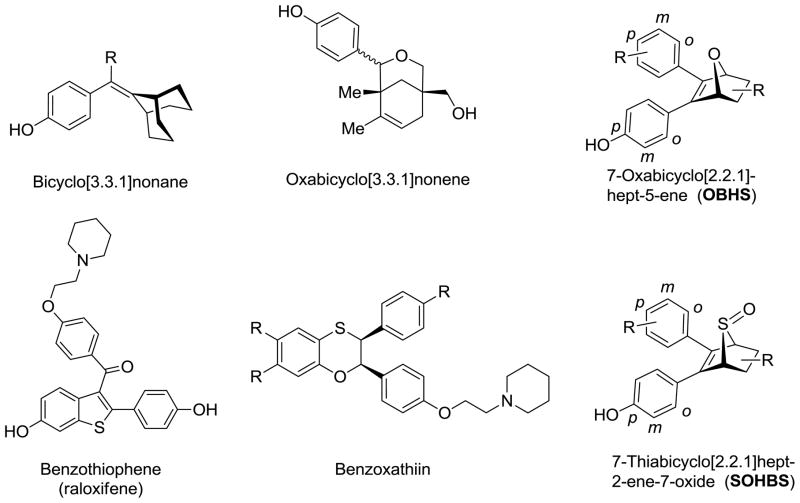
Abstract
To develop estrogen receptor (ER) ligands having novel structures and activities, we have explored compounds in which the central hydrophobic core has a more three-dimensional topology than typically found in estrogen ligands and thus exploit the unfilled space in the ligand-binding pocket. Here, we build upon our previous investigations of 7-oxabicyclo[2.2.1]heptene core ligands, by replacing the oxygen bridge with a sulfoxide. These new 7-thiabicyclo[2.2.1]hept-2-ene-7-oxides were conveniently prepared by a Diels-Alder reaction of 3,4-diarylthiophenes with dienophiles in the presence of an oxidant and give cycloadducts with endo stereochemistry. Several new compounds demonstrated high binding affinities with excellent ERα selectivity, but unlike oxabicyclic compounds, which are transcriptional antagonists, most thiabicyclic compounds are potent, ERα-selective agonists. Modeling suggests that the gain in activity of the thiabicyclic compounds arises from their endo stereochemistry that stabilizes an active ER conformation. Further, the disposition of methyl substituents in the phenyl groups attached to the bicyclic core unit contribute to their binding affinity and subtype selectivity.
Introduction
Estrogens are known to play important roles in the development and maintenance of both reproductive and non-reproductive tissues in both women and men.1, 2 While estrogens are required and can provide some health benefits in some tissues, such as those of the reproductive,3 skeletal,4 cardiovascular5 and central nervous systems,6 the pro-proliferative effect of estrogens can be pathological and promote cancer in the breast and uterus.7–9 The multiple actions of estrogens are mediated by two estrogen receptors (ERα and ERβ) that, although similar, are distinct gene products with non-overlapping and even opposing functions.1 These different functions, combined with the distinct tissue distribution patterns of these two receptors, result in the remarkable tissue-selective effects of estrogens,2 and thus have heightened interest in searching for selective estrogen receptor modulators (SERMs) that are also subtype selective and thus best able to support estrogen health benefits and minimize the risk of cancer.10–15
As part of our long-term interest in the development of ligands for the ERs having novel structures and activities, we have undertaken exploratory studies by preparing new compounds having a central core that has, overall, a more three-dimensional topology than is commonly found in both steroidal and non-steroidal ER ligands. This design strategy was based on structural studies of the ligand binding pockets of both ERα and ERβ: In addition to the obvious flexibility and deformability of the ligand binding pocket,16 it was notable that the cavity of ERα has a probe-accessible size of ca. 450 Å3, whereas estradiol (E2) has a molecular volume of only 245 Å3; though somewhat smaller, the ligand pocket in ERβ is also considerably larger than E2.17 As a result of these marked pocket vs. ligand volume differences, there is substantial unoccupied space on the α face of the B-ring and the β face of the C-ring.18 By incorporating a more three-dimensional hydrophobic bicyclic unit as the core structure of a ligand, we hoped to exploit this unfilled, opportunity space, thereby enhancing the binding affinity and/or ER subtype selectivity, and potentially uncover novel patterns of estrogen responses through the ERs. We and a number of other investigators have prepared some ER ligands having more 3-dimensional character, such as those with ferrocene,19–21 carborane,22, 23 polycyclics,23, 24 and cyclopentadienyl metal tricarbonyls core structures.25
In previous studies, we prepared a series 7-oxabicyclo[2.2.1]hept-5-ene compounds as ER ligands (Scheme 1).26 The best compound, exo-5,6-bis-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene- 2-sulfonic acid phenyl ester (which we named OBHS), exhibited modest ER subtype selectivity, with the relative binding affinity (RBA) values 9.3% and 1.7% for ERα and ERβ, respectively (RBA[estradiol] =100%), and profiled as an antagonist on both ER subtypes, with a modest potency preference for ERβ.26 Bearing some structural relationship to other bicyclic ER ligands, such as bicyclo[3.3.1]nonanes27, 28 and oxabicyclo[3.3.1]nonenes,29–31 the 7-oxabicyclo[2.2.1]hept-5-enes mimic an element of the core of high affinity furan-based ER ligands that we have studied,32, 33 and they also embody a 1,2-diarylethylene unit, a motif found in many high-affinity non-steroidal estrogens.
Scheme 1.
Described three-dimensional, thiophene or sulfur containing ER ligands and the title compounds.
Beyond the oxygen-containing furan and pyran-type heterocycles, sulfur-containing heterocycles also frequently constitute the cores of ER ligands, as the benzothiophene core of raloxifene and the benzoxathiin core of other ER ligands (Scheme 1).34 Some simple aryl-substituted thiophenes can also be ER ligands, as well as inhibitors of certain steroid dehydrogenases.35, 36 It is noteworthy that although some aryl-substituted thiophenes exhibit good ER binding affinities, as thus far reported, they have limited selectivity and bioactivity. In light of these recent reports and in continuation of our interest in non-steroidal estrogens, we extended our previous study of OBHS by replacing the oxabicyclic core with a 7-thiabicyclic core.
Unlike furan, thiophene is not a good diene for the Diels-Alder reaction because of its higher aromaticity.37 In addition, the sulfur or sulfone bridge is not very stable, and such Diels-Alder adducts can spontaneously lose sulfur or sulfur dioxide, respectively, leading to benzene ring formation.38–41 Consequently, after a brief survey, we chose a 7-thiabicyclo[2.2.1]hept-2-ene-7-oxide as the core structure of these novel ER ligands, because of its greater stability and ease of preparation (Scheme 1).
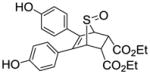
In this report, we describe novel sulfoxide-bridged OBHS analogs constituted of a 7-thiabicyclo[2.2.1]hept-2-ene 7-oxide core that can be prepared conveniently by a Diels-Alder reaction of thiophene with an appropriate dienophile in the presence of an oxidant.42 This bicyclic core system, which we term SOBHS, expands our exploration of ER ligands having an overall three-dimensional topology, and it introduces some new characteristics as well. Therefore, this structure potentially could be further investigated and developed as the basis for new estrogen pharmacological agents. We also evaluate the effect of SOBHS analogs on ER binding affinity and estrogen responsive element (ERE)-driven transcriptional activity.
Results and Discussion
Chemical Synthesis
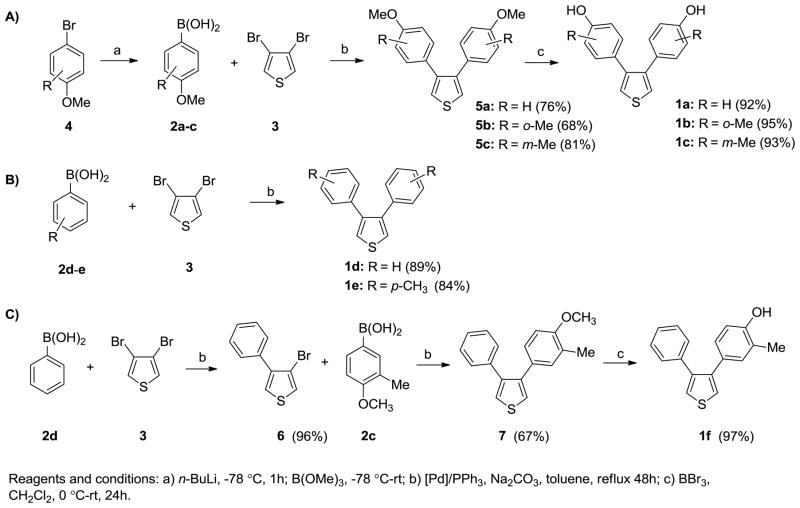
The preparation of the 7-thiabicyclic oxide-bridged compounds involves a Diels-Alder reaction of aryl-substituted thiophenes with various dienophiles. The 3,4-bis(4-hydroxyphenyl)-substituted thiophenes (1a–c) can be conveniently prepared from 3,4-dibromothiophene by a Suzuki coupling sequence that, together with a boronic acid synthesis and a phenol demethylation, proceeds in three steps (Scheme 2A). 3,4-Diphenylthiophene (1d) and 3,4-di-p-tolylthiophene (1e) can be prepared in one step from commercially available boronic acids (Scheme 2B). The unsymmetrical thiophene 1f was obtained by demethylation of 7, which was prepared by two successive cross-coupling reactions (Scheme 2C). In the first step, 1 equivalent of 3,4-dibromothiophene reacted with 1.2 equivalents of phenyl boronic acid using standard conditions. In the second step, the resulting mono-substituted thiophene 6 was subsequently submitted to a second cross-coupling reaction with 1.2 equivalents of aryl boronic acid 2c to yield the intermediate 7, ether cleavage with boron tribromide giving the final compound 1f.
Scheme 2.
Synthesis of thiophenes 1a–f.
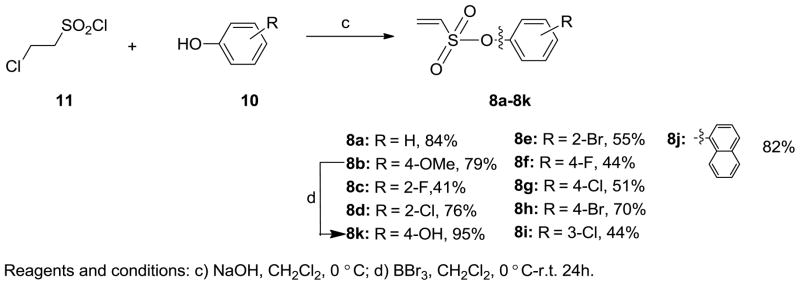
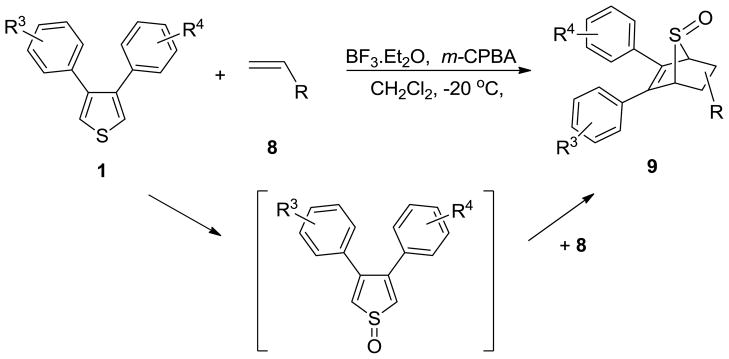
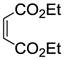
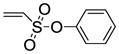
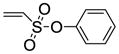
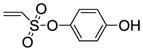
The synthesis of vinyl sulfonates 8a–k was accomplished by the reaction of 2-chloroethanesulfonyl chloride with substituted phenols under basic conditions, as shown below (Scheme 3). The synthesis of 7-thiabicyclic oxide bridged compounds was achieved by a Diels-Alder reaction of thiophene 1 with various dienophiles 8 (2 equiv) (Scheme 4) in the presence of an in situ oxidant (m-CPBA) and a Lewis acid (BF3); the results are summarized in Table 2. This transformation is presumed to proceed via two steps: in situ oxidation of the thiophene to the thiophene S-monoxide, followed by Diels-Alder reaction to produce the 7-thiabicyclic[2.2.1]hept-2-ene-7-oxide structure (Scheme 3).42, 43 A wide range of dienophiles were examined to obtain a broad structure-activity relationship of this series. On the other hand, the dienophiles were restricted to mostly arene esters of vinylsulfonic acid, because earlier work in the OBHS series had indicated that the Diels-Alder products from vinylsulfones and various maleic acid derivatives generally gave products with low affinity for the ERs,26 although we did prepare some of these analogs for comparison purposes.
Scheme 3.
Synthesis of dienophiles 8.
Scheme 4.
Diels-Alder Reaction of Thiophene 1 with Dienophiles 8 to give SOBHS Adducts 9.
Table 2.
Relative Binding Affinity (RBA) of 7-Thiabicyclic-7-oxide Analogs for ERα and ERβa.
| Entry | Compound | ERα | ERβ | α/β ratio |
|---|---|---|---|---|
| 1 |
 10a |
0.956 ±0.16 | 0.110 ±0.03 | 8.33 |
| 2 |
 10b |
0.074 ± 0.002 | 0.080 ± 0.015 | 0.925 |
| 3 |
 10c |
0.077 ± 0.01 | 0.071 ±0.02 | 1.08 |
| 4 |
 10d |
0.017 ±0.005 | 0.013 ±0.001 | 1.31 |
| 5 |
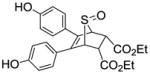 10e |
0.022 ± 0 | 0.048 ± 0.005 | 0.458 |
| 6 |
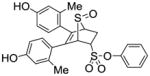 11a |
8.11 ±1.8 | 0.348 ± 0.01 | 23.3 |
| 7 |
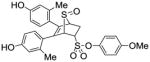 11b |
0.741 ±0.18 | 0.091 ± 0.02 | 8.14 |
| 8 |
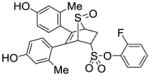 11c |
3.53 ± 0.45 | 0.138 ±0.04 | 25.6 |
| 9 |
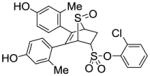 11d |
2.49 ± 0.31 | 0.227 ± 0.03 | 11.0 |
| 10 |
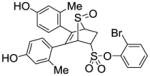 11e |
2.21 ± 0.60 | 0.070 ± 0.02 | 31.6 |
| 11 |
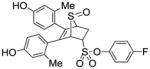 11f |
2.18 ± 0.65 | 0.080 ± 0.02 | 27.2 |
| 12 |
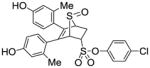 11g |
2.13 ±0.36 | 0.083 ± 0.02 | 25.7 |
| 13 |
 11h |
3.37 ± 0.22 | 0.297 ± 0.08 | 11.3 |
| 14 |
 11i |
1.30 ±0.37 | 0.088 ± 0.021 | 14.8 |
| 15 |
 11j |
0.998 ± 0.22 | 0.180 ±0.05 | 5.54 |
| 16 |
 11k |
2.21 ± 0.48 | 0.812 ±0.06 | 2.72 |
| 17 |
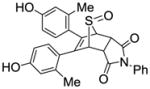 11l |
0.016 ±0.002 | 0.014 ±0.004 | 1.14 |
| 18 |
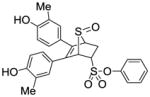 12a |
3.49 ± 0.47 | 0.014 ±0.004 | 249 |
| 19 |
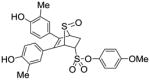 12b |
0.310 ±0.05 | 0.021 ± 0.006 | 14.8 |
| 20 |
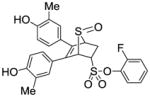 12c |
2.48 ±0.13 | 0.010 ±0.003 | 248 |
| 21 |
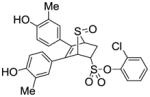 12d |
1.18±0.18 | 0.022 ± 0.005 | 53.6 |
| 22 |
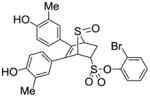 12e |
0.516 ±0.14 | 0.014 ±0.004 | 36.8 |
| 23 |
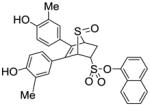 12f |
0.236 ± 0.07 | 0.019 ±0 | 12.4 |
| 24 |
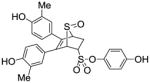 12g |
0.109 ±0.003 | 0.009 ± 0.001 | 12.1 |
| 25 |
 12h |
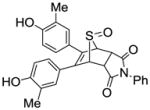
0.009 ± 0.002 | 0.007 ± 0.001 | 1.28 |
| 26 |
 12i |
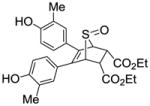
0.026 ± 0.008 | 0.019 ±0.005 | 1.37 |
| 27 |
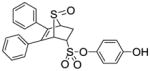 13 |
0.005 ± 0.001 | 0.008 ± 0.001 | 0.62 |
| 28 |
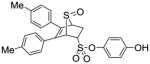 14 |
0.005 ± 0.001 | 0.011 ±0.001 | 0.45 |
| 29 |
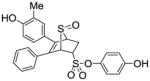 15 |
0.035 ± 0.009 | 0.055 ± 0.001 | 0.64 |
| 30 |
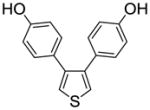 1a |
0.572 ± 0.12 | 1.69 ± 0.45 | 0.338 |
| 31 |
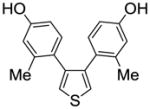 1b |
2.16 ±0.54 | 4.86 ± 1.3 | 0.444 |
| 32 |
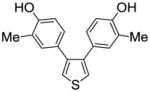 1c |
0.793 ± 0.05 | 0.306 ± 0.01 | 2.59 |
Relative binding affinity (RBA) values are determined by competitive radiometric binding assays and are expressed as IC50estradiol/IC50compound × 100 ± the range or standard deviation (RBA, estradiol = 100%). In these assays, the Kd for estradiol is 0.2 nM on ERα and 0.5 nM on ERβ. For details, see the Experimental Section.
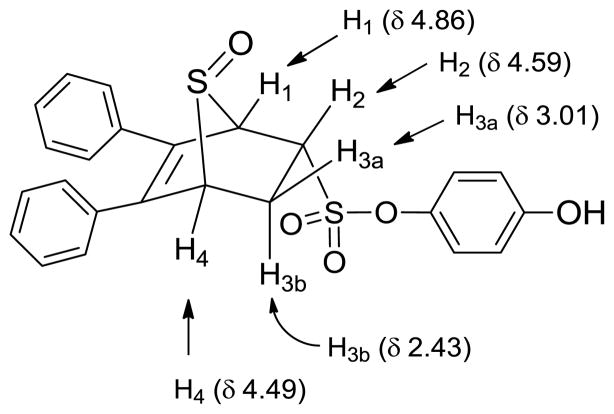
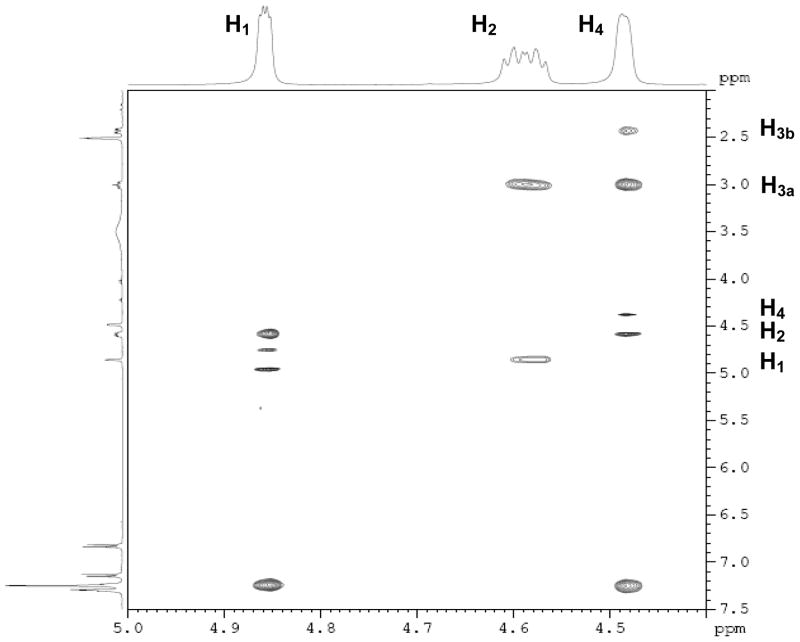
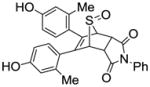
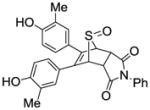
Unlike the high yields obtained in the Diels-Alder reactions with furans, the Diels-Alder reaction with the thiophenes was very sluggish; conversions were typically around 60%, and the yields of the Diels-Alder adducts were moderate. Also, while the exo products predominated in the Diels-Alder reaction the furans, presumably because, as we described previously,26 this very facile cycloaddition is reversible under the conditions used, we observed high endo stereoselectivity in the Diels-Alder reaction with phenolic thiophenes. This endo stereochemistry for one compound (13) (Scheme 5) was verified by the two-dimensional ROESY-NMR (Figure 1, see legend). This observation is in accord with other studies documenting that the Diels-Alder reaction with these systems takes place exclusively in an endo-mode with 100% π-face selectivity, in which dienophiles add in an endo to the thiophene S-monoxide on the syn-π-face relating to the S=O bond, which is the combined result of secondary orbital energy interaction and steric factors.44, 45 It should be noted that all compounds were studied as racemates, and in the one case were an unsymmetrical thiophene was used as a diene, we were unable to separate the regioisomeric products (compound 15), despite our best efforts, although the very low affinity of this compound makes this less of an issue.
Scheme 5.
1H NMR assignments of endo 13.
Figure 1.
ROESY-NMR of endo 13. The peaks at δ 4.86 and δ 4.49 are the hydrogen atoms on the bridgehead carbons (H1 and H4), and the peak between them (at δ 4.59) is the hydrogen attached to the carbon bearing the sulfonate group (H2). It is evident that the H2 interacts with the bridgehead hydrogen H1. Since the bridgehead hydrogen is necessarily at an exo position, this interaction indicates that H2 is also at exo position, and, as a result, the sulfonate group is disposed in an endo configuration.
In our previous work on 7-oxabicyclic[2.2.1]hept-5-ene (OBHS)-core ER ligands,26 we found that compounds bearing a p-hydroxyphenyl group in both the C-5 and C-6 positions, and a phenyl sulfonate in the C-2 position of the bicyclic core, always had the highest ER binding affinities (see below). Therefore, we wondered whether the replacement of the oxygen atom on the bridge with a sulfoxide group might lead to ligands in the SOBHS series with increased binding affinity. Thus, we started our investigation with compound 10a, as shown in Table 1 (entry 1). In addition, we explored the potential binding affinities and estrogenic properties of the SOBHS analogs by varying the substituents on phenol groups in the 5,6-positions, and the phenyl group of sulfonate, while keeping the 7-thiabicyclic[2.2.1]hept-2-ene-7-oxide skeleton intact. Meanwhile, the adducts of diaryl thiophenes with other dienophiles, e.g., naphthyl vinyl sulfonate as well as diethyl maleate and N-phenylmaleimide, were also prepared for comparison with compounds prepared previously in the OBHS series.26 Using molecular modeling as a guiding tool, we designed and synthesized a small array of 29 SOBHS analogs.
Table 1.
Diels-Alder Reaction of Thiophene 1 and Dienophiles 8.
| Entry | Thiophene | Dienophile | Conv.a(%) | Product Yieldb |
|---|---|---|---|---|
| 1 |
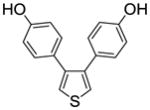 1a |
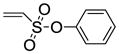 8a |
55 |
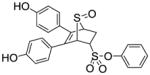 10a (36%) |
| 2 |
8b |
58 |
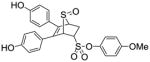 10b (36%) |
|
| 3 |
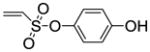 8k |
60 |
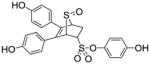 10c (32%) |
|
| 4 |
 8l |
59 |
 10d (36%) |
|
| 5 |
 8m |
59 |
 10e (21%) |
|
| 6 |
 1b |
 8a |
52 |
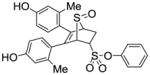 11a (41%) |
| 7 |
8b |
60 |
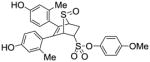 11b (26%) |
|
| 8 |
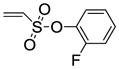 8c |
59 |
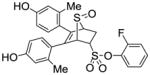 11c (40%) |
|
| 9 |
 8d |
57 |
 11d (31%) |
|
| 10 |
 8e |
57 |
 11e (35%) |
|
| 11 |
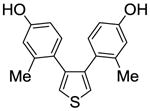 1b |
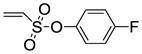 8f |
54 |
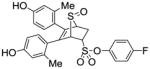 11f (28%) |
| 12 |
 8g |
57 |
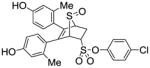 11g (28%) |
|
| 13 |
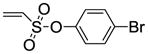 8h |
54 |
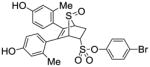 11h (32%) |
|
| 14 |
 8i |
56 |
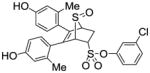 11i (28%) |
|
| 15 |
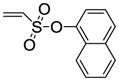 8j |
55 |
 11j (33%) |
|
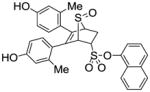
| 16 |
 8k |
64 |
 11k (24%) |
|
| 17 |
 8l |
60 |
 11l (34%) |
|
| 18 |
 1c |
 8a |
56 |
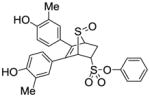 12a (50%) |
| 19 |
8b |
59 |
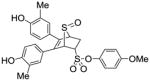 12b (27%) |
|
| 20 |
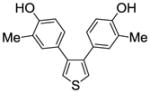 1c |
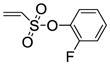 8c |
54 |
 12c (32%) |
| 21 |
 8d |
53 |
 12d (34%) |
|
| 22 |
 8e |
58 |
 12e (32%) |
|
| 23 |
 8j |
56 |
 12f (37%) |
|
| 24 |
 8k |
59 |
 12g (15%) |
|
| 25 |
 8l |
58 |
 12h (39%) |
|
| 26 |
 8m |
59 |
 12i (18%) |
|
| 27 |
 1d |
 8k |
59 |
 13 (55%) |
| 28 |
 1e |
 8k |
54 |
 14 (49%) |
| 29 |
 1f |
 8k |
59 |
 15 (35%) |
The conversion was calculated accounting for the recovered thiophene.
Isolated yield by column chromatography purification based on the thiophene consumed.
Despite the generally moderate yields (30–50%) that we obtained with the diphenolic thiophenes 1a–c and dienophiles, we found that the reaction of 1a–c with the ethenesulfonic acid 4-hydroxyphenyl ester 8k and diethyl maleate 8m gave the corresponding products in lower yields (15–32%) (Table 1, entries 3, 5, 16 and 24). Part of the reduced yield appears to be the sensitivity of the products to purification by silica gel chromatography. In comparison, compounds 1d–e, which have no hydroxyl group on the phenyl ring, reacted well with 8k, giving products 13 and 14 in 55%, and 49% yields, respectively (Table 1, entries 27 and 28).
Binding Affinity for Estrogen Receptors ERα and ERβ
The binding affinities of the SOBHS compounds for both ERα and ERβ were determined using a competitive radiometric assay and are reported in Table 2.46, 47 These affinities are presented as relative binding affinity (RBA) values, where estradiol has an affinity of 100%. At the start, it should be noted that comparisons between compounds in the SOBHS series, presented here, and the OBHS compounds, prepared earlier,26 are between SOBHS endo stereoisomers vs. OBHS exo stereoisomers, although in the one case we investigated previously, there was relatively little difference in the affinity between exo and endo OBHS isomers.26
As a global observation, it is notable first that members of the SOBHS class bind with somewhat lower affinity than the corresponding members of the OBHS class.26 Second, addition of a methyl group in the 5,6-substituted phenol rings, as well as the substituents at the 2 and 3-positions of the 7-thiabicycloheptane 7-oxide core have very significant effects on the binding affinity of the ligands. The series of compounds 11 that have an o-methyl group in both of the core phenyl substituents (o means adjacent to the attachment site to the bicyclic system; see Scheme 1) demonstrate a better binding affinity than the other two series (10 and 12). The compound that has the highest binding affinity for ERα is endo-phenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11a), a compound that possesses a p-hydroxyl group and o-methyl group in both of the core phenyl substituents and a phenyl sulfonate moiety at the 2-position of the bicyclic unit. The RBA values of this compound are 8.11 and 0.348 for ERα and ERβ, respectively (Table 2, entry 6), which is comparable to that of the best compounds we have reported in the original OBHS series.26 However, the compounds in the 12 series, which possesses a m-methyl group instead of an o-methyl group as in 11 series, show the highest ER subtype selective binding affinity. For example, compounds 12a and 12c, which possess a p-hydroxyl group and m-methyl group in both of the core phenyl substituents, have a ERα/ERβ selectivity as high as 249 and 248, respectively, which are the highest selectivity values among the 29 compounds, being more than 10 times greater than 11a and 11c (Table 2, entries 18 vs. 6, and 20 vs. 8). In fact, the ERα binding preference for these compounds approaches that of the most ERα selective ligand of which we are aware, propyl pyrazole triol (PPT), a compound on which we reported some time ago, though the absolute affinity of the SOBHS compounds for ERα is less than that of PPT.48
The position of the hydroxyl group is also of great importance. The hydroxyl group in the 5, 6-substituted phenyl rings is more important than in the phenyl sulfonate moiety at the 2-position, as can be surmised, to some extent, from a comparison of compounds 15 and 12a (Table 2, entries 29 and 18). As it is widely known, the presence of a phenolic ring in ER ligands is crucial to their binding affinity, as this ring is needed to mimic the steroidal “A ring” present in natural estrogens.49 This phenol forms important hydrogen bonds with residues Glu353 and Arg394, and a structured water molecule in ERα or the corresponding residues in ERβ.18 Therefore, this dependence of the RBA value on the position of the hydroxyl group suggests that the hydroxyl group in the 5,6-substituted phenyl rings is better positioned to establish these critical hydrogen bonds with the ERs. Consistent with this required phenolic ring feature, compounds 13 and 14, which lack phenolic hydroxyl groups on the core phenyls, show low affinities (Table 2, entries 27 and 28).
While one might imagine that a single core phenol group, as in compound 15 (Table 2, entry 29), might prove sufficient to engender good binding to the ERs, this is clearly not the case, nor was it the case in the OBHS series studied earlier.26 In the crystal structure of OBHS-like compounds in ERα, one of the core phenols is in the steroidal A-ring position, engaged in the crucial hydrogen bonds, but the second core phenol projects upward in the ligand pocket, roughly in a direction that corresponds to a steroidal 11β substituent.50 This places the second phenolic OH close to Thr347, which aside from Glu353 and Arg394, is the only other polar residue in the ligand-binding pocket. This is very likely an energetically productive interaction, as the second phenol in the bisphenolic ligands of the cyclofenil class, which also greatly enhances the affinity of these ligands, is thought to play the same role.28
In the ligands studied here, the substituents on the C-2 or C-3 position of the bridged core have a significant effect on binding affinity and selectivity. Because many non-steroidal estrogens are triphenols, the introduction of an additional hydroxyl group is also investigated; however, as was the case in the OBHS series,26 the placement of a methoxyl or hydroxyl group at the para position of the phenyl sulfonate ring caused a decrease in affinity for ERα (the trend is not clear for ERβ) and for ER subtype selectivity (Table 2, entries 2, 3, 7, 16, 19 and 24). In fact, the introduction of a third hydroxyl group results in a remarkable drop in affinity for ERα; only compound 11k still shows a moderate binding affinity for ERα; however, its affinity for ERβ increases (RBA ERα 2.21 and ERβ 0.812) (Table 2, entry 16). It is not clear why ERα and ERβ show different responses to these substituent alterations, and this phenomenon is different from that of the OBHS-core ligands.26
Compounds bearing halogens on the sulfonate phenyl group were also evaluated; however, all of them have decreased binding affinity for both ERα and ERβ. With the chlorinated compounds 11d, 11g and 11i (Table 2, entries 9, 12 and 14), the position of the substituent has little effect on binding affinity. For the para halogenated compounds 11f–h (Table 2, entries 11–13), the bromo compound (11h) seems to be superior to the other two. By contrast, for the o-halogenated compounds 11c–e and 12c–e (Table 2, entries 8–10 and 20–22), the fluorine-substituted compounds (11c and 12c) are the best. Other changes to the sulfonate moiety, such as replacing the phenyl with a naphthyl group, as in compound 11j and 12f (Table 2, entries 15 and 23), lower both binding affinity and subtype selectivity
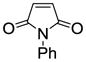
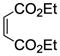
When other dienophiles, like diethyl maleate and N-phenylmaleimide, were used, the Diels-Alder adducts all gave very poor binding affinity and selectivity (compounds 10e and 12i, Table 2, entries 5 and 26, and compounds 10d, 11l and 12h, Table 2, entries 4, 17, and 25). The products from these dienophiles in the furan series also showed very low affinity.26
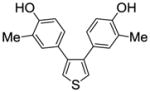
We also measured the ER binding affinities of the three 3,4-bisphenolic thiophenes (1a–c) used for the preparation of the three series of SOBHS compounds in this report (Table 2, entries 30–32). Comparison of the affinities of the three parent thiophenes with members of the three series of SOBHS compounds derived from them is interesting. First, incorporation of a thiophene into the 7-thiabicyclo[2.2.1]heptane 7-oxide phenyl sulfonate system in each case raises ERα binding affinity but lowers ERβ binding affinity (Table 2, entry 30 vs. 1; entry 31 vs. 6; entry 32 vs. 18). Very likely, this has to do with the smaller volume of the ligand-binding pocket in ERβ.17 Second, the highest affinity thiophene (compound 1b, Table 2, entry 31), which has two o-methyl groups, gives rise to, overall, the highest affinity SOBHS series (compounds 11, Table 2, entries 6–17); however, the lowest affinity thiophene (compound 1c, Table 2, entry 32), having two meta methyl groups, gives a series of SOBHS compounds that overall have higher affinity (compounds 12, Table 2, entries 18–26) than those derived from the unsubstituted thiophene (compounds 10, Table 2, entries 1–5). Thus, the sulfoxide bridge structure and other elements of the three-dimensional SOBHS ligand core design make strong contributions to the binding affinity and selectivity of the parent thiophene precursors. Further studies on many other members of the parent thiophene class will be described in a subsequent publication. Overall then, the disposition of the methyl group in the appended phenol substituents in the C-5 and C-6 positions of the bicyclic core unit, and the electron withdrawing group derived from the dienophiles, all prove to be factors in determining the binding affinity and selectivity of these novel SOHBS-core ligands for ERs.
Activation of ERα and ERβ Mediated Transcription
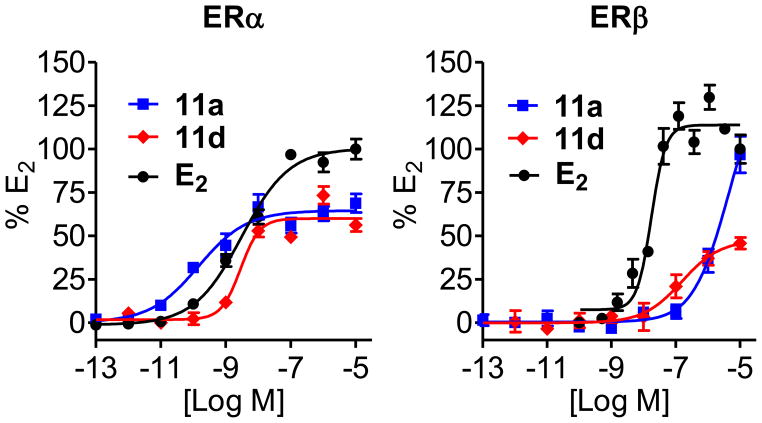
Various SOBHS compounds were tested by luciferase reporter gene assays for their ability to stimulate the transcriptional activities of ERα and ERβ compared to 17β-estradiol (E2). Luciferase assays were conducted in human liver cancer (HepG2) cells transfected with full-length human ERα or ERβ and a widely used estrogen-responsive element (ERE)-driven luciferase reporter.51 These results are summarized in Table 3, and dose-response curves for a few examples are shown in Figure 2.
Table 3.
Transcriptional Activities of 7-Thiabicyclic-7-oxide Analogs through ERα and ERβ
| Entry | Cmpd | ERα EC50 (nM)a | ERα (% E2) | ERβ EC50 (nM) | ERβ (% E2) |
|---|---|---|---|---|---|
| E2 | 2.2 | 100 ± 16 | 11 | 100 ± 6 | |
| 1 | 10a | 670 | 85 ± 20 | - | 3.4 ± 2 |
| 2 | 10b | - | 48 ± 0.2 | - | 19 ± 9 |
| 3 | 10c | 1100 | 48 ± 4 | - | 10 ± 3 |
| 4 | 10d | 7100 | 31 ± 4 | - | 24 ± 9 |
| 5 | 10e | - | 55 ± 8 | - | 27 ± 3 |
| 6 | 11a | 0.14 | 62 ± 7 | 4400 | 66 ± 3 |
| 7 | 11b | 210 | 76 ± 7 | - | 32 ± 7 |
| 8 | 11c | 2.2 | 56 ± 4 | - | 54 ± 2 |
| 9 | 11d | 2.8 | 73 ± 5 | 160 | 46 ± 3 |
| 10 | 11e | 14 | 67 ± 7 | - | 21 ± 2 |
| 11 | 11f | 200 | 74 ± 10 | - | 45 ± 11 |
| 12 | 11g | 110 | 67 ± 5 | - | 50 ± 10 |
| 13 | 11h | 110 | 71 ± 9 | 760 | 100 ± 10 |
| 14 | 11i | n.d. | n.d. | n.d. | n.d. |
| 15 | 11j | 27 | 59 ± 7 | - | 83 ± 3 |
| 16 | 11k | 9.2 | 58 ± 6 | 14000 | 82 ± 6 |
| 17 | 11l | 150 | 65 ± 5 | 800 | 82 ± 20 |
| 18 | 12a | 12 | 65 ± 6 | - | 3.8 ± 1 |
| 19 | 12b | 11000 | 57 ± 4 | - | 9.8 ± 2 |
| 20 | 12c | 1300 | 55 ± 3 | - | 0 |
| 21 | 12d | 2000 | 72 ± 7 | - | 0 |
| 22 | 12e | 1200 | 51 ± 6 | - | 0 |
| 23 | 12f | 770 | 42 ± 2 | - | 10 ± 2 |
| 24 | 12g | n.d. | n.d. | n.d. | n.d. |
| 25 | 12h | - | 29 ± 3 | - | 3.4 ± 2 |
| 26 | 12i | - | 22 ± 3 | - | 3.6 ± 1 |
| 27 | 13 | - | 22 ± 2 | - | 0 |
| 28 | 14 | 5500 | 24 ± 2 | - | 10 ± 4 |
| 29 | 15 | 340 | 32 ± 6 | - | 38 ± 3 |
Luciferase activity was measured in HepG2 cells transfected with 3X-ERE-driven luciferase reporter and expression vectors encoding ERα or ERβ and treated in triplicate with increasing doses (up to 10−5 M) of the compounds. EC50 and average efficacy (mean ± S.E.M.), shown as a percentage of 10−5 M 17β-estradiol (E2), were determined. Effects of 11i and 12g were not determined (n.d.). Omitted EC50 values were too high, while omitted %E2 values were too low to be determined accurately.
Figure 2.
Illustrative dose-response curves for estradiol and two sulfoxide-bridged SOBHS compounds in ERα and ERβ reporter gene assays in HepG2 cells. For details, see experimental section.
Compound 10a, which showed approximately 100-fold weaker binding affinity for ERα than E2 (Table 2, entry 1), stimulated ERα activity with about 300-fold weaker potency than E2, but near full efficacy (Table 3). Within this compound-10 scaffold, modifications of the phenyl sulfonate moiety (i.e., compounds 10a–e, entries 1–5) further reduced the potency and efficacy with which the compounds stimulate ERα activity (Table 3), and the binding affinity for ERα (Table 2).
Compounds having a p-hydroxyl group on the phenyl sulfonate moiety or lacking p-hydroxyl groups on both phenyl substituents, which decreased their binding affinity (Table 2, 10c and 13–15, entries 3, 27–29), also show decreased potency and efficacy as ERα agonists (Table 3). Interestingly, introducing p-hydroxyl and m-methyl groups to one of the phenyl substituents (i.e., compound 15) improved binding affinity (Table 2, entry 29), as well as potency as ER agonists (Table 3).
Compound 11a, which has an o-methyl group in both of the core hydroxyphenyl substituents and about 8-fold improved binding affinity for ERα (Table 2, entry 6), demonstrated about 5,000-fold higher potency but reduced efficacy as an ERα agonist (Figure 2), compared to compound 10a (entry 1) (Table 3). Modifications of the phenyl sulfonate moiety within this compound-11 scaffold (i.e., compounds 11a–l), further reduced potency (Table 3) and affinity (Table 2, entries 6–17) as ERα agonists.
Unlike OBHS or compound 10a (entry 1), both of which do not activate ERβ, compound 11a (entry 6) showed about 3-fold improved binding affinity for ERβ compared to 10a (Table 2), and stimulated ERβ activity, albeit with about 30,000-fold less potency than ERα (Table 3). As ERβ agonists, a 1-naphthyl modification of the phenyl sulfonate moiety (i.e., compound 11j, entry 15) improved efficacy of the compound-11 scaffold (Table 3). Furthermore, introduction of a p-bromine atom to the phenyl sulfonate moiety (i.e., compound 11h) improved ERβ binding affinity as effectively as 11a (Table 2, entries 13 and 6), and increased ERβ agonist efficacy with about 6-fold more potency than 11a (Table 3). In addition, the N-phenylmaleimide adduct, compound 11l, which showed about 24-fold weaker affinity for ERβ (Table 2, entry 17), was more efficacious and at least 5-fold more potent as an ERβ agonist than compound 11a. Therefore, the ability of compounds bearing the compound-11 scaffold to stimulate ERβ activity does not correlate with their relative binding affinities for ERβ.
By contrast, compound 12a which has a m-methyl group in each of the core hydroxyphenyl substituents and shows improved binding affinity for ERα but not ERβ (Table 2, entry 18), exhibits the typical ERα-selective agonist properties of the SOBHS scaffold, with 56-fold higher potency than compound 10a (entry 1)(Table 3). Modifying the phenyl sulfonate moiety within the compound-12 scaffold (i.e., compounds 12a-I, entry 18–26) reduced binding affinity for ERα (Table 2), as well as potency as ERα agonists to various extents (Table 3).
Structure-Activity Relationships
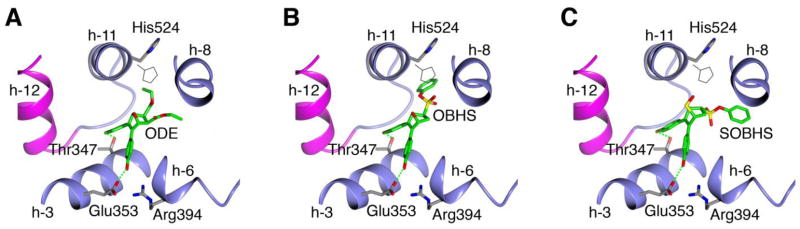
Crystal structures of ERα LBD in complex with oxabicyclic compounds (PBD ID: 2QH6 & 2QR9)50 show that one p-hydroxyphenyl group attached to the oxabicyclic core engages in hydrogen bonding with Glu353 and Arg394, while the other p-hydroxyphenyl group forms a hydrogen bond with Thr347. The ethyl ester moieties of oxabicyclic diethyl ester (ODE, i.e., diethyl 5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene- 2,3-dicarboxylate), which are attached to the oxabicyclic core in the exo position, are accommodated in the pocket at least in part by displacing helix-11 residues, including His524, which engages in hydrogen bonding with 17β-estradiol (E2) (PDB ID: 1ERE) (see the two indicated positions for His524 in Figure 3A).18 Displacement of helix-11 towards the dimer interface is associated with reduced ERα transcriptional activity, a mechanism sometimes termed “passive antagonism,” suggesting a suboptimal ERα LBD conformation for full agonist activity.50, 52
Figure 3.
Structure of ODE and Models of OBHS and SOBHS Binding within the ERα LBD. (A) Crystal structure of transcriptionally active ERα LBD in complex with ODE (PDB ID: 2QH6)50 showing ligand orientation relative to helices 3, 6, 8, 11 and 12 (purple). The ODE hydroxyphenyl groups form hydrogen bonds with Thr347, Glu353 and Arg394, while the exo ethyl ester moiety displaces His524 (shown in bold), compared to its position in the estradiol-bound LBD structure (PDB ID: 1ERE) (gray). (B) Model of OBHS binding within the ERα LBD. Like the ethyl ester moiety of ODE, the exo phenyl sulfonate moiety of OBHS clashes with helix-11 residues including His524. (C) Model of SOBHS binding within the ERα LBD. The endo phenyl sulfonate moiety of SOBHS is accommodated in a different region of the pocket, avoiding the clash with helix-11.
Crystal structures of the ER LBD in complex with OBHS or SOBHS have not yet been reported; therefore, the exact orientation of their phenyl sulfonate moieties in the ER ligand-binding pocket is unclear. Molecular model of OBHS bound to the ERα LBD suggests that its exo phenyl sulfonate moiety will displace helix-11 residues such as His524, at least as severely as the exo ethyl ester moiety of ODE (Figure 3B) and consistent with its greater antagonistic activity. In contrast, the endo phenyl sulfonate moiety of SOBHS is likely accommodated in a different space within the pocket, and therefore is not expected to displace helix-11 as extensively (Figure 3C). Consistent with these models, SOBHS compound 10a, which lacks ERα antagonist activity, is an effective ERα agonist (Table 3), compared to OBHS, which is a potent ERα antagonist.26
An examination of the ODE structure suggests that the o- versus m-methyl substitutions differentially impact how the ligand interacts with the receptor (Figure 3A). The m-methyl groups of compound 12a cannot be accommodated in the pocket without shifting the ligand further away from the hydrogen bonding partners, Glu353, Arg394 and/or Thr347, consistent with the lower affinity and transactivation potency of these compounds. This shift in the ligand is transmitted to the sulfonate phenyl substitution, where it could further shift helix 11 out the position required for agonist activity. This is most apparent with the ERβ compound 12 series, which display very little agonist activity. These differences in the ER subtypes are consistent with our previous findings that ERβ has a smaller pocket, and is more sensitive to ligand induced shifts in helix 11.53 In contrast, the o-methyl substitutions are positioned to make additional hydrophobic contacts, consistent with the higher affinity of these compounds in series 11.
Another factor could be contributing to the increased affinity of the series 11 compounds compared to their unsubstituted analogs (series 10). In an indene system that we studied earlier, we noted that addition of an o-methyl group in a cis-stilbene core structure also raised binding affinity to a considerable degree; we presumed that this was due to an increased twist of the aryl upon methyl substitution, which would increase the molecular surface area.54 Here, we note that according to simple MM2 energy minimization, addition of an o-methyl group to the two hydroxyphenyl groups increases the aryl dihedral angles from ca. 18° in 10a to 30° in 11a, which likely again is responsible for the increased affinity.
Conclusion
To further explore the consequences of expanding ligands for the ERs in the third dimension in terms of ER binding affinity and selectivity, and cellular activity, we have prepared a series of novel ligands for these receptors based on an inherently three-dimensional 7-thiabicyclo[2.2.1]hept-5-ene-7-oxide (SOBHS) core, analogs of the oxa-bridged (7-oxabicyclo[2.2.1]hept-5-ene) OBHS compounds. Ligands in this sulfoxide-bridged series can be readily prepared by an in situ oxidative Diels-Alder reaction between a 3,4-disubstituted thiophene and various dienophiles, which produces exclusively the endo stereoisomers. Careful SAR analysis of ER binding affinities and transcriptional output showed that these novel ligands are largely ERα-selective, and the disposition of methyl groups in the appended phenol substituents has a marked effect on their ER binding affinity and subtype selectivity. The compounds with o-substituted methyl groups show increased binding affinity for ERα, while those in the m-substituted methyl series show significantly enhanced ERα subtype binding-selectivity. The compounds with o-substituted methyl groups also exhibit partial ERβ agonist activity in transcription assays.
Lastly, ER remains an important pharmaceutical target, and the diversity associated with ER ligands provides a strategic platform to improve our understanding of how biological information is encoded within ligand structure and transmitted through ER. Generation of this new series of ER ligands provides key insight into the diversity of structures that can function as ER ligands and specifically as SERMs. Further cellular and in vivo studies on members of this new class of ER ligands, as well as X-ray crystallographic analyses, which are underway, will be reported in due course.
Experimental Section
Materials and Methods
Unless otherwise noted, reagents and materials were obtained from commercial suppliers and were used without further purification. Tetrahydrofuran and toluene were dried over Na and distilled prior to use. Dichloromethane was dried over CaH2 and distilled prior to use. Glassware was oven-dried, assembled while hot, and cooled under an inert atmosphere. Unless otherwise noted, all reactions were conducted in an inert atmosphere. Reaction progress was monitored using analytical thin-layer chromatography (TLC). Visualization was achieved by UV light (254 nm). 1H NMR and 13C NMR spectra were obtain on Bruker Biospin AV400 (400 MHz) instrument. The chemical shifts are reported in ppm and are referenced to either tetramethylsilane or the solvent. Mass spectra were recorded under electron impact conditions at 70 eV. Melting points were obtained on SGW X-4 melting point apparatus and are uncorrected. The purity of all compounds for biological testing was determined by HPLC (see Supporting Information), confirming >95% purity.
General Procedure for Boronic Acid Synthesis
n-BuLi (2.5 M, 2 equiv) was added dropwise to a solution of bromobenzene derivative in THF at −78 °C, resulting in a white suspension. The reaction mixture was stirred for 30 min at −78 °C, then B(OMe)3 (4 equiv) was added. The resulting mixture was stirred at −78 °C for a further 30 min and then allowed to warm to rt. The reaction mixture was acidified with 10% aq. HCl solution and extracted with EtOAc (3 × 30 mL). The organic layer was then dried over anhydrous MgSO4 and concentrated under vacuum. The crude product was purified by chromatography to afford an off-white crystalline material.
General Procedure for Suzuki Coupling Reaction
A solution of deoxygenated toluene was added to a mixture of Pd(OAc)2 (5% mol) (Pd2(dba)3 is used in synthesis of 5b) and PPh3 (25% mol) and stirred at an atmosphere of argon for 15 min at room temperature. Then arylboronic acid (4 equiv) was added to the reaction mixture and stirred for 5 min. A deoxygenated 3,4-dibromothiophene (1 equiv) was added to the mixture and stirred for a further 5 min. A deoxygenated 2M Na2CO3 solution was added to the reaction mixture that was stirred at room temperature for a further 5 min before being heated at reflux for 40 h. The mixture was cooled to rt. and quenched with H2O after which the organic material was extracted with EtOAc (3 × 30 mL), dried over anhydrous MgSO4 and the solvent was evaporated under vacuum. The crude product was subjected to column chromatography and recrystallized in ether to afford the target product.
3,4-Bis(4-methoxyphenyl)thiophene (5a)
Obtained as a white solid (76% yield); 1H NMR (400 MHz, CDCl3) δ 7.23 (s, 2H), 7.12 (d, J = 8.8 Hz, 4H), 6.80 (d, J = 8.8 Hz, 4H), 3.80 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 158.57, 141.32, 130.10, 129.22, 123.08, 113.57, 55.22. MS (ESI) m/z: 297 (M+1)+.
3,4-Bis(4-methoxy-2-methylphenyl)thiophene (5b)
Obtained as a white solid (69% yield); 1H NMR (400 MHz, CDCl3) δ 7.15 (s, 2H), 6.94 (d, J = 8.4 Hz, 2H), 6.64 – 6.58 (m, 4H), 3.75 (s, 6H), 2.03 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 158.46, 141.98, 137.67, 131.61, 128.98, 123.26, 115.26, 110.62, 55.08, 20.52. MS (ESI) m/z: 325 (M+1)+.
3,4-Bis(4-methoxy-3-methylphenyl)thiophene (5c)
Obtained as a white solid (81% yield); 1H NMR (400 MHz, CDCl3) δ 7.21 (s, 2H), 7.05 (s, 2H), 6.93 (dd, J = 8.4, 2.1 Hz, 2H), 6.69 (d, J = 8.4 Hz, 2H), 3.81 (s, 6H), 2.16 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 154.56, 139.23, 130.98, 129.02, 125.14, 124.46, 120.58, 107.17, 53.00, 13.98. MS (ESI) m/z: 325 (M+1)+.
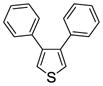
3,4-Diphenylthiophene (1d)
Obtained as a white solid (89% yield) (mp 105–106 °C); 1H NMR (400 MHz, CDCl3) δ 7.25 (dd, J = 5.0, 1.6 Hz, 6H), 7.20 – 7.17 (m, 4H), as previously reported.55 MS (ESI) m/z: 237 (M+1)+.
3,4-Di-p-tolylthiophene (1e)
Obtained as a white solid (84% yield) (mp 68–69 °C); 1H NMR (400 MHz, CDCl3) δ 7.26 (s, 2H), 7.08 (dd, J = 12.1, 8.0 Hz, 8H), 2.33 (s, 6H). MS (ESI) m/z: 265 (M+1)+.
3-(4-Methoxy-3-methylphenyl)-4-phenylthiophene (7)
Obtained as a white solid (73% yield); 1H NMR (400 MHz, CDCl3) δ 7.32–7.23 (m, 7H), 7.05 (d, J = 1.9 Hz, 1H), 6.95 (q, J = 2.2 Hz, 1H), 6.72 (d, J = 8.4 Hz, 1H), 3.84 (s, 3H), 2.18 (s, 3H); 13C NMR δ 156.85, 141.74, 141.61, 136.85, 131.37, 129.06, 128.61, 128.13, 127.45, 126.82, 126.25, 123.88, 123.08, 109.52, 55.30, 16.25. MS (ESI) m/z: 281 (M+1)+.
General Procedure for Demethylation
To a solution of 3,4-diarylthiophene (1 equiv) in dry CH2Cl2 at 0 °C, BBr3 in CH2Cl2 (1 M, 3 equiv per methoxyl function) was added dropwise. The reaction mixture was stirred for 24 h at room temperature under argon. Water was added to quench the reaction, and the aqueous layer was extracted with EtOAc. The combined organic layers were washed with brine, dried over Na2SO4, evaporated to dryness under vacuum, and purified by column chromatography.
3,4-Bis(4-hydroxyphenyl)thiophene (1a)
Obtained as a white solid (92% yield) (mp 198–201 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.41 (s, 2H), 7.40 (s, 2H), 6.93 (d, J = 8.5 Hz, 4H), 6.65 (d, J = 8.5 Hz, 4H); 13C NMR (100 MHz, DMSO-d6) δ 156.14, 140.94, 129.69, 126.98, 123.11, 114.87. MS (ESI) m/z: 269 (M+1)+.
3,4-Bis(4-hydroxy-2-methylphenyl)thiophene (1b)
Obtained as a white solid (95% yield) (mp 214–215 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.28 (s, 2H), 7.41 (s, 2H), 6.80 (d, J = 8.2 Hz, 2H), 6.57 (s, 2H), 6.50 (d, J = 8.2 Hz, 2H), 1.98 (s, 6H); 13C NMR (100 MHz, DMSO-d6) δ 156.09, 141.68, 136.75, 131.19, 126.97, 123.24, 116.38, 112.20, 20.00. MS (ESI) m/z: 319 (M + Na)+.
3,4-Bis(4-hydroxy-3-methylphenyl)thiophene (1c)
Obtained as a white solid (93% yield) (mp 183–184 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.32 (s, 2H), 7.38 (s, 2H), 6.95 (s, 2H), 6.67 (dd, J = 21.5, 8.2 Hz, 4H), 2.06 (s, 6H); 13C NMR (100 MHz, DMSO-d6) δ 154.66, 141.57, 131.36, 127.44, 123.81, 114.48, 16.47. MS (ESI) m/z: 318 (M + Na)+.
3-(4-Hydroxy-3-methylphenyl)-4-phenylthiophene (1f)
Obtained as a white solid (90% yield) (mp 114–115 °C); 1H NMR (400 MHz, DMSO-d6) δ 7.29-7.19 (m, 7H), 7.01 (d, J = 1.6 Hz, 1H), 6.83 (dd, J = 8.2, 2.1 Hz, 1H), 6.63 (d, J = 8.2 Hz, 1H), 4.66 (s, 1H), 2.18 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 152.86, 141.70, 141.47 136.74, 131.67, 129.21, 129.01, 127.83, 126.81, 123.83, 123.42, 123.07, 114.60, 15.70. MS (ESI) m/z: 267 (M+1)+.
General Procedure for Diels-Alder Reaction
To a solution of 3,4-diarylthiophene and dienophile (2 equiv) in dry CH2Cl2 (10 mL) was added slowly BF3·Et2O (10 equiv) under an inert atmosphere and at −20 °C. The reaction mixture was stirred for 10 min at −20 °C, and then a solution of m-CPBA (2 equiv) in dry CH2Cl2 (5 mL) was added slowly. The reaction mixture was stirred for 4 h at −20 °C. Then the suspension was poured into a mixture of conc. aqueous NaHCO3 solution (25 mL) and CH2Cl2 (25 mL) and stirred at room temperature for 20 min. The organic phase was separated, and the aqueous phase was extracted with EtOAc (3 × 30 mL). The combined organic phase was washed with water and brine and dried over anhydrous MgSO4. After removal of the solvent under vacuum, the residue was chromatographed on silica gel to give the target compound.
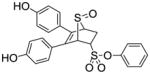
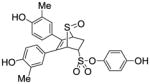
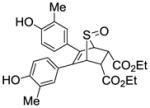
Phenyl-5,6-bis(4-hydroxyphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (10a)
Obtained as a white solid (36% yield) (mp 206–207 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.70 (s, 1H), 9.65 (s, 1H), 7.50 (t, J = 7.8 Hz, 2H), 7.41 (t, J = 7.3 Hz, 1H), 7.33 (d, J = 7.8 Hz, 2H), 7.08 (dd, J = 8.4, 5.5 Hz, 4H), 6.65 (dd, J = 20.1, 8.6 Hz, 4H), 4.74 (s, 1H), 4.59 – 4.55 (m, 1H), 4.39 (s, 1H), 2.96 (ddd, J = 13.2, 9.6, 3.5 Hz, 1H), 2.36 (dd, J = 13.5, 4.3 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 157.39, 157.12, 148.58, 132.97, 130.26, 129.82, 129.50, 129.05, 127.64, 125.09, 124.83, 122.13, 115.33, 114.93, 67.43, 67.34, 58.50, 26.67. HRMS (ESI) calcd for C24H20O6S2H [M + H]+ 469.0780; found 469.0777.
4-Methoxyphenyl-5,6-bis(4-hydroxyphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (10b)
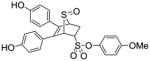
Obtained as a white solid (36% yield) (mp 225–226 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.70 (s, 1H), 9.65 (s, 1H), 7.27 (d, J = 9.1, 2H), 7.08 (dd, J = 8.6, 5.5 Hz, 4H), 7.02 (d, J = 9.2 Hz, 2H), 6.65 (dd, J = 20.0, 8.7Hz, 4H), 4.72 (s, 1H), 4.54 (ddd, J = 9.6, 4.3, 3.8 Hz, 1H), 4.38 (s, 1H), 2.94 (ddd, J = 13.0, 9.4, 3.5 Hz, 1H), 2.33 (dd, J = 13.4, 4.7 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 175.34, 132.37, 132.15, 131.98, 131.19, 129.99, 129.50, 129.11, 128.98, 128.79, 127.03, 115.80, 99.98, 68.87, 65.50, 56.48, 46.62, 30.46. HRMS (ESI) calcd for C25H22NaO7S2 [M + Na]+ 521.0705; found 521.0679.
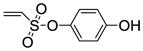
4-Hydroxyphenyl-5,6-bis(4-hydroxyphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (10c)
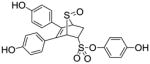
Obtained as a white solid (32% yield) (mp 235–236 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.81 (s, 1H), 9.68 (s, 1H), 9.63 (s, 1H), 7.13 (d, J = 9.0 Hz, 2H), 7.09 – 7.06 (m, 4H), 6.81 (d, J = 9.0 Hz, 2H), 6.64 (dd, J = 20.5, 8.6 Hz, 4H), 6.70 (s, 1H), 4.50 (t, J = 3.8 Hz, 1H), 4.38 (s, 1H), 2.92 (ddd, J = 13.3, 9.8, 3.6 Hz, 1H), 2.32 (dd, J = 13.4, 4.5Hz). 13C NMR (100 MHz, DMSO-d6) δ 167.45, 141.26, 133.48, 132.17, 131.99, 130.33, 129.12, 125.69, 123.61, 116.58, 115.75, 68.00, 65.50, 58.40, 56.43, 30.46, 19.10. HRMS (ESI) calcd for C24H20NaO7S2 [M + Na]+ 507.0548; found 507.0564.
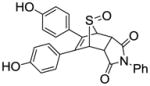
N-Phenyl-5,6-bis(4-hydroxyphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2,3-dicarboxamide-7-oxide (10d)
Obtained as a white solid (36% yield) (mp 264–265 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.74 (s, 2H), 7.52 – 7.33 (m, 3H), 7.02 (d, J = 8.7 Hz, 5H), 6.64 (d, J = 8.8 Hz, 4H), 4.76 (dd, J = 2.8, 1.7 Hz, 2H), 4.21 (dd, J = 2.7, 1.7 Hz, 2H); 13C NMR (100 MHz, DMSO-d6) δ 170.56, 167.43, 131.97, 129.11, 126.42, 99.98, 69.45, 65.50, 61.01, 56.46. HRMS (ESI) calcd for C26H19NNaO5S [M + Na]+ 480.0882; found 480.0893.
Diethyl-5,6-bis(4-hydroxyphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2,3-dicarboxylate-7-oxide (10e)
Obtained as a yellow solid (21% yield) (mp 193–194 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.60 (s, 2H), 7.07 (d, J = 8.8 Hz, 4H), 6.62 (d, J = 8.8 Hz, 4H), 4.46 (t, J = 1.6 Hz, 2H), 3.94 – 3.86 (m, 6H), 0.96 (t, J = 7.1 Hz, 6H); 13C NMR (100 MHz, DMSO-d6) δ 158.60, 142.36, 130.00, 129.56, 129.11, 125.32, 123.70, 115.49, 71.17, 65.52, 56.00, 18.95. HRMS (ESI): calcd for C24H24NaO7S [M + Na]+ 479.1140, found 479.1156.
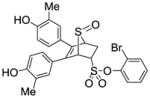
Phenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11a)
Obtained as a white solid (41% yield) (mp 275–276 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.36 (s, 1H), 9.31(s, 1H), 7.43 (t, J = 7.6 Hz, 2H), 7.34 (t, J = 7.3 Hz, 1H), 7.26 (d, J = 7.8 Hz, 2H), 6.90 (d, J = 8.3 Hz, 1H), 6.64 (d, J = 8.4 Hz, 1H), 6.44 – 6.36 (m, 4H), 4.59 (s, 1H), 4.53 (dd, 1H), 4.19 (s, 1H), 2.95 (ddd, 1H), 2.51 (dd, J = 13.5, 4.6 Hz, 1H), 1.89 (s, 3H), 1.84 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 157.02, 156.74, 148.61, 137.22, 136.84, 132.70, 132.19, 130.84, 130.64, 130.41, 130.25, 128.78, 127.88, 125.76, 124.95, 122.09, 116.84, 112.67, 68.26, 68.02, 58.34, 26.73, 20.15. HRMS (ESI) calcd for C26H24NaO6S2 [M + H]+ 497.1103; found 497.10865.
4-Methoxyphenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11b)
Obtained as a white solid (26% yield) (mp 224–226 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.48 (s, 1H), 9.43 (s, 1H), 7.25 (d, J = 9.1 Hz, 2H), 7.00 (d, J = 9.1 Hz, 2H), 6.96 (d, J = 8.3 Hz, 1H), 6.71 (d, J = 8.2 Hz, 1H), 6.48 – 6.41 (m, 4H), 4.62 (s, 1H), 4.55 (dd, J = 7.2, 6.2 Hz, 1H), 4.24 (s, 1H), 3.77 (s, 3H), 3.00 (ddd, 1H), 2.56 (dd, J = 13.9, 4.7 Hz, 1H), 1.95 (s, 3H), 1.90 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 158.13, 157.01, 156.72, 141.93, 137.22, 136.83, 136.76, 132.27, 130.86, 130.40, 125.77, 124.98, 123.11, 116.86, 116.78, 115.02, 112.69, 68.31, 68.12, 58.03, 56.03, 55.54, 26.74, 20.11. HRMS (ESI) calcd for C27H26NaO7S2 [M + Na]+ 549.1018; found 549.1021.
2-Fluorophenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11c)
Obtained as a white solid (40% yield) (mp 210–212 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.46 (s, 1H), 9.40 (s, 1H), 7.51-7.41 (m, 3H), 7.30 (t, J = 7.6 Hz, 1H), 6.94 (d, J = 8.3 Hz, 1H), 6.71 (d, J = 8.3 Hz, 1H), 6.53 – 6.40 (m, 4H), 4.76-4.62 (m, 2H), 4.29 (s, 1H), 3.09 (t, J = 10.6 Hz, 1H), 2.63 (dd, J = 13.7, 3.6 Hz, 1H), 1.95 (s, 3H), 1.91 (s, 3H); 13C NMR (100 MHz, DMSO- d6) δ 157.06, 156.62, 155.05, 152.58, 137.42, 136.76, 135.66, 132.14, 130.39, 129.18, 125.72, 125.56, 124.85, 124.77, 117.48, 117.30, 116.88, 116.81, 112.71, 112.46, 68.35, 59.56, 56.01, 26.84, 20.10, 18.44. HRMS (ESI) calcd for C26H23FO6S2H [M + H]+ 515.0995; found 515.0998.
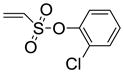
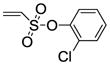
2-Chlorophenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11d)
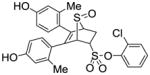
Obtained as a white solid (31% yield) (mp 248–249 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.47 (s, 1H), 9.41 (s, 1H), 7.68 (dd, J = 7.5, 1.8 Hz, 1H), 7.57 – 7.35 (m, 3H), 6.94 (d, J = 8.3 Hz, 1H), 6.71 (d, J = 8.3 Hz, 1H), 6.53 – 6.39 (m, 4H), 4.76 – 4.69 (m, 2H), 4.28 (s, 1H), 3.10 (ddd, J = 13.1, 9.7, 3.7 Hz, 1H), 2.68 (dd, J = 13.4, 3.8 Hz, 1H), 1.95 (s, 3H), 1.91 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 157.03, 156.91, 156.61, 144.40, 137.44, 136.81, 136.77, 132.16, 130.79, 130.40, 128.91, 128.87, 126.13, 125.74, 124.87, 124.27, 116.87, 116.72, 112.70, 112.55, 68.36, 68.15, 60.14, 27.00, 20.13, 19.82. HRMS (ESI) calcd for C26H23ClO6S2H [M + H]+ 531.0703; found 531.0695.
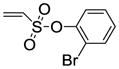
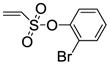
2-Bromophenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11e)
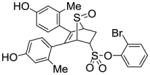
Obtained as a white solid (35% yield) (mp 246–247 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.47 (s, 1H), 9.40 (s, 1H), 7.81 (d, J = 7.9 Hz, 1H), 7.48 (q, J = 8.0 Hz, 2H), 7.38 – 7.29 (m, 1H) 6.94 (d, J = 8.4 Hz, 1H), 6.71 (d, J = 8.3 Hz, 1H), 6.55 – 6.39 (m, 4H), 4.80 – 4.68 (m, 2H), 4.28 (s, 1H), 3.10 (ddd, J = 12.9, 9.4, 3.4 Hz, 1H), 2.70 (dd, J = 13.5, 4.1 Hz, 1H), 1.95 (s, 3H), 1.91 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 157.06, 156.76, 156.61, 137.45, 136.77, 134.01, 132.17, 130.40, 129.45, 129.11, 125.74, 124.88, 124.00, 116.79, 115.46, 112.71, 112.56, 68.39, 68.19, 60.29, 27.08, 20.70, 19.82. HRMS (ESI) calcd for C26H23BrO6S2H [M + H]+ 575.0198; found 575.0199.
4-Fluorophenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11f)
Obtained as a white solid (28% yield) (mp 238–240 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.46 (s, 1H), 9.41 (s, 1H), 7.43 – 7.26 (m, 4H), 6.95 (d, J = 8.2 Hz, 1H), 6.71 (d, J = 8.3 Hz, 1H), 6.55 – 6.39 (m, 4H), 4.69 – 4.59 (m, 1H), 4.26 (s, 1H), 3.03 (ddd, 1H), 2.57 (dd, J = 13.6, 4.6 Hz, 1H), 1.94 (s, 1H), 1.91 (s,1H); 13C NMR (100 MHz, DMSO-d6) δ 162.27, 159.84, 157.52, 157.24, 145.07, 137.33, 137.27, 131.35, 125.42, 124.60, 117.47, 117.24, 113.19, 68.85, 68.58, 60.24, 59.07, 27.27, 20.61, 20.32, 14.52. HRMS (ESI) calcd for C26H23FNaO6S2 [M + Na]+ 537.0818; found 537.0818.
4-Chlorophenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11g)
Obtained as a white solid (28% yield) (mp 218–220 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.47 (s, 1H), 9.41 (s, 1H), 7.55 (d, J = 8.9 Hz, 2H), 7.36 (d, J = 8.9 Hz, 2H), 6.94 (d, J = 8.3 Hz, 1H), 6.71 (d, J = 8.3 Hz, 1H), 6.51–6.41 (m, 4H), 4.67 – 4.60 (m, 2H), 4.26 (s, 1H), 3.03 (ddd, J = 13.3, 9.9, 3.5 Hz, 1H), 2.58 (dd, J = 13.5, 4.0 Hz, 1H), 1.94 (s, 3H), 1.91 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 155.55, 153.08, 136.29, 136.16, 133.55, 131.25, 129.73, 129.26, 127.65, 127.33, 126.06, 125.52, 125.28, 124.36, 123.87, 117.99, 114.98, 114.66, 68.04, 67.93, 60.23, 27.28, 16.41, 16.29. HRMS (ESI) calcd for C26H23ClNaO6S2 [M + Na]+ 533.0522; found 553.0526.
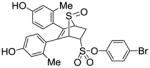
4-Bromophenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11h)
Obtained as a white solid (32% yield) (mp 209–210 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.46 (s, 1H), 9.41 (s, 1H), 7.68 (d, J = 8.9 Hz, 2H), 7.30 (d, J = 8.9 Hz, 2H), 6.93 (d, J = 8.4 Hz, 1H), 6.71 (d, J = 8.3 Hz, 1H), 6.52–6.40 (m, 4H), 4.68 – 4.61 (m, 2H), 4.26 (s, 1H), 3.03 (ddd, J = 13.8, 9.9, 4.1 Hz, 1H), 2.58 (dd, J = 13.5, 4.2 Hz, 1H), 1.94 (s, 3H), 1.91 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 157.53, 157.26, 148.24, 137.78, 137.33, 137.26, 133.57, 132.68, 130.89, 128.37, 126.25, 125.44, 124.84, 120.65, 117.57, 113.04, 68.85, 68.57, 59.33, 27.28, 20.61, 20.32. HRMS (ESI) calcd for C26H23BrNaO6S2 [M + Na]+ 597.0017; found 597.0008.
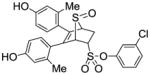
3-Chlorophenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11i)
Obtained as a white solid (28% yield) (mp 217–219 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.46 (s, 1H), 9.40 (s, 1H), 7.56 – 7.45 (m, 2H), 7.41 (s, 1H), 7.32 (d, J = 7.6 Hz, 1H), 6.93 (d, J = 8.4 Hz, 1H), 6.72 (d, J = 8.2 Hz, 1H), 6.56 – 6.38 (m, 4H), 4.76 – 4.62 (m, 1H), 4.27 (s, 1H), 3.05 (ddd, J = 12.5, 9.4, 3.2 Hz, 1H), 2.60 (dd, J = 13.7, 3.8 Hz, 1H), 1.94 (s, 3H), 1.92 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 156.89, 156.79, 148.83, 136.81, 133.91, 132.15, 131.54, 130.84, 130.41, 127.79, 122.46, 121.08, 116.84, 116.74, 112.69, 112.59, 112.54, 68.33, 68.04, 58.96, 26.72, 20.14, 19.85. HRMS (ESI) calcd for C26H23ClNaO6S2 [M + Na]+ 553.0522; found 553.0536.
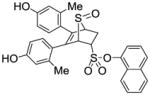
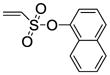
Naphthalen-1-yl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11j)
Obtained as a white solid (33% yield) (mp 273–275 °C); 1H NMR (400 MHz, DMSO) δ 9.60 (s, 1H), 9.55 (s, 1H), 8.06 – 8.00 (m, 2H), 7.96 (d, J = 8.0 Hz, 1H), 7.65 – 7.60 (m, 2H), 7.54 (dt, J = 15.4, 7.7 Hz, 2H), 7.06 (d, J = 15.9 Hz, 2H), 6.88 (ddd, J = 13.2, 8.3, 2.0 Hz, 2H), 6.61 (dd, J = 18.6, 8.4 Hz, 2H), 4.88 – 4.82 (m, 1H), 4.81 (s, 1H), 4.39 (s, 1H), 3.02 (ddd, J = 13.2, 9.6, 3.5 Hz, 1H), 2.01 (s, 3H), 1.95 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 155.44, 155.17, 144.21, 134.36, 132.87, 130.80, 130.42, 128.87, 127.92, 127.37, 127.29, 127.22, 127.11, 126.85, 126.62, 125.67, 123.92, 123.37, 121.35, 118.70, 114.44, 114.12, 80.68, 67.68, 59.57, 26.84, 15.93, 15.81. HRMS (ESI) calcd for C30H26O6S2H [M + H]+ 547.1249; found 547.1260.
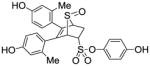
4-Hydroxyphenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (11k)
Obtained as a white solid (24% yield) (mp 240–241 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.86 (s, 1H), 9.49 (s, 1H), 9.43 (s, 1H), 7.17 (d, J = 6.8 Hz, 2H), 7.03 (d, J = 8.1 Hz, 1H), 6.87 (d, J = 6.9 Hz, 2H), 6.76 (d, J = 8.3 Hz, 1H), 6.56-6.48 (m, 4H), 4.67 (s, 1H), 4.59-4.66 (m, 1H), 4.29 (s, 1H), 3.03 (ddd, J = 13.3, 9.8, 3.6 Hz, 1H), 2.57-2.61 (m, 1H), 1.95 (s, 3H), 1.88 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 170.84, 167.44, 156.42, 141.24, 133.34, 132.17, 131.98, 131.32, 130.88, 129.48, 129.12, 127.70, 127.34, 125.66, 124.55, 123.60, 116.59, 114.88, 68.04, 67.90, 65.50, 30.46, 19.10, 18.94. HRMS (ESI) calcd for C26H24NaO7S2 [M + Na]+ 535.0861; found 535.0868.
N-Phenyl-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2,3-dicarboxamide-7-oxide (11l)
Obtained as a white solid (34% yield) (mp 274–276 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.53 (s, 2H), 7.53 (t, J = 7.6 Hz, 2H), 7.44 (t, J = 7.4 Hz, 1H), 7.21 (d, J = 7.5 Hz, 2H), 6.77 (d, J = 8.1 Hz, 2H), 6.48 (d, J = 9.2 Hz, 4H), 4.64 (s, 2H), 4.25 (s, 2H), 1.77 (s, 6H); 13C NMR (100 MHz, DMSO-d6) δ 174.97, 157.05, 136.63, 129.95, 128.99, 128.43, 126.43, 125.19, 117.13, 113.02, 69.35, 46.25, 19.89. HRMS (ESI) calcd for C28H23NNaO5S [M + Na]+ 508.1195; found 508.1168.
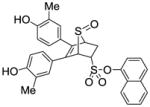
Phenyl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (12a)
Obtained as a white solid (50% yield) (mp 215–216 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.55 (s, 1H), 9.50 (s, 1H), 7.50 (t, J = 7.5 Hz, 2H), 7.40 (t, J = 7.4 Hz, 1H), 7.34 (d, J = 3.2 Hz, 1H), 7.04 (d, J = 6.7 Hz, 2H), 6.86 (t, J = 8.7 Hz, 2H), 6.62 (dd, J = 19.0, 8.4 Hz), 4.72 (s, 1H), 4.56(dd, 1H), 4.37 (s, 1H), 2.95 (ddd, 1H), 2.36(dd, J = 13.4, 4.6 Hz, 1H), 2.02 (s, 3H), 1.99 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 155.44, 155.17, 148.55, 132.82, 130.79, 130.38, 130.25, 128.86, 127.62, 127.20, 126.85, 125.05, 124.77, 123.90, 123.32, 122.12, 114.43, 114.09, 67.46, 67.39, 58.55, 30.66, 15.95, 15.85. HRMS (ESI) calcd for C26H24O6S2H [M + H]+ 497.1093; found 497.1103.
Methoxyphenyl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (12b)
Obtained as a white solid (27% yield) (mp 221–223 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.58 (s, 1H), 9.53 (s, 1H), 7.27 (d, J = 7.0 Hz, 2H), 7.03 (t, J = 6.8 Hz, 4H), 6.86 (t, J = 10.0 Hz, 2H), 6.62 (dd, J = 18.6, 8.4 Hz, 2H), 4.69 (s, 1H), 4.52 (dd, 1H), 4.36 (s, 1H), 2.94 (ddd, 1H), 2.33 (dd, J = 11.6, 4.5 Hz, 1H), 2.02 (s, 1H), 1.99 (s, 1H); 13C NMR (100 MHz, DMSO-d6) δ 158.60, 155.93, 155.64, 142.38, 138.80, 133.33, 131.29, 125.59, 125.30, 123.39, 123.81, 123.66, 115.50, 114.95, 114.61, 81.17, 68.03, 67.89, 56.50, 18.99, 16.46, 16.33. HRMS (ESI) calcd for C27H26NaO7S2 [M + Na]+ 549.1018; found 549.0998.
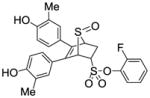
2-Fluorophenyl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (12c)
Obtained as a white solid (32% yield) (mp 203–204 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.58 (s, 1H), 9.53 (s,1H), 7.53 – 7.40 (m, 3H). 7.30 (t, J = 7.6 Hz, 1H), 7.03 (s, 2H), 6.86 (t, J = 8.2 Hz, 2H), 6.62 (dd, J = 19.7, 8.4 Hz, 2H), 4.68 – 4.63 (m, 1H), 4.39 (s, 1H), 3.01 (ddd, J = 13.2, 9.7, 3.3 Hz, 1H), 2.40 (dd, J = 13.4, 4.4 Hz, 1H), 2.02 (s, 3H), 1.98 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 157.54, 157.26, 157.12, 147.75, 137.78, 137.32, 137.25, 132.69, 132.42, 131.34, 130.90, 130.61, 126.28, 125.43, 124.49, 117.28, 113.20, 113.11, 113.03, 112.95, 68.85, 68.58, 59.32, 27.29, 20.62, 20.33. HRMS (ESI) calcd for C26H23FNaO6S2 [M + Na]+ 537.0818; found 537.0809.
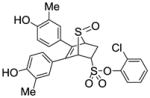
2-Chlorophenyl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (12d)
Obtained as a white solid (34% yield) (mp 219–220 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.58 (s, 1H), 9.53 (s,1H), 7.67 (d, J = 7.7 Hz, 1H), 7.56 – 7.37 (m, 3H), 7.04 (s, 2H), 6.87 (t, J = 8.2 Hz, 2H), 6.62 (dd, J = 20.7, 7.9 Hz, 2H), 4.77 (s, 1H), 4.74-4.68 (m, 1H), 4.39 (s, 1H), 3.03 (ddd, J = 13.0, 9.5, 3.4 Hz, 1H), 2.47 (dd,1H), 2.03 (s, 3H), 1.98 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 155.47, 155.18, 144.38, 133.04, 130.94, 130.76, 130.39, 128.87, 128.72, 127.17, 126.85, 126.13, 124.99, 124.95, 124.80, 124.76, 124.24, 123.91, 123.35, 123.28, 67.58, 67.45, 60.33, 26.90, 15.96, 15.83. HRMS (ESI) calcd for C26H23ClNaO6S2H [M + Na]+ 553.0522; found 553.0513.
2-Bromophenyl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (12e)
Obtained as a white solid (32% yield) (mp 216–217 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.58 (s, 1H), 9.52 (s, 1H), 7.81 (d, J = 7.8 Hz, 1H), 7.48 (q, J = 8.0 Hz, 2H), 7.37-7.27 (m, 1H), 7.04 (s, 2H), 6.86 (t, J = 7.3 Hz, 2H), 6.64 (dd, J = 8.3, 1.8 Hz, 1H), 6.59 (dd, J = 8.4, 1.7 Hz, 1H), 4.77 (s, 1H), 4.75-4.67 (m, 1H), 4.39 (s, 1H), 3.03 (ddd, J = 13.0, 9.5, 3.3 Hz, 1H), 2.48 (dd, 1H), 2.02 (s, 3H), 1.98 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 155.31, 155.18, 145.64, 134.02, 133.04, 130.76, 130.40, 129.46, 129.08, 128.74, 127.17, 126.86, 124.99, 124.77, 123.91, 123.34, 123.27, 67.59, 67.47, 60.45, 26.95, 15.96, 15.81. HRMS (ESI) calcd for C26H23BrNaO6S2H [M + Na]+ 579.0017; found 579.0028.
Naphthalen-1-yl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (12f)
Obtained as a white solid (37% yield) (mp 235–237 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.58 (s, 1H), 9.53 (s, 1H), 8.03 (t, J = 6.3 Hz, 2H), 7.97 (d, J = 7.7 Hz, 1H), 7.63 (t, J = 6.7 Hz, 2H), 7.58 – 7.51 (m, 2H), 7.06 (d, J = 13.7 Hz, 2H), 6.93 – 6.83 (m, 2H), 6.62 (dd, J = 18.5, 8.4 Hz, 2H), 4.89 – 4.83 (m, 1H), 4.82 (s, 1H), 4.40 (s, 1H), 3.02 (ddd, J = 13.1, 9.5, 3.3 Hz, 1H), 2.55 (d, J = 3.5 Hz, 1H), 2.01 (s, 3H), 1.96 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 155.87, 155.60, 144.59, 138.80, 134.81, 133.35, 131.26, 131.16, 130.88, 129.30, 129.23, 128.41, 127.80, 127.63, 127.31, 127.06, 126.14, 125.47, 125.27, 124.49, 123.95, 121.24, 119.19, 114.85, 68.11, 67.98, 60.03, 23.55, 16.41, 16.28. HRMS (ESI) calcd for C30H26O6S2H [M + H]+ 547.1249; found 547.1273.
4-Hydroxyphenyl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (12g)
Obtained as a white solid (15% yield) (mp 211–212 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.80 (s, 1H), 9.55 (s, 1H), 9.50 (s, 1H), 7.1 3(d, J = 8.8 Hz, 2H), 7.03 (d, J = 5.4 Hz, 2H), 6.89 – 6.80 (m, 4H), 6.62 (dd, J = 18.9, 8.2 Hz, 2H), 4.68 (s, 1H), 4.48 (dd, 1H), 4.35 (s, 1H), 2.93 (ddd, 1H), 2.34 (dd, 1H), 2.02 (s, 3H), 1.99 (s, 3H); 13C NMR (100 MHz, DMSO-d6) δ 157.37, 157.09, 156.85, 141.30, 137.71, 137.26, 133.20, 131.98, 130.90, 129.28, 129.12, 128.38, 126.32, 125.55, 123.57, 117.26, 116.66, 113.09, 68.79, 68.62, 65.50, 30.47, 20.63, 20.34. HRMS (ESI) calcd for C26H24NaO7S2 [M + Na]+ 535.0861; found 535.0837.
N-Phenyl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2,3-dicarboxamide-7-oxide (12h)
Obtained as a white solid (39% yield) (mp 260–261 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.62 (s, 2H), 7.47 – 7.38 (m, 3H), 7.03 – 6.97 (m, 4H), 6.84 (dd, J = 8.3, 2.0 Hz, 2H), 6.62 (d, J = 8.4 Hz, 2H), 4.74 (s, 2H), 4.19 (s, 1H), 1.98 (s, 6H); 13C NMR (100 MHz, DMSO-d6) δ 175.35, 156.02, 132.40, 131.05, 130.92, 129.47, 128.95, 127.35, 124.99, 124.29, 114.93, 68.90, 46.65, 19.00. HRMS (ESI) calcd for C28H23NNaO5S [M + Na]+ 508.1195; found 508.1179.
Diethyl-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2,3-dicarboxylate-7-oxide (12i)
Obtained as a white solid (18% yield) (mp 200–201 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.48 (s, 2H), 7.00 (s, 2H), 6.85 (dd, J = 8.3, 2.0 Hz, 2H), 6.59 (d, J = 8.4 Hz, 2H), 4.43 (s, 2H), 3.92 – 3.87 (m, 6H), 2.00 (s, 6H), 0.97 (t, J = 7.1 Hz, 6H); 13C NMR (100 MHz, DMSO-d6) δ 170.56, 155.47, 138.80, 131.62, 126.38, 123.75, 114.57, 81.17, 69.48, 60.98, 44.93, 18.99, 14.04. HRMS (ESI) calcd for C26H28NaO7S [M + Na]+ 507.1453; found 407.1464.
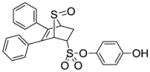
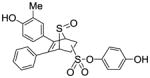
4-Hydroxyphenyl-5,6-diphenyl-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (13)
Obtained as a white solid (55% yield) (mp 194–195 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.86 (s, 1H), 7.33 – 7.21 (m, 9H), 7.14 (d, J =9.6 Hz, 2H), 6.83 (d, J = 8.9 Hz 2H), 4.86 (s, 1H), 4.59 (dd, 1H), 4.49 (s, 1H), 3.01 (ddd, J = 13.3, 9.7, 3.6 Hz, 1H), 2.43 (dd, J = 13.5, 4.4 Hz, 1H); 13C NMR (100 MHz, DMSO-d6) δ 156.54, 140.67, 135.49, 134.17, 134.12, 131.53, 128.60, 128.55, 128.39, 128.16, 128.13, 128.07, 123.11, 116.15, 67.55, 67.40, 57.62, 26.50. HRMS (ESI): calcd for C24H20NaO5S2 [M + Na] + 475.0650, found 475.0652.
4-Hydroxyphenyl-5,6-di-p-tolyl-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (14)
Obtained as a yellow solid (49% yield) (mp 164–165 °C); 1H NMR (400 MHz, DMSO-d6) δ 9.81 (s, 1H), 7.16 – 7.12 (m, 9H), 7.05 (d, J = 8.1 Hz, 2H), 6.81 (d, J = 8.9 Hz, 2H), 4.80 (s, 1H), 4.43-4.56 (m, 1H), 4.44 (s, 1H), 2.95 (ddd, J = 13.2, 9.8, 3.6 Hz, 1H), 2.39 (dd, J = 13.5, 4.4 Hz, 1H), 2.26 (dd, J = 6.6 Hz, 6H); 13C NMR (100 MHz, DMSO-d6) δ 165.98, 140.73, 137.85, 137.51, 134.66, 133.33, 132.72, 131.33, 130.64, 129.16, 128.41, 127.88, 123.11, 116.10, 67.52, 57.72, 26.53, 20.72. HRMS (ESI) calcd for C26H24NaO5S2 [M + Na]+ 503.0963; found 503.0963.
4-Hydroxyphenyl-6-(4-hydroxy-3-methylphenyl)-5-phenyl-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate-7-oxide (15, Mixture of 1:1 isomers)
Obtained as a brown solid (35% yield); 1H NMR (400 MHz, DMSO-d6) δ 9.84 (s, 1H), 9.65 (s, 0.5H), 9.60 (s, 0.5H), 7.72 (d, J = 7.8 Hz, 1H), 7.56 (t, J = 8.1 Hz, 1H), 7.33 – 7.21 (m, 4H), 7.14 (t, J = 8.6 Hz, 2H), 7.02 (s, 1H), 6.88 – 6.78 (m, 2H), 6.62 (dd, J = 20.2, 8.2 Hz, 1H), 4.78 (s, 1H), 4.54 (dd, J = 7.6, 2.0 Hz, 1H), 4.43 (s, 1H), 2.96 (t, J = 11.7 Hz, 1H), 2.42 – 2.29 (m, 1H), 2.01 (s, 1.5H), 1.97 (s, 1.5H); 13C NMR (100 MHz, DMSO-d6) δ 170.83, 167.44, 157.37, 157.09, 156.85, 141.30, 137.71, 137.34, 137.26, 133.20, 132.77, 131.98, 131.36, 131.13, 130.90, 129.28, 129.12, 128.38, 126.32, 125.55, 123.57, 117.26, 116.66, 113.09, 68.79, 68.62, 65.50, 60.23, 58.25, 30.47, 27.20, 20.63, 20.34, 19.11, 18.93. HRMS (ESI) calcd for C25H22NaO6S2 [M + Na]+ 505.0755; found 505.0763.
Estrogen Receptor Binding Affinity
Relative binding affinities were determined by a competitive radiometric binding assay, as previously described,46, 47 using 2 nM [3H]estradiol as tracer ([2,4,6,7-3H] estra-1,3,5(10)-triene-3,17-β-diol, 70–115 Ci/mmol, Perkin Elmer, Waltham, MA), and purified full-length human ERα and ERβ, purchased from PanVera/Invitrogen (Carlsbad, CA). Incubations were for 18–24 h at 0 °C. Hydroxyapatite (BioRad, Hercules, CA) was used to absorb the receptor-ligand complexes, and free ligand was washed away. The binding affinities are expressed as relative binding affinity (RBA) values with the RBA of estradiol set to 100%. The values given are the average ± range or SD of two to three independent determinations. Estradiol binds to ERα with a Kd of 0.2 nM and to ERβ with a Kd of 0.5 nM.
Luciferase Assay
Assays were performed as previously described with a few modifications.51, 56 HepG2 cells were cultured in growth media containing Dulbecco’s minimum essential medium (DMEM) (Cellgro by Mediatech, Inc. Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Hyclone by Thermo Scientific, South Logan, UT), and 1% non-essential amino acids (Cellgro), Penicillin-Streptomycin-Neomycin antibiotic mixture and Glutamax (Gibco by Invitrogen Corp. Carlsbad, CA), and maintained at 37 °C and 5% CO2. The cells were transfected with 10.0 ug of 3XERE-luciferase reporter plus 1.6 ug of ER expression vector per 10 cm dish using FugeneHD reagent (Roche Applied Sciences, Indianapolis IN). The next day, the cells were re-suspended in phenol red-free growth media containing 10% charcoal-dextran sulfate-treated FBS, transferred to 384-well plates at a density of 20,000 cells/well, incubated overnight at 37 °C and 5% CO2, and treated in triplicate with increasing doses of ER ligands. After 24 hours, luciferase activity was measured using BriteLite reagent (PerkingElmer Inc., Shelton, CT) according to manufacturer’s protocol.
Molecular Modeling
Crystal structures of ER LBD in complex with E2 and ODE were downloaded from the protein data bank (PDB IDs: 1ERE and 2QH6).57 OBHS or SOBHS was docked into the electron density of ODE using the molecular graphic program, COOT.58 The models were transferred to CCP4MG and superposed for presentation.59
Supplementary Material
Acknowledgments
We are grateful to the NSFC (20872116, 20972121, 91017005), the Program for New Century Excellent Talents in University (NCET-10-0625), the National Mega Project on Major Drug Development (2009ZX09301-014-1), and the Research Fund for the Doctoral Program of Higher Education of China (20100141110021) for support of this research. Research support from the National Institutes of Health (PHS R37 DK015556 to J.A.K. and R01 DK077085 to K.W.N.) is gratefully acknowledged.
Footnotes
Abbreviations: E2, estradiol; ER, estrogen receptor; HepG2, human liver cancer cells; RBA, relative binding affinity; OBHS, 7-oxabicyclo[2.2.1]hept-5-ene; ODE, diethyl 5,6-bis(4-hydroxyphenyl)- 7-oxabicyclo[2.2.1]hept-5-ene-2,3-dicarboxylate; SAR, structure-activity relationship; SERMs, selective estrogen receptor modulators; SOBHS, 7-thiabicyclo[2.2.1]hept-5-ene-7-oxide; THF, tetrahydrofuran; m-CPBA, m-chloroperbenzoic acid.
Supporting Information Available: HPLC results and HPLC spectra for compounds 10–14. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Stroem A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: how do they signal and what are their targets? Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 2.Gustafsson JA. What pharmacologists can learn from recent advances in estrogen signalling. Trends Pharmacol Sci. 2003;24:479–485. doi: 10.1016/S0165-6147(03)00229-3. [DOI] [PubMed] [Google Scholar]
- 3.Hess RA. Estrogen in the adult male reproductive tract: a review. Reprod Biol Endocrinol. 2003;1(52):1–14. doi: 10.1186/1477-7827-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syed F, Khosla S. Mechanisms of sex steroid effects on bone. Biochem Biophys Res Commun. 2005;328:688–696. doi: 10.1016/j.bbrc.2004.11.097. [DOI] [PubMed] [Google Scholar]
- 5.Mendelsohn ME. Protective Effects of Estrogen on the Cardiovascular System. Am J Cardiol. 2002;89(suppl):12E–18E. doi: 10.1016/s0002-9149(02)02405-0. [DOI] [PubMed] [Google Scholar]
- 6.Behl C. Oestrogen as a neuroprotective hormone. Nat Rev Neurosci. 2002;3:433–442. doi: 10.1038/nrn846. [DOI] [PubMed] [Google Scholar]
- 7.Carruba G. Estrogens and mechanisms of prostate cancer progression. Ann NY Acad Sci. 2006;1089:201–217. doi: 10.1196/annals.1386.027. [DOI] [PubMed] [Google Scholar]
- 8.Zumoff B. Does Postmenopausal Estrogen Administration Increase the Risk of Breast Cancer? Proc Soc Exp Biol Med. 1998;217:30–37. doi: 10.3181/00379727-217-44202. [DOI] [PubMed] [Google Scholar]
- 9.Vergote I, Neven P, vanDam P, Serreyn R, De Prins F, De Sutter P, Albertyn G. The estrogen receptor and its selective modulators in gynecological and breast cancer. Eur J Cancer. 2000;36:S1–S9. doi: 10.1016/s0959-8049(00)00203-3. [DOI] [PubMed] [Google Scholar]
- 10.Katzenellenbogen BS, Katzenellenbogen JA. Estrogen receptor transcription and transactivation: Estrogen receptor alpha and estrogen receptor beta: regulation by selective estrogen receptor modulators and importance in breast cancer. Breast Cancer Res. 2000;2:335–344. doi: 10.1186/bcr78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 12.Katzenellenbogen BS, Katzenellenbogen JA. Defining the “S” in SERMs. Science. 2002;295:2380–2381. doi: 10.1126/science.1070442. [DOI] [PubMed] [Google Scholar]
- 13.Zhou HB, Nettles KW, Bruning JB, Kim Y, Joachimiak A, Sharma S, Carlson KE, Stossi F, Katzenellenbogen BS, Greene GL, Katzenellenbogen JA. Elemental isomerism: A boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands. Chem Biol. 2007;14:659–669. doi: 10.1016/j.chembiol.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Zhou HB, Sheng SB, Compton DR, Kim Y, Joachimiak A, Sharma S, Carlson KE, Katzenellenbogen BS, Nettles KW, Greene GL, Katzenellenbogen JA. Structure-guided optimization of estrogen receptor binding affinity and antagonist potency of pyrazolopyrimidines with basic side chains. J Med Chem. 2007;50:399–403. doi: 10.1021/jm061035y. [DOI] [PubMed] [Google Scholar]
- 15.Zhou HB, Carlson KE, Stossi F, Katzenellenbogen BS, Katzenellenbogen JA. Analogs of methyl-piperidinopyrazole (MPP): Antiestrogens with estrogen receptor alpha selective activity. Bioorg Med Chem Lett. 2009;19:108–110. doi: 10.1016/j.bmcl.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nettles KW, Bruning JB, Gil G, O’Neill EE, Nowak J, Guo Y, Kim Y, DeSombre ER, Dilis R, Hanson RN, Joachimiak A, Greene GL. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 2007;8:563–568. doi: 10.1038/sj.embor.7400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engstrom O, Ljunggren J, Gustafsson JA, Carlquist M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 19.Top S, Vessieres A, Leclercq G, Quivy J, Tang J, Vaisserman J, Huche M, Jaouen G. Synthesis, biochemical properties and molecular modelling studies of organometallic specific estrogen receptor modulators (SERMs), the ferrocifens and hydroxyferrocifens: evidence for an antiproliferative effect of hydroxyferrocifens on both hormonedependent and hormone-independent breast cancer cell lines. Chem Eur J. 2003;9:5223–5236. doi: 10.1002/chem.200305024. [DOI] [PubMed] [Google Scholar]
- 20.Plazuk D, Vessieres A, Le Bideau F, Jaouen G, Zakrzewski J. Synthesis of benzyl- and benzhydrylferrocenes via Friedel-Crafts alkylation of ferrocene. Access to ferrocenyl bisphenols with high affinities for estrogen receptors. Tetrahedron Lett. 2004;45:5425–5427. [Google Scholar]
- 21.Pigeon P, Top S, Zekri O, Hillard EA, Vessieres A, Plamont MA, Buriez O, Labbe E, Huche M, Boutamine S, Amatore C, Jaouen G. The replacement of a phenol group by an aniline or acetanilide group enhances the cytotoxicity of 2-ferrocenyl-1,1-diphenyl-but-l-ene compounds against breast cancer cells. J Organomet Chem. 2009;694:895–901. [Google Scholar]
- 22.Causey PW, Besanger TR, Valliant JF. Synthesis and Screening of Mono- and Di-Aryl Technetium and Rhenium Metallocarboranes. A New Class of Probes for the Estrogen Receptor. J Med Chem. 2008;51:2833–2844. doi: 10.1021/jm701561e. [DOI] [PubMed] [Google Scholar]
- 23.Endo Y, Yoshimi T, Ohta K, Suzuki T, Ohta S. Potent Estrogen Receptor Ligands Based on Bisphenols with a Globular Hydrophobic Core. J Med Chem. 2005;48:3941–3944. doi: 10.1021/jm050195r. [DOI] [PubMed] [Google Scholar]
- 24.Yamakoshi Y, Otani Y, Fujii S, Endo Y. Dependence of estrogenic activity on the shape of the 4-alkyl substituent in simple phenols. Biol Pharm Bull. 2000;23:259–261. doi: 10.1248/bpb.23.259. [DOI] [PubMed] [Google Scholar]
- 25.Mull ES, Sattigeri VJ, Rodriguez AL, Katzenellenbogen JA. Aryl cyclopentadienyl tricarbonyl rhenium complexes: novel ligands for the estrogen receptor with potential use as estrogen radiopharmaceuticals. Bioorg Med Chem. 2002;10:1381–1398. doi: 10.1016/s0968-0896(01)00406-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhou HB, Comninos JS, Stossi F, Katzenellenbogen BS, Katzenellenbogen JA. Synthesis and Evaluation of Estrogen Receptor Ligands with Bridged Oxabicyclic Cores Containing a Diarylethylene Motif: Estrogen Antagonists of Unusual Structure. J Med Chem. 2005;48:7261–7274. doi: 10.1021/jm0506773. [DOI] [PubMed] [Google Scholar]
- 27.Muthyala RS, Carlson KE, Katzenellenbogen JA. Exploration of the bicyclo[3.3.1]nonane system as a template for the development of new ligands for the estrogen receptor. Bioorg Med Chem Lett. 2003;13:4485–4488. doi: 10.1016/j.bmcl.2003.08.061. [DOI] [PubMed] [Google Scholar]
- 28.Muthyala RS, Sheng S, Carlson KE, Katzenellenbogen BS, Katzenellenbogen JA. Bridged bicyclic cores containing a 1,1-diarylethylene motif are high-affinity subtype-selective ligands for the estrogen receptor. J Med Chem. 2003;46:1589–1602. doi: 10.1021/jm0204800. [DOI] [PubMed] [Google Scholar]
- 29.Sibley R, Hatoum-Mokdad H, Schoenleber R, Musza L, Stirtan W, Marrero D, Carley W, Xiao H, Dumas J. A Novel Estrogen Receptor Ligand Template. Bioorg Med Chem Lett. 2003;13:1919–1922. doi: 10.1016/s0960-894x(03)00307-x. [DOI] [PubMed] [Google Scholar]
- 30.Hamann LG, Meyer JH, Ruppar DA, Marschke KB, Lopez FJ, Allegretto EA, Karanewsky DS. Structure-activity relationships and subtype selectivity in an oxabicyclic estrogen receptor alpha/beta agonist scaffold. Bioorg Med Chem Lett. 2005;15:1463–1466. doi: 10.1016/j.bmcl.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh RW, Rajan SS, Sharma SK, Guo Y, DeSombre ER, Mrksich M, Greene GL. Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity. J Biol Chem. 2006;281:17909–17919. doi: 10.1074/jbc.M513684200. [DOI] [PubMed] [Google Scholar]
- 32.Mortensen DS, Rodriguez AL, Carlson KE, Sun J, Katzenellenbogen BS, Katzenellenbogen JA. Synthesis and biological evaluation of a novel series of furans: ligands selective for estrogen receptor alpha. J Med Chem. 2001;44:3838–3848. doi: 10.1021/jm010211u. [DOI] [PubMed] [Google Scholar]
- 33.Mortensen DS, Rodriguez AL, Sun J, Katzenellenbogen BS, Katzenellenbogen JA. Furans with basic side chains: synthesis and biological evaluation of a novel series of antagonists with selectivity for the estrogen receptor alpha. Bioorg Med Chem Lett. 2001;11:2521–2524. doi: 10.1016/s0960-894x(01)00488-7. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Wu JY, Birzin ET, Frisch K, Chan W, Pai LY, Yang YT, Mosley RT, Fitzgerald PM, Sharma N, Dahllund J, Thorsell AG, DiNinno F, Rohrer SP, Schaeffer JM, Hammond ML. Estrogen Receptor Ligands. II. Discovery of Benzoxathiins as Potent, Selective Estrogen Receptor α Modulators. J Med Chem. 2004;47:2171–2175. doi: 10.1021/jm034243o. [DOI] [PubMed] [Google Scholar]
- 35.Bey E, Marchais-Oberwinkler S, Negri M, Kruchten P, Oster A, Klein T, Spadaro A, Werth R, Frotscher M, Birk B, Hartmann RW. New Insights into the SAR and Binding Modes of Bis(hydroxyphenyl)thiophenes and -benzenes: Influence of Additional Substituents on 17β-Hydroxysteroid Dehydrogenase Type 1 (17β-HSD1) Inhibitory Activity and Selectivity. J Med Chem. 2009;52:6724–6743. doi: 10.1021/jm901195w. [DOI] [PubMed] [Google Scholar]
- 36.Bey E, Marchais-Oberwinkler S, Werth RNM, Al-Soud YA, Kruchten P, Oster AFM, Birk B, Hartmann RW. Design, Synthesis, Biological Evaluation and Pharmacokinetics of Bis(hydroxyphenyl) substituted Azoles, Thiophenes, Benzenes, and Aza-Benzenes as Potent and Selective Nonsteroidal Inhibitors of 17β-Hydroxysteroid Dehydrogenase Type 1 (17β-HSD1) J Med Chem. 2008;51:6725–6739. doi: 10.1021/jm8006917. [DOI] [PubMed] [Google Scholar]
- 37.Katritzky AR, Rees CW, editors. Comprehensive Heterocyclic Chemistry. 1–8 John Wiley and Sons; New York: 1974. [Google Scholar]
- 38.Kumar AS, Balasrubrahmanyam SN. Reactivity of Some Tetra Substituted Furans and Thiophenes Towards BF3-Et2O Catalysed Diels-Alder Reaction. Tetrahedron Lett. 1997;38:1900–1911. [Google Scholar]
- 39.Nakayama J, Kurida K. Synthesis and Reactivities of a Highly Strained Thiophene with Two Fused Four-Membered Rings, 1,2,4,5-Tetahydrodicyclobuta[b,d]thiophene. J Am Chem Soc. 1993;115:4612–4617. [Google Scholar]
- 40.Nakayama J, Hasemi R, Yoshimura K, Suguhara Y, Yamaoka S. Preparation of Congested Thiophenes Carrying Bulky Substituents on the 3- and 4-Positions and Their Conversion to the Benzene Derivatives. J Org Chem. 1998;63:4912–4924. [Google Scholar]
- 41.Bailey D, Williams VE. An efficient synthesis of substituted anthraquinones and naphthoquinones. Tetrahedron Lett. 2004;45:2511–2513. [Google Scholar]
- 42.Li Y, Thiemann T, Sawada T, Mataka S, Tashiro M. Lewis Acid Catalysis in the Oxidative Cycloaddition of Thiophenes. J Org Chem. 1997;62:7926–7936. doi: 10.1021/jo961985z. [DOI] [PubMed] [Google Scholar]
- 43.Thiemann T, Ohira D, Li Y, Sawada T, Mataka S, Rauch K, Noltemeyer M, de Meijere A. [4+2] Cycloaddition of thiophene S-monoxides to activated methylenecyclopropanes. J Chem Soc, Perkin Trans. 2000;1:2968–2976. [Google Scholar]
- 44.Otani T, Takayama J, Sugihara Y, Ishii A, Nakayama J. π-Face-Selective Diels-Alder Reactions of 3,4-Di-tert-butylthiophene 1-Oxide and 1-Imide and Formation of 1,2-Thiazetidines. J Am Chem Soc. 2003;123:8255–8263. doi: 10.1021/ja029867i. [DOI] [PubMed] [Google Scholar]
- 45.Takayama J, Yoshiaki S, Takayanagi T, Nakayama J. syn-π-Face- and endo-selective, inverse electron-demand Diels–Alder reactions of 3.4-di-tert-butylthiophene 1-oxide with electron-rich dienophiles. Tetrahedron Lett. 2005;46:4165–4169. [Google Scholar]
- 46.Katzenellenbogen JA, Johnson HJ, Jr, Myers HN. Photoaffinity labels for estrogen binding proteins of rat uterus. Biochemistry. 1973;12:4085–4092. doi: 10.1021/bi00745a010. [DOI] [PubMed] [Google Scholar]
- 47.Carlson KE, Choi I, Gee A, Katzenellenbogen BS, Katzenellenbogen JA. Altered ligand binding properties and enhanced stability of a constitutively active estrogen receptor: evidence that an open pocket conformation is required for ligand interaction. Biochemistry. 1997;36:14897–14905. doi: 10.1021/bi971746l. [DOI] [PubMed] [Google Scholar]
- 48.Stauffer SR, Coletta CJ, Tedesco R, Nishiguchi G, Carlson K, Sun J, Katzenellenbogen BS, Katzenellenbogen JA. Pyrazole ligands: structure-affinity/activity relationships and estrogen receptor-alpha-selective agonists. J Med Chem. 2000;43:4934–4947. doi: 10.1021/jm000170m. [DOI] [PubMed] [Google Scholar]
- 49.Anstead GM, Carlson KE, Katzenellenbogen JA. The estradiol pharmacophore: ligand structure-estrogen receptor binding affinity relationships and a model for the receptor binding site. Steroids. 1997;62:268–303. doi: 10.1016/s0039-128x(96)00242-5. [DOI] [PubMed] [Google Scholar]
- 50.Nettles KW, Bruning JB, Gil G, Nowak J, Sharma SK, Hahm JB, Kulp K, Hochberg RB, Zhou H, Katzenellenbogen JA, Katzenellenbogen BS, Kim Y, Joachmiak A, Greene GL. NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat Chem Biol. 2008;4:241–247. doi: 10.1038/nchembio.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun J, Meyers MJ, Fink BE, Rajendran R, Katzenellenbogen JA. Novel ligands that function as selective estrogens or antiestrogens for estrogen receptor-alpha or estrogen receptor-beta. Endocrinology. 1999;140:800–804. doi: 10.1210/endo.140.2.6480. [DOI] [PubMed] [Google Scholar]
- 52.Shiau AK, Barstad D, Radek JT, Meyers MJ, Nettles KW, Katzenellenbogen BS, Katzenellenbogen JA, Agard DA, Greene GL. Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism. Nat Struct Biol. 2002;9:359–364. doi: 10.1038/nsb787. [DOI] [PubMed] [Google Scholar]
- 53.Nettles KW, Sun J, Radek JT, Sheng S, Rodriguez AL, Katzenellenbogen JA, Katzenellenbogen BS, Greene GL. Allosteric control of ligand selectivity between estrogen receptors alpha and beta: implications for other nuclear receptors. Mol Cell. 2004;13:317–327. doi: 10.1016/s1097-2765(04)00054-1. [DOI] [PubMed] [Google Scholar]
- 54.Anstead GM, Peterson CS, Pinney KG, Wilson SR, Katzenellenbogen JA. Torsionally and hydrophobically modified 2,3-diarylindenes as estrogen receptor ligands. J Med Chem. 1990;33:2726–2734. doi: 10.1021/jm00172a008. [DOI] [PubMed] [Google Scholar]
- 55.Tamao K, Kodama S, Nakajima I, Kumada M, Minato A, Suzuki K. Nickel-phosphine complex-catalyzed Grignard coupling. II. Grignard coupling of heterocyclic compounds. Tetrahedron. 1982;38:3347–3354. [Google Scholar]
- 56.Mclnerney EM, Tsai MJ, O’Malley BW, Katzenellenbogen BS. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc Natl Acad Sci U S A. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J. The Protein Data Bank and the challenge of structural genomics. Nat Struct Biol. 2000;7(Suppl):957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.