Abstract
The ability of the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm (BOADICEA) model to predict BRCA1 and BRCA2 mutations and breast cancer incidence in women with a family history of breast cancer was evaluated. Observed mutations in 263 screened families were compared to retrospective predictions. Similarly, observed breast cancers in 640 women were compared to retrospective predictions of breast cancer incidence. The ratios of observed to expected number of BRCA1- , BRCA2- and BRCA(1 or 2) mutations were 1.43 (95% CI 1.05–1.90), 0.63 (95% CI 0.34–1.08), and 1.12 (95% CI 0.86–1.44), showing a significant underestimation of BRCA1 mutations. Discrimination between carriers and non-carriers as measured by area under the receiver operating characteristic (ROC) curve was 0.83 (95% CI 0.76–0.88). The ratio of observed to expected number of invasive breast cancers was 1.41 (0.91–2.08). The corresponding area under the ROC curve for prediction of invasive breast cancer at individual level was 0.62 (95% CI 0.52–0.73). In conclusion, the BOADICEA model can predict the total prevalence of BRCA(1 or 2) mutations and the incidence of invasive breast cancers. The mutation probability as generated by BOADICEA can be used clinically as a guideline for screening, and thus decrease the proportion of negative mutation analyses. Likewise, individual breast cancer risks can be used for selecting women whose risk of breast cancer indicates follow-up. Application of local mutation frequencies of BRCA1 and BRCA2 could improve the ability to distinguish between the two genes.
Keywords: Risk assessment, BOADICEA, Breast cancer, BRCA1, BRCA2
Introduction
Women with a family history of breast- and/or ovarian cancer, early age at breast cancer onset or men with breast cancer can be referred for genetic counselling. The procedure involves investigation of family history, if indicated, BRCA1 and BRCA2 mutation screening and estimation of and information on cancer risk. A personal risk-reducing strategy might be recommended, such as increased surveillance or prophylactic surgery. It is important for patients and caregivers alike, that the separation of high risk individuals, eligible for follow-up action, from individuals with population risk, is as sensitive and specific as possible.
A number of risk assessment tools for familial breast- and ovarian cancer have been described. These include the Claus model [1–3], the BRCAPRO computer program [4] and web-based programs such as IBIS [5] and the Breast and Ovarian Analysis of Disease Incidence and Carrier Estimation Algorithm (BOADICEA) [6, 7].
BOADICEA is used in Swedish cancer genetic clinics. It was developed using women/families mainly from the UK but also including a few families from other countries, among them Sweden [8].
BOADICEA’s prediction of BRCA1 and BRCA2 mutations in different populations has been investigated in several studies [9–13] and found to perform similarly or better than other risk assessment tools. The frequencies of BRCA1 and BRCA2 mutations in Sweden are likely to differ from the default values given by the model due to the presence of several BRCA1 founder mutations. Thus, a need to evaluate the model in Swedish women was identified. Regarding breast cancer risk, BOADICEA accurately predicted the age-specific familial relative risk (FRR) for an individual with an affected mother, when compared to the observed FRR in epidemiological studies [7]. To our knowledge, no studies of the model’s ability to predict breast cancer in a specified group of women represented by extended pedigrees have been published. The model was developed for invasive breast cancer [6, 7] and the program’s homepage [14] states that the accuracy of the program is less clear for ductal cancer in situ (DCIS). Consequently there was also a need to investigate the effect of including DCIS diagnoses in the calculations.
The present aims were to investigate BOADICEA’s ability to predict BRCA1 and BRCA2 mutations, and its ability to predict breast cancer within a specified observation period, in a cohort of Swedish women.
Methods
Study material
The material consisted of pedigrees and medical records belonging to 652 women (index persons) who consecutively attended the cancer genetic clinic for hereditary breast- and ovarian cancer at any of three hospitals in Stockholm (Karolinska University Hospital, Södersjukhuset, Danderyd Hospital) during January 2002 to June 2006.
The women had been assigned at least 17% life time risk for breast cancer using Claus tables which is an almost doubled risk compared to the general population. They also fulfilled age criteria for being eligible for annual breast imaging (mammography ± ultrasound). Earliest age for initializing breast controls was either 10 years before the youngest age of breast cancer onset in respective family, or from 25 years of age in families with a known BRCA1 or BRCA2 mutation. Highest age was 60 years. Furthermore, individuals with an identified mutation other than in BRCA1 or BRCA2 were not included. Women with a personal history of ovarian cancer before start of observation were not included in the breast cancer risk prediction study but her pedigree could be included in the study of mutation risk predictions. All 652 women had had a normal mammogram within 1 year before the start of observation in order to avoid inclusion of women with a current breast cancer. Most of the women had been included in a prospective study on breast cancer diagnostic methods (PSDM) including mammography, ultrasound and, in the case of BRCA1 or BRCA2 mutation carriers, breast magnetic resonance imaging. The design and result of that study will be presented elsewhere. Pedigrees provided information about cancer diagnoses verified through medical records and/or death certificates, age at cancer onset, year of birth for affected individuals with a cancer diagnosis, and screening status for BRCA1 and BRCA2.
Mutation screening on genomic DNA had been offered in families fulfilling the clinical criteria regarding family history of breast- and/or ovarian cancer [15]. The pre-screening techniques protein truncation test and denaturizing high-performance liquid chromatography (WAVE DNA Fragment Analysis System, Transgenomic Inc., San Jose, CA, USA) were used in combination with direct sequencing and multiplex ligation-dependent probe amplification (MLPA, MRC Holland, The Netherlands). If no fresh material was available, genomic DNA was extracted from paraffin-embedded tissue and analyzed using the Sequenom MassARRAY system (Sequenom, San Diego, CA, USA) or the pyrosequencing PSQ HS96 system (Biotage AB, Uppsala, Sweden). All mutation analyses were performed at Lund University Hospital.
A minor proportion of cancer diagnoses had not been verified from medical records due to insufficient data. These diagnoses were included if diagnosis as well as age at onset were considered reliable by the oncologist performing the family-history investigation. Most pedigrees covered four generations including siblings and offspring in each. Year of birth was sometimes noted but not for the majority of healthy individuals drawn in the pedigrees. However, in general, information on premature death had been documented. Three oncologists and one genetic counsellor had been involved in the family history investigation.
In the present study, pedigrees were transformed into text files that were uploaded for risk calculations according to BOADICEA version 1 guidelines. Since information on ages for healthy family members frequently was lacking, the parameters “year of birth”, “age” or “age at death” were set according to: (1) year of birth in agreement with 25 years between each generation and 2 years between siblings. (2) 75 years as age at death. Spouses not drawn in the pedigree but required for BOADICEA analyses were consistently censored at age zero. If age at death for a deceased individual with cancer (breast-, ovarian-, prostate-, pancreatic-) was unknown, it was set to 2 years after cancer diagnosis. When known, Jewish ancestry was recorded in the text files prepared for BOADICEA.
The risk analyses were performed retrospectively during 2010 using BOADICEA version 1. The non optional values of mutation frequencies were for BRCA1 0.0006394 and for BRCA2 0.00102.
The present study did not cause any interventions in the care of the included women. The risk calculations used data about family history, incidence of breast cancer and mutation screening status that had been collected as part of the clinical routine and/or the PSDM.
Prediction of BRCA1 and BRCA2 mutations
The 652 available pedigrees included 288 mutation screening results, documented before 1 September 2010. The screened individual, for whom the mutation risks were calculated, was most often not the index person in the family but a close relative with breast or ovarian cancer. When several individuals in the same family had been screened, only the mutation risks of the first screened person were calculated and later gained information about family history and screening events were not taken into account. Twenty-five of the screening events were not included due to difficulties in interpreting records of family data at the date of screening (18), screened individual common for two families (6), or screening of only one gene (1). The remaining 263 individuals (representing 263 distinct families) were included, regardless of screening result, for the retrospective calculation of mutation probabilities prior to BRCA1 and BRCA2 mutation screening. All included individuals were of Scandinavian origin except for five with Iranian (1), Iraqi (1) and Ashkenazi Jewish (3) ancestry respectively.
The screening events represented 246 analyses on fresh material and 17 analyses on paraffin-embedded tissue. Among the 246 analyses there were 25 analyses without the MLPA technique. The sensitivities of the mutation screening methods were taken into account when calculating the expected number of mutations. The sensitivities were estimated to; 75% for analyses that were performed on DNA from paraffin-embedded tissue; 80% for analyses on DNA from fresh material prior to the introduction of the MLPA technique; 90% for DNA analyses from fresh material that included MLPA. The BRCA1 and BRCA2 mutation risks for all screened individuals were multiplied with the corresponding mutation detection sensitivity. The sum of the individual risks represented the expected number of mutations.
Observed BRCA1 and BRCA2 mutations
Individuals were classified as BRCA1 or BRCA2 mutation carriers if they carried a pathogenic mutation according to internationally recognized criteria [16].
Prediction of breast cancer
The 652 pedigrees included eleven index persons with ovarian cancer and one index person who underwent prophylactic mastectomy before start of observation. Exclusion of these twelve women resulted in 640 index persons for whom breast cancer risk during observation period were calculated. The 640 women represented 622 distinct families since there were 18 pairs of relatives (sisters/mother/aunt). All 640 individuals were of Scandinavian origin except for six with Iranian (1), Iraqi (1) or Ashkenazi Jewish (5) ancestry respectively. Approximately one-fifth of all diagnoses included were unverified. In total 27 included index persons had had a first breast cancer, all invasive, prior to start of observation. Results of mutation screening that were known prior to start of observation were recorded in the text files for BOADICEA. The pre-set values for mutation search sensitivities were 0.7 and 0.8 for BRCA1 and BRCA2 respectively. These values were in accordance with the estimated sensitivity of the mutation screening techniques used until June 2006. There were 95 pedigrees that included a negative mutation analysis and among the index persons there were 36 known mutation carriers, 29 for BRCA1 and 7 for BRCA2. Among the 640 pedigrees 12 diagnoses were solely DCIS. These were also included in the calculations.
The risk of a first or contralateral breast cancer for each index person during the observation period was calculated. The date when the family history was summarized in the medical records and the woman was offered annual breast examinations was defined as the start of the observation period for each individual in the present study. Cancer diagnoses or mutation screening for any family member after that date were not included. The end of the observation period was defined as the date of PM/death/migration/1 September 2010, whichever took place first. Each index person’s risk was obtained from the BOADICEA result file that lists annual risks for the first 5 years, followed by risks for every fifth year. For periods equalling 6, 7, 8 or 9 years the resulting risk was obtained as the sum of the 5-year risk and the term “10-year risk minus 5-year risk, divided by 5, multiplied by 1, 2, 3 or 4 respectively”.
Subsequently the breast cancer risks for all index persons during the observation period were summarized to obtain the expected number of breast cancers within the period.
Observed breast cancers at end of observation period
Information on all incidental breast cancer cases among the index persons during the observation period was found in medical records. Women no longer living in the catchment area were checked for cancer status until 1 September 2010 in the Swedish Cancer Registry.
Statistical methods
The overall accuracy in terms of the expected number of BRCA1 and BRCA2 mutation carriers, and of incidental breast cancers (first or contralateral), was evaluated by calculating the ratio of the observed and expected number of events. This ratio was presented together with its 95% exact Poisson confidence interval.
To evaluate the ability of the model—at individual level—to distinguish between carrier and non-carrier, and between those who would develop breast cancer and those who would not, the area under the receiver operating characteristic (ROC) curve was used. The larger the area under the curve the better the test, with a value of 1 indicating perfect discrimination and a value of 0.5 no better than chance discrimination. The ROC analysis results are presented as the area under the curve together with 95% bootstrap confidence intervals.
Results
Prediction of BRCA1 and BRCA2 mutations
The expected numbers of BRCA1-, BRCA2-, and BRCA(1 or 2) mutations were 33, 21 and 54 respectively. The observed numbers were 47 BRCA1- and 13 BRCA2 mutations, altogether 60 BRCA(1 or 2) mutations. The ratios of observed to expected numbers of mutations were 1.43, 0.63 and 1.12 for BRCA1-, BRCA2-, and BRCA(1 or 2) respectively. The ratios demonstrated that the number of BRCA2 mutations and the total number of BRCA1 and BRCA2 mutations could be predicted by the model, whereas the specific number of BRCA1 mutations was significantly underestimated (Table 1). There were 203 negatively screened samples, of these 25 had not yet been supplemented with the MLPA method.
Table 1.
Observed and expected numbers of BRCA1 and BRCA2 mutations in a subset of individuals (n = 263)
| Observed (O) | Expected (E) | O/E (95% CI) | ROC area (95% CI) |
|---|---|---|---|
| BRCA1 | BRCA1 versus non-BRCA1 | ||
| 47 (2 P) | 33 | 1.43 (1.05–1.90)* | 0.86 (0.79–0.91) |
| BRCA2 | BRCA2 versus non-BRCA2 | ||
| 13 (1 P) | 21 | 0.63 (0.34–1.08) | 0.69 (0.58–0.78) |
| BRCA(1 or 2) | BRCA1 or BRCA2 versus non-carriers | ||
| 60 (3 P) | 54 | 1.12 (0.86–1.44) | 0.83 (0.76–0.88) |
Discrimination between carriers and non-carriers of mutations in BRCA1 and/or BRCA2 as measured by area under the ROC curve
P, analyses of paraffin embedded tissue. * Significant deviation from 1
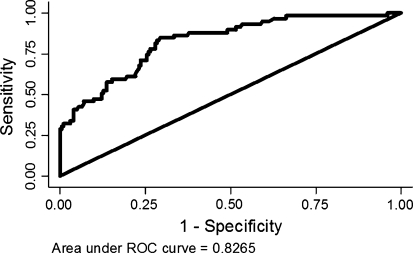
BOADICEA’s ability to discriminate between carriers and non-carriers at individual level as measured by the area under the ROC curve (AUC) was 0.86, 0.69 and 0.83 for BRCA1, BRCA2 and BRCA(1 or 2) respectively (Table 1, Fig. 1). All ROC areas were significantly different from 0.5.
Fig. 1.
ROC curve showing the discrimination between BRCA(1 or 2) mutation carriers and non-carriers
The sensitivities and specificities corresponding to the carrier probability cut-off points 4, 5, 10 and 15% ranged from 98.3 to 81.7% and 34.0–72.4% respectively (Table 2).
Table 2.
BRCA1/2 carrier probability thresholds and corresponding sensitivity and specificity
| Threshold, % | Sensitivity, % (95% CI) | Specificity, % (95% CI) |
|---|---|---|
| ≥ 4 | 98.3 (91.1–100.0) | 34.0 (27.5–41.0) |
| ≥ 5 | 95.0 (86.1–99.0) | 40.9 (34.1–48.0) |
| ≥ 10 | 88.3 (77.4–95.2) | 62.6 (55.5–69.2) |
| ≥ 15 | 81.7 (69.6–90.5) | 72.4 (65.7–78.4) |
Prediction of breast cancer
The sum of all breast cancer risks during the observation periods (in total 4,507 years), corresponding to the expected number of breast cancers, was calculated for the 640 index persons. In total, 613 risk estimates corresponded to a first breast cancer and 27 to a contralateral disease. The total number of expected breast cancers was 17.7.
The observed diagnoses among the 640 women during the observation period were 25 invasive breast cancers and five DCIS. Two DCIS cases were diagnosed at PM. The ratio of “observed” to “expected” for invasive cancers was 1.41. When DCIS cases were included the corresponding ratio was 1.69 (Table 3). The ratio did not deviate significantly from 1 if only invasive cancers were counted. In contrast, when DCIS cases were included, the model significantly under predicted the breast cancer incidence.
Table 3.
Observed and expected numbers of breast cancers
| Observed (O) | Expected (E) | O/E (95% CI) | ROC area (95% CI) |
|---|---|---|---|
| Invasive BCs | |||
| 25 | E: 17.74 | 1.41 (0.91–2.08) | 0.62 (0.52–0.73) |
| All BCsa | |||
| 30 | E: 17.74 | 1.69 (1.14–2.41)* | 0.63 (0.51–0.78) |
Discrimination between individuals diagnosed with breast cancer and individuals without diagnosis measured by area under the ROC curve
BC breast cancer. a Including 5 DCIS. * Significant deviation from 1
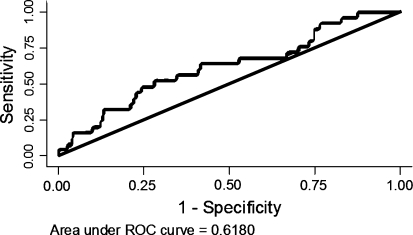
The ability of the model to discriminate between individuals who had invasive breast cancer and those that did not during the observation period as measured by AUC was 0.62 (Table 3, Fig. 2). When DCIS was accounted for among the observed cases the corresponding areas was 0.63 (Table 3). Both ROC areas were significantly different from 0.5.
Fig. 2.
ROC curve showing the discrimination between women diagnosed with breast cancer and women without breast cancer diagnosis
Discussion
The ability of BOADICEA to predict BRCA1 and BRCA2 mutations
The model’s ability to predict BRCA1 and BRCA2 mutations has previously been investigated in several different populations here exemplified by 195 French-Canadian families [9] and 1934 British families [10]. Both studies demonstrated that BOADICEA was well calibrated at group level, as reflected by ratios O/E ~1. Regarding discrimination between carriers and non-carriers, ROC curve areas ranged from 0.77 to 0.81.
The present study evaluated BOADICEA by using family data corresponding to the time of mutation screening. The model’s prediction of the total number of BRCA1 and BRCA2 mutations among the 263 individuals that had been screened for mutations was close to the observed number. This result indicated agreement between the data used to develop the model and the total prevalence of BRCA(1 or 2) mutations in our cohort. Further, the ability to discriminate between carriers and non-carriers was measured to an AUC of 0.83 which is similar to the reported value for British families.
The model performed worse in predicting the respective numbers of BRCA1 and BRCA2 mutations, which was to be expected since its pre-set values corresponded to mutation frequencies in the UK. In Sweden BRCA1 mutations are more frequent than BRCA2 mutations due to several well-known BRCA1-founder mutations [15], the opposite from the situation in the UK. In BOADICEA version 2 the parameter mutation frequency can be changed manually in order to fit a certain population.
All of the 263 individuals had met the clinical criteria regarding family history for mutation screening. Applying the present results in this group by setting screening thresholds based on carrier probabilities demonstrated that substantial proportions of mutation screening analyses with a negative outcome can be avoided. At a threshold of 10% carrier probability the model had 88.3% sensitivity and 62.6% specificity in the present study, but as a result seven mutation carriers would not have been offered mutation screening. For comparison, at the same threshold (10%), BOADICEA had a sensitivity of 90.4% and a specificity of 39.5% when investigated in British families [10]. In our cohort, we observed a sensitivity of 98.3% and a specificity of 34.0% at a threshold of 4% carrier probability. As a consequence, had mutation screening been offered only to individuals with a carrier probability of at least 4%, a quarter of all 263 individuals had not been screened for mutations. Still we would have identified all but one of the 60 confirmed carriers. We conclude that one missed mutation carrier versus 69 saved screening analyses is an acceptable trade-off.
The predictive ability may increase further in the next version of BOADICEA, which is reported to include tumour pathology data such as estrogen, progesterone and HER2 receptor status, and the expression of ‘basal’ markers [17]. According to the model’s homepage, other planned improvements are the addition of mutations in other breast cancer susceptibility genes such as TP53, ATM, CHEK2, PALB2, BRIP1, FGFR2, TNRC9 and MAP3K1 [18].
The present evaluation of the models ability to predict BRCA1 and BRCA2 mutations was based on data that was collected before BOADICEA was in use. Healthy individuals were included even if detailed information regarding age was lacking (further discussed below). It is possible that this approach influenced our results regarding overall accuracy and discrimination.
The ability of BOADICEA to predict breast cancer
The present study calculated breast cancer risk for women who at the start of observation were free from cancer. Consequently, no woman was under treatment for breast cancer at the start of observation, which would have decreased the risk of a second breast cancer during observation time. However, we did not account for the reduction in breast cancer risk conferred by the prophylactic salpingo-oophorectomy that 18 women had undergone.
Although a slight underestimation was found, the breast cancer prediction of 17.7 did not significantly deviate from the 25 observed invasive breast cancers. Similarly, the Tyrer–Cuzick model was reported to predict the risk of breast cancer in 1933 women, recruited from a family history clinic and surveyed by annual mammography, with the same degree of underestimation as BOADICEA’s in the present study. It was also demonstrated that another model, the Manual model, predicted more closely the observed number of breast cancers in that subgroup of women, whereas the Claus model (an algorithm that calculates remaining risk as opposed to the Claus tables which give lifetime risks) and BRCAPRO statistically significantly underestimated the risk [19].
There are factors that might have contributed to that the estimated number of breast cancers were lower than the observed. For example, the BRCA1 mutation frequency found in the present cohort was higher than predicted by the model. Since the average cumulative breast cancer risk at younger ages is higher for BRCA1 than for BRCA2 mutation carriers, this discrepancy will result in an underestimation of breast cancer risk [7]. In addition, BOADICEA performed better in predicting BRCA1 and BRCA2 mutations when larger pedigrees were used [9] although including uncertain data made the model perform worse [10]. When we calculated breast cancer risks with BOADICEA, the effects of including healthy family members became obvious. The average individual lifetime risk decreased from 24.9 to 23.1%, a reduction of 7%, when year of birth and life length were assessed for healthy family members. The information collected at the start of the observation period regarding healthy family members would have been more detailed if BOADICEA guidelines had been in use at that time. Nevertheless, information of putative premature death among family members without any of the cancer diagnoses relevant for BOADICEA was routinely collected. Therefore we believe that the occasional underestimation of risk as a result of assigning longer lives to individuals who died prematurely would be smaller than the corresponding overestimation of risk if all healthy individuals that lacked detailed birth and death data had been censored at age zero. A person was censored at age zero if lack of knowledge was indicated in the pedigree. Likewise, we chose to censor a deceased individual with a relevant cancer diagnosis by setting death to 2 years after diagnosis unless date of death was known. This is a minor deviation from BOADICEA version 1 guidelines that suggest that deceased individuals with cancer diagnosis and unknown date of death should be censored at the age of cancer diagnosis.
Finally, 96% of the women had annual breast examinations, 3% had mammography every second year and 1% were not monitored due to migration or death. Intensive surveillance and breast cancer diagnosis at PM confer a lead time bias that contributes to a higher number of observed than expected invasive breast cancers and to the significant underestimation of breast cancer when DCIS are included among the observed cases. It has been suggested that the incidence of DCIS can be adjusted by adding 10 years to age at diagnosis, assuming that the DCIS would have become invasive during that time [20]. If so, none of ‘our’ five observed DCIS would have been accounted for, since observation periods were shorter than 10 years. The demonstrated underestimation of breast cancer risk when DCIS was included among observed breast cancers is in accordance with that the model was developed for invasive breast cancers, not DCIS.
The ability of BOADICEA to distinguish individuals who were diagnosed with breast cancer (invasive or DCIS) from those who were not—on the basis of each individual’s risk for the observation period—was investigated. An AUC of 0.63 indicated a clinical usefulness. This value was lower than the corresponding values reported for the Gail (0.74), Claus (0.72), Ford (0.74) and Tyrer–Cuzick (0.76) risk assessment methods, which were evaluated in a British cohort [19].
In Swedish oncogenetic clinics there are ongoing discussions regarding at what breast cancer risk level surveillance beyond general mammography screening should be offered. It has been suggested that a life time risk of 20% according to BOADICEA should indicate follow-up and that a life time risk of 17–19.9% should lead to an individualized assessment. In the present cohort BOADICEA was used to retrospectively calculate life time risks (20–80 years). It was demonstrated that the proportion of breast cancer in the groups with <17% and 17–19.9% life time risk was 1.8 and 5.4% respectively, compared to 6.7% in the group “ ≥20% life time risk/previous breast cancer”. The corresponding age-adjusted risk in Swedish women in general with a life time risk of 10% [21], was 0.9%, or 5.5 breast cancer cases. Four of the observed breast cancers were diagnosed in women with a life time breast cancer risk of less than 17%.
Taken together, the estimated background risk and the observed high incidence of breast cancer cases in women with 17–19.9% life time risk indicate that a cut-off point of 17% for follow-up could be considered.
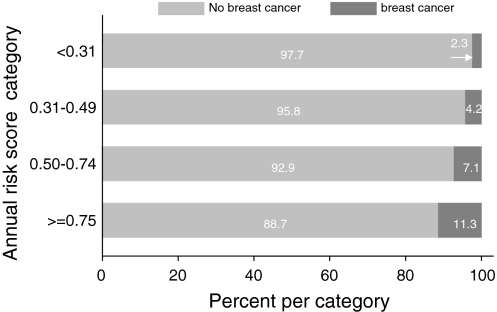
As opposed to life time risk, each woman’s risk for the observation period depended on her age at start of observation. To identify an alternative cut-off point for follow-up, based on the risks for the observation periods, we needed to compensate for that the observation period length differed between individuals. Therefore, an annual risk was calculated for all index persons by dividing each risk for the observation period by the corresponding number of years. Here, we allowed the observation period to be uninterrupted by PM/death/migration in order to mimic a counselling situation at the clinic. An analysis of the annual risk score regarding discrimination between breast cancer and no breast cancer resulted in an AUC of 0.68. If an annual risk score of 0.3% had been used as cut-off point for breast cancer preventive measures in the present study, the numbers of breast cancers in the group below the threshold would have been five, similar to the estimated background risk. The distribution of observed breast cancers, including DCIS, over four intervals of annual risk is shown in Fig. 3 and the number of women per interval in Table 4. It was demonstrated that with a cut-off point of 0.3% annual risk 210 women would not have been offered follow-up at the time of risk assessment. An annual risk level of 0.33% coincides with a five-year risk of 1.67% (as estimated by the Gail model 2 [22]) which is accepted by the US Food and Drug Administration as a cut-off point for segregating high- and low-risk individuals [23].
Fig. 3.
Percentage of women per category of annual risk score and percentage of breast cancer incidence in each category
Table 4.
Numbers of individuals and observed breast cancers per interval of annual risk
| Breast cancer | Annual risk % | Total | |||
|---|---|---|---|---|---|
| <0.31 | 0.31–0.49 | 0.50–0.74 | ≥0.75 | ||
| No | 210 | 254 | 91 | 55 | 610 |
| Yes | 5 | 11 | 7 | 7 | 30 |
| Total | 215 | 265 | 98 | 62 | 640 |
Contralateral breast cancer and ovarian cancer
There were four cases of contralateral disease among the 25 invasive breast cancers. Due to the small sample size statistical analyses of the ability of BOADICEA to predict contralateral breast cancer were not presented. Likewise, the model’s ability to predict ovarian cancer could not be evaluated since 18 of the 36 women that were known BRCA1 or BRCA2 mutation carriers at the start of observation underwent salpingo-oophorectomy prior to or during the observation period. Among the 36 known carriers, the predicted number of ovarian cancers was 1.3. No ovarian cancer was observed in the study population.
Summary
The findings in the present study population support that individuals with a mutation probability of 4%, as calculated by BOADICEA, could be offered mutation screening. In addition, a threshold of 17% life time risk or 1.67% five-year risk could be considered for increased breast surveillance.
Acknowledgments
We thank Bodil Edman Ahlbom, Clara Wäppling and Ulla Platten for valuable contributions to the preparation of the study material. This study was supported by grant from the Swedish Cancer Society.
Conflict of interest
The authors have no conflict of interest.
Ethical standards
The study was approved by the Regional Ethics Committee in Stockholm, no 2010/1864-31/2. No interventions were performed.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Claus EB, Risch N, Thompson WD. Genetic analysis of breast cancer in the cancer and steroid hormone study. Am J Hum Genet. 1991;48:232–242. [PMC free article] [PubMed] [Google Scholar]
- 2.Claus EB, Risch N, Thompson WD. The calculation of breast cancer risk for women with a first degree family history of ovarian cancer. Breast Cancer Res Treat. 1993;28:115–120. doi: 10.1007/BF00666424. [DOI] [PubMed] [Google Scholar]
- 3.Claus EB, Risch N, Thompson WD. Autosomal dominant inheritance of early-onset breast cancer. Implications for risk predictions. Cancer. 1994;73:643–651. doi: 10.1002/1097-0142(19940201)73:3<643::AID-CNCR2820730323>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Parmigiani G, Berry D, Aguilar O. Determining carrier probabilities for breast cancer-susceptibility genes BRCA1 and BRCA2. Am J Hum Genet. 1998;62:145–158. doi: 10.1086/301670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial, personal risk factors. Stat Med. 2004;23:1111–1130. doi: 10.1002/sim.1668. [DOI] [PubMed] [Google Scholar]
- 6.Antoniou AC, Pharoah PP, Smith P, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancer. Br J Cancer. 2004;91:1580–1590. doi: 10.1038/sj.bjc.6602175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA, Risch HA, Eyfjord JE, Hopper JL, Southey MC, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tryggvadottir L, Syrjakoski K, Kallioniemi OP, Eerola H, Nevanlinna H, Pharoah PD, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antoniou AC, Pharoah PDP, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoniou AC, Durocher F, Smith P, Simard J, INHERIT BRCAs program members. Easton DF. BRCA1 and BRCA2 mutation predictions using the BOADICEA and BRCAPRO models and penetrance estimation in high-risk French-Canadian families. Breast Cancer Res. 2006;8:R3. doi: 10.1186/bcr1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniou AC, Hardy R, Walker L, Evans DG, Shenton A, Eeles R, Shanley S, Pichert G, Izatt L, Rose S, Douglas F, Eccles D, Morrison PJ, Scott J, Zimmern RL, Easton DF, Pharoah PD. Predicting the likelihood of carrying a BRCA1 or BRCA2 mutation: validation of BOADICEA, BRCAPRO, IBIS, Myriad and the Manchester scoring system using data from UK genetics clinic. J Med Genet. 2008;45:425–431. doi: 10.1136/jmg.2007.056556. [DOI] [PubMed] [Google Scholar]
- 11.Barcenas CH, Hosain GM, Arun B, Zong J, Zhou X, Chen J, Cortada JM, Mills GB, Tomlinson GE, Miller AR, Strong LC, Amos CI. Assessing BRCA carrier probabilities in extended families. J Clin Oncol. 2006;24:354–360. doi: 10.1200/JCO.2005.02.2368. [DOI] [PubMed] [Google Scholar]
- 12.Thirthagiri E, Lee SY, Kang P, Lee DS, Toh GT, Selamat S, Yoon S-Y, Mohd Taib NA, Thong MK, Yip CH, Teo SH. Evaluation of BRCA1 and BRCA2 mutations and risk-prediction models in a typical Asian country (Malaysia) with a relatively low incidence of breast cancer. Breast Cancer Res. 2008;10:R59. doi: 10.1186/bcr2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurian AW, Gong GD, John EM, Miron A, Felberg A, Phipps A, West DW, Whittenmore AS. Performance of prediction models for BRCA mutation carriage in three racial/ethnic groups: findings from the Northern California breast cancer family registry. Cancer Epidemiol Biomarkers Prev. 2009;18:1084–1091. doi: 10.1158/1055-9965.EPI-08-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BOADICEA http://www.srl.cam.ac.uk/genepi/boadicea/boadicea_home.html. Accessed 7 July 2011
- 15.The National Oncogenetic Group, Arver B, Einbeigi Z, Loman N, Malander S. 20080901. State-of-the-art. Utredning, uppföljning och omhändertagande av personer med misstänkt ärftlig ökad risk för tumörsjukdom. Bröstcancer och äggstockscancer
- 16.National Human Genome Research Institute http://research.nhgri.nih.gov/bic/. Accessed 7 July 2011
- 17.Mavaddat N, Rebbeck TR, Lakhani SR, Easton DF, Antoniou AC. Incorporating tumour pathology information into breast cancer risk prediction algorithms. Breast Cancer Res. 2010;12:R28. doi: 10.1186/bcr2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.BOADICEA http://www.srl.cam.ac.uk/genepi/boadicea/boadicea_faqs.html#q32. Accessed 12 July 2011
- 19.Amir E, Evans DG, Shenton A, Lalloo F, Moran A, Boggis C, Wilson M, Howell A. Evaluation of breast cancer risk assessment packages in the family history evaluation and screening programme. J Med Genet. 2003;40:807–814. doi: 10.1136/jmg.40.11.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domchek SM, Eisen A, Calzone K, Stopfer J, Blackwood A, Weber BL. Application of breast cancer risk prediction models in clinical practice. J Clin Oncol. 2003;21:593–601. doi: 10.1200/JCO.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Nationella riktlinjer för bröstcancer (national guidelines for breast cancer) http://www.socialstyrelsen.se/Lists/Artikelkatalog/Attachments/9085/2007-114-51_200711453.pdf. Accessed 7 July 2011
- 22.Constantino J, Gail MH, Pee D, Anderson S, Redmond CK, Benichou J, Wieand HS. Validation studies for models projecting the risk of invasive and total breast cancer incidence. J Natl Cancer Inst. 1999;91:1541–1548. doi: 10.1093/jnci/91.18.1541. [DOI] [PubMed] [Google Scholar]
- 23.Rockhill B, Spiegelman D, Byrna C, Hunter DJ, Colditz GA. Validation of the Gail et al model of breast cancer risk prediction and implications for chemoprevention. J Natl Cancer Inst. 2001;93:358–366. doi: 10.1093/jnci/93.5.358. [DOI] [PubMed] [Google Scholar]