Abstract
Cellular differentiation and developmental programs require changing patterns of gene expression. Recent experiments have revealed that chromatin organization is highly dynamic within living cells, suggesting possible mechanisms to alter gene expression programs, yet the physical basis of this organization is unclear. In this article, we contrast the differences in the dynamic organization of nuclear architecture between undifferentiated mouse embryonic stem cells and terminally differentiated primary mouse embryonic fibroblasts. Live-cell confocal tracking of nuclear lamina evidences highly flexible nuclear architecture within embryonic stem cells as compared to primary mouse embryonic fibroblasts. These cells also exhibit significant changes in histone and heterochromatin binding proteins correlated with their distinct epigenetic signatures as quantified by immunofluorescence analysis. Further, we follow histone dynamics during the development of the Drosophila melanogaster embryo, which gives an insight into spatio-temporal evolution of chromatin plasticity in an organismal context. Core histone dynamics visualized by fluorescence recovery after photobleaching, fluorescence correlation spectroscopy, and fluorescence anisotropy within the developing embryo, revealed an intriguing transition from plastic to frozen chromatin assembly synchronous with cellular differentiation. In the embryo, core histone proteins are highly mobile before cellularization, actively exchanging with the pool in the yolk. This hyperdynamic mobility decreases as cellularization and differentiation programs set in. These findings reveal a direct correlation between the dynamic transitions in chromatin assembly with the onset of cellular differentiation and developmental programs.
Introduction
Recent experiments have revealed that the three-dimensional organization of chromatin is central to its function, whether in transcription, replication, or repair mechanisms (1,2). The fundamental unit of chromatin is the nucleosome, which consists of ∼200 basepairs of DNA wrapped round a histone octamer core, stabilized primarily by electrostatic interactions and tuned by posttranslational modifications (3,4). Chromatin assembly is further stabilized by interactions with the nuclear lamina (5). Live-cell imaging combined with fluorescence recovery after photobleaching (FRAP) and fluorescence correlation spectroscopy (FCS) experiments reveal that chromatin binding proteins exhibit distinct dynamics within differentiated cells (6,7). Core histone proteins (H2A, H2B, H3, and H4) and lamin (A and B1) stably bind chromatin with a residence time of several hours (8,9). However, linker histones and other chromatin binding proteins such as high mobility group proteins undergo transient interactions with residence timescales ranging from a few seconds to minutes (10–12). The differential dynamics of chromatin related proteins elicits a spatio-temporal heterogeneity in higher-order chromatin rigidity, as revealed by fluorescence anisotropy imaging (13). In contrast, undifferentiated cells in culture exhibit hyperdynamic mobility of both core and linker histones, suggesting a highly flexible chromatin structure (14,15). In addition, embryonic stem (ES) cells exhibit altered histone modifications (16,17), suggesting a possible functional correlation with the hyperdynamic mobility of chromatin binding proteins.
While plasticity at the level of higher-order chromatin assembly is functionally important, how it relates to the structural dynamics of nuclear architecture is poorly understood. More importantly, the coupling between the nuclear architecture proteins and histone protein dynamics are critical to determining three-dimensional architecture of the cell nucleus and thus genome function. In this work, we have explored the dynamics of nuclear proteins in mouse ES cells and contrasted that with lineage-restricted primary cells using live-cell fluorescence imaging and spectroscopy methods. The corresponding changes in epigenetic modifications in ES cells are also mapped. Further, the temporal evolution of translational and rotational dynamics of histone proteins during development of the Drosophila embryo is investigated. These results directly track the changing chromatin structure during development and differentiation, which could provide an insight into the transcription-level control of these processes.
Materials and Methods
Cell culture
R1 ES cells were cultured on a layer of feeder cells (primary mouse embryonic fibroblasts (PMEF)) with DMEM-F12 supplemented with 15% fetal bovine serum (HyClone, Logan, UT), 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 2 mM L-Glutamine, 0.1 mM β-mercaptoethanol (Sigma Aldrich, St. Louis, MO), and 500 U/ml leukemia inhibitory factor (Sigma Aldrich) and penicillin-streptomycin (All cell culture reagents, unless noted otherwise, are from Gibco Invitrogen,Carlsbad,CA). PMEF were cultured with DMEM-F12 supplemented with 5% fetal bovine serum, penicillin-streptomycin. Cells were maintained at 37°C in a 5% CO2 incubator. PMEF cells up to third passage were used in experiments. Both cell types grown on coverslip dishes for one day were transfected with 500 ng of DNA using Lipofectamine 2000 and Opti-MEM. The cells were imaged 24 h later for FRAP and FCS experiments. Details about the plasmids are included in the Supporting Material.
Fly lines
Early embryo of transgenic Drosophila melanogaster, where one of the core histones H2B is tagged to EGFP (H2B-EGFP), is used as a model system. Flies were kept for 1 h on a sucrose plate for egg laying. The embryo was then mounted on a #1 coverslip and covered with halocarbon oil 700. Alexa-488 (Molecular Probes, Eugene, OR) labeled linker histones (H1-Alexa488) were microinjected into wild-type CS embryos before the 11th nuclear division.
Confocal imaging
A model No. LSM510-Meta/Confocor2 fluorescence microscope equipped with an FCS module (Carl Zeiss, Oberkochen, Germany) was used in our experiments. Imaging and FRAP experiments on ES cells and PMEF cells were carried out using a C-Apochromat 63×/1.4 NA oil immersion objective with identical acquisition settings. FCS experiments in both the model systems were done using a C-Apochromat 40×/1.2 NA water-corrected objective. The same objective was used to acquire confocal images and FRAP/fluorescence-loss-in-photobleaching (FLIP) experiments inside the live Drosophila embryo. EGFP- and Alexa-488 tagged proteins were excited with the 488-nm line of an Argon-ion laser (LASSO) and the emission collected with a 500–530 nm bandpass filter. Confocal images (512 × 512 pixels, 12-bit images, with optimal pinhole aperture sizes) were acquired. In FCS experiments, the intensity timeseries I(t) values were collected at an optimum experimental setting to avoid artifacts arising due to photobleaching and to ensure high counts/particle. The pinhole size was kept at 70 μm for 488-nm laser line excitation (confocal diameter of ∼300 nm). From the intensity time series, the autocorrelation function, , was calculated online, where τ is the correlation time. For this, the data was collected for a period of 10-s intervals and averaged over 10 runs (18).
Fluorescence anisotropy
Anisotropy images were captured on an Olympus microscope (Melville, NY) with 60×/1.4 NA objective using an intensified charge-coupled device camera (Roper Scientific, Duluth, GA). Mercury arc source was used for the excitation light, which was then selected for vertical polarization using a sheet polarizer (Melles Griot, Carlsbad, CA). The light was collected by swapping between two polarizers (Melles Griot) parallel and perpendicular to the excitation. Images were captured using V++ Digital Optics (Auckland, New Zealand) software and analyzed using LabVIEW as described before (13). Parallel and perpendicular images were acquired at two regions of an embryo at the indicated time points. Anisotropy is given by , where I‖ is the parallel component of the emission intensity with respect to the excitation polarization direction and I⊥ is the perpendicular component.
Results
Undifferentiated mouse ES cells show significant fluidity in their lamin scaffolds compared to differentiated PMEF
Lamin proteins are type-V intermediate filaments that form the scaffold for nuclear organization (5). To probe lamin-mediated structural stability in the differentiated and undifferentiated states, FRAP experiments were carried out by strip-bleaching 6 μm in nuclear membrane regions of cells transiently expressing Lamin-B1 tagged with EGFP in both ES cells and PMEFs. The images of EGFP-LaminB1 before and after photobleaching are shown in Fig. 1 A for ES cells. In Fig. 1 B, the FRAP data indicate a higher recovery of nuclear lamin (∼20% in 200 s) in ES cell nuclei compared to PMEF nuclei. To interrogate whether nuclear lamin exists in a preassembled state in the nucleus, we carried out FCS experiments in these cells. The autocorrelation curves for Lamin-B1 proteins show a single timescale with subdiffusive behavior as parameterized by β. The value β is an anomalous scaling factor depicting the degree of confinement for protein diffusion within a heterogeneous medium as described previously (18). Diffusion under conditions of macromolecular crowding or through an anisotropic mesh, is characterized by not merely an increase in effective viscosity, but also a change of the exponent of the time dependence, β. For pure diffusion β = 1, while values of β < or > 1 characterize subdiffusive and superdiffusive processes, respectively. The mean β-value between ES cells and PMEF were within experimental error. Fitting the autocorrelation curves with free β for 18 correlation curves for PMEF cells, we find the mean β-factor to be 0.64. Using β = 0.64 as a fixed parameter, the mean correlation timescale for PMEF nuclei was found to be 729 ± 269 μs and ES cell nuclei as 758 ± 348 μs, suggesting that the free fraction of Lamin-B1 undergoes similar translation diffusion in both cell types (inset to Fig. 1 B).
Figure 1.
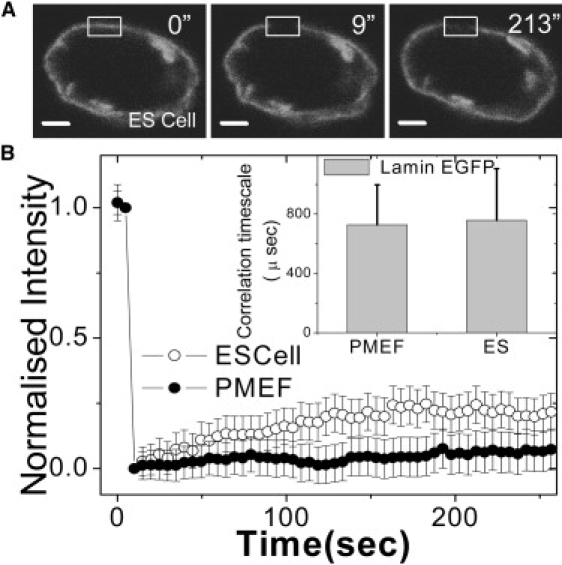
Plasticity in nuclear organization I: FRAP/FCS of lamin proteins in mouse ES and PMEF cells. (A) Fluorescent images of an ES cell nucleus transiently expressing EGFP-LaminB1. Images indicate prebleach (0 s), immediately after bleach (9 s), and 213 s after bleach. The scale bar is 2 μm. The bleach region is shown by the rectangle. (B) Normalized fluorescence intensities of EGFP-LaminB1 transiently expressed in both ES cells and in PMEF cells. PMEF cells show lower recovery in the shown time interval compared to ES cells (n = 20). (Inset) Diffusion timescales of EGFP-Lamin-B1 in PMEF and ES cell nuclei obtained using FCS show no significant difference in the two systems. The error bars are standard deviations.
Antibody staining against Lamin-B1 reveals an ill-defined nuclear lamina in ES cells, which becomes distinct and well defined with the progression of differentiation (15,19). To see whether this structural difference revealed in fixed cells correlates with a more fluidic lamin scaffold in living cells, we carried out time-lapse fluorescence imaging of the EGFP-LaminB1. These indicated a highly dynamic nuclear lamina in the ES cells (see Movie S1 and Movie S2 in Supporting Material) and Fig. 2 A. Here EGFP-LaminB1 shows significant nucleoplasmic intensity in PMEF cells as compared to ES cells where it is restricted to the envelope. In Fig. 2 B, the time series of the mean-square fluctuation [〈(δr)2〉 = Σ(δri)2/N] of the nuclear radius, computed over all angles from the centroid position (see Fig. S1 in Supporting Material), reveals large fluctuations in the ES cells compared to the PMEF cells, indicative of a more plastic organization of the lamina architecture in the ES cells. Thus, undifferentiated ES cells show significant fluidity in their lamin scaffolds compared to differentiated PMEFs. To investigate whether the plasticity in nuclear lamina is reflected in histone dynamics in ES cells, we carried out FRAP and FCS experiments as described in the next section.
Figure 2.
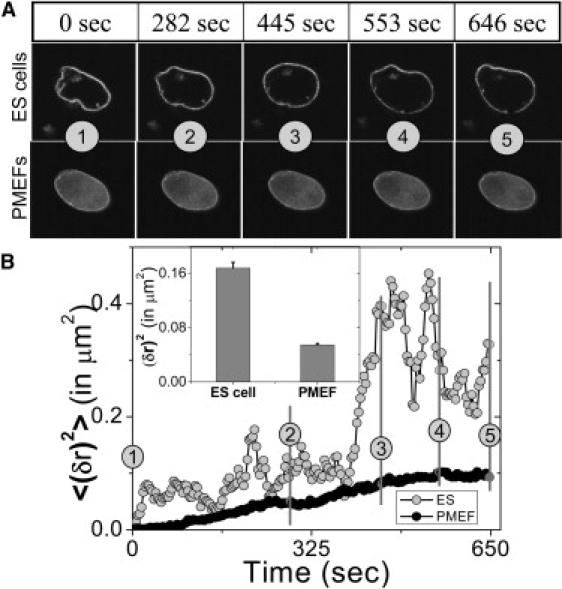
Plasticity in nuclear organization II: nuclear envelope fluctuations in mouse ES and PMEF cells. (A) Nuclear envelope fluctuations of ES cell and PMEF expressing EGFP-LaminB1 are shown for the time points indicated above the panel. The numbers in circles indicate corresponding time points. (B) Time series of the mean-square fluctuation of the nuclear membrane (EGFP-LaminB1) in ES cells and in PMEFs. (Inset) The mean and the standard error of mean-square fluctuations of the nuclear membrane, showing a more fluidic structure in ES cells compared to PMEFs.
Mouse ES cells exhibit hyperdynamic mobilities of histone and chromatin binding proteins compared to PMEFs
The dynamic nature of nuclear proteins is emerging as a crucial determinant of genome regulation (6,7,14). To assess the dynamic nature of nuclear proteins in both cell types, FRAP experiments with core histone H2B tagged with EGFP (H2B-EGFP) transiently expressed in both cell types was performed. The nucleus of both cell types was bleached in a region of interest of ∼1.8-μm diameter and recovery was plotted as a function of time. We observe in undifferentiated ES cells ∼50% recovery in ∼250 s. However, in the case of PMEF, negligible recovery in this time interval was observed as shown in Fig. 3 A (see also Movie S3 and Movie S4). Representative images of H2B-EGFP in ES cells and PMEFs before and after photobleaching are shown in the insets of Fig. 3 A and Fig. S2 A, respectively. The mean correlation timescale for core histone (H2B-EGFP) diffusion in PMEF nuclei was found to be 892 ± 165 μs and in ES cell nuclei was 731 ± 262 μs with the subdiffusive parameter β ∼ 0.64 (Fig. S2 C), indicating that core histone H2B diffuses as a multimer regardless of the differentiation status of the cell (20).
Figure 3.
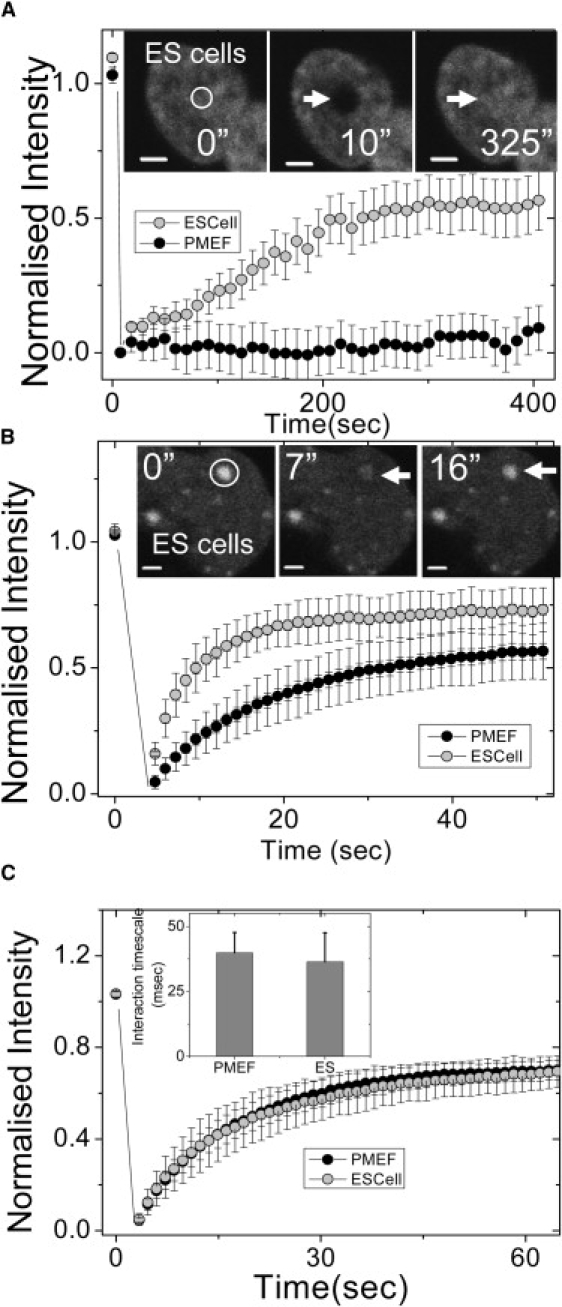
Dynamics of core, linker histones, and HP1 proteins in mouse ES and PMEF cells. (A) Normalized fluorescence intensities of core histone (H2B-EGFP) in ES and PMEF cells. A high fluorescence recovery was observed in the nuclei of ES cells, with negligible recovery in PMEFs in similar timescales (n = 20). (Inset) Images of H2B-EGFP expressing ES cells prebleach (0 s), after bleach (10 s), and after recovery of fluorescence (325 s) are shown. The scale bar is 2 μm. (B) FRAP of heterochromatin protein-1 (HP1α-EGFP) transiently expressed in ES cells and PMEFs showed a higher and faster fluorescence recovery in the nuclei of ES cells compared to PMEFs (n = 18). (Inset) Images of HP1α-EGFP expressing ES cells prebleach (0 s), after bleach (7 s), and after recovery of fluorescence (16 s) are shown. The scale bar is 2 μm. (C) Normalized fluorescence intensities of linker histone H1.5-EGFP in ES cells and PMEFs show no difference in dynamics (n = 15). (Inset) The graph indicates similar interaction timescales of H1.5-EGFP in ES cells and PMEFs revealed by FCS experiments.
In addition to specific epigenetic signatures, the identity of the heterochromatin nodes is maintained by nonhistone proteins like Heterochromatin Protein 1 (HP1). Moreover, the heterochromatin structure is inherently different in undifferentiated cells—with fewer and larger heterochromatin nodes/areas (15), thought to be exclusively constitutive in nature. Previous studies have explored the mobility of the HP1 proteins in mouse ES and T cells (14,21). HP1 protein dynamics in undifferentiated ES cells and terminally differentiated primary cells would offer insight into chromatin architecture during differentiation. FRAP experiments in ES cells transiently expressing HP1α-EGFP fusion protein showed transient dynamics, in line with previous experiments (14). With a bleach diameter of ∼1.8 μm at heterochromatin foci, fluorescence recovered to 70% in case of ES cells as compared to 50% in PMEFs, indicating a higher free fraction in undifferentiated cells. In addition, the recovery time was faster in ES cells (∼20 s) as compared to PMEF (∼45 s), as shown in Fig. 3 B (see also Movie S5 and Movie S6). This indicates that HP1α protein is highly dynamic in undifferentiated cells as compared to terminally differentiated cells. The images of HP1α-EGFP in ES cells and PMEFs before and after photobleaching are shown in insets to Fig. 3 B and Fig. S2 B, respectively.
Linker histone proteins stabilize higher order chromatin assembly. Widespread changes in chromatin structure with the onset of differentiation (15) could reflect on the diffusion and chromatin interaction properties of the linker histones. To investigate this, FRAP studies on both cell types expressing H1.5-EGFP–N1 were performed. In both ES cells and PMEFs, a similar percentage of recovery (∼70%) was observed (Fig. 3 C). This was reaffirmed by FCS measurements on H1.5-EGFP, which also shows similar behavior in both ES cells (36.4 ± 11.1 ms) and in PMEF (39.9 ± 7.7 ms) nuclei (see Fig. 3 C, inset). While FRAP experiments suggest that the dynamic recovery fraction and timescales of nuclear proteins are distinct in ES cells and PMEFs, FCS experiments reveal that their diffusion properties remain unaltered in both cell types.
To investigate whether the differences in the recovery dynamics of nuclear proteins correlate with a change at the level of histone modification, we carried out quantitative immunostaining for different histone modifications in ES cells and PMEFs. Posttranslational histone modifications play a central role in chromatin compaction and mobility of nuclear proteins (3,22). Several studies have established distinct epigenetic signatures of ES cells compared to differentiated cells (16,17). We compared the level of various histone marks between ES and PMEF cells using antibody staining to specific histone modifications (Fig. S3, A–D). H3 acetylation shows a higher level in ES cells, as shown in Fig. S3, A and B. Recently bivalent states of chromatin in ES cells have been reported, where both activating marks like H3K4 trimethylation and repressive marks like H3K27 trimethylation are present simultaneously on transcriptional start sites of a number of developmental genes (16,17,23). In close agreement with this, higher levels of facultative repressive mark H3K27 trimethylation in ES cells are observed (as shown in Fig. S3, A and B). This indicates perhaps that high level of H3K27 trimethylation prevents activation of genes, yet the hyperacetylation state in ES cells keeps a decondensed chromatin structure. Supporting this, linker histones that participate in higher order chromatin folding show a uniform pattern in ES cell nuclei compared to PMEF (Fig. S3, A and B). Enhanced level of acetylation in ES cells is also complemented by higher levels of the RNA pol II phosphorylated subunit, which is indicative of active transcriptional machinery (Fig. S3, C and D). However, the LaminB1 levels are not statistically significant in both cell types, although their structural dynamics are dramatically different as in Fig. S3, C and D. To address whether the plastic chromatin structure evidenced in undifferentiated ES cells in culture is observed in a developmental context, we further studied the dynamics of histone proteins in a Drosophila embryo using H2B-EGFP transgenic lines.
Developing Drosophila embryo nuclei, before cellularization, evidence dynamic exchange of histone proteins with maternal pool
Apart from the differentiation of ES cells, another context that is marked by widespread changes in transcription programs, and possibly therefore chromatin states, is the development of an organism. Thus, we carried out similar mobility experiments in the early embryo of a transgenic Drosophila melanogaster line expressing the core histone H2B tagged to EGFP. The images of H2B-EGFP in whole nuclei of a developing Drosophila syncytial blastoderm, before and after photobleaching, are shown in Fig. 4 A (see also Movie S7, Movie S8, and Movie S9). To test whether core histone H2B-EGFP exchanged between the nucleus and the yolk, the whole nucleus was bleached in FRAP experiments. The exchange of H2B-EGFP through the nuclear pores is calculated by normalizing the fluorescent intensities to the intensity of the whole nucleus before photobleaching. Fig. 4 B indicates a high fluorescence recovery of the core histones in the photobleached nuclei before cellularization, which decreases significantly with subsequent nuclear divisions and shows minimal recovery after cellularization. The recovery fraction in 400 s was ∼94% after the 11th division, ∼65% after the 12th division, ∼25% after the 13th division, and ∼8% 1 h postcellularization. Since there is no zygotic gene expression until the beginning of the 14th interphase cycle, these data indicate the exchange rates of maternally expressed H2B-EGFP. Fig. 4 illustrates the fact that depletion of maternal pool results in the reduction of histone recovery. To further delineate whether the source of the recovered fluorescence is the yolk, or the neighboring nuclei, a FLIP experiment was carried out. To this end, all the adjacent nuclei surrounding a given nucleus were photobleached, and the intensity followed with time. After photobleaching, the intensity of all the adjacent bleached nuclei increase without affecting the intensity of the unbleached nucleus (see Fig. 4 C). This clearly indicates an exchange of the core histones between the embryonic yolk and the nuclei. To determine whether the observed recovery was due to movements in the embryo yolk, we conducted experiments in long bleach regions of the embryo (see Fig. S4, A and B, and Movie S10). These indicate that the exchange of the core histones between the yolk and the nuclei is independent of the positioning of the nucleus in the embryo as well as the direction of flow in the yolk.
Figure 4.
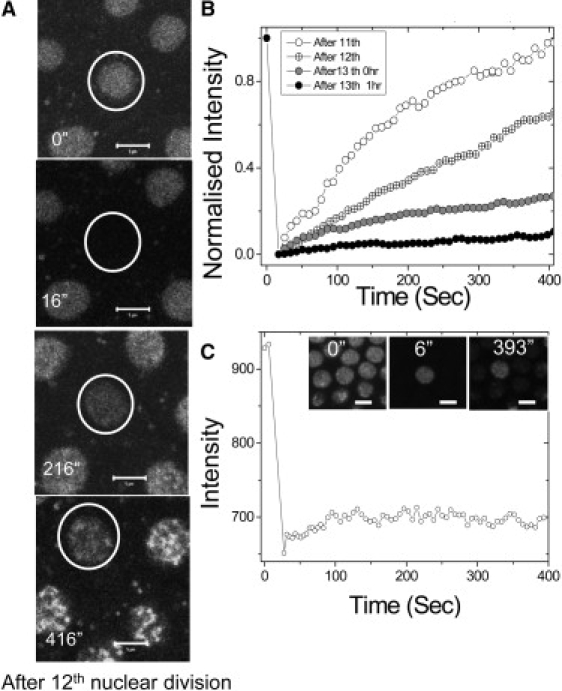
Large exchange of core histones between the nuclei and the yolk in the Drosophila melanogaster syncytial blastoderm. (A) A nucleus before photobleaching and subsequent frames after photobleaching (0 s, 200 s, and 400 s) before cellularization (after the 12th nuclear division) is shown. The scale bar is 5 μm. (B) Normalized fluorescence intensities of typical nuclei at different time points (after the 11th, 12th, and 13th nuclear divisions and 1 h from the 13th nuclear division), showing progressive fall in recovery. (C) Quantified fluorescence intensity indicates no fluorescence loss in a nucleus in the time after photobleaching of its adjacent nuclei, indicating exchange of the core histones between the yolk and the nucleus. (Inset) FLIP images after the 13th nuclear division (before cellularization) at indicated time points. The scale bar is 5 μm.
To understand the exchange mechanism of the linker histones between the nuclei and the yolk before cellularization, Alexa-488 labeled linker histones were microinjected into wild-type CS Drosophila embryos and these were used for FRAP and FCS studies. Nuclei were followed through the 12th and 13th mitotic divisions and the incorporation of the linker histones in the chromatin fibers was observed. Whole nuclei bleach experiments show that the exchange dynamics of linker histones recapitulated the behavior of core histones in the early embryo before cellularization (Fig. S4 C).
Developing Drosophila embryos evidence reduction in histone protein dynamics postcellularization
To determine the exchange characteristics of free and bound core histones within the cell nucleus after cellularization and isolation from the yolk, FRAP measurements were carried out on Drosophila embryos after cellularization set in. For this, a region of diameter of ∼2 μm inside the cell nucleus was photobleached at different time points, after the 13th nuclear division (0 h), 1 h postcellularization, and 5 h postcellularization, Fig. 5 A (see also Movie S11 and Movie S12). FRAP data show ∼40% fluorescence recovery in 150 s, for H2B-EGFP at the 1 h time point, indicating plasticity in chromatin organization even when cellularization is complete. Importantly, at the ∼5 h timepoint, recovery was significantly lowered, possibly indicating that the chromatin architecture had become more well defined and compacted at this stage. Confocal fluorescent images show distinct patches of condensed chromatin in the nucleus at the 5 h timepoint. FRAP curves show minimal fluorescence recovery of H2B-EGFP both in the euchromatin and heterochromatin regions (Fig. S5 B), reminiscent of core-histone diffusion characteristics inside the nuclei of differentiated systems such as HeLa cells, Drosophila SR2+ cells, and PMEFs (Fig. 5 B).
Figure 5.
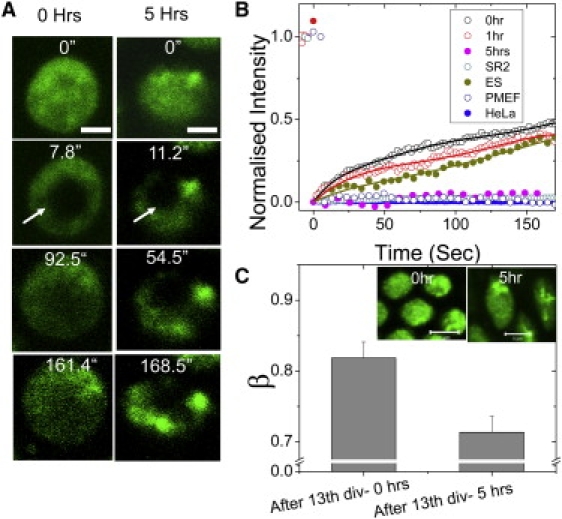
Plasticity in chromatin assembly in the early Drosophila embryo, postcellularization. (A) Time-lapse images of a small bleached region inside a cell nucleus at two stages of development (0 and 5 h after the 13th nuclear division). (B) Quantitation of images as in panel A, showing a high fluorescence recovery in the nuclei at 0 h from the 13th nuclear division and after 1 h from the 13th nuclear division (after completion of cellularization) and is similar to the recovery observed in ES cells. This fractional recovery comes down 5 h from the 13th nuclear division, and is comparable to FRAP recovery of H2B-EGFP in human HeLa cells, Drosophila SR2+ cells, and PMEFs. (C) The mean and the standard deviation of the β-factor inside cell nuclei at 0 h from the 13th nuclear division and 5 h from the 13th nuclear division. The decrease in β-factor indicates the condensation of the chromosomal mesh at 5 h from the 13th nuclear division (n = 30, each). (Inset) Confocal fluorescence images of the nuclei at 0 h from the 13th nuclear division and 5 h after the 13th nuclear division. Heterogeneous patches of chromatin condensation after 5 h from the 13th nuclear division can be seen.
To investigate the nature of core histones in the developing Drosophila embryo, FCS experiments were carried out at various locations in the embryo at different time points. The autocorrelation timescales indicate that the diffusion timescales of the H2B-EGFP does not diffuse as a monomeric species in line with earlier findings (20). The typical correlation timescales obtained for the H2B-EGFP did not change significantly during embryogenesis (Fig. S5 A). Importantly the diffusion behavior of linker histones measured by Alexa-488 labeled linker histones showed no secondary interaction timescales in Drosophila nuclei even 1 h postcellularization in Drosophila embryos, while in terminally differentiated HeLa cells, they show both a diffusion and interaction timescale (Fig. S6, A and B).
The autocorrelation function curves obtained for the multimeric core histones inside the nucleus at all experimental time points (t = 0 h and 5 h postcellularization) do not fit to a model of unhindered single species diffusion, but can be adequately described by a model of anomalous diffusion, with β as a scaling parameter defining the diffusive behavior. Fitting the correlation curves with β as a free parameter, it was found that the correlation curves fit well with β < 1, indicating subdiffusive transport. The mean β-value was used to estimate the condensation of the chromosomal mesh structure in the nucleus before cellularization (0 h after the 13th nuclear division) and after 5 h postcellularization. The mean value for β obtained over 30 such nuclei was 0.82 ± 0.08 for the nuclei at t = 0 h, whereas the β changed to 0.71 ± 0.08 inside the nucleus at t = 5 h (Fig. 5 C). The distinct patches in the fluorescent images of the nucleus and the change in β-factor correlate with the observation of a distinct heterogeneous chromatin compaction in the nucleus after the completion of cellularization.
Emergence of chromatin rigidity with progression of cellularization in the Drosophila embryo
Given that, within the first hour postcellularization, of the syncytial blastoderm of the Drosophila embryo there is a reduction in the diffusion of the core histones, we wanted to investigate whether these events coincided with an increased compaction of the chromatin, which is a characteristic of differentiated cells. To this end, we used fluorescence anisotropy imaging (13) to map the rotational freedom of the bound H2B-EGFP in these early nuclei reflecting chromatin compaction. An increase in anisotropy of H2B-EGFP emission implies a decreased rotational freedom of the protein and hence an increased chromatin rigidity (13). Fluorescence anisotropy images of H2B-EGFP expressing Drosophila embryos showed that, concomitant with the impeded mobility of core histone proteins, there was an overall rise in chromatin rigidity. This is also reflected in the standard deviation of anisotropy, indicating the emergence of heterogeneity in chromatin rigidity states (Fig. 6 A). The graph in Fig. 6 B shows the rise in anisotropy of these nuclei and the small changes in standard deviation over pixels are represented in the inset. Thus, with the onset of cellularization and lowered core histone mobility, there is an increase in chromatin rigidity states in different nuclei within a given embryo.
Figure 6.
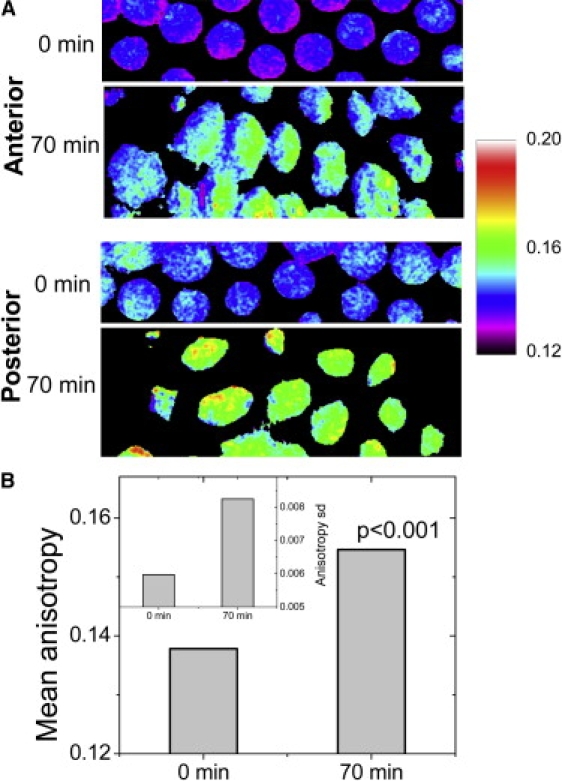
Evolution of rigidity maps of the chromatin in the Drosophila embryo using fluorescence anisotropy. (A) Color-coded anisotropy maps are shown for typical nuclei in the anterior and posterior of the embryo at 0 min (defined at 5 min after the completion of the 13th mitotic cycle), and at 70 min when nuclei of higher and more variegated chromatin rigidity states have emerged. The bar to the right shows anisotropy values corresponding to specific colors. (B) The anisotropy rise at the indicated time points are computed at each pixel and the plot shows the rise in the mean anisotropy (p < 0.001). (Inset) Corresponding small changes in standard deviation of anisotropy over pixels.
Discussion
In this work, we follow changes in chromatin architecture and structural proteins of the nucleus that accompany the onset of differentiation and development. We find negligible recovery of EGFP-LaminB1 in PMEF cells in ∼200 s, which is in line with previous studies (8). Interestingly, we also note a substantial (∼20%) recovery of fluorescence in undifferentiated ES cells on similar short timescales. Corresponding to the faster recovery dynamics of EGFP-LaminB1, for the first time, to our knowledge, we visualize large-scale fluctuations of the nuclear boundary in living cells. This suggests a less structured, fluidic state of the nuclear lamina in ES cells. This is consistent with recent micromanipulation studies that also indicate that nuclei in human ES cells are highly deformable and stiffen considerably through terminal differentiation (24). Taken together, these findings suggest that in the undifferentiated state, not just the chromatin, even the nuclear lamina is maintained in a fluidic state.
Although experiments by us, and others, clearly demonstrate a plastic state of both the chromatin and the nuclear lamin scaffold in an undifferentiated state, it is not clear how the freezing-down of these structures is brought about with differentiation. An interesting possibility is that the fluidic nature of these structures in ES cells is maintained by their unique epigenetic signatures (16,17). Indeed, previous work from our lab has demonstrated that mobility of histone proteins is sensitive to the acetylation states of the chromatin (22). Thus, with immunofluorescence microscopy, we explored the global levels of certain histone modifications. We found a hyperacetylated state of the chromatin in the undifferentiated state, and also found higher levels of H3K27-trimethylation—an epigenetic mark explored in other studies on ES cells, although these studies (16,17) had not discussed the global levels of these modifications. Thus, these studies suggest a possible coupling between histone modifications and structural plasticity of the nucleus in ES cells, which changes dramatically upon differentiation.
Taking further our studies on ES cells in culture, and validating the emergent principles in an organismal context, we first demonstrate a dynamic exchange mechanism of core histones in the precellular Drosophila embryo. This is unique compared to diffusion studies on core histones in terminally differentiated systems whether in mammalian, or Drosophila cells, which show little turnover over long periods of time (20). Before cellularization and the onset of zygotic transcription, the progressively lower recovery might be explained by doubling of the nuclei number with each division, with the histones derived from a fixed maternally laid pool in the embryo yolk. Here again, the rapid turnover of histones is a unique signature of these early nuclei. Progression of cellularization in the hour after the 13th mitotic cycle is also accompanied by an increase in chromatin rigidity as revealed by fluorescence anisotropy experiments. Even postcellularization enhanced core-histone mobility is observed in the nuclei of the early embryo, which seems analogous to observations in undifferentiated ES cells in culture (our studies and (14)). FRAP experiments complemented by FCS studies indicate a rise in the resistance offered by a chromosomal mesh to the diffusion of nuclear proteins with the emergence of different tissue types, as envisaged by a significant fall in the subdiffusive parameter β. Thus, with the emergence of cell identity, in processes of differentiation or development, chromatin structure loses its plasticity. This could have vital implications for the maintenance of tissue-specific transcriptional programs and thus, serve as a means for the emergence of cellular memory in three-dimensional organization of chromatin assembly. The transition between plastic to frozen configuration in chromatin dynamics may facilitate rapid response to cues for differentiation into specific lineages.
The molecular mechanisms underlying the highly fluidic organization of the nucleus in an undifferentiated cell and how it transits into defined conformations during the processes of differentiation and development are poorly understood. Firstly, Lamin A/C proteins are absent in ES cells (19), perhaps thus rendering the nuclear envelope highly dynamic. Moreover, since posttranslational modifications on histones are known to regulate higher order chromatin structure (23), it is possible that the higher acetylation levels in undifferentiated ES cells, maintain the chromatin in a decompacted state, thus also effecting rapid turnover of nuclear architecture proteins. It is thought that bivalent chromatin marks in such cells (16,17) keep them poised for differentiation into a variety of pathways, while maintaining tissue-specific genes in repressed states. Ongoing work in our lab is focused at understanding the mechanisms for maintaining a plastic chromatin structure in undifferentiated cells, and its significance in regulating cell fate decisions.
Acknowledgments
We thank Azim Surani, Maneesha Inamdar, Apurva Sarin, Yamuna Krishnan, and K. Vijay Raghavan for useful discussions. Mouse R1 ES cells were a kind gift from Dr. Maneesha Inamdar (Jawaharlal Nehru Centre for Advanced Scientific Research, India).
We also thank the Nanoscience Initiative of Department of Science and Technology for funding, and The National Centre for Biological Sciences Common Imaging and Flow Facility, supported in part by the Wellcome Trust.
Supporting Material
References
- 1.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Lanctot C., Cheutin T., Cremer M., Cavalli G., Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 3.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 4.Luger K., Hansen J.C. Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Gruenbaum Y., Margalit A., Goldman R.D., Shumaker D.K., Wilson K.L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 6.Gasser S.M. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 7.Spector D.L. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- 8.Moir R.D., Yoon M., Khuon S., Goldman R.D. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura H., Cook P.R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misteli T., Gunjan A., Hock R., Bustin M., Brown D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 11.Lever M.A., Th'ng J.P., Sun X., Hendzel M.J. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 12.Phair R.D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee B., Bhattacharya D., Shivashankar G.V. Chromatin structure exhibits spatio-temporal heterogeneity within the cell nucleus. Biophys. J. 2006;91:2297–2303. doi: 10.1529/biophysj.105.079525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meshorer E., Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 16.Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Weiss M., Hashimoto H., Nilsson T. Anomalous protein diffusion in living cells as seen by fluorescence correlation spectroscopy. Biophys. J. 2003;84:4043–4052. doi: 10.1016/S0006-3495(03)75130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Constantinescu D., Gray H.L., Sammak P.J., Schatten G.P., Csoka A.B. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya D., Mazumder A., Miriam S.A., Shivashankar G.V. EGFP-tagged core and linker histones diffuse via distinct mechanisms within living cells. Biophys. J. 2006;91:2326–2336. doi: 10.1529/biophysj.105.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Festenstein R., Pagakis S.N., Hiragami K., Lyon D., Verreault A. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 22.Rao J., Bhattacharya D., Banerjee B., Sarin A., Shivashankar G.V. Trichostatin-A induces differential changes in histone protein dynamics and expression in HeLa cells. Biochem. Biophys. Res. Commun. 2007;363:263–268. doi: 10.1016/j.bbrc.2007.08.120. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 24.Pajerowski J.D., Dahl K.N., Zhong F.L., Sammak P.J., Discher D.E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


