Abstract
During the course of an infection, viruses take advantage of a variety of mechanisms to travel in cells, ranging from diffusion within the cytosol to active transport along cytoskeletal filaments. To study viral motility within the intrinsically heterogeneous environment of the cell, we have developed a motility assay that allows for the global and unbiased analysis of tens of thousands of virus trajectories in live cells. Using this assay, we discovered that poliovirus exhibits anomalously rapid intracellular movement that was independent of microtubules, a common track for fast and directed cargo transport. Such rapid motion, with speeds of up to 5 μm/s, allows the virus particles to quickly explore all regions of the cell with the exception of the nucleus. The rapid, microtubule-independent movement of poliovirus was observed in multiple human-derived cell lines, but appeared to be cargo-specific. Other cargo, including a closely related picornavirus, did not exhibit similar motility. Furthermore, the motility is energy-dependent and requires an intact actin cytoskeleton, suggesting an active transport mechanism. The speed of this microtubule-independent but actin-dependent movement is nearly an order of magnitude faster than the fastest speeds reported for actin-dependent transport in animal cells, either by actin polymerization or by myosin motor proteins.
Introduction
The interior of eukaryotic cells is an extremely crowded environment consisting of a densely packed network of organelles, macromolecules, and the cytoskeleton. Whereas small molecules are able to passively diffuse in the cytoplasm, larger complexes, vesicles, and organelles with molecular masses >500 kDa typically require active transport during fundamental cellular processes such as endocytosis, membrane trafficking, and cytokinesis (1). Both actin filaments and microtubules contribute to intracellular motility by serving as tracks for cargo transport by motor proteins, including kinesins and dyneins, which travel along microtubules, and myosins, which travel along actin filaments (2–4). In addition, actin polymerization itself can propel cargo resulting in characteristic actin “comet-tails” (5). Each of these active intracellular transport mechanisms is reported to exhibit a characteristic range of speeds. Actin polymerization has been shown to propel cargo in mammalian cells with speeds of up to ∼0.4 μm/s (6). The processive human myosins (Va, VI, and X) travel at speeds as high as 0.3–0.9 μm/s in vitro and 0.1–0.4 μm/s in live cells (7–12). Kinesin- and dynein-based cargo transport typically occurs at much faster speeds of up to several μm/s (13). Recent studies on various cargos including viruses have extended our understanding of transport in the complex environment of the cell and have revealed the emerging role of the cargo itself in directing intracellular transport and cytoskeletal dynamics (14,15).
Given the fact that viruses need to enter and exit cells, it comes as no surprise that viruses have adapted to use the cell's many transport systems to efficiently infect their hosts (16,17). Many viruses have been observed to use active transport along microtubule and/or actin tracks. For example, murine leukemia virus, avian leukosis virus, human immunodeficiency virus (HIV), papillomavirus, and vesicular stomatitis virus use the cortical actin cytoskeleton, together with myosin II, for directed movement along actin rich filopodia (18,19). After entering the cell, many viruses, such as adenovirus, herpes simplex virus, HIV, and influenza virus are able to travel along microtubules to reach a specific site for genome release (20–23). In addition to using motor proteins to travel along existing cytoskeletal tracks, viruses are also able to manipulate the actin polymerization machinery to propel themselves deeper into the cell, as in the case of simian virus 40 (24), or to exit the cell, as in the case of vaccinia virus (25). Because viruses widely exploit diverse cellular mechanisms for motility, they are natural probes of intracellular transport, prompting investigation of viral transport both at a fundamental level and as a potential target for antiviral therapies (17,26–28).
Here, we examine the intracellular transport of poliovirus (PV), a ∼9000 kDa model pathogen for the study of nonenveloped RNA viruses such as rhinoviruses, enteroviruses, echoviruses, and hepatoviruses (29,30). By tracking individual viral particles in live cells, we discovered anomalously rapid, microtubule-independent but actin-dependent motility of internalized PV with speeds as high as 5 μm/s. This type of motility was observed in multiple types of cell lines but was cargo specific. Remarkably, the observed speed is nearly an order of magnitude higher than the fastest speeds reported for actin-dependent transport mechanisms in animal cells.
Materials and Methods
Cell culture, viruses, reagents, and plasmids
Additional information regarding cell culture, viruses, reagents, and plasmids is described in the Supporting Material.
Drugs
All inhibitors were purchased from Sigma Aldrich (St. Louis, MO) and used at the following concentrations: nocodazole (60 μM), sodium azide (20 μM), cytochalasin D (5–20 μM), latrunculin B (25 μM), latrunculin A (10 μM), jasplakinolide (1 μM), and blebbistatin (200 μM). Drugs were dissolved in a minimum volume of DMSO to make a concentrated stock solution, and the concentrated stock solution was then diluted in culture media during experiments. For all tracking experiments, cells were incubated with the appropriate concentration of inhibitor at 37°C for at least 50 min before data acquisition unless otherwise indicated. Because some drugs inhibit PV internalization, for all experiments except those shown in Figs. 1 and 2, PV was allowed to enter cells first for 10 min followed by a wash step and then incubation with the drug. Data acquisition in this case was started 60 min past PV infection, unless otherwise indicated, ensuring the full effect of the chosen drug. All experiments using sodium azide treatment of cells also included treatment with 25 μM deoxyglucose that helps to reduce cellular levels of ATP. The intensity of green 514 nm laser light used during the blebbistatin experiments (every tenth frame) was <1.6 × 10−4 W/cm2, which is much less than the intensity regime reported by Sakamoto et al. to be capable of photoinactivating blebbistatin (Fig. S13 in the Supporting Material) (31).
Figure 1.
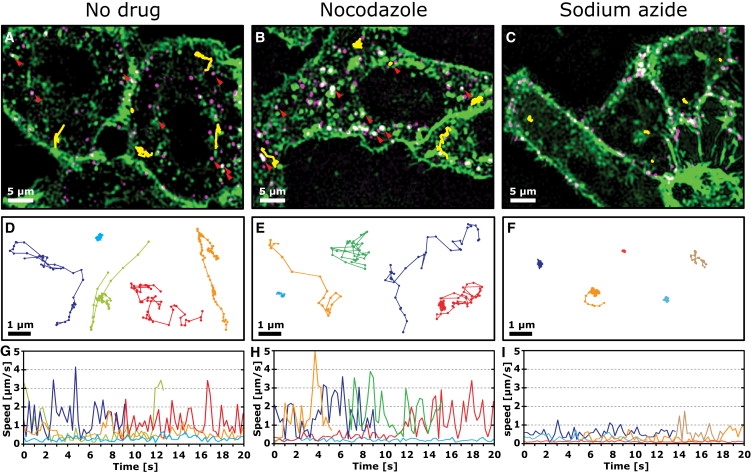
Motion of PV in untreated and nocodazole or sodium azide-treated cells. Confocal images (A–C) of PV (magenta) in live cells labeled with R18 (green; see also Movie S1, Movie S3, and Movie S4). Representative trajectories are overlaid in yellow. Magnified views of these trajectories are shown in D–F, and their instantaneous speed as a function of time are shown in G–I. Cells were pretreated with drug (60 μM nocodazole or 20 μM sodium azide) for 50 min before incubation with Cy5-labeled PV for 10 min at a low multiplicity of infection (MOI = 1 pfu/cell). Just before imaging, cells were briefly incubated with R18, which nonspecifically stained the plasma membrane and allowed the determination of the boundary of the cell and the observation of R18-stained vesicles. The red arrowheads in A and B indicate examples where PV colocalizes with R18-labeled vesicles. Successive points in trajectories are separated by 0.25 s.
Figure 2.
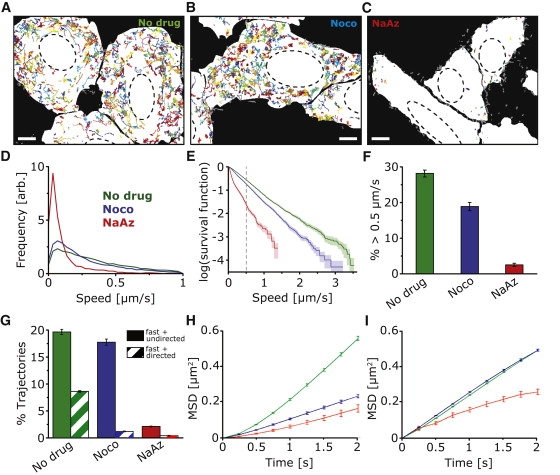
Quantitative analysis of PV motion. (A–C) Color overlays of all PV trajectories identified in representative movies of untreated, nocodazole-treated (Noco), and sodium azide-treated (NaAz) cells (see also Movie S1, Movie S3, and Movie S4). The corresponding still frames of the movies are shown in Fig. 1, A–C. The white cell maps indicate the interior of the cells as identified by confocal images of the plasma membrane marker R18, and the dashed lines represent the outlines of the nuclei. Trajectories not contained entirely within the white cell maps are rendered in gray and were excluded from further analysis. All movies were 400–500 frames (100–125 s) long. Scale bars in A–C = 5 μm; ∼40,000 trajectories from ∼150 cells and 61 movies were used for the analysis. (D) PV peak speed distributions in untreated (green), nocodazole-treated (blue), and NaAz-treated (red) cells. The speeds shown here and subsequently are calculated from the viral trajectories after four-point moving average. (E) Survival function plot of the observed peak speed distribution. The survival function plot shows along the y axis the fraction of trajectories with peak speed higher than the corresponding threshold value given in the x axis. (F) Percentage of trajectories with peak speeds above 0.5 μm/s. (G) Percentage of trajectories that are categorized as “fast and directed” and “fast and undirected” as defined in the text. Error bars = SE. (H) Mean-square displacement (MSD) analysis of the fast and directed subgroup and (I) of the fast and undirected subgroup for untreated (green), nocodazole (blue), and NaAz (red)-treated cells. Shown in the plots is the average MSD of all trajectories within the indicated subgroup.
Fluorescence microscopy
Two-color live cell fluorescence imaging was carried out using a customized Yokogawa spinning disk confocal scan head attached to an Olympus IX-71 microscope (Olympus, Center Valley, PA) with an Olympus 60×, 1.35 NA oil-immersion objective. The 514 nm line of an argon ion laser and the 647 nm line of a krypton ion laser were used to excite the lipophilic membrane dye R18 and cargo (Cy5 labeled PV, Cy5 labeled Seneca Valley Virus, or Alexa 647 labeled transferrin), respectively. The fluorescence from the two fluorophores was separated using a 650 nm longpass dichroic beamsplitter (Chroma, Rockingham, VT) and imaged onto two separate regions of an Andor 885 electron-multiplying charge coupled device camera for simultaneous two-color imaging. Because the R18 fluorescence has a weak crosstalk (that was subtracted, see below) into the 647 nm channel, we typically exposed only one out of every 10 frames with 514 nm laser light, whereas the 647 nm laser continuously illuminated the sample. R18 signals were filtered using a 570/80 nm bandpass emission filter (Chroma). Cy5 or Alexa 647 signals from cargo were filtered using a 665 nm longpass emission filter (Chroma). Except where otherwise indicated, movies were collected at a frame rate of 4 Hz for 400–500 frames. In all movies, the confocal microscope focal plane was positioned above the bottom of the cells in a plane that optically cut though the nucleus as shown in Fig. S2.
All experiments were conducted at 37°C using ∼75% confluent cells in Ibidi (Munich, Germany) μ-slide VI chambers. Imaging was carried out in phenol red free Dulbecco's modified Eagle's medium containing 50 mM Hepes and one of two oxygen scavenging systems depending on the experiment. All experiments except for those involving treatment by sodium azide/deoxyglucose used a glucose oxidase/catalase based oxygen scavenging system (32) consisting of 1% glucose, 0.5 mg/mL glucose oxidase (Sigma-Aldrich), and 40 μg/mL catalase (Roche Applied Science, Indianapolis, IN). The glucose oxidase system provided superior photostability, but was not suitable for experiments involving sodium azide/deoxyglucose treatment because glucose counteracts the effectiveness of deoxyglucose. Instead, for all experiments involving sodium azide/deoxyglucose, we used a glucose-free oxygen scavenging system based on the enzyme protocatechuate dioxygenase (PCD) (33,34). This alternative oxygen scavenging system uses PCD (Sigma-Aldrich) added to a final activity of 0.5 U/mL, and its substrate protocatechuate (Sigma-Aldrich) added to a final concentration of 50 mM. Control experiments on nocodazole-treated cells (not shown) using the PCD oxygen scavenging system showed identical motility to that measured in nocodazole-treated cells when using the glucose oxidase/catalase system.
Particle tracking and data analysis
Image processing and data analysis were carried out using custom written MATLAB (The MathWorks, Natick, MA) and IDL software (ITT Visual Information Solutions, Boulder, CO). Initial steps included the application of a spatial Fourier bandpass filter to remove high and low frequency background noise, the registration of dual-channel movies to account for optical aberrations or differences in magnification between the two channels, and the subtraction of a weak (4%) crosstalk of the R18 signal from the Cy5/Alexa 647 channel. Particles were tracked using custom-written software based on an algorithm described previously (35) that was modified to reliably obtain the trajectories for all particles identified in large batches of movies in an automated fashion. In some movies, there was a slow lateral drift of the microscope stage, which was corrected by subtracting off the average motion of many (mostly) nonmoving particles from all trajectories. Cell boundary maps were drawn by hand based on R18 membrane signals. The cell boundary maps were used to select out only those trajectories that occurred entirely within the interior of the cell whereas ambiguous trajectories that traversed the edge of the cell were discarded. Although rare, trajectories from cells that moved significantly on the timescale of the movies (∼2 min) were discarded, such that it was sufficient for all movies to use the cell map generated in one frame for all successive frames. A four-frame vectorial moving average filter was applied to the obtained trajectories before analyzing the speed of any trajectory. The minimum trajectory length was eight frames.
Results
Rapid microtubule-independent motility of PV
To observe the movement of PV in cells, we infected cells with fluorescently labeled PV and tracked individual virus particles in live cells using a spinning-disk confocal microscope. Fig. 1 A and Movie S1 in the Supporting Material show representative examples of PV motility in Hela S3 (human cervical cancer) cells. The great majority of particles entered cells by 20 min postinfection (mpi) and exhibited high motility inside the cells. Various types of movements were observed. Although some particles showed comparatively little motion and appeared to be confined to a small area (e.g., the cyan trajectory in Fig. 1, D and G), others were able to achieve high transport speeds (e.g., the green, blue, orange, and red trajectories in Fig. 1, D and G). Some of these rapidly moving particles exhibited highly directed movements (e.g., the green and orange trajectories in Fig. 1, D and G). Interestingly, the static or slowly moving particles were often localized to distinct vesicular structures nonspecifically stained by the lipophilic dye octadecyl rhodamine B (R18) after its internalization from the plasma membrane (Fig. 1, A and B, Fig. S1, and Movie S2), whereas the rapidly moving virus particles rarely colocalized with R18. Similar results were observed with other lipid markers, including DiI and NBDPS (data not shown), suggesting that the rapidly moving viruses were either sorted to highly specific endocytic vesicles that were not labeled by general purpose membrane markers, or were free inside the cytosol. The optical sectioning power of confocal microscopy allows us to rule out the possibility that these rapidly moving particles were on the cell surface—only the trajectories from particles inside the cell significantly far away from the plasma membrane, as marked by the lipophilic dyes, were analyzed (Fig. S2). Note that because the movements of virus particles inside the cell typically occur in all three dimensions, we only measured a two-dimensional projection of three-dimensional motion. The speeds calculated from the two-dimensional projection therefore represent lower bounds to the actual speeds.
Given the high motility of PV observed inside the cell during infection, we tested whether all movements were caused by microtubule-based motor proteins, which are known to transport cargo at speeds of up to several μm/s (13). Surprisingly, treatment of cells with nocodazole (that has been shown previously to have no affect on the efficiency of infection (36)) only moderately reduced the motility of PV (Fig. 1 B, Movie S3), despite the fact that nocodazole treatment completely depolymerized microtubules (Fig. S3). Virus particles were still able to achieve high speeds of several micrometers per second (up to 5 μm/s), often for multiple consecutive frames (e.g., orange, blue, and green trajectories, Fig. 1 E and H). The observation that these high speeds are sustained for several consecutive frames suggests that they are unlikely due to artifacts arising from frame-to-frame noise. To further illustrate this point, we carried out additional measurements at a higher frame rate of 20 Hz (instead of the 4 Hz frame rate used in recording data in Fig. 1). We then carried out a vectorial moving average of four consecutive points to generate smoothed virus trajectories that are used to calculate the speed of movement. Even after treatment with such a low-pass filter, speeds as high as 5 μm/s sustained for several frames were observed for PV in nocodazole-treated cells (Fig. S4).
To test whether the observed rapid, microtubule-independent motility of PV resulted from passive diffusion, we depleted cells of ATP by incubating the cells with sodium azide such that active processes in the cell were suppressed (37). We have shown previously that this treatment inhibits poliovirus infection (36). In contrast to untreated and nocodazole-treated cells, PV particles in sodium azide-treated cells were overwhelmingly static and were mainly found at the periphery of cells (Fig. 1, C, F, and I, and Movie S4), suggesting that passive diffusion cannot account for the rapid intracellular motility of PV. It should be noted that pretreatment of cells with sodium azide also prevented a large fraction of virus particles from being internalized. Here we selectively analyzed those trajectories that were inside the cells, as indicated by their significant separation from the R18-labeled cell membrane in the confocal image. As control experiments, we added sodium azide to cells after PV had been already internalized. These experiments yielded a substantially larger number of virus trajectories inside the cell, among which the great majority were also static (Fig. S5 and Movie S5).
Quantitative analysis of PV motility
The study of viral transport is often complicated by the fact that the intracellular environment is complex and heterogeneous and that viruses can exploit multiple cellular mechanisms simultaneously (28). To characterize and compare the full spectrum of viral motility, it is therefore necessary to track a large number of virus particles in cells and quantitatively analyze their behavior such that specific types of motion can be characterized, the fractions of viruses showing each type of motion can be determined, and the effects of various inhibitors on each type of motion can be assessed with confidence. To this end, we developed a motility assay that recorded the motion of many virus particles in cells using a confocal microscope and analyzed their movements using an automated particle tracking algorithm. The obtained viral trajectories were then compared with the cell boundaries derived from the fluorescence image of a plasma membrane marker to ensure that only viruses inside the cell were analyzed. Cells were infected with PV with an MOI of 1, a physiologically relevant condition in which there are ∼100–300 viruses per cell and a similar number of observed trajectories. All of the trajectories observed in the example movies corresponding to Fig. 1, A–C, (Movie S1, Movie S3, and Movie S4) are displayed in Fig. 2, A–C, in different colors on top of a white background indicating the interior of the cells. The viral trajectories in untreated and nocodazole-treated cells showed that PV particles not only moved rapidly but were also able to access nearly all regions of the interior of the cell except for the nucleus. We note that although the density of trajectories seems to be high in Fig. 2, not all trajectories were recorded simultaneously. The trajectories that appear to overlap in space did not overlap in time and thus did not cause ambiguity in tracking.
Statistical analyses were then carried out on tens of thousands of PV trajectories taken from ∼150 cells in many independent experiments. One of the parameters we extracted from each trajectory is the peak speed, defined as the maximum speed observed during the trajectory. To faithfully track rapidly moving virus particles, we imaged the sample at a relatively high frame rate (4 Hz). Before calculating the peak speed of each trajectory, a four-frame vectorial moving average was applied to all trajectories to reduce noise in the speed analysis and to avoid artifactual high-speed excursions that typically lasted for only one frame resulting from frame-to-frame noise. The peak speed distributions were then constructed from ∼20,000, ∼20,000, and ∼3000 viral trajectories in untreated, nocodazole-treated, and sodium azide-treated cells, respectively (Fig. 2 D). Plotting this data in the form of a survival function allows comparison of motility over the full range of observed speeds (Fig. 2 E). In addition to these full speed distributions, we also present the fraction of trajectories with speeds above 0.5 μm/s to offer a simpler comparison for the motility of rapidly moving particles under different conditions (Fig. 2 F). For instance, the percentage of trajectories with peak speeds >0.5 μm/s is 28% in untreated cells, 19% in nocodazole-treated cells, and 2.5% in sodium azide-treated cells. This threshold value of 0.5 μm/s was chosen because it represents the upper speed limit of previously reported actin-dependent movements in mammalian cells. We note that the trend of how the fraction of rapidly moving particles changes under different drug conditions does not substantially depend on the choice of the threshold value because the survival functions appear roughly linear on a semilogarithmic plot and the curves under different conditions rarely cross each other (Fig. 2 E). Time course experiments revealed that the fast, microtubule-independent PV motility inside the cell was achieved after 15 mpi and lasted until at least 80 mpi without any significant decrease (Fig. S6). This allowed us to probe PV motility at a variety of time points past infection.
In addition to speed, we also analyzed the shape of trajectories to elucidate what type of cytoskeletal tracks the viruses might be moving on. According to the speed and trajectory shape, we categorized trajectories as either: i), “slow”—with a peak speed below 0.5 μm/s; ii), “fast and directed”—having a peak speed ≥0.5 μm/s and possessing 5 or more consecutive steps with <30° turns between steps; or iii), “fast and undirected”—having a peak speed ≥0.5 μm/s but not satisfying the directedness criterion given above. Many approaches, such as mean-square displacement (MSD) analysis, can be used for the identification of directed and undirected trajectories (38–40). The simple trajectory sorting criteria used here facilitates global analysis of an extremely large number of trajectories (on the order of tens to hundreds of thousands), which is important for drawing unbiased and statistically significant conclusions of viral motility in a heterogeneous environment like the interior of a cell. Despite involving relatively arbitrary thresholds for discriminating fast versus slow and directed versus undirected trajectories, these criteria were nonetheless adequate to allow statistically significant comparisons between different drug treatment conditions. In addition, these criteria also allow us to sort out the small fraction with trajectories with directed movement among a relatively large population of undirected trajectories, or the short, transient bursts of directed movements in a long trajectory, both of which tend to be masked by ensemble or full-trajectory MSD analysis. According to these simple criteria, 9% of the PV trajectories in untreated cells were fast and directed, 19% were fast but undirected, whereas the remaining were slow. In contrast, only 1% of the trajectories in nocodazole-treated cells were fast and directed, whereas the fast and undirected fraction (18%) remained at a similar level to that observed in untreated cells (Fig. 2 G). These results indicated that the fast and directed movements obtained in untreated cells were largely due to microtubule-dependent transport, consistent with the facts that microtubules have a persistence length of ∼5 μm (41) and that kinesins and dyneins in cells often exhibit highly processive motion with run lengths of up to several micrometers (42). Sodium azide-treated cells showed a nearly undetectable amount of fast and directed trajectories and a drastic reduction of the fast and undirected trajectories (to only 2%), suggesting that the fast and undirected movements observed in untreated and nocodazole-treated cells required energy and were thus unlikely to be the result of passive diffusion despite its undirected appearance. We have also calculated the false positive rate of identifying a fully undirected trajectory as being directed using the above criteria. This rate (∼3%) is much lower than the fast and directed subpopulation identified in untreated cells but comparable to the small fraction of fast and directed trajectories identified in nocodazole-treated cells (Fig. S7), indicating that the identified directed movements were statistically significant only in untreated cells but not in nocodazole-treated cells. By varying the parameters used in the trajectory sorting criteria, we found these qualitative conclusions to be insensitive to the parameter choice (Fig. S7).
As expected, MSD analysis of the fast and directed subgroup shows a clear superlinear dependence on time (Fig. 2 H), whereas the MSD of the fast and undirected subgroup shows an approximately linear or slightly sublinear dependence on time (Fig. 2 I). The MSD analysis lends further support to our trajectory sorting procedure. Interestingly, the average MSD for the fast and directed subgroup decreased substantially on nocodazole treatment whereas the average MSD for the fast and undirected subgroup are nearly identical for untreated and nocodazole-treated cells, suggesting that the apparently fast and undirected movements observed in untreated and microtubule-disrupted cells share a similar mechanism. As anticipated earlier, the ensemble average MSD of all trajectories show a linear dependence on time (Fig. S8), masking the small fraction of trajectories with directed movement. Trajectories with directed movements transiently appearing as short bursts often exhibit a linear MSD versus time behavior too (data not shown).
Comparison of PV motion to other cargos and in different cell types
The above results show that PV is likely transported actively with speeds up to several micrometers per second in cells lacking intact microtubules. This is surprising considering that such high speeds are typically associated with transport by motor proteins on microtubules. To verify the absence of microtubules in nocodazole-treated cells we not only stained for tubulin as shown in Fig. S3 but also carried out experiments in green fluorescent protein (GFP)-tubulin expressing and nocodazole-treated cells (data not shown) and in neither case were microtubular structures observed. Next, we tested whether this type of motility was specific to the cargo or cell type used. In addition to the HeLa S3 cell lines used above, we studied PV motility in two additional cell lines, H446 (human lung carcinoma) and Hep 3B (human hepatocellular carcinoma). In these two cell lines, 36% and 26% of PV trajectories showed a peak speed of ≥0.5 μm/s, respectively, which is comparable to the 28% observed in HeLa S3 cells (Fig. 3 and Fig. S9). On treatment with nocodazole to disrupt microtubules, the fraction was only moderately reduced (by 1.7 and 1.8-fold), also comparable to the 1.4-fold reduction observed in HeLa S3 cells (Fig. 3 and Fig. S9).
Figure 3.
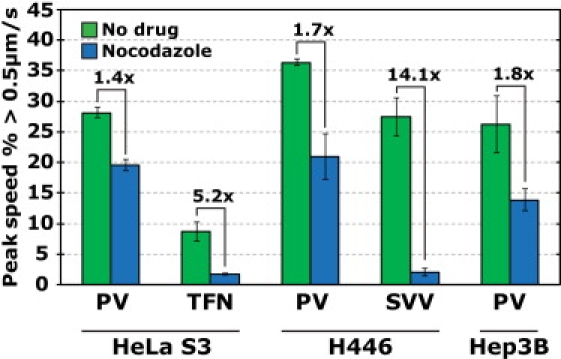
Comparison of motility between different cell types and different cargos. To examine the motility of various cargos in microtubule-disrupted cells, cells were first incubated with cargo (PV, transferrin (TFN), or Seneca Valley Virus (SVV)) for 10 min to allow internalization and were then treated with 60 μM nocodazole for 50 min to disrupt microtubules. More than 190,000 trajectories were analyzed. Error bars = SE. The comparison between PV and SVV were not carried out in HeLa S3 cells, but in H446 cells as the HeLa S3 cells cannot be infected by SVV. Survival function plots of the full speed distributions for the data shown here may be found in Fig. S9.
To test the cargo specificity, we chose two other cargoes for comparison: transferrin, a well characterized ligand responsible for iron delivery to the cell (14,43), and Seneca Valley Virus, a virus that belongs to the same family as PV (Picornaviridae) and is identical in size to PV (44,45). In contrast to PV, the fraction of fast transferrin trajectories was drastically reduced (by 5.2-fold) on depolymerization of microtubules (Fig. 3, Fig. S9, and Fig. S10). The comparison between Seneca Valley Virus and PV was even more dramatic. In H446 cells, which can be infected by both PV and Seneca Valley Virus, the viruses are highly dynamic with 36% of PV trajectories and 27% of Seneca Valley Virus trajectories exhibiting a peak speed ≥0.5 μm/s. However, the treatment of cells with nocodazole caused the fraction of fast Seneca Valley Virus trajectories to plummet to 2%, whereas 20% of PV trajectories remained fast (Fig. 3, Fig. S9, and Movie S6). Despite being members of the same family and equal in size, the two viruses apparently use different means for cellular transport. The rapid, microtubule-independent transport is therefore cargo-specific but not cell type-dependent.
The fact that the motility of transferrin and Seneca Valley Virus was strongly inhibited by nocodazole treatment further supports the notion that our nocodazole treatment successfully depolymerized microtubules and that the rapid motion of PV in nocodazole-treated cells is not attributable to microtubule-based motor proteins. Additionally, because PV and Seneca Valley Virus are of the same size and would be expected to diffuse with similar speeds, the fact that Seneca Valley Virus particles appeared largely static in nocodazole-treated cells lends further credence to the idea that the rapid microtubule-independent motility of PV did not result from diffusion, but was due to active transport.
Actin dependence of PV motility
Given that the rapid motility of PV is most likely active, independent of microtubules, and present in multiple human-derived cell lines, we next sought to explore the mechanism of this motility by testing whether it depends on the actin cytoskeleton. To this end, cells were treated with the pharmacological drugs cytochalasin D, latrunculins A and B, and jasplakinolide, which perturb the actin cytoskeleton in different ways. To focus only on the microtubule-independent movement, we treated the cells studied here also with nocodazole to eliminate microtubule-dependent transport. Because the uptake of PV itself requires an intact actin cytoskeleton (36), we first incubated the cells with PV for 10 min to allow virus entry before drug treatment. Interestingly, the microtubule-independent motility of PV was strongly reduced by cytochalasin D treatment (Fig. 4 A and Fig. S11). This significant reduction is dependent on the concentration of cytochalasin D as shown in Fig. S12. Treatment with latrunculins A and B or jasplakinolide yielded a more moderate reduction in PV motility. Given that cytochalasin D binds to the barbed, fast growing end of actin and depolymerizes actin filaments, the above results suggest that the rapid motility of PV requires an intact actin cytoskeleton but depends, to a lesser extent, on the dynamic behavior of actin, which is inhibited either by sequestering actin monomers, as in the case of latrunculins A and B, or by stabilizing actin filaments, as in the case of jasplakinolide. Such actin-dependent motility of PV was also observed in H446 cells (Fig. 4 C).
Figure 4.
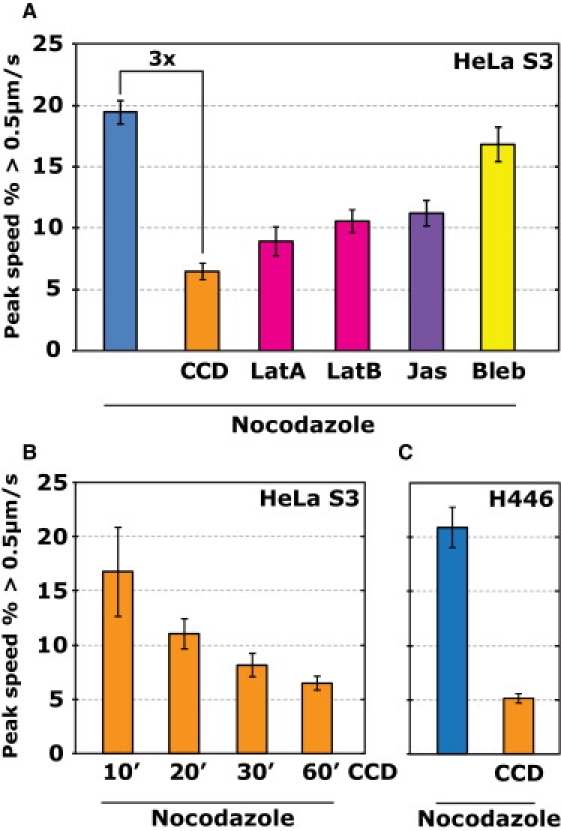
Actin dependence of PV motility. (A) Effects of the actin perturbing drugs cytochalasin D (CCD, see also Movie S7), latrunculin A (LatA), latrunculin B (LatB), and jasplakinolide (Jas), and the myosin II inhibitor blebbistatin (Bleb) on the observed motility of PV in HeLa S3 cells. Survival function plots of the full speed distributions for the data shown here may be found in Fig. S11. (B) Inhibition of PV motility in H446 cells by treatment of CCD. (C) CCD inhibition of PV motility over time. HeLa S3 cells were pretreated for 50 min with nocodazole, infected with PV for 10 min, and then incubated with CCD. Movies were recorded at different time points (10–60 min) after CCD addition, indicating that significant inhibition of PV motility did not occur until at least 20 min after CCD incubation. Error bars = SE.
Given that the formin-induced polymerization of actin filaments can achieve speeds of up to 3 μm/s and that the propulsion of cargo by actin comet tails was observed at speeds of up to 0.4 μm/s, respectively (6,46), we tested for the possibility that actin polymerization is responsible for the observed PV motility using two approaches. First, we assayed the motility of PV at various time points after incubation with cytochalasin D (Fig. 4 B), which is known to almost immediately inhibit actin polymerization but only completely disrupt the actin cytoskeleton after relatively long treatment. In both cases of polymerization of actin filaments and cargo propulsion by actin comet tails, cytochalasin D fully inhibited motility within 2 min of drug treatment (46,47). Here, we do not observe such a quick effect—significant inhibition of PV motility by cytochalasin D was only observed after at least 20 min of treatment (Fig. 4 B). Second, we infected cells stably expressing YFP-actin to search for comet-tail like actin structures colocalizing with moving PV but saw none. Thus, known mechanisms of actin polymerization do not seem to explain the observed motility of PV. Additionally, treatment of cells with the nonmuscle myosin II inhibitor blebbistatin (48,49) also caused little inhibition of the microtubule-independent PV motion (Fig. 4 A), suggesting that myosin II is not required for the motility. Although blebbistatin can be inactivated by irradiation with blue and ultraviolet light (≤488 nm), this effect is unlikely to be present here because the weak 514 nm light that we used to excite the plasma membrane marker R18 has an average power density that is five orders of magnitude lower than the reported photoinactivation regime (31). Indeed, exploiting a commonly used actin stress fiber assay to test the activity of blebbistatin, we found the drug to be active under our experimental conditions (Fig. S13) (50).
Last, we tested whether the rapid, actin-dependent motility of PV may be associated with myosin motors, we coimaged PV with a small panel of myosin proteins. To this end, GFP tagged myosins IIc, Va, Vb, VI, or X (9,51–54) were expressed in HeLa cells, which were subsequently infected with fluorescently labeled PV. The results are largely negative. None of these five myosins tested showed statistically significant colocalization with rapidly moving PV particles. A fraction of slow or nonmoving PV particles showed substantial colocalization with the GFP-labeled myosin VI or Vb punctae (Fig. S14, A and B, and Movie S8). Some PV particles were also observed to travel along actomyosin rich filopodia outlined by GFP-labeled myosin Va and X (Fig. S13, C and D, and Movie S9). Note that a lack of significant colocalization between the GFP tagged myosins and moving PV particles does not rule out the possibility that a myosin may be involved in the rapid motility of PV, considering that only a small subset of the 40 known human myosins were tested here and that the myosin signal colocalized with PV may be too weak to detect if only a small number of GFP-labeled myosin molecules were associated with the particle.
Discussion
Various mechanisms, both active and passive, give rise to the different modes of intracellular transport essential to the proper function of living cells. Viruses are able to exploit these mechanisms to infect cells. The study of viral motility can thus reveal valuable insights not only into virus-cell interactions but also into endogenous intracellular transport mechanisms. Here, we report the surprising finding that PV is capable of rapidly exploring the interior of cells with unusually high speeds, independent of microtubules. This was observed by imaging and tracking individual PV particles in live cells using spinning-disk confocal microscopy. Considering the wide spectrum of motility observed for virus particles, due to the presence of multiple transport mechanisms and a heterogeneous intracellular environment, we developed a quantitative motility assay to track tens of thousands of cargo particles in an automated and unbiased manner to allow for the systematic characterization of cargo motility.
Using this assay, we observed that a significant fraction of PV trajectories exhibits rapid motility in cells lacking intact microtubules, with ∼20% of trajectories showing sustained speeds above 0.5 μm/s and many particles achieving speeds of up to 5 μm/s. This motility is both ATP- and actin-dependent, suggesting an active transport mechanism. The peak speed of the actin-dependent movement observed here in various human cell lines is an order of magnitude faster than the highest speed of myosin motors observed in mammalian cells, although myosin-based transport of several micrometers per second has been reported for myosin V in yeast and for myosin XI isolated from plants (55,56). Cargo transport by actin polymerization also occurs at a much slower speed (6,25). The anomalously high microtubule-independent but actin-dependent PV motility observed here also seems to be cargo-specific: microtubule depolymerization induces near complete inhibition of motion for Seneca Valley Virus, a virus that is of the same physical size and belongs to the same family as PV. The same inhibition was also found for transferrin as shown here and many other cargoes as reported in the literature, including vaccinia virus (57), adenovirus (39), peroxisomes (58), lipid droplets (59), and endosomes (60).
Although it is presently unclear what exact mechanism underlies this rapid transport of PV, we propose a few possible scenarios consistent with our observations. Treatments with cytochalasin D at long times showed a substantial inhibition of PV motility, whereas those at short times showed little inhibition. This evidence established that the motility requires an intact actin cytoskeleton, but that known forms of actin polymerization for cargo transport, which are highly sensitive to short cytochalasin D incubations, are unlikely to be responsible. Furthermore, the nonmuscle myosin II inhibitor blebbistatin did not inhibit the rapid motility of PV, suggesting that cortical actomyosin dynamics are also unlikely to be responsible, owing to the prominent role of nonmuscle myosin II in actomyosin based motility.
One possible scenario that may account for the observed motility is the upregulation of the activity/speed of a myosin motor protein by PV either directly, or indirectly through the PV receptor or other proteins residing on the PV-harboring vesicles. The putative myosin may transport PV along a zigzagged path with frequent turns on the highly cross-linked actin network, explaining the observation that the trajectories do not appear directed (61). Though we tested a few myosin proteins and did not find measurable colocalization between these myosins and the rapidly moving PV, these results do not exclude the possibility that a myosin may be involved in the rapid motility of PV. Alternatively, PV may influence cytoskeletal dynamics (15) to achieve such high speeds. Drastic actin remodeling induced by PV has been observed during early and late stages of infection (62,63). It has also been shown recently that Vaccinia virus induces rapid and massive rearrangement of the cortical actin cytoskeleton shortly after binding to the cell (64). A fast induction of cytoskeleton remodeling after virus binding and internalization potentially provides an alternative mechanism for the rapid motility of PV. Future work will seek to further clarify the molecular mechanism of the rapid PV motility by isolating PV (free or vesicle bound) shortly after internalization and identifying the proteins associated with the virus or virus harboring vesicles.
Acknowledgments
The authors thank Dr. Michael Way (Cancer Research UK) for providing the GFP-tubulin and GFP-actin HeLa cell lines, Dr. P. Seshidhar Reddy (Neotropix) for samples of Seneca Valley Virus and the H446 cell line, James Spudich, James Goldenring, Richard Cheney, and Robert Adelstein for the GFP-myosin plasmids, and Yajun Zhou for helpful discussions. X.Z. is a Howard Hughes Medical Investigator. J.C.V. is a recipient of a Burroughs-Wellcome Career Award at the Scientific Interface and is a National Institutes of Health Ruth L. Kirschstein National Research Service Award fellow.
This work is supported in part by the National Institutes of Health (GM068518 to X.Z. and AI20566 to J.M.H).
Footnotes
Joshua C. Vaughan and Boerries Brandenburg contributed equally to this work.
Supporting Material
References
- 1.Dix J.A., Verkman A.S. Crowding effects on diffusion in solutions and cells. Annu. Rev. Biophys. 2008;37:247–263. doi: 10.1146/annurev.biophys.37.032807.125824. [DOI] [PubMed] [Google Scholar]
- 2.Spudich J.A. How molecular motors work. Nature. 1994;372:515–518. doi: 10.1038/372515a0. [DOI] [PubMed] [Google Scholar]
- 3.Vale R.D., Milligan R.A. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 4.Soldati T., Schliwa M. Powering membrane traffic in endocytosis and recycling. Nat. Rev. Mol. Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- 5.Gouin E., Welch M.D., Cossart P. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Gouin E., Gantelet H., Egile C., Lasa I., Ohayon H. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 1999;112:1697–1708. doi: 10.1242/jcs.112.11.1697. [DOI] [PubMed] [Google Scholar]
- 7.Dunn A.R., Spudich J.A. Dynamics of the unbound head during myosin V processive translocation. Nat. Struct. Mol. Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 8.Wu X., Bowers B., Rao K., Wei Q., Hammer J.A., 3rd Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function In vivo. J. Cell Biol. 1998;143:1899–1918. doi: 10.1083/jcb.143.7.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg J.S., Cheney R.E. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 10.Park H., Li A., Chen L.Q., Houdusse A., Selvin P.R. The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc. Natl. Acad. Sci. USA. 2007;104:778–783. doi: 10.1073/pnas.0610066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakata S., Watanabe Y., Usukura J., Mabuchi I. Characterization of native myosin VI isolated from sea urchin eggs. J. Biochem. 2007;142:481–490. doi: 10.1093/jb/mvm161. [DOI] [PubMed] [Google Scholar]
- 12.Nagy S., Ricca B.L., Norstrom M.F., Courson D.S., Brawley C.M. A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard J. Sinauer Associates; Sunderland, MA: 2001. Mechanics of Motor Proteins and the Cytoskeleton. [Google Scholar]
- 14.Lakadamyali M., Rust M.J., Zhuang X. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell. 2006;124:997–1009. doi: 10.1016/j.cell.2005.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hehnly H., Stamnes M. Regulating cytoskeleton-based vesicle motility. FEBS Lett. 2007;581:2112–2118. doi: 10.1016/j.febslet.2007.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sodeik B. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 2000;8:465–472. doi: 10.1016/s0966-842x(00)01824-2. [DOI] [PubMed] [Google Scholar]
- 17.Greber R.F., Way M. A superhighway to virus infection. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann M.J., Sherer N.M., Marks C.B., Pypaert M., Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J. Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schelhaas M., Ewers H., Rajamaki M.L., Day P.M., Schiller J.T. Human papillomavirus type 16 entry: retrograde cell surface transport along actin-rich protrusions. PLoS Pathog. 2008;4:e1000148. doi: 10.1371/journal.ppat.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suomalainen M., Nakano M.Y., Keller S., Boucke K., Stidwill R.P. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohner K., Wolfstein A., Prank U., Echeverri C., Dujardin D. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell. 2002;13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald D., Vodicka M.A., Lucero G., Svitkina T.M., Borisy G.G. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakadamyali M., Rust M.J., Babcock H.P., Zhuang X. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelkmans L., Puntener D., Helenius A. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science. 2002;296:535–539. doi: 10.1126/science.1069784. [DOI] [PubMed] [Google Scholar]
- 25.Cudmore S., Cossart P., Griffiths G., Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 26.Pelkmans L., Helenius A. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 2003;15:414–422. doi: 10.1016/s0955-0674(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 27.Dohner K., Nagel C.H., Sodeik B. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 2005;13:320–327. doi: 10.1016/j.tim.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Brandenburg B., Zhuang X. Virus trafficking—learning from single-virus tracking. Nat. Rev. Microbiol. 2007;5:197–208. doi: 10.1038/nrmicro1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogle J.M. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitton J.L., Cornell C.T., Feuer R. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 2005;3:765–776. doi: 10.1038/nrmicro1284. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto T., Limouze J., Combs C.A., Straight A.F., Sellers J.R. Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry. 2005;44:584–588. doi: 10.1021/bi0483357. [DOI] [PubMed] [Google Scholar]
- 32.Payne C.K., Jones S.A., Chen C., Zhuang X. Internalization and trafficking of cell surface proteoglycans and proteoglycan-binding ligands. Traffic. 2007;8:389–401. doi: 10.1111/j.1600-0854.2007.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patil P.V., Ballou D.P. The use of protocatechuate dioxygenase for maintaining anaerobic conditions in biochemical experiments. Anal. Biochem. 2000;286:187–192. doi: 10.1006/abio.2000.4802. [DOI] [PubMed] [Google Scholar]
- 34.Crawford D.J., Hoskins A.A., Friedman L.J., Gelles J., Moore M.J. Visualizing the splicing of single pre-mRNA molecules in whole cell extract. RNA. 2008;14:170–179. doi: 10.1261/rna.794808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakadamyali M., Rust M.J., Brandenburg B., Zhuang X. Single-virus tracking in live cells. In: Selvin P.R., Ha T., editors. Single-Molecule Techniques. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2007. pp. 171–192. [DOI] [PubMed] [Google Scholar]
- 36.Brandenburg B., Lee L.Y., Lakadamyali M., Rust M.J., Zhuang X. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5:1543–1555. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwoebel E.D., Ho T.H., Moore M.S. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J. Cell Biol. 2002;157:963–974. doi: 10.1083/jcb.200111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ewers H., Smith A.E., Sbalzsarini I.F., Lilie H., Koumoutsakos P. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc. Natl. Acad. Sci. USA. 2005;102:15110–15115. doi: 10.1073/pnas.0504407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helmuth J.A., Burckhardt C.J., Koumoutsakos P., Greber U.F., Sbalzarini I.F. A novel supervised trajectory segmentation algorithm identifies distinct types of human adenovirus motion in host cells. J. Struct. Biol. 2007;159:347–358. doi: 10.1016/j.jsb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janson M.E., Dogterom M. A bending mode analysis for growing microtubules: evidence for a velocity-dependent rigidity. Biophys. J. 2004;87:2723–2736. doi: 10.1529/biophysj.103.038877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross S.P., Welte M.A., Block S.M., Wieschaus E.F. Coordination of opposite-polarity microtubule motors. J. Cell Biol. 2002;156:715–724. doi: 10.1083/jcb.200109047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dautry-Varsat A., Ciechanover A., Lodish H.F. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy P.S., Burroughs K.D., Hales L.M., Ganesh S., Jones B.H. Seneca Valley virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J. Natl. Cancer Inst. 2007;99:1623–1633. doi: 10.1093/jnci/djm198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkataraman S., Reddy S.P., Loo J., Idamakanti N., Hallenbeck P.L. Crystallization and preliminary X-ray diffraction studies of Seneca Valley virus-001, a new member of the Picornaviridae family. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008;64:293–296. doi: 10.1107/S1744309108006921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Higashida C., Miyoshi T., Fujita A., Oceguera-Yanez F., Monypenny J. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- 47.Sanger J.M., Sanger J.W., Southwick F.S. Host cell actin assembly is necessary and likely to provide the propulsive force for intracellular movement of Listeria monocytogenes. Infect. Immun. 1992;60:3609–3619. doi: 10.1128/iai.60.9.3609-3619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 49.Limouze J., Straight A.F., Mitchison T., Sellers J.R. Specificity of blebbistatin, an inhibitor of myosin II. J. Muscle Res. Cell Motil. 2004;25:337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- 50.Brzeska H., Szczepanowska J., Matsumura F., Korn E.D. Rac-induced increase of phosphorylation of myosin regulatory light chain in HeLa cells. Cell Motil. Cytoskeleton. 2004;58:186–199. doi: 10.1002/cm.20009. [DOI] [PubMed] [Google Scholar]
- 51.Wei Q., Adelstein R.S. Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol. Biol. Cell. 2000;11:3617–3627. doi: 10.1091/mbc.11.10.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golomb E., Ma X., Jana S.S., Preston Y.A., Kawamoto S. Identification and characterization of nonmuscle myosin II–C, a new member of the myosin II family. J. Biol. Chem. 2004;279:2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- 53.Fan G.H., Lapierre L.A., Goldenring J.R., Sai J., Richmond A. Rab11-family interacting protein 2 and myosin Vb are required for CXCR2 recycling and receptor-mediated chemotaxis. Mol. Biol. Cell. 2004;15:2456–2469. doi: 10.1091/mbc.E03-09-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altman D., Goswami D., Hasson T., Spudich J.A., Mayor S. Precise positioning of myosin VI on endocytic vesicles in vivo. PLoS Biol. 2007;5:e210. doi: 10.1371/journal.pbio.0050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schott D.H., Collins R.N., Bretscher A. Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J. Cell Biol. 2002;156:35–39. doi: 10.1083/jcb.200110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tominaga M., Kojima H., Yokota E., Orii H., Nakamori R. Higher plant myosin XI moves processively on actin with 35 nm steps at high velocity. EMBO J. 2003;22:1263–1272. doi: 10.1093/emboj/cdg130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carter G.C., Rodger G., Murphy B.J., Law M., Krauss O. Vaccinia virus cores are transported on microtubules. J. Gen. Virol. 2003;84:2443–2458. doi: 10.1099/vir.0.19271-0. [DOI] [PubMed] [Google Scholar]
- 58.Kural C., Kim H., Syed S., Goshima G., Gelfand V.I. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 59.Nan X., Potma E.O., Xie X.S. Nonperturbative chemical imaging of organelle transport in living cells with coherent anti-stokes Raman scattering microscopy. Biophys. J. 2006;91:728–735. doi: 10.1529/biophysj.105.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loubery S., Wilhelm C., Hurbain I., Neveu S., Louvard D. Different microtubule motors move early and late endocytic compartments. Traffic. 2008;9:492–509. doi: 10.1111/j.1600-0854.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- 61.Klumpp S., Lipowsky R. Active diffusion of motor particles. Phys. Rev. Lett. 2005;95:268102. doi: 10.1103/PhysRevLett.95.268102. [DOI] [PubMed] [Google Scholar]
- 62.Kang G., Desikan P., Mathan M. Cytoskeletal changes during poliovirus infection in an intestinal cell line. Indian J. Med. Res. 2002;115:37–45. [PubMed] [Google Scholar]
- 63.Coyne C.B., Kim K.S., Bergelson J.M. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J. 2007;26:4016–4028. doi: 10.1038/sj.emboj.7601831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mercer J., Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


