Abstract
Membrane protein stability is a key parameter with important physiological and practical implications. Inorganic salts affect protein stability, but the mechanisms of their interactions with membrane proteins are not completely understood. We have undertaken the study of a prototypical G-protein-coupled receptor, the α-helical membrane protein rhodopsin from vertebrate retina, and explored the effects of inorganic salts on the thermal decay properties of both its inactive and photoactivated states. Under high salt concentrations, rhodopsin significantly increased its activation enthalpy change for thermal bleaching, whereas acid denaturation affected the formation of a denatured loose-bundle state for both the active and inactive conformations. This behavior seems to correlate with changes in protonated Schiff-base hydrolysis. However, chromophore regeneration with the 11-cis-retinal chromophore and MetarhodopsinII decay kinetics were slower only in the presence of sodium chloride, suggesting that in this case, the underlying phenomenon may be linked to the activation of rhodopsin and the retinal release processes. Furthermore, the melting temperature, determined by means of circular dichroism and differential scanning calorimetry measurements, was increased in the presence of high salt concentrations. The observed effects on rhodopsin could indicate that salts favor electrostatic interactions in the retinal binding pocket and indirectly favor hydrophobic interactions at the membrane protein receptor core. These effects can be exploited in applications where the stability of membrane proteins in solution is highly desirable.
Introduction
Membrane proteins comprise ∼30% of all proteins; however, in contrast to globular proteins, membrane proteins represent <0.2% of the solved high-resolution protein structures because of difficulties—such as low stability and poor expression levels—of applying classical methods of structural determination to proteins of this class (1–6). G-protein-coupled receptors (GPCRs) are the largest family of membrane-bound receptors and also the target of many drugs under investigation (7). Vision is one of the many processes mediated by GPCRs, in particular the photoreceptor protein rhodopsin (Rho), which captures light and converts it into an electrical signal. The visual pigment Rho is the most extensively studied member of the superfamily of GPCRs due to the fact that it constitutes >90% of the disk membrane protein in the outer segments of rod photoreceptor cells (8). Rho consists of seven transmembrane α-helices connected by loop domains (9). Light sensitivity is conferred by the presence of the 11-cis-retinal chromophore, which upon light absorption undergoes isomerization to its all-trans configuration. The stability of Rho is determined by several factors, such as structural features (10–12), ligands (13–15), membrane composition (16), detergents (17), and salts (18–21). Understanding the molecular interactions that stabilize or destabilize membrane proteins is essential to our understanding of their function (11,22,23). High stability of GPCRs in solution is desirable for biotechnological applications (24), biochemical assays, and structural studies. It has been previously shown that inorganic salts play a key role in the stability of proteins (25–35), and whereas a large number of studies are available for globular proteins, less information is available in the case of membrane proteins. Thus, we have undertaken a detailed analysis of the effect of several inorganic salts on the stability and conformational properties of Rho in both its inactive and active states. We observe that some salts clearly slow the thermal bleaching processes of this retinal protein and that this effect is proportional to salt concentration. Specifically, thermal bleaching results in the presence of high sodium chloride concentrations would reflect an increase in favorable tertiary interactions—in the retinal binding pocket environment—that may help to stabilize the correctly folded conformation of this GPCR. Ultraviolet-visible (UV-vis) spectroscopic measurements suggest that the presence of high salt could affect the hydrolysis rate of the protonated Schiff base. Chromophore regeneration kinetics and MetarhodopsinII (MetaII) decay experiments (measured by following the retinal release process by means of fluorescence spectroscopy) suggest a specific behavior for sodium chloride. Furthermore, differential scanning calorimetry (DSC) and circular dichroism (CD) analysis indicate that the secondary structure of dark-state inactive Rho is more resistant to denaturation in the presence of high salt concentrations, which may be related to improved interactions at the photoreceptor transmembrane core.
Experimental Procedures
Materials
Bovine retinas were from J. A. Lawson (Lincoln, NE). n-Dodecyl-β-D maltoside (DM) detergent was purchased from Anatrace (Maumee, OH). Water used for buffer preparation was Milli-Q (Millipore, Billerica, MA) quality (16–18 mΩ). Rod outer segment (ROS) membranes from frozen bovine retinas were prepared under dim red light using a sucrose gradient method. Briefly, the membranes were suspended in 70 mM potassium phosphate, 1 mM MgCl2, and 0.1 mM EDTA, pH 6.9, and pelleted by centrifugation. Then, membrane pellets were resuspended in 5 mM Tris-HCl (pH 7.5) and 0.5 mM MgCl2. Two alternating washes with these buffers were carried out to decrease the amount of any further contaminating proteins. Finally, ROS membranes were split into several aliquots and stored in the dark at −20°C. A 1-mg sample of ROS membranes was solubilized in 2 ml of Milli-Q water containing 1% DM in the dark for 1 h at 4°C. The solubilized sample was subjected to centrifugation for 30 min in a T865 Sorvall rotor (Thermo Scientific, Waltham, MA) at 30,000 rpm (to remove insoluble material), and the clear supernatant was used for the different assays perfomed. Samples were usually stored in the dark at 4°C, or at −80°C for long-term storage. Protein concentration was determined using a molar extinction coefficient value at λ = 500 nm of 40,600 M−1 cm−1, and a molecular mass of 40 kDa. Preparations showed UV-vis A280/A500 ratios in the 1.8–2.2 range.
Sample preparation
Rho was diluted in 100 mM HEPES buffer, pH 7.4, containing 0.05% DM, and the corresponding salt was added from a concentrated stock solution to the spectrophotometric cuvette to obtain the desired final salt concentration. The sample was then incubated in the dark for 10 min at room temperature, and UV-vis spectra were recorded at 2-min intervals over cycle mode to ensure that there was no spectral change with time. Control samples were used and treated according to the same protocol, but without salt addition.
Absorbance and fluorescence spectroscopy
UV-vis spectra measurements were carried out with a Cary 100 Bio spectrophotometer (Varian, Palo Alto, CA) equipped with water-jacketed cuvette holders connected to a Digiterm circulating water bath (Selecta, Barcelona, Spain) to control the temperature by a dual-cell Peltier accessory (Varian). All spectra were recorded in the 250–650 nm range, with a bandwidth of 2 mm, a response time of 0.1 s, and a scan speed of 250 nm min−1. First, salt incubation of Rho was carried out in the spectrophotometric cuvette for 10 min at room temperature so that spectral changes over time due to salt addition could be monitored. These samples were then used for photobleaching, thermal bleaching, regeneration, and acidification experiments.
Fluorescence assays were performed using a Photon Technologies QM-1 steady-state fluorescence spectrophotometer (PTI Technologies, Birmingham, NJ). Sample temperature was controlled with a cuvette holder Peltier accessory TLC 50 (Quantum Northwest, Liberty Lake, WA) connected to a hybrid liquid coolant system Reserator XT (Zalman, Garden Grove, CA). All fluorescence spectra were carried out by exciting the samples for 2 s at λ295 nm and a bandwidth slit of 0.5 nm and blocking the excitation beam for 28 s with a beam shutter to avoid photobleaching of the sample. Tryptophan emission was monitored at λ330 nm and a bandwidth slit of 5 nm. All assays were performed in triplicate.
Acidification kinetics (photoactivated and dark Rho)
Rho samples were illuminated for 30 s by using a 150-watt power source with a 495-nm cutoff filter and acidified by immediate addition of HCl to a final concentration of 100 mM. Spectra were recorded every 5 min at 15°C according to the experimental conditions described in the previous section. Dark samples were treated as described above, but without Rho illumination.
Thermal bleaching assay of Rho
Dark-state Rho thermal stability was followed in the absence and in the presence of 3.5 M salt, using two different spectroscopic techniques. In a first assay, the loss of absorbance at λ500 nm over time was monitored by UV-vis spectroscopy at 54°C. Thermodynamic parameters for the dark thermal bleaching process were determined as a function of temperature. This method regards thermal bleaching as an irreversible process (36). The thermodynamic parameters Ea, Δ‡Go, Δ‡Ho, and Δ‡So were calculated from the decay rate constants versus T−1 (Arrhenius plot) using the values for the slopes (k), which were derived from least-squares analysis. For this purpose, Rho samples in the dark, and in the absence or presence of 3.5 M salt, were thermally bleached at different temperatures in the range 48–58°C. Our calculations are based on the equations
and
where Ea is the activation energy and A is a collision factor, both obtained from the Arrhenius plot; R is the ideal gas constant; kB is Boltzmann's constant; and h is Planck's constant. In a complementary approach, fluorescence spectroscopy was also used to follow specific changes in Trp fluorescence from Rho due to retinal release (retinal acts as a quencher of Trp fluorescence in dark-state Rho) when treated at 55°C in the absence or presence of 3.5 M salt. Briefly, 0.5 μM Rho in 100 mM Hepes, pH 7.4, containing 0.05% DM was kept at 55°C and Trp fluorescence changes were measured over time. The t1/2 values for Trp fluorescence increase were determined by fitting the experimental data to a single-exponential curve using Sigma Plot version 11.0 computer software (Systat Software, Chicago, IL).
Salt effect on the Schiff-base linkage hydrolysis
Stability of the opsin-retinal Schiff-base linkage, in the absence and presence of the studied salts, was determined by means of an acidification assay. Briefly, Rho, in the absence or presence of salts (NaCl, KCl, and NH4Cl at 3.5 M), was treated at 55°C and aliquots were collected at 0, 5, 10, and 20 min in the absence of salt and at 0, 20, 40, 60, and 100 min in the presence of salt. These collected samples were immediately acidified with 2 N H2SO4 and absorbance at λ = 440 nm was measured. T1/2 values were determined by fitting the experimental data to an exponential decay curve using the Sigma Plot version 11.0 computer software (Systat Software).
Regeneration of Rho
Regeneration of Rho was carried out at room temperature in the dark. Briefly, 11-cis-retinal in a methanolic stock solution was added to a dark Rho sample in either the absence or presence of 3.5 M salt. Then, the sample was illuminated for 30 s by using a 150-watt power source with a 495-nm cutoff filter, and absorption spectra were immediately recorded at 2.0-min cycles over 2 h. Rho regeneration rate was monitored by the increase of absorbance at λ = 500 nm. Samples were eventually acidified to confirm the presence of Schiff-base-linked species.
Stability of the active and inactive state against acid denaturation
These assays were carried out with Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM. The reaction was started by adding 10% (v/v) of 1 M HCl, in the dark, to the thermostated sample and following spectral changes over time at 1-min intervals. In the case of the active-state samples, the same procedure was used, but the Rho samples were illuminated with a 150-watt power source equipped with an optic fiber guide and using a 495-nm cutoff filter for 30 s at maximum intensity before HCl addition.
Effect of salt on the active state (MetaII) by fluorescence spectroscopy
MetaII-state stability was determined by the increase in Trp fluorescence from opsin that occurs over time as the Schiff-base linkage is hydrolyzed in MetaII and the retinal is subsequently released. Briefly, 0.5 μM Rho in 100 mM Hepes, pH 7.4, containing 0.5% DM, in the absence or presence of 3.5 M NaCl, was kept at 20°C for 10 min and illuminated for 30 s. The t1/2 values, derived from curves measuring Trp fluorescence increase due to retinal release, were determined by fitting the experimental data to single-exponential curves using Sigma Plot version 11.0 (Systat Software).
Measuring thermal stability of Rho by DSC
DSC experiments were performed using an MC-II differential scanning calorimeter (Microcal, Amherst, MA) at a scan rate of 60°C/h. Rho samples (0.5 mg·ml−1) in 100 mM HEPES, pH 7.4, containing 0.05% DM, with and without salt, were used for calorimetric analysis. The excess heat capacity was obtained by subtracting the baseline, which is a thermal scan of the buffer alone. The melting temperatures (Tm) reported were derived as the temperatures corresponding to the maximum (baseline corrected) excess heat capacity curve.
Measuring thermal stability of Rho by CD spectroscopy
Thermal stability was also assessed with an 810 CD spectropolarimeter (Jasco, Great Dunmow, UK), and a single wavelength was chosen (222 nm) as a specific probe of the secondary-structure changes for dark-state Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, in the absence and presence of salt. Molar ellipticity was followed continuously as the temperature was raised in the range 30–80°C, with a scan rate of 60°C/h. The melting data, θ222 nm, were recorded in the range 35–85°C, and data collection was carried out at equilibrium. The equilibrium constant can be calculated from the fraction of unfolded protein at each temperature near the transition point (37). Since this is an equilibrium-based calculation, the absolute amount or concentration of the sample is not needed for the calculation. Also, the ΔG° for unfolding of Rho at each temperature can be calculated using , where R is the ideal gas constant and T the specific temperature. ΔG° is then plotted versus temperature, and using the slope, one can calculate the transition temperature, Tm, because at the Tm, the unfolded/folded species ratio is 1 and ΔG° = 0.
Results
Influence of salts on Rho thermal bleaching process
One way to examine the influence of salts on the dark ground-state conformation of Rho is thermal bleaching. This assay can provide information about changes in tertiary interactions within the retinal chromophore environment, after thermally induced isomerization of 11-cis-retinal to all-trans-retinal, caused by helical movements and molecular rearrangements during the semidenaturation process (38). The influence of salts on dark-state Rho thermal decay was determined at 50°C in the dark in the presence of different salts at 3.5 M concentration. Kinetic profiles of the thermal bleaching processes of Rho in the dark were obtained by using UV-vis spectroscopy. Absorbance changes in the visible chromophoric band at λ500 nm were monitored, at 50°C, as a function of time (Fig. 1 A). At this temperature, a slower decay of this band was observed in the presence of high salt concentration relative to the control sample (Fig. 1 B).
Figure 1.
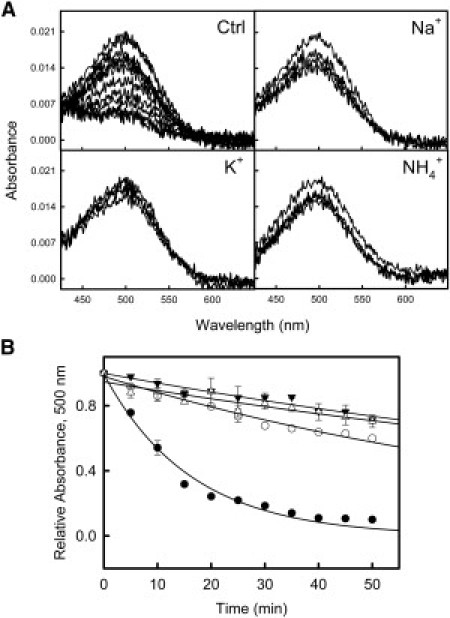
Rho thermal bleaching at 50°C. Rho samples, in 100 mM HEPES, pH 7.4, containing 0.05% DM, were spectroscopically analyzed in the presence of 3.5 M NaCl, KCl, and NH4Cl. (A) Spectra obtained from the thermal stability of the retinal chromophore environment tested in the presence of these salts. Ctrl refers to the control sample in the absence of high salt concentration. Spectra were recorded at 5-min intervals for a total time of 35 min. (B) Absorbance decay at λ500 nm was plotted versus time in the absence and presence of the different salts for the control sample (●), Na+ (○), K+ (▾), and NH4+(▵).
Rho thermal bleaching was also studied by monitoring the retinal release kinetics of three different Rho samples, treated at 55°C, by means of fluorescence spectroscopy. In this case, the t1/2 for the Trp fluorescence increase process is 31 ± 2 min in the presence of NaCl compared to the value of 14 ± 1 min for the control sample (Fig. S1 a in Supporting Material). These results were compared to the decrease of A500 nm for Rho at this temperature of 55°C. t1/2 for the thermal process was 28 ± 2 min in the presence and 7 ± 1 min in the absence of 3.5 M NaCl (Fig. S1 b). Similar results were obtained for KCl and NH4Cl by means of this fluorescence approach (33 ± 2 min and 35 ± 2 min, respectively).
We observed a linear dependence between thermal bleaching rates and salt concentration, with higher concentrations of salt increasing the t1/2 for the respective processes (Fig. 2A). Thus, the thermal process was slower in the presence of 4 M NaCl, with a rate constant at 50°C of k = −0.000399 ± 8·10−6 min−1, compared with the process in the presence of 2 M NaCl, k = −0.0085 ± 0.0003 min−1, or in the absence of salt, k = −0.0978 ± 0.002 min−1 observed. Thermodynamic parameters derived from Arrhenius plots obtained from experiments performed with different salts at different temperatures (Fig. 2 B), were calculated and are shown in Table 1. Here, Δ‡Ho and Δ‡Socomponents were significantly increased in all salts tested.
Figure 2.
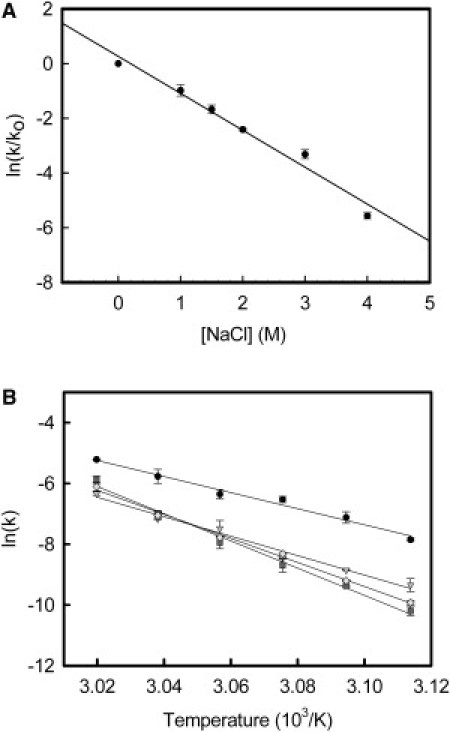
Dependence of thermal bleaching rates on salt concentration and temperature. (A) Variations on the decay rate constant values (k) of Rho thermal bleaching at different NaCl concentrations. Thermal bleaching of Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, at 55°C, was studied in the presence of 0, 1, 1.5, 2, 3, and 4 M NaCl. A linear dependence was found for rate constant k versus [NaCl] expressed in molar concentration. Rate constants of thermal bleaching were determined from the corresponding experimental data fit to a single-exponential function. The same tendency was observed in the cases of K+- and NH4+-containing samples (data not shown). In general, k values followed the equation , where k0 is the k value in the absence of salts, A+ can be Na+, K+, or NH4+, and κ is a constant dependent on the type of salt and the temperature. Values are the mean ± SE (n = 3). (B) Arrhenius plots of the dark-state thermal decay rates (k) for Rho in the presence of different salts. k was determined from thermal decay curves in the dark state of Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, in the absence (●), or presence of 3.5 M NaCl (▾), KCl (♦) and NH4Cl (■) at different temperatures between 48°C and 58°C. Symbols for salt-containing samples are in different shades of gray. Values represent the mean ± SE (n = 3).
Table 1.
Thermodynamic parameters for the thermal bleaching kinetics of Rho
| Salt | Δ‡Ho (kJ mol−1) | Δ‡So (kJ mol−1 K−1)103 | Δ‡Go (kJ mol−1) |
|---|---|---|---|
| Ctrl | 217.27 ± 0.19 | 366.78 ± 0.07 | 98.00 ± 0.01 |
| Na+ | 263.36 ± 0.17 | 495.90 ± 0.03 | 102.11 ± 0.02 |
| K+ | 326.40 ± 0.15 | 688.16 ± 0.06 | 102.64 ± 0.01 |
| NH4+ | 369.60 ± 0.20 | 819.69 ± 0.05 | 103.07 ± 0.01 |
These parameters were determined from Rho thermal stability assays. Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM was treated at different temperatures between 48°C and 58°C, in the absence and presence of 3.5 M NaCl, KCl, or NH4Cl. The Ea parameter was determined using values from the slopes of the Arrhenius plots (Fig. 3), which were derived from least-squares analysis. , , , and , where Ea is the activation energy and A is a collision factor, both obtained from the Arrhenius plot; R is the ideal gas constant, 8.314 J mol−1 K−1; kB is the Boltzmann constant, 1.38 J K−1; and h is Planck's constant, 6.26 × 10−34 J s. The values were derived for T = 52°C. These parameters were calculated from three independent experiments (p < 0.05).
Effect of salts on Schiff-base-linkage hydrolysis
Rho Schiff-base-linkage stability in the absence and presence of salt was determined by acidification, with the assay performed in triplicate. We found that salts stabilized the Schiff-base linkage by slowing down its hydrolysis (t1/2 = 6.5 ± 1 min, 14 ± 2 min, 15 ± 1 min, and 14 ± 2 min in the absence and presence of NaCl, KCl, and NH4Cl, respectively).
Chromophore regeneration and MetaII decay measurements
The effect of high salt concentrations on pigment regeneration with 11-cis-retinal after Rho photobleaching was also analyzed (Fig. 3 A). A slower regeneration kinetics was observed in the presence of NaCl, with regard to KCl or NH4Cl, which showed a behavior similar to that of the control sample without added salt (data not shown). t1/2 for the Rho regeneration process in the presence of 3.5 M NaCl was 18 ± 3 min, compared to 11.5 ± 1 min for the control sample. MetaII stability was studied by fluorescence spectroscopy, which measures retinal release upon sample illumination. The results showed that NaCl also increased the stability of the Rho active state (Fig. 3 B). The t1/2 for the MetaII assay in the presence of 3.5 M NaCl was determined to be 21 ± 2 min, compared to the value of 13 ± 3 min obtained in the absence of added salt. In contrast, KCl and NH4Cl gave similar behavior to that of the control sample (data not shown).
Figure 3.
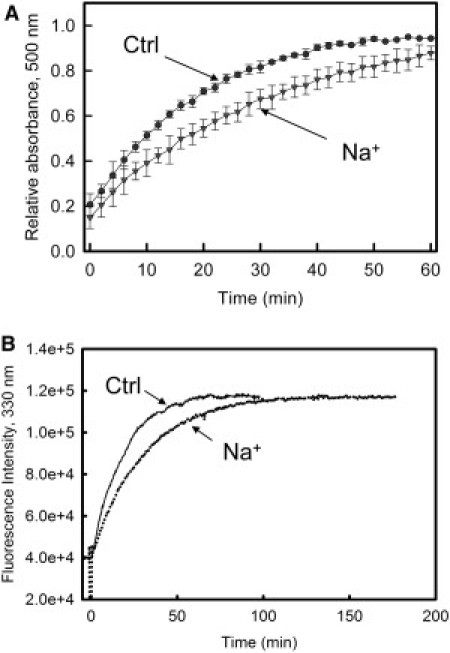
Effect of NaCl on Rho regeneration and MetaII stability. (A) Rho regeneration in the presence of NaCl. Rho in 100 mM HEPES, pH 7.4, containing 0.02% DM, with a 1.5 molar excess of 11-cis-retinal, was preincubated with or without 3.5 M NaCl and then illuminated for 30 s. The plot shows the time course of Rho regeneration by monitoring the absorbance increase at 500 nm at room temperature. Values are the mean ± SE (n = 3). (B) MetaII stability was determined by monitoring Trp fluorescence at λ330nm to measure retinal release upon Schiff-base hydrolysis. Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, in the absence (−) or in the presence of 3.5 M NaCl (⋅⋅⋅), was illuminated for 30 s and Trp fluorescence was measured by fluorescence spectroscopy at 20°C. Experiments were performed in triplicate.
Influence of salts on the acid denaturation of the active and inactive states of Rho
Addition of 100 mM HCl to detergent-solubilized Rho in the dark leads to denaturation of the dark state and formation of a 440-nm protonated-Shiff-base species. The transition from native dark state to the 440-nm denatured species proceeds via a slightly blue-shifted intermediate (λmax = 495 nm), with a reduced absorption coefficient, which is formed immediately after acidification and whose formation precedes the irreversible transition to the 440-nm denatured species (18). Acid denaturation of Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, was performed at 15°C, and the shift of the visible absorbance band to 440 nm was monitored for 15 min (Fig. 4). Total denaturation time was ∼12 min in the absence of salt, but a different behavior was observed depending on the type of salt used. NH4Cl increased Rho stability against denaturation, with a total denaturation time of ∼80 min. In the case of the NaCl- and KCl-treated samples, their total denaturation time was >200 min. Acid denaturation of the active conformation was performed as in the case of dark-state Rho, but an additional 30s illumination step was carried out before HCl addition. A stabilizing effect elicited by these salts was also observed for the active state (Fig. 5). In the case of the NaCl-containing sample, an absorbance increase at 475 nm could be detected (a semidenatured intermediate), and the kinetics of this increase was found to be proportional to the absorbance decrease at 380 nm, with an isosbestic point at 418 nm. After 70 min, we observed a gradual displacement of the maximum absorbance to 440 nm, which is interpreted as the complete denaturation of the intermediate formed at 475 nm. Acid denaturation of the active state in the presence of KCl was similar to that observed for NaCl, with the difference that the lifetime of the semidenatured intermediate at 475 nm was only 35 min, and as a consequence, the kinetics of the acid denaturation for the active state was slightly faster than in the presence of NaCl. Finally, acid denaturation in the presence of NH4Cl, was characterized by the nongradual appearance of the intermediate (as in the case of NaCl and KCl). However, the kinetics of the transition from the semidenatured intermediate (λmax = 475 nm) to complete denaturation (λmax = 440 nm) was similar to those for the sodium and potassium cases.
Figure 4.
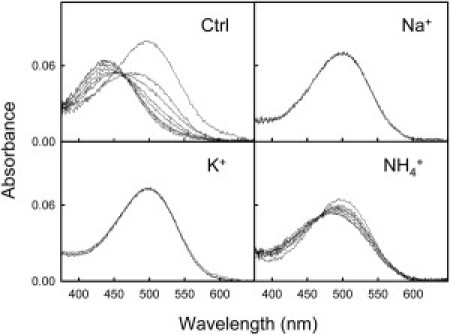
Influence of salts on the conformation of acid-denatured Rho. Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, was measured by Uv-vis spectrophotometry at 15°C in the absence and presence of 3.5 M NaCl, KCl, and NH4Cl. Samples in dark conditions were acidified by the addition of 1 M HCl up to 100 mM final concentration. Spectral changes were followed for a total time of 15 min for each condition.
Figure 5.
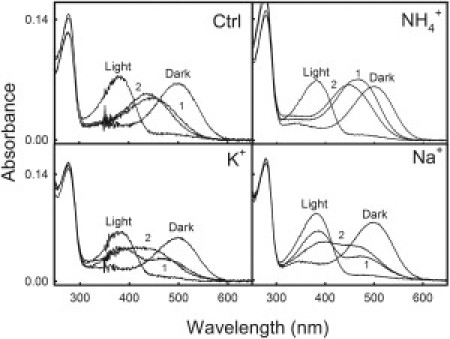
Influence of high salt concentrations on the acid denaturation of the active state of Rho. Acidification to 100 mM HCl final concentration of the active state of Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, was measured at 15°C. Spectral changes were recorded in the absence (Ctrl) and presence of NaCl, KCl, and NH4Cl. Sample spectra 1 and 2 were obtained 0 min and 2 min (control), 5 and 85 min (NH4+), 10 and 85 min (K+), and 15 and 85 min (Na+) after acidification.
DSC and CD analysis of Rho denaturation process
The influence of high salt concentration on Tm was determined by using DSC for unbleached Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM in the absence and in the presence of NaCl, KCl, and NH4Cl at 3.5 M. Individual thermal scans at fixed salt concentrations are shown in Fig. 6 A. The shape of the endotherm in the case of the control sample, as well as for samples containing NaCl or NH4Cl, was more symmetric than the endotherm for the KCl case. This asymmetry is characterized by the region below Tm exhibiting more gradual temperature dependence than the region above Tm. We could determine an increase in Tm for the different salts when compared with the control sample with no added salt (Fig. 6 A).
Figure 6.
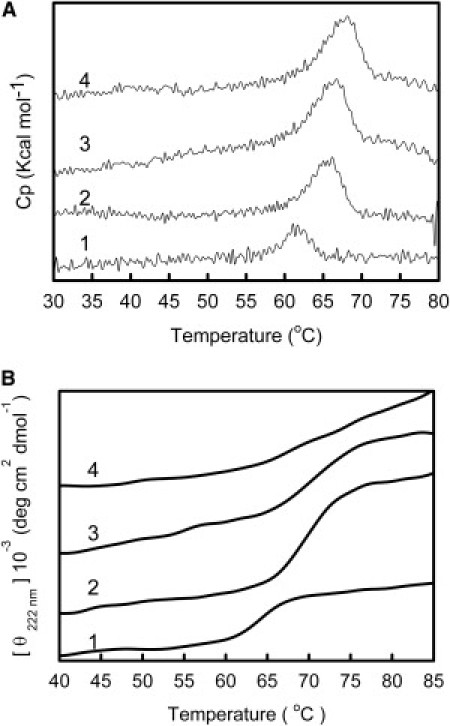
Effects of salts on Rho dark-state stability monitored by DSC and CD. (A) In the DSC assays, heat capacity (Cp) was recorded within the range of 20–90°C, with a scan rate of 60°C/h. Protein concentration was 0.5 mg ml−1 in 100 mM HEPES, pH 7.4, containing 0.05% DM, in the presence of 3.5 M NaCl, KCl, and NH4Cl. (B) Thermal unfolding of Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, in the absence and presence of NaCl, KCl, and NH4Cl, was monitored by CD following the ellipticity at 222 nm as a function of temperature, with a scan rate of 60°C/h. For DSC and CD in panels A and B, line 4 corresponds to the NH4+, line 3 to the K+, line 2 to the Na+, and line 1 to the control samples.
The effects of salt on the secondary structure—particularly on α-helical structure, which is better determined by CD analysis—of Rho were investigated by CD spectroscopy. The ellipticity at 222 nm was plotted versus temperature to obtain a thermal denaturation profile for Rho in 100 mM HEPES, pH 7.4, containing 0.05% DM, in the absence and presence of NaCl, KCl, or NH4Cl at 3.5 M (Fig. 6 B). The values of Tm determined from the calculations described in the Materials and Methods section are summarized in Table 2. Tm values increased for the samples containing NH4Cl, NaCl, and KCl, when compared with those for the control sample (without salt addition).
Table 2.
Determination of Rho stability by CD spectroscopy and DSC
| Sample | Tm (°C) CD | Tm (°C) DSC |
|---|---|---|
| Ctrl | 63.8 ± 0.6 | 61.8 ± 1 |
| Na+ | 68.8 ± 0.4 | 66.3 ± 0.5 |
| K+ | 68.4 ± 0.3 | 67.0 ± 0.6 |
| NH4+ | ND | 68.5 ± 0.4 |
Thermal stability of Rho in the presence of NaCl, KCl, and NH4Cl at 3.5 M, and in its absence, was measured by CD and DSC. For CD spectroscopy, the change in molar ellipticity at 222 nm was measured as a function of temperature for Rho samples (0.2 mg ml−1) in 5 mM HEPES buffer, pH 7.4, containing 0.05% DM, with a scan rate of 60°C/h and a cell of 0.1-cm path length. For DSC, the heat capacity of Rho was determined in 100 mM HEPES, pH 7.4, containing 0.05% DM, in the 20–90°C range, with a scan rate of 60°C/h. In both cases, Tm (°C) values were derived from the denaturation curves shown in Fig. 6, A and B. Values represent the average of three independent experiments. ND, not determined.
Discussion
A number of studies have been published that describe the effects of salt on globular soluble proteins, but much less information is available with regard to the stability of membrane proteins under high salt concentrations. In this context, the main aim of our study was to acquire further insights into the physicochemical nature of the effects of salt on dark-state Rho secondary (especially at the helical structure level) and tertiary structures (within the retinal chromophore helical environment). The Δ‡Ho increase (Table 1 and Fig. 3) observed in the Rho thermal bleaching process in the presence of the salts studied can be interpreted as a stabilizing effect, presumably due to favoring tertiary contacts within the transmembrane domain in the vicinity of the retinal chromophore, similar to what has been described in previous reports (28). In contrast, Δ‡So increase would have a different interpretation, pointing to a destabilizing effect arising from some degree of structural disorder, presumably associated with other domains of the protein. It is likely that the stabilizing effect seen at high salt concentrations of Na+, K+, or NH4+ on Rho dark-state chromophoric stability is the result of two contributions acting in opposite directions, as has been previously proposed (18).
Rho thermal bleaching processes were also investigated for other inorganic salts (i.e., NaI, LiCl, Na2SO4, MgCl2, and NaBr) to further analyze the effect of other anions and cations, and a clearly different behavior was observed (Fig. S2). All salts tested showed a stabilizing effect toward thermal bleaching except for NaI and LiCl. NaI was the only sodium salt that showed a destabilizing effect even at much lower concentrations. LiCl is a special case showing a destabilizing effect at low concentrations which we associate with specific effects on Rho structure (A. Reyes-Alcaraz, M. Martínez-Archundia, M. Morillo, E. Ramon, and P. Garriga, unpublished results). The results obtained for the thermal bleaching behavior of the different salts analyzed suggest that specific anions play an important role in salt behavior that cannot be overlooked. In our case, however, our study focuses on chloride salts, so we are mainly following the effect of the associated cation. These findings highlight the difficulty of proposing a unifying mechanism that integrates all the results reported in this study.
The stability of Rho in its dark inactive state is often inferred from hydroxylamine reactivity assays, which reflect the accessibility to the Schiff base of this reagent, thus providing an indirect measure of the structural status of the protein around the Schiff-base linkage. Hydroxylamine is a reagent used to study Rho tertiary structure stability indirectly by measuring its access to the Schiff base and subsequent breakage of the Schiff-base linkage. Previously reported results showed that the presence of increasing concentrations of NaCl progressively increased the reactivity of hydroxylamine with dark-state Rho, suggesting that exogenous NaCl addition decreases the thermal stability of the Rho dark state (18). In our case, we found only a small effect of high salt concentrations on the hydroxylamine reactivity of Rho (data not shown), which could not account for the stabilizing effect observed in the thermal bleaching assays. Thus, our results are in apparent disagreement with those previously reported (18). It should be noted that different buffer and detergent concentrations were used in that study. Furthermore, we carried out a detailed acidification analysis at high temperatures, finding a good correlation with the thermal bleaching results that suggests that the presence of the salt stabilized the Schiff-base linkage by slowing down its breakage rate. This result is consistent with the thermal bleaching results and is in agreement with our interpretation that high salt concentrations—under the specific experimental conditions studied—favor tertiary interactions that make the protein more resistant to high temperatures.
Previous studies have shown that under given specific experimental conditions—which are different from ours in both the detergent concentration and the molar Rho/detergent ratios used—there should be no loss of secondary structure in the thermal bleaching UV-vis assays at temperatures <60°C (38). Therefore, the decrease in absorbance at 500 nm would reflect thermal isomerization of the chromophore. This could lead to changes at the tertiary structure level, such as helical translocations, which could be the cause of Rho denaturation (38). This proposal is in agreement with our previous findings concerning thermally induced retinal isomerization (39).
In the case of the chromophore regeneration experiments, we found to our surprise that only NaCl—and not the other chloride salts analyzed—had a significant influence on this process, increasing the regeneration half-lifetime without affecting the maximal Rho regeneration. This could be interpreted as reflecting that, immediately after light exposure, all-trans-retinal remains attached to the chromophoric site longer when this salt is present than in the case of the control sample. In this case, our observations could be related to effects that salts may have on the aqueous environment, particularly on those molecules in the hydrating water shell in the external milieu of Rho. Therefore, the regeneration kinetic results could be interpreted as NaCl's effect on the properties of the surrounding aqueous environment of the protein, suggesting a role that may be related to that described in the case of the Hofmeister ion series (40–42). These results are consistent with the retinal release assay perfomed by fluorescence spectroscopy (which closely parallels the MetaII decay process), where a delay of the release was observed in the presence of 3.5 M NaCl, but not in the case of KCl and NH4Cl. This is in agreement with the slower hydrolysis of the Schiff-base linkage observed with the acidification assay. This specific effect caused by sodium ions may reflect direct binding to the protein, similar to that recently observed in the case of the D2 dopaminergic receptor (43). The increase in the regeneration half-lifetime (slower regeneration), and the delay in the retinal release, in the NaCl case may be two aspects of an underlying common phenomenon that would be more likely related to the photoactivation process of rhodopsin than to the stability of the dark conformation of the receptor observed in the thermal bleaching assays.
In a different set of experiments, we investigated the effect of salts on the overall thermal stability and particularly on secondary structure, which cannot be studied by the thermal bleaching analysis. Our DSC results indicate a stabilizing effect, for all tested salts, in the case of dark-state Rho, indicating that this stabilizing effect occurs not only at the tertiary interaction level (thermal bleaching) but also at the level of the secondary structure. The stabilizing effect—particularly for the α-helical secondary structure—could also be observed by CD spectroscopy. An additional observation is that the difference between the initial and final ellipticities for the denaturing process in the case of NaCl and KCl is larger than that observed for the control sample. To determine whether this change could have specific significance would require further study.
In conclusion, we have shown that several inorganic salts, i.e., NaCl, KCl, and NH4Cl, slow down the thermal bleaching of dark-state Rho and its Schiff-base hydrolysis rate. This can be interpreted as improved tertiary contacts in the chromophoric binding-site environment and favoring of electrostatic network interactions in the vicinity of the Schiff-base linkage. However, the specific effect seen for NaCl on chromophore regeneration and MetaII decay suggests that these effects are likely related to the activation mechanism of the receptor and to the associated retinal release process. On the other hand, salts had a stabilizing effect on the secondary structure of Rho, as determined by our complementary CD and DSC analyses. Tm values derived from these analyses were higher in the presence of the studied salts, suggesting an effect at structural levels beyond those of the retinal binding pocket. Our results represent a contribution to Rho stability studies—with potential extrapolation to other GPCRs—that may be useful either for structural analysis or, potentially, in nanobiotechnological applications.
Acknowledgments
We thank all the members of our laboratory and Prof. Juan Jesús Pérez for helpful discussions. Help from Elodia Serrano, Michael Orel, and Prof. Joan Manyosa (Autonomous University of Barcelona) with the calorimetric measurements is acknowledged. We also thank Dr. Núria Ferrer from the Barcelona Scientific Park for her help with the CD measurements.
This research was supported by grants from Ministerio de Investigación, Ciencia e Innovación (SAF2008-04943-C02-02) and from Comissionat per a Universitats i Recerca del DIUE de la Generalitat de Catalunya (2009 SGR 1402) to P.G. and predoctoral fellowships from CONACYT (Mexico City, Mexico) to A.R.A. and M.M.A.
Supporting Material
References
- 1.Unger V.M., Schertler G.F.X. Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys. J. 1995;68:1776–1786. doi: 10.1016/S0006-3495(95)80354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palczewski K., Kumasaka T., Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 3.Cherezov V., Rosenbaum D.M., Stevens R.C. High-resolution crystal structure of an engineered human β2-adrenergic G-protein coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warne T., Serrano-Vega M.J., Schertler G.F. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaakola V.-P., Griffith M.T., Stevens R.C. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu B., Chien E.Y.T., Stevens R.C. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malin C.L., Schiöth H.B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Natl. Rev. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 8.Landin J.S., Katragadda M., Albert A.D. Thermal destabilization of rhodopsin and opsin by proteolytic cleavage in bovine rod outer segment disk membranes. Biochemistry. 2001;40:11176–11183. doi: 10.1021/bi0100539. [DOI] [PubMed] [Google Scholar]
- 9.Ji T.H., Grossmann M., Ji I. G protein-coupled receptors. I. Diversity of receptor-ligand interactions. J. Biol. Chem. 1998;273:17299–17302. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- 10.Okada T., Fujiyoshi Y., Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl. Acad. Sci. USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanuj Sapra K., Park P.S.-H., Palczewski K. Detecting molecular interactions that stabilize native bovine rhodopsin. J. Mol. Biol. 2006;358:255–269. doi: 10.1016/j.jmb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Liu M.Y., Yan E.C. Thermal decay of rhodopsin: role of hydrogen bonds in thermal isomerization of 11-cis retinal in the binding site and hydrolysis of protonated Schiff base. J. Am. Chem. Soc. 2009;131:8750–8751. doi: 10.1021/ja903154u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S.M., Bolen W., McDowell J.H. Differential scanning calorimetry of bovine rhodopsin in rod-outer-segment disk membranes. Eur. J. Biochem. 1991;200:53–59. doi: 10.1111/j.1432-1033.1991.tb21047.x. [DOI] [PubMed] [Google Scholar]
- 14.Ahuja S., Crocker E., Smith S.O. Location of the retinal chromophore in the activated state of rhodopsin. J. Biol. Chem. 2009;284:10190–10201. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Malley M.A., Naranjo A.N., Robinson A.S. Analysis of adenosine A2a receptor stability: effects of ligands and disulfide bonds. Biochemistry. 2010;49:9181–9189. doi: 10.1021/bi101155r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soubias O., Niu S.-L., Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J. Am. Chem. Soc. 2008;130:12465–12471. doi: 10.1021/ja803599x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramon E., Marron J., Garriga P. Effect of dodecyl maltoside detergent on rhodopsin stability and function. Vision Res. 2003;43:3055–3061. doi: 10.1016/j.visres.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Vogel R., Siebert F. Conformation and stability of α-helical membrane proteins. 2. Influence of pH and salts on stability and unfolding of rhodopsin. Biochemistry. 2002;41:3536–3545. doi: 10.1021/bi016024f. [DOI] [PubMed] [Google Scholar]
- 19.Vogel R. Influence of salts on rhodopsin photoproduct equilibria and protein stability. Curr. Opin. Colloid Interface Sci. 2004;9:133–138. [Google Scholar]
- 20.del Valle L.J., Ramon E., Garriga P. Zinc-induced decrease of the thermal stability and regeneration of rhodopsin. J. Biol. Chem. 2003;278:4719–4724. doi: 10.1074/jbc.M210760200. [DOI] [PubMed] [Google Scholar]
- 21.Park P.S.-H., Sapra K.T., Muller D.J. Stabilizing effect of Zn2+ in native bovine rhodopsin. J. Biol. Chem. 2007;282:11377–11385. doi: 10.1074/jbc.M610341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thévenin D., Lazarova T. Stable interactions between the transmembrane domains of the adenosine A2A receptor. Protein Sci. 2008;17:1188–1199. doi: 10.1110/ps.034843.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komolov K.E., Aguilà M., Koch K.W. On-chip photoactivation of heterologously expressed rhodopsin allows kinetic analysis of G-protein signaling by surface plasmon resonance spectroscopy. Anal. Bioanal. Chem. 2010;397:2967–2976. doi: 10.1007/s00216-010-3876-4. [DOI] [PubMed] [Google Scholar]
- 24.Reyes-Alcaraz A., Tzanov T., Garriga P. Stabilization of membrane proteins: the case of G-protein coupled receptors. Eng. Life Sci. 2008;8:207–217. [Google Scholar]
- 25.Schwartz C.P., Uejio J.S., Saykally R.J. Investigation of protein conformation and interactions with salts via X-ray absorption spectroscopy. Proc. Natl. Acad. Sci. USA. 2010;107:14008–14013. doi: 10.1073/pnas.1006435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pegram L.M., Wendorff T., Record M.T., Jr. Why Hofmeister effects of many salts favor protein folding but not DNA helix formation. Proc. Natl. Acad. Sci. USA. 2010;107:7716–7721. doi: 10.1073/pnas.0913376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sedlák E., Stagg L., Wittung-Stafshede P. Effect of Hofmeister ions on protein thermal stability: roles of ion hydration and peptide groups? Arch. Biochem. Biophys. 2008;479:69–73. doi: 10.1016/j.abb.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 28.Boström M., Williams D.R., Ninham B.W. Hofmeister effects in membrane biology: the role of ionic dispersion potentials. Phys. Rev. E. 2003;68:041902. doi: 10.1103/PhysRevE.68.041902. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Cremer P.S. Interactions between macromolecules and ions: the Hofmeister series. Curr. Opin. Chem. Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H.-X. Interactions of macromolecules with salt ions: an electrostatic theory for the Hofmeister effect. Proteins. 2005;61:69–78. doi: 10.1002/prot.20500. [DOI] [PubMed] [Google Scholar]
- 31.Gloss L.M., Placek B.J. The effect of salts on the stability of the H2A-H2B histone dimer. Biochemistry. 2002;41:14951–14959. doi: 10.1021/bi026282s. [DOI] [PubMed] [Google Scholar]
- 32.Lavelle L., Fresco J.R. Stabilization of nucleic acid triplexes by high concentrations of sodium and ammonium salts follows the Hofmeister series. Biophys. Chem. 2003;105:681–699. doi: 10.1016/s0301-4622(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 33.Ramos C.H.I., Baldwin R.L. Sulfate anion stabilization of native ribonuclease A both by anion binding and by the Hofmeister effect. Protein Sci. 2002;11:1771–1778. doi: 10.1110/ps.0205902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright D.B., Banks D.D., Gloss L.M. The effect of salts on the activity and stability of Escherichia coli and Haloferax volcanii dihydrofolate reductases. J. Mol. Biol. 2002;323:327–344. doi: 10.1016/s0022-2836(02)00916-6. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu S., McLaren W.M., Matubayasi N. The Hofmeister series and protein-salt interactions. J. Chem. Phys. 2006;124:234905. doi: 10.1063/1.2206174. [DOI] [PubMed] [Google Scholar]
- 36.Davidson F.F., Loewen P.C., Khorana H.G. Structure and function in rhodopsin: replacement by alanine of cysteine residues 110 and 187, components of a conserved disulfide bond in rhodopsin, affects the light-activated metarhodopsin II state. Proc. Natl. Acad. Sci. USA. 1994;91:4029–4033. doi: 10.1073/pnas.91.9.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creighton T.E. Oxford University Press; New York: 1997. Protein Structure: A Practical Approach. [Google Scholar]
- 38.Liu J., Liu M.Y., Yan E.C. Thermal decay of rhodopsin: role of hydrogen bonds in thermal isomerization of 11-cis retinal in the binding site and hydrolysis of protonated Schiff base. J. Am. Chem. Soc. 2009;131:8750–8751. doi: 10.1021/ja903154u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Valle L.J., Ramon E., Garriga P. Specific isomerization of rhodopsin-bound 11-cis-retinal to all-trans-retinal under thermal denaturation. Cell. Mol. Life Sci. 2003;60:2532–2537. doi: 10.1007/s00018-003-3113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geisler M., Pirzer T., Hugel T. Hydrophobic and Hofmeister effects on the adhesion of spider silk proteins onto solid substrates: an AFM-based single-molecule study. Langmuir. 2008;24:1350–1355. doi: 10.1021/la702341j. [DOI] [PubMed] [Google Scholar]
- 41.Baldwin R.L. How Hofmeister ion interactions affect protein stability. Biophys. J. 1996;71:2056–2063. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broering J.M., Bommarius A.S. Evaluation of Hofmeister effects on the kinetic stability of proteins. J. Phys. Chem. B. 2005;109:20612–20619. doi: 10.1021/jp053618+. [DOI] [PubMed] [Google Scholar]
- 43.Selent J., Sanz F., de Fabritiis G. Induced effects of sodium ions on dopaminergic G-protein coupled receptors. PLoS Comput. Biol. 2010;6:e1000884. doi: 10.1371/journal.pcbi.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrens D.L., Khorana H.G. Structure and function in rhodopsin: measurement of the rate of the metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


