Abstract
We have observed the assembly of the staphylococcal pore-forming toxin α-hemolysin using single-molecule fluorescence imaging. Surprisingly, assembly from the monomer to the complete heptamer is extremely rapid, occurring in <5 ms. No lower order oligomeric intermediates are detected. Monte Carlo simulation of our experiment shows that assembly is diffusion limited, and pore formation is dependent on the stability of intermediate species. There are close similarities between bacterial pore-forming toxins, such as staphylococcal α-hemolysin, the anthrax protective antigen, and the cholesterol-dependent cytolysins, and their eukaryotic analogs, such as the complement pore membrane attack complex and perforin domain. The assembly mechanism we have observed for α-hemolysin provides a simple model that aids our understanding of these important pore formers.
Introduction
Pore-forming proteins play a vital role in the eukaryotic immune response, where immune surveillance can lead to the targeted-attack of infected cells and bacteria via pore generation (1–4). Conversely, many bacteria use similar protein pores to kill target cells and acquire nutrients from their host (5). Interest in these pore-forming proteins has increased because of their use as tools in nanotechnology, including biosensing and single-molecule DNA sequencing (6). However, the mechanisms by which many of these multimeric membrane proteins assemble to form pores are not well understood. Fortunately, the close structural similarities between prokaryotic β-barrel pore-forming toxins (βPFTs) (7,8) and their eukaryotic analogs have shown that much can be learned about the general mechanisms of βPFT assembly by studying simple prokaryotic pore formers (9). For example, recent high-resolution crystal structures have demonstrated a large degree of homology between the cholesterol-dependent cytolysins and the complement pore membrane attack complex and perforin domain (3–5,9,10).
Staphylococcal α-hemolysin, is the archetypal βPFT. Its assembly requires a lipid bilayer: First, α-hemolysin monomers bind to the bilayer surface (5). Phosphocholine (11,12) and the disintegrin and metalloprotease 10 (13) have been identified as potential low- and high-affinity receptors for the protein. Second, protein diffusion on the lipid bilayer results in lateral contact between monomers. Monomers assemble on the membrane by an as yet uncharacterized process to form a heptameric ring-shaped intermediate termed a prepore (14–17). Third, prepores spontaneously insert a membrane-spanning β-barrel domain into the lipid bilayer, forming a water-filled nanopore (7). Pore formation subsequently affects the osmotic balance of the target cell and permits small molecule outflow (18).
Here, we follow the assembly of staphylococcal α-hemolysin using single-molecule fluorescence imaging. To image rapidly diffusing α-hemolysin monomers and observe their assembly into higher order oligomers requires bilayer longevity, high signal to noise, high time resolution and control of lipid composition. We have recently developed a new, to our knowledge, synthetic mimic of the cell membrane that fulfills these requirements; we form a Droplet Interface Bilayer by contact of two lipid monolayers between a nanoliter aqueous droplet and a hydrogel support immersed in a solution of phospholipid in hexadecane (Fig. 1 A) (19–22). To monitor the entire assembly process we introduced Cy3b-labeled α-hemolysin monomers into the droplet using a piezo-driven nanoinjector. The labeled α-hemolysin species were imaged on the bilayer using total internal reflection fluorescence microscopy (Fig. 1).
Figure 1.
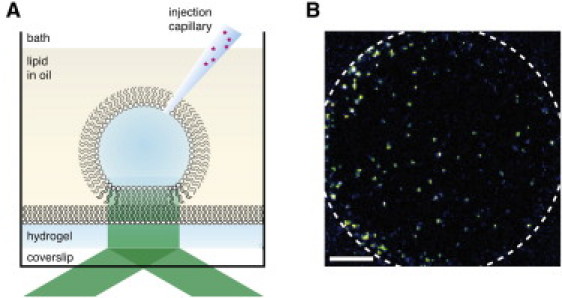
Diagram of experiment. (A) Singly labeled α-hemolysin monomers are injected into a droplet that contains an interface bilayer and are imaged by total internal reflection fluorescence microscopy. (B) A single frame (5 ms) from an acquisition showing fluorescent α-hemolysin monomers diffusing on a droplet interface bilayer. The white dashed line illustrates the edge of the lipid bilayer (scale bar 10 μm).
Materials and Methods
Single labeling of monomers was achieved by introduction of a C-terminal mutant cysteine and subsequent reaction with Cy3b maleimide (Fig. S1 in the Supporting Material). Absorption measurements and mass spectrometry confirmed a 1:1 labeling ratio while hemolytic activity and electrical conductance of the labeled monomer were unaffected with respect to the wild-type protein. Single-channel recording also confirmed that the monomer forms functional pores in the droplet interface bilayers used for imaging (see the Supporting Material).
Droplet interface bilayers were prepared following our previously reported methods (19–22), which are fully described in the Supporting Material. Briefly, an agarose substrate on a glass coverslip and an aqueous droplet are allowed to equilibrate separately for 15 min in a solution of (1,2-diphytanoyl-sn-glycero-3-phosphatidylchlorine; DPhPC) lipid in hexadecane (∼5 mM) to form lipid monolayers at the oil-water interface. The aqueous droplet is then pipetted onto the agarose and a lipid bilayer forms spontaneously where the monolayers of the droplet and the agarose contact.
Monte Carlo simulations modeled 10 particles in a two-dimensional box of side length 900 nm. The particle radius was 1 nm. The time step of simulation was 51.34 ns and was chosen to ensure the root mean-square motion of the particle in one dimension within the step size of the order of a particle radius per step given the known diffusion coefficient of the monomer (20). The spatial resolution of the simulation was 0.155 nm or 1/10th of a step size.
Results and Discussion
Unexpectedly, we did not observe the stepwise growth of a complex during oligomer formation; rather we observed the rapid formation of individual fully assembled oligomers from α-hemolysin monomers in an apparent single step that occurred in <5 ms (Fig. 2 A and Movie S1). Only monomers and fully assembled oligomers were detected, and no intermediates were detected at any point during assembly, regardless of monomer concentration. The time resolution of these experiments was 5 ms. Assembled oligomers were stable after formation and diffused more slowly than monomers (Fig. 2 B and Movie S1).
Figure 2.
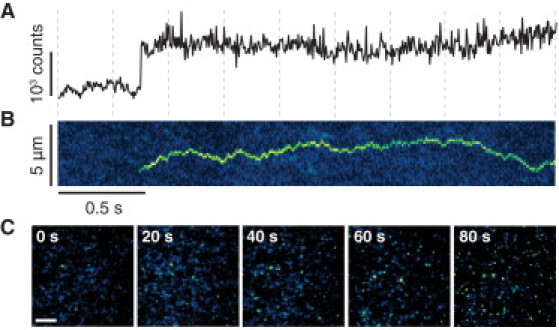
Rapid assembly of α-hemolysin oligomers. (A) Intensity trace for the appearance of a single oligomer (5 ms resolution). (B) Kymograph showing the same oligomer appearing and diffusing. Each pixel in the horizontal axis represents a single 5 ms frame, the vertical axis represents the lateral diffusion in one arbitrarily chosen dimension on the lipid bilayer. (C) Time-lapse sequence showing the appearance of oligomers at 20 s intervals (imaging commences 30 s after injection), bright oligomers can be seen above the intensity of the monomeric protein (scale bar 10 μm).
To establish the stoichiometry of the observed α-hemolysin oligomers, we allowed assembly to occur in darkness before analyzing all diffraction-limited fluorescent spot intensities (Fig. 3 A). An intensity distribution with two clear peaks was observed. The higher intensity peak of 7.0 ± 1.2 multiples brighter than the lower intensity peak (see Fig. 3 C). In a separate experiment, photobleaching at higher laser intensities enabled determination of the stoichiometries of the complexes by counting the stepwise loss of fluorescence intensity from diffraction-limited spots as we had previously observed for purified preassembled heptamers ((23) and Supporting Material). By this method, we assigned the lower intensity distribution to the fluorescence from single fluorophores (3,821 photobleaching trajectories). Analysis of the photobleaching trajectories of the oligomeric species revealed a mean of seven steps per spot, as expected (23) (see the Supporting Material for details of 152 oligomeric photobleaching trajectories) (Fig. 3, B and D). We therefore attribute the peaks in the intensity distribution to monomers and heptamers. Lower order oligomeric intermediates were not detected (Fig. 3 C). Because we are able to detect monomers, if oligomeric intermediates are formed in the assembly of α-hemolysin they must persist for less than the temporal resolution of this experiment. This rapid assembly is consistent with the current consensus from biochemical evidence (17) where nontime-resolved biochemical cross-linking experiments have not detected intermediates other than heptameric prepores.
Figure 3.
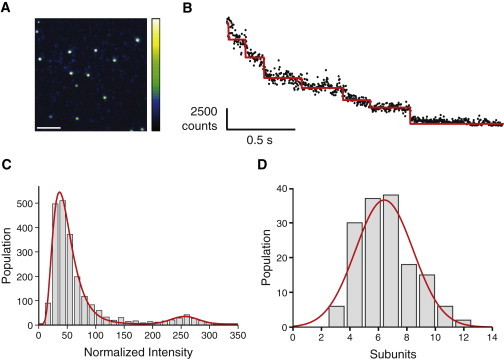
Oligomer stoichiometry. (A) Single frame (5 ms) from a droplet interface bilayer acquired 30 min after injection of labeled protein. Bright oligomers and monomers are resolved as separate fluorescent spots (scale bar 5 μm). (B) An example oligomer photobleaching trajectory. The intensity is plotted (black circles) with detected steps overlaid in red. Steps were detected automatically as jumps in intensity greater than three standard deviations above the average noise in the unfiltered data. Seven stepwise decreases in fluorescence intensity are detected corresponding to complete photobleaching of a heptameric oligomer. (C) A normalized (see the Supporting Material) fluorescent spot intensity histogram showing two intensity distributions. The histogram was fitted with a sum of log-normal and normal probability density functions by least-squares minimization. The lower intensity peak (monomers) has a mean value of 36.4 ± 0.3, and the higher intensity peak (heptamers) has a mean value of 256.1 ± 5.9, corresponding to 7.0 ± 1.2 multiples of the monomer intensity. (D) Histogram of oligomer stoichiometries. This distribution is fitted with a Gaussian with a mean value of 6.4 ± 0.2 steps.
To confirm that the observed complexes were related to pore-formation, we performed simultaneous measurements of complex appearance, by time-lapse imaging, and pore insertion, by single-channel recording (Fig. 4). The number of appearing oligomers and the number of pores inserting into the bilayer were correlated. Simultaneous measurements that allowed correlation of single oligomer appearances and insertion events were not feasible, because at the concentrations where it was possible to observe appearance events there were too many pores inserted and inserting into the bilayer to resolve individual events. Conversely, at concentrations where individual insertion events could be observed electrically, complex assembly could not be observed continuously due to photobleaching. Although not definitive, these data (Fig. 4) suggest that the prepore intermediate is short lived.
Figure 4.
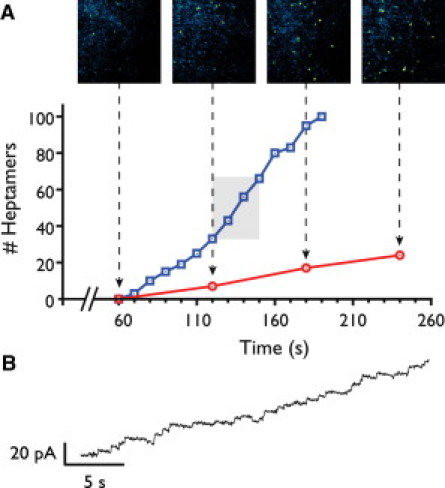
Comparison of single molecule fluorescence with single-channel recording. (A) Pore insertions as measured by increases in electrical current (blue line) are compared with a simultaneous time-lapsed fluorescence acquisition (solid red line). The fluorescent events are observed from the inset panels with imaging area 4.9 × smaller than the total bilayer area. (B) Section of electrical trace corresponding to the period between 120 and 150 s.
To further understand the assembly process, we calculated the number of monomer-monomer interactions that can occur per unit area within the timescale of heptamer formation. Following Smoluchowski's theory developed for bimolecular reactions (see review (24)), we calculate a rate of collisions between monomers of 9,401 μm−2 s−1 (corresponding to ∼107 collisions in each image per second). This number is far greater than the observed rate of heptamer appearance, 0.199 μm−2 s−1, indicating that the vast majority of monomer-monomer collisions do not result in oligomerization (see Supporting Material for details).
We also used a two-dimensional Monte Carlo simulation of our experiment (Fig. 5) to compare the relative probability of oligomer formation and monomer-monomer collision events. If purely diffusive encounters are modeled, where there are no persistent interactions between monomers, our simulations show that only two and three body collisions occur in any significant number (P = 0.003 for a triple monomer collision relative to the monomer-monomer collision probability). As might be expected, this cannot explain the experimentally observed heptameric complex formation rate.
Figure 5.
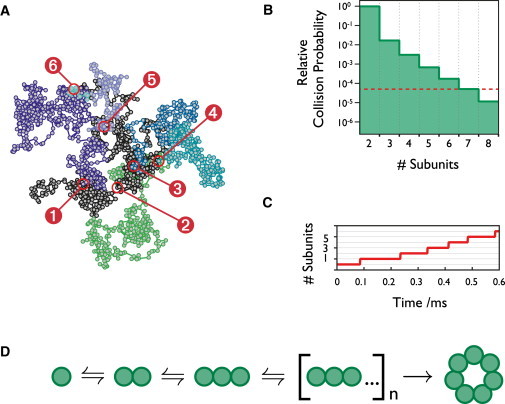
Monte Carlo simulation of assembly. (A) Example trajectory of monomer addition events during the formation of an individual α-hemolysin complex (51 ns simulation step). (B) Probability of a monomer colliding with a forming complex, relative to the probability of a monomer-monomer collision (n = 868, 372) vs. the size of the complex formed upon collision. Assembly steps are reversible . Red dotted line indicates the experimentally observed probability of heptamer formation. (C) Time-dependent heptamer assembly for the example trajectory shown in (A), each step is the stochastic addition of a monomer to a nascent complex. (D) Proposed assembly mechanism. All steps during complex formation are reversible, except the ring formation step, which is irreversible.
We can only account for the observed probability of heptamer formation (Fig. 5 B, red dotted line) by including the possibility that collisions between monomers result in the formation of a transiently stable complex. By allowing monomer-monomer collisions to be sticky, we reproduce the observed heptamer formation rate when the mean persistence time following a collision of two species on the membrane is 51.6 μs for the monomer surface densities present in our experiment. For this persistence, we calculate the mean time taken for a heptameric complex to assemble as 361 μs. This is consistent with our experimental observations (Fig. 2) that show complexes must assemble within 5 ms.
Conclusion
On the basis of this evidence, we propose the following model for the cooperative assembly of α-hemolysin (Fig. 5 D): 1), Monomers first bind to phosphocholine lipids and diffuse on the lipid bilayer; 2), Monomers undergo many collisions, which result in transient dimer formation. These dimers have a lifetime of ∼50 μs after which they dissociate to monomers; 3), Monomer collisions with the transient complex result in reversible stepwise growth; 4), Formation of the heptamer yields a stable complex that does not dissociate. This mechanism very rarely results in complete heptamer formation as almost all of the intermediate complexes rapidly dissociate.
The intensity distribution after assembly is complete (Fig. 3 C) shows only monomer and heptamer populations; smaller oligomers were notably absent. Although our biochemical evidence suggests a labeling ratio of 1:1.08, even if the labeling efficiency was lower than predicted, given our signal to noise, we would still observe single step changes in intensity when complexes are formed. Poor labeling efficiency would merely affect the accuracy of fit to the intensity and photobleaching step distributions. The absence of lower order oligomers is consistent with our model: Initially, when monomer concentrations are high, the assembly rate of monomers to oligomers, a sequence of bimolecular reactions, is greater than any disassembly rate of lower order oligomers back to monomers, a sequence of unimolecular reactions. As assembly continues, the surface monomer concentration is depleted as stable heptamers form irreversibly. This results in a decreasing assembly rate until the competing disassembly dominates, leading to a final population consisting only of heptamers and monomers. The fact that we do not observe oligomers with greater than seven subunits implies that either the final terminating step in oligomerization is either very rapid, or the disassembly rate of higher order (>7) oligomers is itself rapid. In this case the terminating step may be the ring formation of the heptamer, in an analogous way to other cyclic complexes (25). This all or none process has been suggested previously for the mechanism of assembly of perfringolysin O (26).
The rotational correlation time for lipids is around 10 μs, whereas integral membrane proteins have rotational correlation times between 10 μs and 1 ms (27). As the α-hemolysin monomer diffuses on the membrane and does not penetrate through the lipid bilayer, it is likely that it has a rotational correlation time closer to 10 μs. This is consistent with our calculated lifetime for intermediate steps (∼50 μs) as the monomers could undergo several rotations before the next monomer collision, enabling effective sampling of the binding interface.
It is interesting to consider why the assembly of α-hemolysin might need to proceed by a mechanism with reversible intermediate steps. Conceivably, the mechanism may have evolved to overcome limitations posed by low concentrations present on target cell surfaces; where the stalled assembly of lower order intermediates would inhibit the formation of functional pores.
This proposed mechanism of assembly is not an exclusive model consistent with our experimental observations. Another possible mechanism would require a rare monomer activation step, followed by rapid and successful accumulation of monomers into the activated complex at each subsequent diffusive encounter with a monomer. However, given the lack of biochemical evidence to support such an activation step, we favor a model with rapid reversible intermediate steps in assembly. Our results provide an interesting comparison to previous pioneering work by Higuchi and co-workers on the assembly mechanism of the closely related bicomponent γ-hemolysin pores (28,29), where the authors concluded that lower order oligomer intermediates were detected in the assembly pathway. Droplet interface bilayers yield a higher signal/noise ratio for single molecule fluorescence than could be expected when monitoring protein assembly on a cell membrane, and gives us confidence in our proposed model for α-hemolysin (30).
The experimental approach we describe here is applicable to the study of the assembly of other multimeric membrane protein complexes (31). It will be interesting to apply our methods to other pore-forming proteins, particularly those with much larger subunit stoichiometries (3,4,32). Whereas α-hemolysin assembly is extremely rapid, we would expect to resolve assembly intermediates for larger pore complexes.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council and the Engineering and Physical Sciences Research Council. Bríd Cronin is an EPSRC Postdoctoral Research Fellow.
Supporting Material
References
- 1.Iacovache I., Bischofberger M., van der Goot F.G. Structure and assembly of pore-forming proteins. Curr. Opin. Struct. Biol. 2010;20:241–246. doi: 10.1016/j.sbi.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Voskoboinik I., Smyth M.J., Trapani J.A. Perforin-mediated target-cell death and immune homeostasis. Nat. Rev. Immunol. 2006;6:940–952. doi: 10.1038/nri1983. [DOI] [PubMed] [Google Scholar]
- 3.Rosado C.J., Buckle A.M., Whisstock J.C. A common fold mediates vertebrate defense and bacterial attack. Science. 2007;317:1548–1551. doi: 10.1126/science.1144706. [DOI] [PubMed] [Google Scholar]
- 4.Hadders M.A., Beringer D.X., Gros P. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science. 2007;317:1552–1554. doi: 10.1126/science.1147103. [DOI] [PubMed] [Google Scholar]
- 5.Bayley H. Membrane-protein structure: piercing insights. Nature. 2009;459:651–652. doi: 10.1038/459651a. [DOI] [PubMed] [Google Scholar]
- 6.Branton D., Deamer D.W., Schloss J.A. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song L., Hobaugh M.R., Gouaux J.E. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 8.Katayama H., Janowiak B.E., Fisher M.T. GroEL as a molecular scaffold for structural analysis of the anthrax toxin pore. Nat. Struct. Mol. Biol. 2008;15:754–760. doi: 10.1038/nsmb.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukoyanova N., Saibil H.R. Friend or foe: the same fold for attack and defense. Trends Immunol. 2008;29:51–53. doi: 10.1016/j.it.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Law R.H.P., Lukoyanova N., Whisstock J.C. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature. 2010;468:447–451. doi: 10.1038/nature09518. [DOI] [PubMed] [Google Scholar]
- 11.Galdiero S., Gouaux E. High resolution crystallographic studies of α-hemolysin-phospholipid complexes define heptamer-lipid head group interactions: implication for understanding protein-lipid interactions. Protein Sci. 2004;13:1503–1511. doi: 10.1110/ps.03561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valeva A., Hellmann N., Bhakdi S. Evidence that clustered phosphocholine head groups serve as sites for binding and assembly of an oligomeric protein pore. J. Biol. Chem. 2006;281:26014–26021. doi: 10.1074/jbc.M601960200. [DOI] [PubMed] [Google Scholar]
- 13.Wilke G.A., Bubeck Wardenburg J. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus α-hemolysin-mediated cellular injury. Proc. Natl. Acad. Sci. USA. 2010;107:13473–13478. doi: 10.1073/pnas.1001815107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker B., Krishnasastry M., Bayley H. Assembly of the oligomeric membrane pore formed by Staphylococcal α-hemolysin examined by truncation mutagenesis. J. Biol. Chem. 1992;267:21782–21786. [PubMed] [Google Scholar]
- 15.Walker B., Braha O., Bayley H. An intermediate in the assembly of a pore-forming protein trapped with a genetically-engineered switch. Chem. Biol. 1995;2:99–105. doi: 10.1016/1074-5521(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 16.Fang Y., Cheley S., Yang J. The heptameric prepore of a staphylococcal α-hemolysin mutant in lipid bilayers imaged by atomic force microscopy. Biochemistry. 1997;36:9518–9522. doi: 10.1021/bi970600j. [DOI] [PubMed] [Google Scholar]
- 17.Kawate T., Gouaux E. Arresting and releasing Staphylococcal α-hemolysin at intermediate stages of pore formation by engineered disulfide bonds. Protein Sci. 2003;12:997–1006. doi: 10.1110/ps.0231203 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husmann M., Beckmann E., Bhakdi S. Elimination of a bacterial pore-forming toxin by sequential endocytosis and exocytosis. FEBS Lett. 2009;583:337–344. doi: 10.1016/j.febslet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 19.Bayley H., Cronin B., Wallace M. Droplet interface bilayers. Mol. Biosyst. 2008;4:1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson J.R., Heron A.J., Wallace M.I. Enhanced stability and fluidity in droplet on hydrogel bilayers for measuring membrane protein diffusion. Nano Lett. 2007;7:3875–3878. doi: 10.1021/nl071943y. [DOI] [PubMed] [Google Scholar]
- 21.Heron A.J., Thompson J.R., Wallace M.I. Direct detection of membrane channels from gels using water-in-oil droplet bilayers. J. Am. Chem. Soc. 2007;129:16042–16047. doi: 10.1021/ja075715h. [DOI] [PubMed] [Google Scholar]
- 22.Heron A.J., Thompson J.R., Wallace M.I. Simultaneous measurement of ionic current and fluorescence from single protein pores. J. Am. Chem. Soc. 2009;131:1652–1653. doi: 10.1021/ja808128s. [DOI] [PubMed] [Google Scholar]
- 23.Das S.K., Darshi M., Bayley H. Membrane protein stoichiometry determined from the step-wise photobleaching of dye-labelled subunits. ChemBioChem. 2007;8:994–999. doi: 10.1002/cbic.200600474. [DOI] [PubMed] [Google Scholar]
- 24.Keizer J. Diffusion effects on rapid bimolecular chemical reactions. Chem. Rev. 1987;87:167–180. [Google Scholar]
- 25.Hunter C.A., Anderson H.L. What is cooperativity? Angew. Chem. Int. Ed. 2009;48:7488–7499. doi: 10.1002/anie.200902490. [DOI] [PubMed] [Google Scholar]
- 26.Hotze E.M., Heuck A.P., Tweten R.K. Monomer-monomer interactions drive the prepore to pore conversion of a β-barrel-forming cholesterol-dependent cytolysin. J. Biol. Chem. 2002;277:11597–11605. doi: 10.1074/jbc.M111039200. [DOI] [PubMed] [Google Scholar]
- 27.Murray E.K., Restall C.J., Chapman D. Monitoring membrane protein rotational diffusion using time-averaged phosphorescence. Biochim. Biophys. Acta. 1983;732:347–351. doi: 10.1016/0005-2736(83)90050-0. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen V.T., Kamio Y., Higuchi H. Single-molecule imaging of cooperative assembly of γ-hemolysin on erythrocyte membranes. EMBO J. 2003;22:4968–4979. doi: 10.1093/emboj/cdg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen A.H., Nguyen V.T., Higuchi H. Single-molecule visualization of environment-sensitive fluorophores inserted into cell membranes by staphylococcal γ-hemolysin. Biochemistry. 2006;45:2570–2576. doi: 10.1021/bi0514156. [DOI] [PubMed] [Google Scholar]
- 30.Joo C., Balci H., Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 31.Groves J.T., Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tilley S.J., Orlova E.V., Gilbert R.J., Andres P.W., Saibil H.R. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell. 2005;121:247–256. doi: 10.1016/j.cell.2005.02.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


