Abstract
Urodele amphibians are unique amongst adult vertebrates in their ability to regenerate missing limbs. The process of limb regeneration requires several key tissues including a regeneration-competent wound epidermis called the regeneration epithelium (RE). We used microarray analysis to profile gene expression of the RE in the axolotl, a Mexican salamander. A list of 125 genes and expressed sequence tags (ESTs) showed a ≥1.5 fold expression in the RE than in a wound epidermis covering a lateral cuff wound. A subset of the RE ESTs and genes were further characterized for expression level changes over the time-course of regeneration. This study provides the first large scale identification of specific gene expression in the RE.
Keywords: limb regeneration, urodele amphibian, gene expression microarray, regeneration epithelium
INTRODUCTION
Limb regeneration is a unique ability of urodele amphibians, which are the only vertebrates able to replace such a complex structure throughout their adult life. The process of regenerating an amputated limb proceeds from the early phase of wound healing to digit development through formation of a mass of cells called the blastema. Until recently it was believed that the blastema was comprised of dedifferentiated mesenchymal cells with pluripotent properties (Lo et al., 1993). On the contrary, the blastema appears to be a heterogeneous mass of cells that retain memory of their tissue origin and replenish their respective missing structures (Kragl et al., 2009) through a recapitulation of signaling molecules and pathways used in development (Gardiner et al., 1999). Nevertheless, it is unclear how blastemal cells are recruited to the amputation plane and how urodele amphibians, such as newts and axolotls, are able to proceed past wound healing to replace missing limbs.
The nerve has been extensively studied as the source of regeneration signaling molecules since it was first described as required for regeneration (Singer, 1952). Recently, the newt AG protein (nAG) was identified as a secreted nerve factor that rescues regeneration in denervated newt limbs (Kumar et al., 2007). nAG is expressed in Schwann cells of the nerve sheath as the severed axon regrows and it has been shown to promote proliferation of blastemal cells in culture. Later in regeneration, nAG appears in the gland cells of the regeneration epithelium (RE). The RE is an epithelial structure covering the distal part of the regenerate and is also required for successful regeneration (Stocum, 2004). The RE is unique to the amputation wound and forms as a result of epidermal migration over the wound from around the circumference of the amputation plane (Repesh and Oberpriller, 1978). In addition to nAG, genes such as Sp9 (Satoh et al., 2008) and Dlx-3 (Mullen et al., 1996) display nerve-dependent expression patterns in the RE. Historically, the RE has been referred to as the wound epidermis (WE) and the apical epithelial cap (AEC), however it has recently been suggested that the structure be termed regeneration epithelium (RE) (Satoh et al., 2008) to distinguish it as a specialized structure that communicates with the nerve to promote regeneration. It has been demonstrated that the RE is required for successful regeneration since removal of the structure delays the process (Thornton, 1957) and preventing formation of the structure inhibits regeneration (Mescher, 1976). Markers for the RE are limited (Campbell and Crews, 2008), but include transcription factors such as Msx-2 (Carlson et al., 1998), Dlx-3 (Mullen et al., 1996), and Sp9 (Satoh et al., 2008), FGF signaling molecules (Han et al., 2001; Christensen et al., 2002), and matrix metalloproteinases (Yang et al., 1999; Kato et al., 2003). Of these, Msx-2 is expressed the earliest, within hours after amputation, but is not RE-specific since it is also expressed during healing of a lateral wound (Carlson et al., 1998). Sp9, another early marker, is RE-specific and expressed within 24 hours after amputation (Satoh et al., 2008). As more investigations focus on exploring the molecular pathways involved in regeneration such as with the Accessory Limb Model (Endo et al., 2004) or with in vitro work (Ferris et al., 2010), a larger set of RE-specific markers will be needed.
The discovery and characterization of molecules and signaling pathways involved in limb regeneration has been improved by the development of genomic tools for salamanders. Significant effort has been put towards sequencing and organizing expressed sequence tags (ESTs) from Ambystoma mexicanum and Ambystoma tigrinum (Habermann et al., 2004; Putta et al., 2004). The Sal-Site at http://www.ambystoma.org (Smith et al., 2005) provides an Ambystoma gene collection and EST database that has allowed for approaches such as microarray analysis and high-throughput 454 cDNA sequencing to investigate aspects of limb regeneration on a broader level (Monaghan et al., 2007; Monaghan et al., 2009). These studies have introduced many new candidate genes that will enable future regeneration studies.
In the present study we utilized the publicly available collections of Ambystoma genes and ESTs to compare, by microarray analysis, the expression profiles of the RE and the wound epidermis covering a lateral cuff wound. From these results we focused on a list of Ambystoma ESTs and genes that are significantly more highly expressed in the RE. A subset of this list was further characterized using quantitative polymerase chain reaction (qPCR) to show the temporal expression pattern during the time-course of regeneration. We discuss the characterized ESTs and genes in terms of best hit candidates discovered through BLAST searches and with respect to previously published regeneration studies.
RESULTS AND DISCUSSION
Identification of RE-Specific Gene Expression
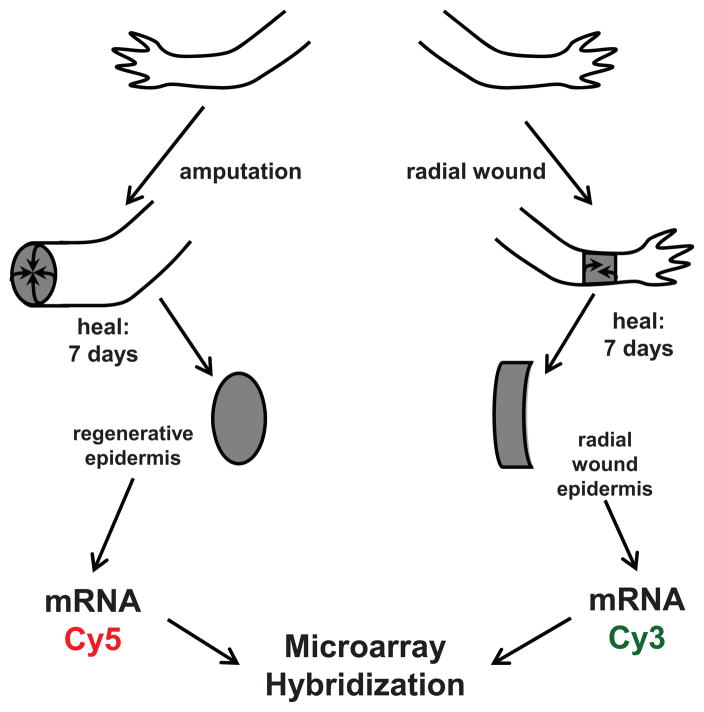
It is well documented that the RE is necessary for successful regeneration in urodele amphibians (Thornton, 1957; Mescher, 1976), however, there are few markers for RE-specific gene expression. In order to identify RE-specific genes, we collected both the epidermis from a lateral cuff wound and from a RE after 7 days of healing, which represented an early stage of regeneration between wound healing and early bud formation (Figure 1). Messenger RNA (mRNA) from the two populations were labeled and hybridized to oligonucleotide arrays containing 42,553 elements that represented 16,257 sequences from A. mexicanum and A. tigrinum and 396 sequences from other salamander species.
Fig. 1.
Schematic depicting tissue dissection and RNA isolation for microarray hybridization. Axolotl limbs were amputated or wounded with a lateral cuff wound. After 7 days of healing, the RE and the LE were dissected and RNA was isolated. mRNA was labeled with Cy3 and Cy5 dyes and hybridized to custom microarray slides designed from EST assemblies for A. mexicanum, A. tigrinum, and other salamander species.
A total of 698 probe sets demonstrated significantly different (p≤0.0016) hybridization intensities between the RE and the lateral cuff wound epidermis (LE) (266 in the RE; 432 in the LE; Additional file 1). We focused on the significant probe sets that exhibited at least a 1.5 fold intensity increase in the RE over the LE (n=195; Additional file 2). The Sal-Site (http://www.ambystoma.org, (Smith et al., 2005)) was used to BLAST the probe sets and identify sequences targeted by the probes. Best hits were identified with NCBI BLAST (Table 1; Additional file 2).
TABLE 1.
List of Interesting Hits Up-regulated in Regeneration Epithelium as Compared to Radial Lateral Wound Epidermis
| Fold change (RE/LE) | Sal ID/accession no. | Gene name | BLASTX (E) | Best hit accession no. |
|---|---|---|---|---|
| 7.79 | M003080 | Similar to methyltransferase 24 (Gallus gallus) | 3E-66 | XM_421871 |
| 6.47 | D82576 | Msx2 (Ambystoma mexicanum) | ||
| 5.73 | M011831 | N/A | N/A | No hit |
| 5.30 | M064466 | N/A | N/A | No hit |
| 5.18 | M002949 | Sodefrin precursor-like factor (Desmognathus monticola) | 3E-26 | DQ097063 |
| 5.11 | M006889 | hyaluronan and proteoglycan link protein 3 (Xenopus laevis) | 8E-42 | NM_001086162 |
| 5.02 | M065735 | N/A | N/A | No hit |
| 4.53 | M061758 | Dynein, cytoplasmic 1, intermediate chain 1 (Homo sapiens) | 2E-39 | NM_004411 |
| 4.08 | Z14047 | Wnt-5a (Ambystoma mexicanum) | ||
| 3.53 | M065526 | Similar to desmoglein 4 preproprotein (Gallus gallus) | 2E-05 | XM_426082 |
| 3.38 | M003433 | Apolipoprotein C-I (Hemibarbus mylodon) | 6E-06 | FJ170109 |
| 3.26 | M002254 | Uromodulin-like (Xenopus tropicalis) | 1E-16 | XM_002934636 |
| 2.86 | M008800 | laminin, beta 1 (Homo sapiens) | 9E-67 | NM_002291 |
| 2.66 | M003964 | Prostate stem cell antigen (Homo sapiens) | 8E-16 | NM_005672 |
| 2.25 | M004510 | Mal, T-cell differentiation protein-like (Homo sapiens) | 2E-27 | NM_005434 |
| 2.13 | M062282 | EGF-like-domain, multiple 6 (Homo sapiens) | 7E-26 | NM_015507 |
| 2.06 | AY326272 | BMP2/4-like (Ambystoma mexicanum) | ||
| 1.88 | U59480 | Distal-less (Dlx-3) (Ambystoma mexicanum) | ||
| 1.68 | M061881 | LY6/PLAUR domain containing 2 (Homo sapiens) | 6E-17 | NM_205545 |
| 1.58 | M062365 | Krüppel-like factor 2 (Homo sapiens) | 3E-19 | NM_016270 |
Best hit identification revealed that blood-specific genes, such as globins, were over-represented in the RE list. Due to the extent of injury during amputation it is not surprising to see this upregulation in the RE with respect to the LE. It is expected that there was carryover of blood cells in the dissection and RNA isolation of the RE samples. Additionally, the best hit identification revealed that genes in the list are targeted by multiple probe sets. In order to reduce the list of RE genes we averaged the multiple hits and achieved a list of 125 genes that are upregulated in the RE (Additional file 3).
Validation of RE Gene Overexpression by qPCR
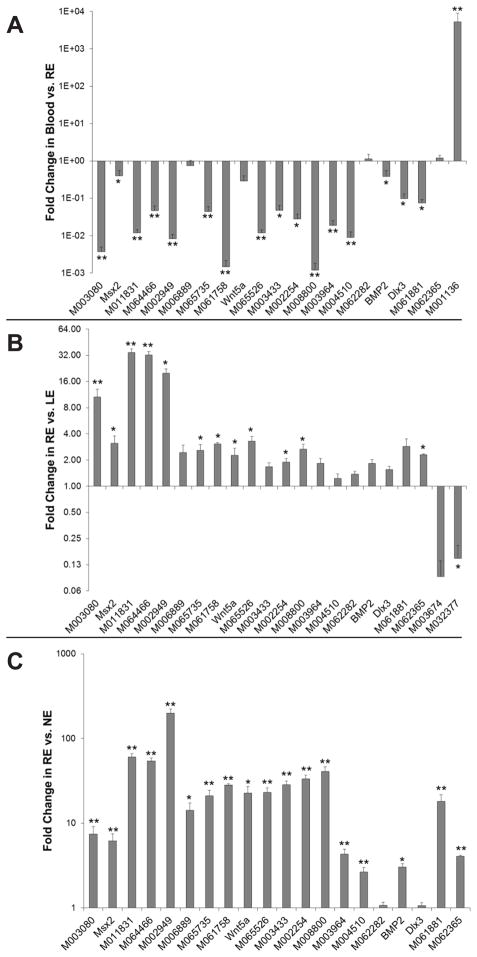
For the purposes of validation and further investigation we focused on the top ten genes as well as ten additional genes of interest (Table 1). Since we observed several blood-specific genes in the RE list it was necessary to confirm that these selected genes are specific to RE and not to blood. Total RNA was isolated from RE at 7 days post amputation, as well as from blood cells, and qPCR was used to examine expression levels of the genes of interest. The geometric mean of four endogenous genes, GAPDH, β-Actin, EF1α, and L27, was used for data normalization. A delta globin gene with Sal ID M001136 (Table 2) was used as an internal control. The results confirmed a >5,000 fold level of expression for M001136 in blood over RE, while 16 of the 20 RE genes were significantly more highly expressed in RE than in blood (Figure 2A). Two genes, M006889 and Wnt-5a, tended towards higher expression levels in RE than in blood but did not have significant p-values. QPCR analysis with primers for M062282 and M062365 showed expression levels of 1.1 and 1.2 fold higher, respectively, in blood over RE, suggesting that M062282 and M062365 may be expressed by both blood and RE. Some of the fold difference in expression level of M062282 and M062365 in RE over LE may be due to blood cell contamination.
TABLE 2.
Summary of Fold Level Changes in Expression Between Tissuesa
| Sal ID/gene name (best hit gene name) | RE/LE (microarray) | RE/LE (qPCR) | Blood/RE (qPCR) | RE/NE (qPCR) |
|---|---|---|---|---|
| M003080 (methyltransferase) | 7.79 | 10.64 | 0.0037 | 7.44 |
| Msx2 | 6.47 | 3.11 | 0.3995 | 6.17 |
| M011831 | 5.73 | 34.50 | 0.0118 | 60.33 |
| M064466 | 5.30 | 32.37 | 0.0468 | 54.15 |
| M002949 (sodefrin precursor-like factor) | 5.18 | 19.91 | 0.0083 | 199.06 |
| M006889 (hyaluronan and proteoglycan link protein 3) | 5.11 | 2.43 | 0.7441 | 14.23 |
| M065735 | 5.02 | 2.58 | 0.0441 | 20.97 |
| M061758 (dynein) | 4.53 | 3.07 | 0.0015 | 28.15 |
| Wnt-5a | 4.08 | 2.28 | 0.2899 | 22.65 |
| M065526 (similar to desmoglein 4 preprotein) | 3.53 | 3.29 | 0.0119 | 23.07 |
| M003433 (apolipoprotein C-I) | 3.38 | 1.68 | 0.0474 | 28.49 |
| M002254 (uromodulin-like) | 3.26 | 1.89 | 0.0278 | 33.23 |
| M008800 (laminin, beta 1) | 2.86 | 2.68 | 0.0012 | 40.71 |
| M003964 (prostate stem cell antigen) | 2.66 | 1.84 | 0.0190 | 4.32 |
| M004510 (mal, T-cell differentiation protein-like) | 2.25 | 1.23 | 0.0089 | 2.66 |
| M062282 (EGF-like domain, multiple 6) | 2.13 | 1.38 | 1.1405 | 1.08 |
| BMP2/4-like | 2.06 | 1.83 | 0.3872 | 3.03 |
| Dlx-3 | 1.88 | 1.55 | 0.0974 | 1.06 |
| M061881 (LY6/PLAUR domain containing 2) | 1.68 | 2.88 | 0.0756 | 18.05 |
| M062365 (Kruppel-like factor 2) | 1.58 | 2.31 | 1.2014 | 4.05 |
|
| ||||
| M001136 (hemoglobin, delta) | 2.09 | N/A | 5225 | N/A |
|
| ||||
| M003674 (myosin, heavy chain 3) | 0.05 | 0.09 | N/A | N/A |
| M032377 | 0.04 | 0.15 | N/A | N/A |
RE, regeneration epithelium; LE, lateral cuff wound epidermis; qPCR, quantitative polymerase chain reaction (qPCR).
Fig. 2.
qPCR validation of fold change gene expression of RE upregulated hits. A: Fold change expressions in blood over RE. B: Fold change expressions in RE over LE. C: Fold change expressions in RE over NE. Genes are represented by Sal ID or gene name as in Table 1. Data represents the mean of three biological replicates and standard error of the mean. *p≤0.05; **p≤0.01.
QPCR was used as an independent verification method of RE gene expression levels as compared to LE. The 20 genes of interest were tested in RE and LE at 7 days post wounding and also in normal unwounded epidermis. As with the tissue collection for the microarray data set, the RE tissues were collected at a time-point corresponding to a phase between wound healing and early bud. RNA was isolated only from the epidermal tissue of the regenerate. Again, the analysis was performed using the geometric mean of four endogenous controls, GAPDH, βActin, EF1α, and L27, as reference. Two genes, M003674 and M032377, which were identified in the microarray as more highly expressed in LE, were used as internal controls (Table 2). The qPCR analysis confirmed overexpression of all 20 genes of interest in the RE with respect to LE (Figure 2B). The two LE genes were validated with higher fold levels in the lateral cuff wound than the amputation wound. M011831, M064466, and M002949 demonstrated much higher fold level differences in RE than in LE by qPCR analysis than by microarray (Table 2), while the remaining genes displayed relatively similar fold differences by both methods. These data suggest that the five genes with the greatest fold difference between RE and LE are M011831, M064466, M002949, M003080, and M065526, respectively.
Normal epidermis (NE) was also compared to RE using qPCR. The skin removed from the limb to make the lateral cuff wound was soaked in dispase I, which promoted the separation of epidermis from the underlying dermis (Kitano and Okada, 1983). As before, analysis was performed using the geometric mean of four endogenous controls, GAPDH, βActin, EF1α, and L27. QPCR analysis confirmed that all but two genes were significantly overexpressed in RE over NE (Figure 2C). M002949 demonstrated the greatest fold difference at nearly 200 fold (Table 2). The two genes that did not show significant difference were M062282 and Dlx-3, with fold level differences of 1.08 and 1.06, respectively.
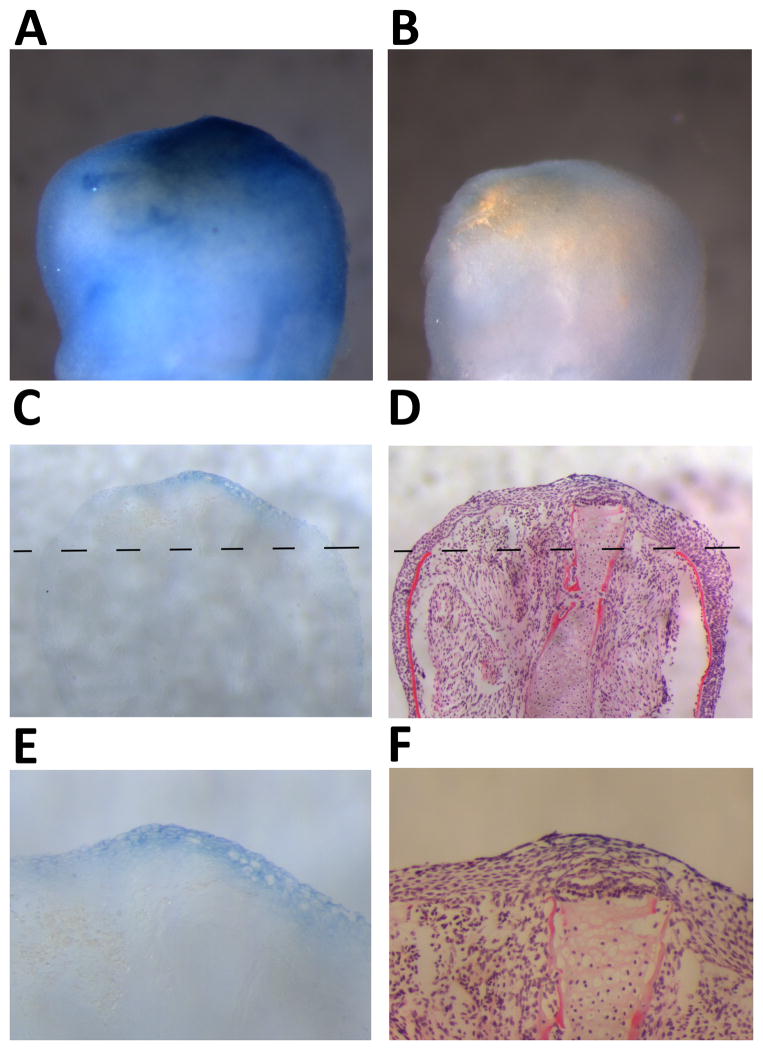
Given the technical difficulty of isolating epidermal tissue from the underlying stump and blastemal tissue, we cannot eliminate the possibility that gene expression differences between RE and LE or RE and NE may be partially due to blastemal cell contamination. Whole mount in situ hybridization analysis shows that M002949, one of the genes with the greatest difference between RE and LE, as well as RE and NE, is expressed over the distal tip of the regenerating limb at 7 days post amputation (Figure 3A). The sense control showed no stain (Figure 3B). Sectioning of the tissue showed that expression is limited to the RE, with no expression in the underlying stump tissue (Figure 3C, D). The expression of M002949 in the top layers of the RE (Figure 3E, F) confirms that the identification of this gene by microarray analysis was due to tissue-specific expression in the RE as opposed to contamination of blastemal or underlying stump tissues. It is important to note that this does not exclude the possibility that some microarray-identified RE genes may be expressed by the mesenchymal tissue of the regenerate, however RE-specific expression of M002949 demonstrates that mesenchymal cell contamination did not contribute to the identification of one of the most highly expressed RE genes as compared to LE. The microarray results in combination with the qPCR data demonstrate that the RE is unique in terms of gene expression as compared to the LE or NE. These results provide a set of regeneration markers for the identification of the RE in regeneration studies.
Fig. 3.
Whole mount in situ hybridization analysis of M002949 (sodefrin precursor factor) expression in 7 day post amputation regenerating axolotl limbs. All panels are forelimbs with distal at the top. A: M002949 expression covering distal tip of regenerating limb indicated in blue as determined with anti-sense probe. B: Negative control using M002949 sense probe. C, D: Sister sections of tissue from panel A. Dashed line represents level of amputation. Tissue sectioned at 14 μm. C: Expression in the regeneration epithelium indicated in blue. D: Hematoxylin and eosin. E, F: Higher magnification of the distal tip of the regenerating limb shows that the M002949 expression is localized to the top layers of the regeneration epidermis.
RE-specific Genes Display Changes in Expression Levels throughout the Regeneration Process
The temporal expression patterns of the RE genes of interest were characterized using qPCR. Regenerating limbs representing six stages of regeneration were processed for RNA isolation and analyzed for each gene with two endogenous controls for normalization: GAPDH and EF1α. Regenerates were collected from slightly smaller animals than those used for the microarray and for the qPCR validation data in Table 2. Therefore, the 7 day post amputation time-point used in the microarray study corresponds with the 2–4 day time-points used for the time-course expression profiling. Since the entire regenerate was collected and assayed, expression level changes in the time-course experiments, unlike the microarray and qPCR validation experiments, account for gene expression levels in the RE as well as possible expression in the blastema tissue. Therefore it should be noted that expression level changes could account for gene expression in different or multiple tissues of the regenerate during the various phases of limb regeneration. In the following sections we present the temporal expression patterns of all 20 genes of interest and discuss their identity as determined by BLAST search or previous characterization in limb regeneration.
Temporal Expression Patterns of Genes Previously Described in Limb Regeneration
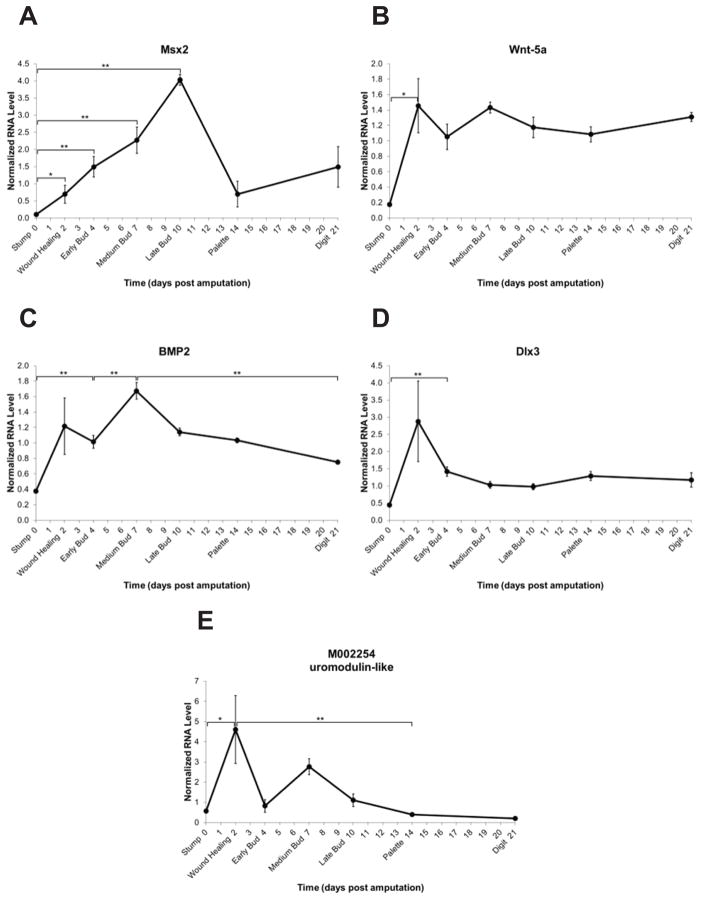
Five genes identified in the microarray have been previously described to play a role during limb regeneration. Msx2 encodes a transcription factor that has been described to be expressed in the RE and distal mesenchyme of the regenerating limb (Carlson et al., 1998; Koshiba et al., 1998).
The time course data show that the expression level gradually increases over time from the wound healing stage through late bud (Figure 4A). This result concurs with the previous report that Msx2 is re-expressed after amputation and is highly expressed in the late bud stage (Carlson et al., 1998).
Fig. 4.
Expression level time-courses of genes previously reported to play a role in limb regeneration. The y-axis represents normalized RNA level and the x-axis represents days post amputation. Genes are represented by Sal ID and/or gene name as in Table 1. Each dark circle represents the mean of three samples ± standard error of the mean. *p≤0.05; **p≤0.01.
Wnt-5a, a member of the Wnt family of secreted proteins, has been previously described to play an important role in the early stages of limb regeneration during dedifferentiation (Ghosh et al., 2008). The temporal expression pattern described by the qPCR time course agrees with this previous report that expression was detected from very early stages and throughout regeneration (Figure 4B).
BMP2 has been described as an important factor in the onset of condensation during limb regeneration (Guimond et al., 2010). The qPCR time-course data show an increase in expression from the stump through medium bud stages and a significant decrease from medium bud to digit stage (Figure 4C). This concurs with the in situ hybridization patterns that show strong staining in the medium to late bud and then a distinct localization to the interdigital regions (Guimond et al., 2010).
Dlx-3, or distal-less 3, has been previously described to play a nerve-dependent role in regeneration with an expression pattern that is very low at early stages and peaks at the late bud stage of regeneration (Mullen et al., 1996). Contrary to the previous report, qPCR time course analysis indicates that Dlx-3 significantly increases early in expression level as compared to stump tissue and tends to be expressed at a constant level throughout regeneration with no significant increase or decrease over time (Figure 4D).
M002254 shows a significant increase in expression upon wound healing and decreases over time from the wound healing stage to palette (Figure 4E). This gene shows similarity to Xenopus tropicalis uromodulin-like. It has been linked to nerve-dependent blastema outgrowth in axolotl (Monaghan et al., 2009) and has been shown to be down-regulated in response to thyroid hormone-induced metamorphosis in Xenopus (Brown et al., 1996).
Temporal Expression Patterns of Three-Finger Protein Family Members
Three genes (M002949, M003964, and M061881) are structurally similar to Prod1 (Garza-Garcia et al., 2009). Prod1 is a three-finger protein (TFP) family member that is involved in positional identity of the proximal-distal axis in the newt limb (da Silva et al., 2002) and binds to nAG, the nerve factor that rescues regeneration in denervated newt limbs (Kumar et al., 2007). The structural similarity suggests that these factors may be secreted or anchored to the cell membrane by glycosylphosphatidylinositol (GPI) linkage.
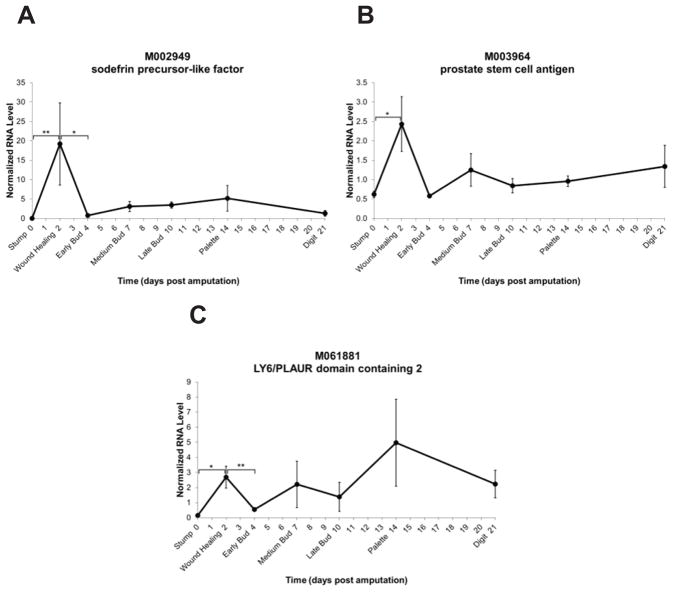
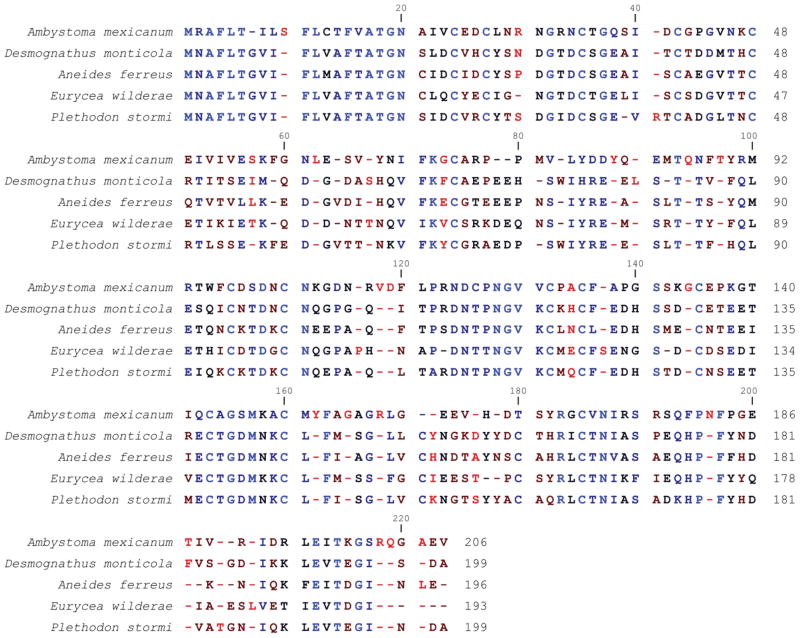
M002949 shows high expression level at the wound healing stage and takes a drop in expression by the early bud stage (Figure 5A). The tendency for lower expression during the remaining stages of limb regeneration suggests that it may have a very early role in regeneration. M002949 shows similarity to sodefrin precursor-like factor from several salamander species, particularly at the N-terminal end and amongst the cysteine residues (Figure 6).
Fig. 5.
Expression level time-courses of three finger protein family members. The y-axis represents normalized RNA level and the x-axis represents days post amputation. Genes are represented by Sal ID and gene name as in Table 1. Each dark circle represents the mean of three samples ± standard error of the mean.
Fig. 6.
Alignment of axolotl (A. mexicanum) M002949 amino acid sequence with sodefrin precursor-like factor from four species of salamander: seal salamander (D. monticola; accession: AAZ06333), clouded salamander (A. ferreus; accession: AAZ06336), Blue Ridge two-lined salamander (E. wilderae; accession: AAZ06337), and Siskiyou Mountain salamander (P. stormi, accession: AAZ06325). Conserved residues are indicated by blue and non-conserved residues are indicated by red. Gaps are indicated by a dash (−).
M003964 shows similarity to prostate stem cell antigen (PSCA), a GPI-anchored cell membrane protein. The expression pattern of this PSCA-like gene takes a significant increase immediately upon wound healing and then a tendency towards low expression levels throughout regeneration (Figure 5B).
Similarly, the M061881 gene shows a significant increase upon wound healing and then a significant drop in expression level at the early bud stage (Figure 5C). This gene shows similarity to the LY6/PLAUR domain containing 2.
Temporal Expression Patterns of Genes with Roles in Cell Adhesion and Organization
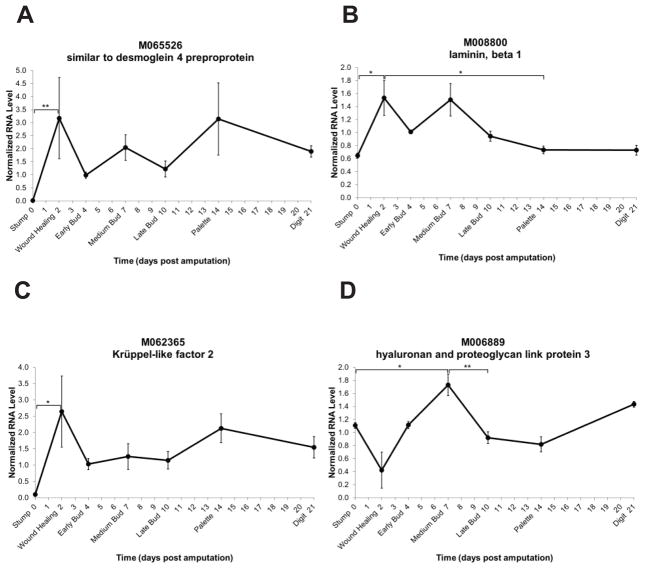
Four genes have similarities with cell adhesion factors. The M065526 pattern shows higher expression in regenerating tissue as compared to stump tissue with fluctuating expression levels during regeneration (Figure 7A). This EST shows similarity to desmoglein 4 preprotein, a cadherin family member that is involved in the formation of desmosomes. More specifically it has been shown to localize to desmosomes in the human hair follicle (Bazzi et al., 2006), a regenerating dermal appendage.
Fig. 7.
Expression level time-courses of genes involved in cell adhesion and organization. The y-axis represents normalized RNA level and the x-axis represents days post amputation. Genes are represented by Sal ID and gene name as in Table 1. Each dark circle represents the mean of three samples ± standard error of the mean. *p≤0.05; **p≤0.01.
M008800 shows similarity to laminin, beta 1, which has been localized to epithelial basement membranes (Virtanen et al., 2003). During limb regeneration its expression increases upon wound healing and significantly decreases by the palette stage (Figure 7B).
M062365 encodes a gene with strong similarity to the Krüppel-like zinc-finger transcription factor 2. This gene is also described as lung KLF (LKLF). Knockout mouse studies have shown that loss of LKLF results in a change in smooth muscle cell morphology and loss of organization in the blood vessel wall (Kuo et al., 1997). The temporal expression pattern shows a significant increase upon wound healing and a tendency towards relatively stable expression level during regeneration (Figure 7C).
The M006889 gene shows similarity to a hyaluronan and proteoglycan link protein family member. Alignment with Xenopus and human sequences shows high conservation in the second hyaluronan and proteoglycan binding link domain (Figure 8). This family of proteins functions in cell adhesion and migration by binding hyaluronan, a glycosaminoglycan, with proteoglycans to modify the extracellular matrix or cell surfaces (Fraser et al., 1997). The temporal expression pattern demonstrates an increase in expression level at the medium bud stage as compared to stump tissue and then a decrease in expression at the late bud stage (Figure 7D). The expression pattern suggests an interesting initial decrease in expression level upon wound healing.
Fig. 8.
Alignment of axolotl (A. mexicanum) M006889 amino acid sequence with hyaluronan and proteoglycan link protein 3 from X. laevis (accession: NP_001079631) and human (H. sapiens; accession: NP_839946). Conserved residues are indicated in blue and non-conserved residues are indicated by red. Gaps are indicated by a dash (−). The shaded boxes identify the hyaluronan and proteoglycan link domains.
Temporal Expression Patterns of Genes with Lipid-Associated and Neurite Regeneration Roles
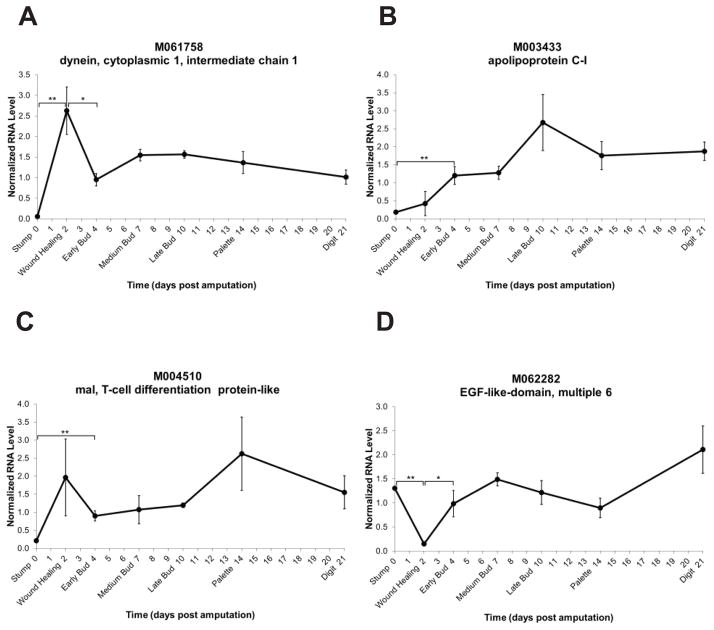
M061758 shows similarity to dynein, a minus-end directed motor protein (Figure 9A). Dynein has been described to play an important role in axonal regeneration by signaling to the cell body that an axon has been injured (Tuck and Cavalli, 2010). The qPCR data for temporal limb regeneration expression shows that expression for this gene peaks at the wound healing stage.
Fig. 9.
Expression level time-courses of genes with lipid-associated and neurite regeneration roles. The y-axis represents normalized RNA level and the x-axis represents days post amputation. Genes are represented by Sal ID and gene name as in Table 1. Each dark circle represents the mean of three samples ± standard error of the mean. *p≤0.05; **p≤0.01.
M003433 (Figure 9B) and M004510 (Figure 9C) are two lipid-associated proteins. Lipid rafts have been described as crucial for the signaling that drives neurite outgrowth and regeneration (Zhao et al., 2009). M003433 shows similarity to apolipoprotein C-I, which is a lipid-binding protein believed to be involved in lipid transport. M004510 shows similarity to mal, T-cell differentiation protein-like, which is a protein that appears to localize in lipid rafts. Both of these genes show a significant increase in expression by the early bud stage.
It has also been suggested that an epidermal growth factor-like molecule and lipid rafts are involved in cutaneous wound healing (Mathay et al., 2007). The M062282 sequence shows similarity to an epidermal growth factor repeat superfamily member, EGFL6. Genes of this type have been shown to be expressed during early development (Buchner et al., 2000b) and in the anterior part of the dermal placode, which induces hair follicle formation (Buchner et al., 2000a). The qPCR temporal expression pattern for EGFL6 shows a very low expression level after amputation and then a significant increase at each time point thereafter, with the highest expression at the digit stage (Figure 9D). Taken in concert with the comparison data that suggested nearly equal expression levels between RE and NE (Figure 2C), M062282 may play a role in the maturation of the epidermis covering the regenerate.
Temporal Expression Patterns of Sequences with Unknown or Hypothetical Identity
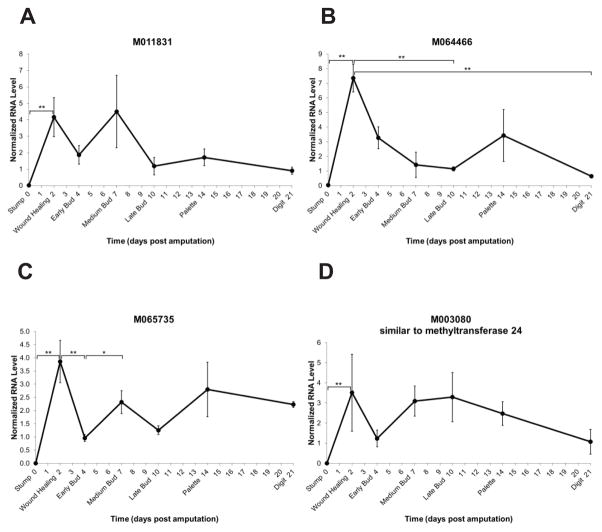
The M011831 sequence does not show strong similarity to any known genes but demonstrates a high fold difference of expression level in RE over LE (Table 2, Figure 2B). The temporal expression pattern shows a significant increase in expression at the wound healing stage and then a tendency for higher expression through medium bud and lower expression from late bud through digit stage (Figure 10A).
Fig. 10.
Expression level time-courses of genes with unknown or hypothetical identity. The y-axis represents normalized RNA level and the x-axis represents days post amputation. Genes are represented by Sal ID and gene name as in Table 1. Each dark circle represents the mean of three samples ± standard error of the mean. *p≤0.05; **p≤0.01.
The expression pattern of M064466 is highest at the wound healing stage and then significantly decreases by the late bud and digit stages (Figure 10B), suggesting that the gene product plays a role in wound healing and early blastema formation.
M065735, another sequence with no identifiable similarities, shows a significant increase upon wound healing followed by a decrease at early bud and then an increase at medium bud stage, suggesting a dynamic expression pattern (Figure 10C).
M003080, which presented in the microarray with the highest fold difference in RE over LE, demonstrated an increase in expression upon wound healing followed by a tendency towards higher expression during medium and late bud stages and then a decrease to the digit stage (Figure 10D). The gene shows similarity to a putative S-adenosylmethionine-dependent methyltransferase (AdoMet-MTase), class I (Figure 11). The class I family of AdoMet-MTases is the largest and most diverse including members with substrate specificity to small molecules, lipids, proteins, and nucleic acids (Martin and McMillan, 2002; Schubert et al., 2003). Further investigation is needed to identify the substrate of this potential axolotl AdoMet-MTase and its role in limb regeneration.
Fig. 11.
Alignment of axolotl (A. mexicanum) M003080 amino acid sequence with putative methyltransferase sequences from chicken (G. gallus; accession: XP_001232694) and X. laevis (accession: NP_001136263). Conserved residues are indicated by blue and residues that are not conserved are indicated by red. Gaps are indicated by a dash (−). The shaded boxes identify the S-adenosylmethionine-dependent methyltransferase, class I regions in chicken and Xenopus.
CONCLUSIONS
This study provides the first expression profiling of the RE in urodele amphibian limb regeneration. The analysis identified 125 genes that demonstrate higher expression in the regenerative epithelium than in wound epidermis covering a lateral cuff wound, suggesting that the expression is specific to the regeneration response of an amputation wound as opposed to general wound healing. QPCR data for a subset of the genes support the microarray findings and show that they are significantly more highly expressed in RE than in NE. Additional qPCR data show interesting expression changes for the genes during the time-course of regeneration. These markers will provide an important tool for studying the early events in limb regeneration. Further study into the function of the individual genes will illuminate the role of the RE for successful limb regeneration.
EXPERIMENTAL PROCEDURES
Animal procedures
Axolotls (Ambystoma mexicanum) were spawned at Yale University or obtained from the Ambystoma Genetic Stock Center at the University of Kentucky. Amputations and tissue collections were performed on animals measuring 8–15 cm from snout to tip of tail. All animals were anesthetized in 0.1% MS222 solution (Ethyl 3-aminobenzoate methanesulfonate salt, Sigma-Aldrich, St. Louis, MO, USA). Radial lateral wounds were created by cutting through full thickness skin around the circumference of the limb with spring scissors and peeling away the full thickness skin from the underlying stump tissue. Animal care and use protocols were approved by the Yale University Institutional Animal Care and Use Committee.
Tissue collection
Amputations and lateral wounds were made in the zeugopod region of the limbs (between the wrist/ankle joint and the elbow/knee joint). RE and LE were collected at 7 days post amputation/wounding. NE was collected by soaking full thickness skin from the radial lateral wounding in a 1% solution of dispase I (Sigma-Aldrich, St. Louis, MO, USA) in 0.8x PBS for 5 hours at room temperature, washing with 0.8x PBS, and then gently peeling the epidermis from the dermis layer. Dispase treatment was only performed on the NE tissue sample. Blood was collected from the radial lateral and amputation wounds at the time of RE and LE collection. Limb regenerates for qPCR were collected at 0, 2, 4, 7, 10, 14, and 21 days post amputation. Tissues were soaked in RNAlater® (Ambion, Foster City, CA, USA) prior to RNA isolation. For microarray analysis, 3 pools of 7 animals each were used. For qPCR validation studies, 3 pools of 4 animals each were used. For regeneration time-course qPCR studies, 3 pools of 3 animals each were used.
RNA isolation and microarray analysis
RNA was isolated using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). Following isolation, RNA was purified and DNase treated using RNeasy minicolumns (Qiagen, Valencia, CA, USA). RNA quality was assessed by spectrophotometry using a NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). RNA samples were also analyzed on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Microarray analysis was performed on custom eArrays (Agilent Technologies, Santa Clara, CA, USA) using 60-mer probes designed against genes and ESTs from Ambystoma mexicanum, Ambystoma tigrinum and other salamander species. Hybridization and data collection were performed by the Whitehead Institute Genome Technology Core (Cambridge, MA, USA). RE samples were labeled with Cy5; LE samples were labeled with Cy3. Each of the three pools of tissues collected for RE and LE were hybridized to three arrays as biological replicates, while a fourth array was used as a technical replicate of one of the biological samples. Analysis of the arrays was done using the limma (Smyth, 2005) package in Bioconductor (Gentleman et al., 2004). Multiple probes for the same clone were averaged. Arrays were normalized with the loess routine (Yang et al., 2001; Yang et al., 2002; Smyth and Speed, 2003) without background correction (Zahurak et al., 2007). A linear model was fit to the expression values, and an empirical Bayes routine was used to moderate the t-statistic (Smyth, 2004). Genes were selected as differentially expressed after adjusting p-values for repeated tests by controlling the false discovery rate (Benjamini and Hochberg, 1995).
Quantitative PCR
Reverse transcription was performed with iScript™ Reverse Transcription Supermix (Bio-Rad, Hercules, CA, USA). Quantitative PCR assays were run using Power SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) on a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) and analyzed with the CFX96 Real-Time System. Primer sequences and annealing temperatures for the genes assayed are listed in Additional File 4.
Whole mount in situ hybridization and Histology
Whole mount in situ hybridization was performed as previously described (Gardiner et al., 1995) with modifications. Tissues were fixed overnight at 4°C with gentle rocking in freshly-made MEMFA (0.1 M MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, 3.7% formaldehyde) and then dehydrated and stored at −20°C in 100% methanol. Proteinase K (New England BioLabs, Ipswich, MA, USA) treatment was performed at 10 μg/mL for 30 minutes at 4°C followed by 15 minutes at 37°C to permeabilize tissue. Prehybridization was performed at 60°C overnight. Probes were prepared from pBluescript SK- vector containing the 1062 bp M002949 fragment. The vector was linearized with PstI and anti-sense probe was transcribed with T7 RNA polymerase (New England BioLabs, Ipswich, MA, USA) and digoxigenin RNA labeling mix (Roche Applied Science, Indianapolis, IN, USA). Sense probe was transcribed with T3 RNA polymerase (Roche Applied Science, Indianapolis, IN, USA) and digoxigenin RNA labeling mix after linearization with XhoI. Hybridization was performed at 60°C for 72 hours. Alkaline-phosphatase (AP) conjugated anti-digoxigenin antibody was obtained from Roche Applied Science (Indianapolis, IN, USA). The colorimetric alkaline-phosphatase reaction was developed with BM Purple (Roche Applied Science, Indianapolis, IN, USA). Tissues from whole mount in situ hybridization were cryoembedded and sectioned at 14 μm. Hematoxylin and eosin stains (Sigma-Aldrich, St. Louis, MO, USA) were used to stain nuclei blue and cytoplasm and extracellular proteins shades of pink.
Supplementary Material
Acknowledgments
We thank the Crews laboratory for helpful discussion. We thank Randall Voss for providing the vector containing M002949. This work was supported by the NIH (GM094944) and the ARO (Army US #W911NF-07-1-0252). HOZ is supported, in part, by P20RR016470. We acknowledge the services of the Ambystoma Genetic Stock Center, which is supported by NSF-DBI-0443496, and the resources available at the Sal-Site, which are supported by NIH-NCRR-R24RR16344.
RFERENCES
- Bazzi H, Getz A, Mahoney MG, Ishida-Yamamoto A, Langbein L, Wahl JK, III, Christiano AM. Desmoglein 4 is expressed in highly differentiated keratinocytes and trichocytes in human epidermis and hair follicle. Differentiation. 2006;74:129–140. doi: 10.1111/j.1432-0436.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Brown DD, Wang Z, Furlow JD, Kanamori A, Schwartzman RA, Remo BF, Pinder A. The thyroid hormone-induced tail resorption program during Xenopus laevis metamorphosis. Proc Natl Acad Sci U S A. 1996;93:1924–1929. doi: 10.1073/pnas.93.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner G, Broccoli V, Bulfone A, Orfanelli U, Gattuso C, Ballabio A, Franco B. MAEG, an EGF-repeat containing gene, is a new marker associated with dermatome specification and morphogenesis of its derivatives. Mech Dev. 2000a;98:179–182. doi: 10.1016/s0925-4773(00)00462-7. [DOI] [PubMed] [Google Scholar]
- Buchner G, Orfanelli U, Quaderi N, Bassi MT, Andolfi G, Ballabio A, Franco B. Identification of a New EGF-Repeat-Containing Gene from Human Xp22: A Candidate for Developmental Disorders. Genomics. 2000b;65:16–23. doi: 10.1006/geno.2000.6146. [DOI] [PubMed] [Google Scholar]
- Campbell LJ, Crews CM. Wound epidermis formation and function in urodele amphibian limb regeneration. Cell Mol Life Sci. 2008;65:73–79. doi: 10.1007/s00018-007-7433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson MR, Bryant SV, Gardiner DM. Expression of Msx-2 during development, regeneration, and wound healing in axolotl limbs. J Exp Zool. 1998;282:715–723. doi: 10.1002/(sici)1097-010x(19981215)282:6<715::aid-jez7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Christensen RN, Weinstein M, Tassava RA. Expression of fibroblast growth factors 4, 8, and 10 in limbs, flanks, and blastemas of Ambystoma. Dev Dyn. 2002;223:193–203. doi: 10.1002/dvdy.10049. [DOI] [PubMed] [Google Scholar]
- da Silva SM, Gates PB, Brockes JP. The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev Cell. 2002;3:547–555. doi: 10.1016/s1534-5807(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Endo T, Bryant SV, Gardiner DM. A stepwise model system for limb regeneration. Dev Biol. 2004;270:135–145. doi: 10.1016/j.ydbio.2004.02.016. [DOI] [PubMed] [Google Scholar]
- Ferris DR, Satoh A, Mandefro B, Cummings GM, Gardiner DM, Rugg EL. Ex vivo generation of a functional and regenerative wound epithelium from axolotl (Ambystoma mexicanum) skin. Dev Growth Differ. 2010;52:715–724. doi: 10.1111/j.1440-169X.2010.01208.x. [DOI] [PubMed] [Google Scholar]
- Fraser JRE, Laurent TC, Laurent UBG. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Blumberg B, Komine Y, Bryant SV. Regulation of HoxA expression in developing and regenerating axolotl limbs. Development. 1995;121:1731–1741. doi: 10.1242/dev.121.6.1731. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Carlson MR, Roy S. Towards a functional analysis of limb regeneration. Semin Cell Dev Biol. 1999;10:385–393. doi: 10.1006/scdb.1999.0325. [DOI] [PubMed] [Google Scholar]
- Garza-Garcia A, Harris R, Esposito D. Solution Structure and Phylogenetics of Prod1, a Member of the Three-Finger Protein …. 2009 doi: 10.1371/journal.pone.0007123. pubmedcentral.nih.gov. [DOI] [PMC free article] [PubMed]
- Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini A, Sawitzki G, Smith C, Smyth G, Tierney L, Yang J, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Roy S, Séguin C, Bryant SV, Gardiner DM. Analysis of the expression and function of Wnt-5a and Wnt-5b in developing and regenerating axolotl (Ambystoma mexicanum) limbs. Dev Growth Differ. 2008;50:289–297. doi: 10.1111/j.1440-169X.2008.01000.x. [DOI] [PubMed] [Google Scholar]
- Guimond J-C, Levesque M, Michaud P-L, Berdugo J, Finnson K, Philip A, Roy S. BMP-2 functions independently of SHH signaling and triggers cell condensation and apoptosis in regenerating axolotl limbs. BMC Developmental Biology. 2010;10:15. doi: 10.1186/1471-213X-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann B, Bebin A-G, Herklotz S, Volkmer M, Eckelt K, Pehlke K, Epperlein H, Schackert H, Wiebe G, Tanaka E. An Ambystoma mexicanum EST sequencing project: analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries. Genome Biology. 2004;5:R67. doi: 10.1186/gb-2004-5-9-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MJ, An JY, Kim WS. Expression patterns of Fgf-8 during development and limb regeneration of the axolotl. Dev Dyn. 2001;220:40–48. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1085>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Kato T, Miyazaki K, Shimizu-Nishikawa K, Koshiba K, Obara M, Mishima HK, Yoshizato K. Unique expression patterns of matrix metalloproteinases in regenerating newt limbs. Dev Dyn. 2003;226:366–376. doi: 10.1002/dvdy.10247. [DOI] [PubMed] [Google Scholar]
- Kitano Y, Okada N. Separation of the epidermal sheet by dispase. Br J Dermatol. 1983;108:555–560. doi: 10.1111/j.1365-2133.1983.tb01056.x. [DOI] [PubMed] [Google Scholar]
- Koshiba K, Kuroiwa A, Yamamoto H, Tamura K, Ide H. Expression of Msx genes in regenerating and developing limbs of axolotl. J Exp Zool. 1998;282:703–714. doi: 10.1002/(sici)1097-010x(19981215)282:6<703::aid-jez6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009 doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kumar A, Godwin J, Gates P, Garza-Garcia A, Brockes J. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murineembryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci U S A. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Mathay C, Giltaire S, Minner F, Bera E, Herin M, Poumay Y. Heparin-Binding EGF-Like Growth Factor Is Induced by Disruption of Lipid Rafts and Oxidative Stress in Keratinocytes and Participates in the Epidermal Response to Cutaneous Wounds. J Invest Dermatol. 2007;128:717–727. doi: 10.1038/sj.jid.5701069. [DOI] [PubMed] [Google Scholar]
- Mescher AL. Effects on adult newt limb regeneration of partial and complete skin flaps over the amputation surface. J Exp Zool. 1976;195:117–128. doi: 10.1002/jez.1401950111. [DOI] [PubMed] [Google Scholar]
- Monaghan JR, Epp LG, Putta S, Page RB, Walker JA, Beachy CK, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol. 2009;7:1. doi: 10.1186/1741-7007-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Walker JA, Page RB, Putta S, Beachy CK, Voss SR. Early gene expression during natural spinal cord regeneration in the salamander Ambystoma mexicanum. J Neurochem. 2007;101:27–40. doi: 10.1111/j.1471-4159.2006.04344.x. [DOI] [PubMed] [Google Scholar]
- Mullen LM, Bryant SV, Torok MA, Blumberg B, Gardiner DM. Nerve dependency of regeneration: the role of Distal-less and FGF signaling in amphibian limb regeneration. Development. 1996;122:3487–3497. doi: 10.1242/dev.122.11.3487. [DOI] [PubMed] [Google Scholar]
- Putta S, Smith J, Walker J, Rondet M, Weisrock D, Monaghan J, Samuels A, Kump K, King D, Maness N, Habermann B, Tanaka E, Bryant S, Gardiner D, Parichy D, Voss SR. From biomedicine to natural history research: EST resources for ambystomatid salamanders. BMC Genomics. 2004;5:54. doi: 10.1186/1471-2164-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repesh LA, Oberpriller JC. Scanning electron microscopy of epidermal cell migration in wound healing during limb regeneration in the adult newt, Notophthalmus viridescens. Am J Anat. 1978;151:539–555. doi: 10.1002/aja.1001510408. [DOI] [PubMed] [Google Scholar]
- Satoh A, Graham GMC, Bryant SV, Gardiner DM. Neurotrophic regulation of epidermal dedifferentiation during wound healing and limb regeneration in the axolotl (Ambystoma mexicanum) Dev Biol. 2008;319:321–335. doi: 10.1016/j.ydbio.2008.04.030. [DOI] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. The influence of the nerve in regeneration of the amphibian extremity. Q Rev Biol. 1952;27:169–200. doi: 10.1086/398873. [DOI] [PubMed] [Google Scholar]
- Smith J, Putta S, Walker J, Kump D, Samuels A, Monaghan J, Weisrock D, Staben C, Voss S. Sal-Site: Integrating new and existing ambystomatid salamander research and informational resources. BMC Genomics. 2005;6:181. doi: 10.1186/1471-2164-6-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Stocum DL. Amphibian regeneration and stem cells. Curr Top Microbiol Immunol. 2004;280:1–70. doi: 10.1007/978-3-642-18846-6_1. [DOI] [PubMed] [Google Scholar]
- Thornton CS. The effect of apical cap removal on limb regeneration in Amblystoma larvae. J Exp Zool. 1957;134:357–381. doi: 10.1002/jez.1401340209. [DOI] [PubMed] [Google Scholar]
- Tuck E, Cavalli V. Roles of membrane trafficking in nerve repair and regeneration. Commun Integr Biol. 2010;3:209–214. doi: 10.4161/cib.3.3.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen I, Korhonen M, Petajaniemi N, Karhunen T, Thornell L-E, Sorokin LM, Konttinen YT. Laminin Isoforms in Fetal and Adult Human Adrenal Cortex. J Clin Endocrinol Metab. 2003;88:4960–4966. doi: 10.1210/jc.2003-030418. [DOI] [PubMed] [Google Scholar]
- Yang EV, Gardiner DM, Carlson MR, Nugas CA, Bryant SV. Expression of Mmp-9 and related matrix metalloproteinase genes during axolotl limb regeneration. Dev Dyn. 1999;216:2–9. doi: 10.1002/(SICI)1097-0177(199909)216:1<2::AID-DVDY2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Speed TP. Normalization for cDNA microarray data. In: Bittner ML, Chen Y, Dorsel AN, Dougherty ER, editors. Microarrays: Optical Technologies and Informatics: Proc SPIE. 2001. pp. 141–152. [Google Scholar]
- Zahurak M, Parmigiani G, Yu W, Scharpf R, Berman D, Schaeffer E, Shabbeer S, Cope L. Pre-processing Agilent microarray data. BMC Bioinformatics. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Cao X, Wu G, Loh HH, Law PY. Neurite Outgrowth is Dependent on the Association of c-Src and Lipid Rafts. Neurochem Res. 2009 doi: 10.1007/s11064-009-0016-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.