Abstract
Randomized trials with dropouts or censored data and discrete time-to-event type outcomes are frequently analyzed using the Kaplan–Meier or product limit (PL) estimation method. However, the PL method assumes that the censoring mechanism is noninformative and when this assumption is violated, the inferences may not be valid. We propose an expanded PL method using a Bayesian framework to incorporate informative censoring mechanism and perform sensitivity analysis on estimates of the cumulative incidence curves. The expanded method uses a model, which can be viewed as a pattern mixture model, where odds for having an event during the follow-up interval , conditional on being at risk at , differ across the patterns of missing data. The sensitivity parameters relate the odds of an event, between subjects from a missing-data pattern with the observed subjects for each interval. The large number of the sensitivity parameters is reduced by considering them as random and assumed to follow a log-normal distribution with prespecified mean and variance. Then we vary the mean and variance to explore sensitivity of inferences. The missing at random (MAR) mechanism is a special case of the expanded model, thus allowing exploration of the sensitivity to inferences as departures from the inferences under the MAR assumption. The proposed approach is applied to data from the TRial Of Preventing HYpertension.
Keywords: Clinical trials, Hypertension, Ignorability index, Missing data, Pattern-mixture model, TROPHY trial
1. INTRODUCTION
Randomized control trials where the primary outcome is time-to-event are subject to censoring. Censoring can occur due either to dropout or violations of the study protocol. The resulting censoring mechanism could be considered noninformative if the time of censoring is independent of the time of event and informative otherwise. To analyze these data, where the censoring mechanism is potentially informative, additional data or assumptions are required to make valid inferences (Zheng and Klein, 1995).
There are 3 general approaches for analyzing data with dropouts or censoring. Rotnitzky and others (2001), Scharfstein and others (2001), and Scharfstein and Robins (2002) adopt a selection model approach where an explicit censoring mechanism is formulated and related to the unobserved outcome. On the other hand, Little (1995), Little and Rubin (2002), and Birmingham and others (2003) used a pattern-mixture (PM) approach where the distribution of the outcome across different patterns formed by censoring times are related by some identifiability constraints. Wu and Carroll (1988) used a shared-parameter model. This model uses common random effects to relate the response to the missing-data indicator (Guo and others, 2004). All 3 approaches involve untestable assumptions about the censoring mechanism, therefore must be followed with sensitivity analysis (Molenberghs and Kenward, 2007).
In a PM setting, Daniels and Hogan (2000) and Kenward and others (2003) identified parameters using constraints. Roy and Daniels (2008) use latent class PM models with Bayesian model averaging. Previously, we developed a model for binary outcomes with missing data (Kaciroti and others, 2009) and used Bayesian estimation methods for sensitivity analysis. Here, we propose a Bayesian model using a parameterization within the PM approach to analyze time-to-event data with informative censoring.
The product limit (PL) estimator is a general purpose nonparametric approach used to analyze data from clinical trials with time-to-event outcomes and noninformative censoring. Using a Bayesian methodology, PS83 and Tsai (1986) extend the method of Susarla and Van Ryzin (1976) by introducing a bivariate exponential distribution on the parameters of the Dirichlet process to allow the possibility of dependent censoring. However, choosing a distribution that captures the correct dependence between censoring and event times is not easy to specify as it is not estimatable from the observed data. Ibrahim and others (2001) give a comprehensive review on Bayesian survival analysis. More recently, Scharfstein and others (2003) propose a fully Bayesian methodology that accounts for informative censoring by introducing prior restrictions on the selection mechanism and follow with sensitivity analysis. Alternatively, we propose here a fully Bayesian methodology within the PM setting, using a nonparametric approach for the censoring mechanism. Our goal is to use the appealing methodology of PL estimator but expand it to incorporate informative censoring. We first formulate the PL estimator using a Bayesian framework. When the censoring mechanism is informative, we introduce intuitive parameters that are used for sensitivity analysis. These parameters capture the difference between the censored and the observed data.
To reduce the large number of the sensitivity parameters, we consider them random from a prior distribution. The prior distribution links all these parameters together around an average missing-data mechanism (mean parameter) while simultaneously accommodating possible differences among them by introducing random variation. The parameterization and the prior distribution associated with it are easy-to-use and accommodate modeling of different types of missing data, including those with informative censoring or missing not at random (MNAR). It also contains MAR as a special case enabling a local sensitivity analysis for departures in a neighborhood of MAR (Ma and others, 2005). We incorporate a probabilistic range on the sensitivity parameters by introducing a prior distribution on them and then applying Bayesian strategies to derive inferences (Kaciroti and others, 2006, 2008, 2009; Daniels and Hogan, 2007).
In Section 2, we develop the Bayesian approach for PL estimate with no censoring and expand it to include informative censoring in Section 3. In Section 4, we describe the Trial Of Preventing Hypertension (TROPHY) study, which motivated and provided the context for the methods developed here. In Section 5, we apply the proposed models to the TROPHY data and in Section 6, we give results from comparative studies and offer conclusions in Section 7.
2. BAYESIAN FORMULATION OF PL ESTIMATOR
First, we begin with the formulation of a version of a PL method using a Bayesian framework without censoring. Let be a vector of binary responses, indicating the presence of the event at baseline () and K follow-up visits . Let denote the time from baseline for the follow-up visit , and . Let T be the measure of time when the event occurred, ; with if and only if and , and if no event occurred by . The focus here is to compare the cumulative incidence, , between groups.
Let be the probability of an event in for a subject who is at risk at in the treatment group g. The cumulative incidence function up to time is
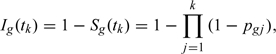 |
(2.1) |
where .
Let be the number of individuals at risk at time ; and be the number of events occurring in among these subjects for and . In absence of censoring, , where is the number of subjects at the beginning of the study and . The likelihood function of the parameter , conditional on , , and treatment indicator is
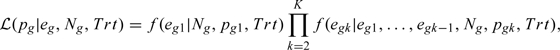 |
where and for ,
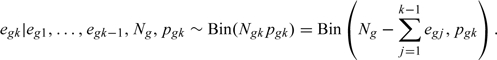 |
Framing the inference problem from a Bayesian perspective, the posterior distribution of assuming a noninformative prior is
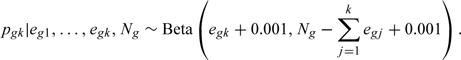 |
(2.2) |
The inference on the cumulative incidence is derived from (2.1) and using Monte Carlo (MC) simulations, with the posterior mean of estimated by
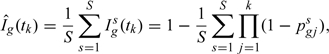 |
where is the sth draw from (2.2), and simulations. The posterior standard deviation of is also estimated using MC simulations.
Our main hypothesis is that and (or equivalently and ) are different over the study period and also at the 4-year mark. We compare these curves between 2 groups by using the average distance on a log scale:
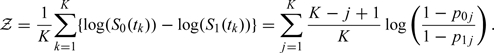 |
(2.3) |
If , i.e., no group differences, then 𝒵 should be 0. In a Bayesian approach, we calculate the posterior distribution of 𝒵 and the Bayesian p value using MC simulations. The p value is equal to 0), Pr(𝒵< 0)} and is calculated as the smaller of the proportion of 0 or 0 times 2 for a 2-sided test. Similarly, the incidence rates at the end of the study are compared using .
3. EXPANDED MODEL
When censored data occurred, the estimates, described in Section 2, are modified to account for censoring mechanism. Let be the random variable indexing the missing-data patterns, which is equal to k if subject i was censored in , . Let +1 when subject i had no event and completed the study. If the censoring mechanism is nonignorable, then would depend on the unobserved , and additional assumptions or constraints are needed to identify the model.
In a longitudinal setting, Little (1993) used identifying restrictions by setting the density of missing components, conditional on the observed components, to be equal to the corresponding distribution of the subgroup of completers or some other relevant pattern. For monotone missing data, Molenberghs and others (1998) extended Little's approach to available case missing value (ACMV) restriction by setting the density of missing components at visit k, conditioned on the previous components, to be equal to the corresponding distribution of the observed data at visit k. That is, for 0, . They showed that ACMV restriction is equivalent to MAR. Here, we extend the ACMV approach to identify PM models when missing data are MNAR: the extension also provides a necessary and sufficient condition for MAR, which is used for local sensitivity analysis around MAR. For MNAR, the identifying constraints relate the distribution of the missing data to that of the observed data. Because there are no data to estimate the identifying constraints, it is useful to have intuitive and easy-to-use parameters, which can then be evaluated using a sensitivity analysis. We propose the following parameterization.
Let be the probability of an event in for subjects who are at risk and observed in . Let be the number of subjects at risk and observed in , and be the number of events that occurred in among these subjects. Next, let be the probability of an event in for subjects censored in who would be at risk at for . Last, let be the number of subjects censored in who would have been at risk at , and be the number of events occurring in among these subjects. Then,
and for ,
| (3.1) |
where and for . There are no data to estimate the parameters of the distribution (3.4). To overcome this problem, we relate the parameters of the distribution for the missing data to the parameters of the distribution for the observed data, using the following parameterization:
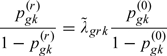 |
(3.2) |
for any , , and g. Here, is the odds ratio of having an event in , being at risk at , between those subjects in missing pattern r (censored in ), and the observed subjects in for group g. The parameters measure the departure from MAR and cannot be estimated from the data. We assume that has a log-normal prior distribution with mean and variance , where c is the coefficient of variation. The and c parameters are used for sensitivity analysis, where and correspond to MAR, satisfying the ACMV constraint. Note that for time-to-event outcomes (where ), the ACMV restriction is equivalent to , , . Indeed, ; and because , the only requirement for ACMV is or equivalently , , , and g.
In the longitudinal setting, with K follow-up measures, up to missing-data patterns can occur, resulting in a maximum of constraints ( indices) for each group required to identify the model. As K increases, the number of identifying parameters become too large to be feasible for sensitivity analysis. Therefore, it is important to reduce the dimension of the space for the sensitivity parameters. For example, Scharfstein and others (2001) introduced a parametric function for their sensitivity parameters; Birmingham and others (2003) reduced the dimension of their sensitivity parameters by assuming all to be the same.
Here, we reduce the dimension of the sensitivity space by introducing a prior distribution for the identifying parameters with mean, , yet we introduce random variation that allows each to vary within a range for different r and k. Thus, are consider random variables from a log-normal prior distribution 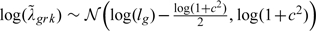 , (this corresponds to and ). Here, and c are second-order sensitivity parameters: indicates the average difference from MAR, and c indicates the degree of the random variation (potential dissimilarities) across different patterns and over time. A coefficient of corresponds to a deterministic constraint, whereas corresponds to a stochastic constraint that becomes less informative as c increases. As in complete case analysis, we test the null hypothesis of no difference between 2 curves using the 𝒵 distance (2.3).
, (this corresponds to and ). Here, and c are second-order sensitivity parameters: indicates the average difference from MAR, and c indicates the degree of the random variation (potential dissimilarities) across different patterns and over time. A coefficient of corresponds to a deterministic constraint, whereas corresponds to a stochastic constraint that becomes less informative as c increases. As in complete case analysis, we test the null hypothesis of no difference between 2 curves using the 𝒵 distance (2.3).
3.1. Fitting the model
We fit our model following a full Bayesian approach, where the quantities of interest are the cumulative incidence curves for each group as if no subject dropped out: . Here, is the probability of having an endpoint in , interval for subjects who would have been at risk at , this includes both observed and censored subjects. Under PM specification , where is the weight (proportion) of the dropout pattern r at time j for each group. Here, are not observed and thus are unknown parameters.
Under a fully Bayesian approach, the inferences are derived from the joint posterior distribution . Where Data=; is the set of parameters for the observed data; are the unobserved data for the censored subjects; are the proportions of subjects who would be at risk at a given time for each missing-data pattern; and .
Draws from this posterior distribution are obtained using the following partition:
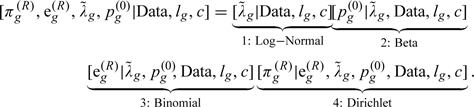 |
We sequentially draw values based on this partition as described in the following steps.
1. Draw from . In a PM setting, the data provide no evidence for sensitivity parameters, (Little, 1995). Thus, are drawn from
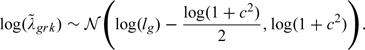 |
2. Draw from . First note that within the PM approach, the fit of the model to the observed data is identical for all choices of sensitivity parameters or (Little, 1995). Let be the initial number of subjects at , let and be the number of the observed events and the censored subjects in the interval (). Then,
where is the number of subjects at risk and observed in the : for , and . We assume noninformative independent priors from which
3. Draw from . When and , let be the number of subjects censored at visit r who would have been at risk for an event at visit k (no event by ). Let be the number among these subjects who would have had the event in the interval. Then,
and for ,
where from (3.2),
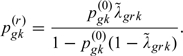 |
(3.3) |
Here, , and for . Within this step, the draw of is sequential, specifically, is drawn first, and then up to are drawn next, sequentially conditioned on the previous draws. If , then .
4. Draw from . Here, , , is the proportion of subjects who belong to missing-data pattern r at time among subjects who would be at risk at the beginning of interval; corresponds to the proportion of subjects who are at risk and observed in interval. Then for each k,
Assuming a noninformative Dirichlet prior distribution for with parameters , are drawn from the corresponding posterior Dirichlet distribution 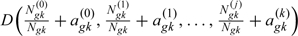 .
.
5. Derive draws of marginal parameters . For each draw of , , and , the marginal parameters are calculated as , where for is derived from (3.3).
5A. Draws of marginal parameters . Instead of computing in Step (5), we can simulate draws of from the completed data. This can be viewed as a multiple imputation version of Step 5 and has been proposed in Demirtas and Schafer (2003). Thus, under this approach, once are drawn/imputed, the draws for are obtained based on the imputed data set as if there were no censoring. For each , we assume a noninformative prior distribution, , independent of each other, which results in the following posterior distributions:
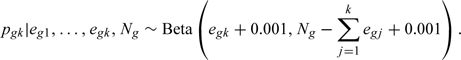 |
Here, , where are observed and are draws from Step 3. Though Step 5 is probably most computationally efficient, but this alternative step is much easier to implement in available software packages such as WinBUGS. Applying either Step 5 or 5A to the smaller data set analyzed in Section 6, we obtained identical results up to the third decimal. For the TROPHY data, which has a larger number of missing-data patterns, we used the Step 5A approach. Note that, for MAR case (, ), the (based on the full imputed data) was the same as (based on the observed data) as expected.
Sensitivity analysis. We vary and c to perform sensitivity analysis. When the missing data are MAR, then all or and ; thus and c can be considered ignorability indices. When the missing data are MNAR, then not all equal to 1. Finding the best combination of sensitivity parameters is usually not possible, but reasonable choices for such parameters can often suffice. For instance, trying different values of sensitivity parameters in a neighborhood of the MAR (, small c) could often provide reassuring information about whether the inferences are robust to moderately informative censoring mechanisms. In addition, finding values for which group comparisons are just significant is also important. That is, finding how much different the censored data must be from the observed data for treatment differences to be just significant.
Here, are interpretable and easy-to-use for sensitivity analysis. By considering them random, we (1) provide a probabilistic range for differences among censored and observed subjects, (2) allow for random variation on such differences across different missing-data patterns and over time, (3) reduce the dimension of the sensitivity parameters' space by using the shrinkage parameters ( and c) for sensitivity analysis, and (4) account for the overall uncertainty related to censoring by using Bayesian methods.
4. MOTIVATION: TROPHY STUDY
The TROPHY (Julius and others, 2006) was an investigator-initiated study to examine whether early treatment of prehypertension might prevent or delay the development of hypertension. Our primary objective was to determine whether for patients with prehypertension, 2 years of treatment with candesartan would reduce the incidence of hypertension for up to 2 years after active treatment was discontinued.
The study consisted first of a 2-year double-blind placebo-controlled phase; followed by a 2-year phase in which all study patients received a placebo. Subjects were examined every 3 months as well as 1 month after the beginning of each phase.
4.1. Primary event
The development of hypertension was chosen as the primary study event. It was defined as the first occurrence of any one of the following: (i) systolic blood pressure (SBP) ≥140 and/or diastolic blood pressure (DBP) ≥90 mmHg at any 3 visits during the 4 years of the study; (ii) SBP ≥160 and/or DBP ≥100 mmHg at any visit during 4 years; (iii) SBP ≥140 and/or DBP ≥90 mmHg at the end of the study; and (iv) patients requiring treatment as decided by the attending physician. After an event occurred, antihypertension treatment with metoprolol, hydrochlorothiazide, or other medications, with the exception of angiotensin-receptor blockers, was offered at no cost.
The “3 times in 4 years” definition of hypertension differs from more contemporary accepted definition of hypertension based on the guidelines published in the Seventh Report of the Joint National Committee on Hypertension (JNC 7). Following the new guidelines, patients with an average clinic reading of systolic 140 mmHg or higher and/or diastolic of 90 mmHg or higher on 2 consecutive clinic visits are now considered to need treatment for hypertension. TROPHY results are published for both definitions (Julius and others, 2006), (Julius and others, 2008). We focus here on the sensitivity analysis for the definition of hypertension that follows the current JNC 7 guidelines.
4.2. Censored data
Data in TROPHY are censored for 2 reasons: (1) if subjects dropout before the event or (2) if subjects violated the protocol. The violation is related to a post hoc change in the definition of the hypertension. That is, a subject who satisfied the 3 times in 4 years criteria of hypertension may not have had 2 consecutive readings of SBP ≥140 and /or DBP ≥90 and so would not be considered as hypertensive based on the new definition. Following the protocol, treatment of blood pressure (BP) was initiated for such subjects an thus the follow-up BP data are affected and cannot be used.
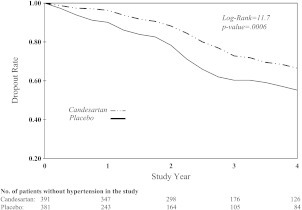
The intention-to-treat population for the study consisted of 772 patients randomly assigned to candesartan (391) or placebo (381). Among the 772 participants, 109 (54 in placebo) dropped out before developing hypertension. When the new definition of hypertension was used, 92 subjects, classified with hypertension based on the original definition, did not satisfy the new definition (JNC 7). Following the protocol, these subjects received antihypertension treatment, therefore, they are censored at the time the treatment was initiated for the analysis using the new event definition. In addition, 2 subjects who had dropped out satisfy the new event definition. Thus, the number of subjects with a missing event per the new definition is increased to 199 (101 in placebo). Figure 1 shows the rate of censoring, which is higher for subjects in the placebo group.
Fig. 1.
PL estimate for time-to-dropout in placebo and candesartan group.
5. APPLICATION
In this section, we apply the method developed in Section 3 to the data from the TROPHY study using WinBUGS1.4 (Spiegelhalter and others, 2003). The effect of candesartan compared with a placebo is estimated over the entire study period as well as its end. Different scenarios about the censoring mechanism are considered to assess the effect of the censored data on the final results. We perform a local sensitivity analysis around the region where missing-data mechanism is MAR (, ). We consider , , and with varying between 0.8 and 1.2. A value of indicates that, on average, the censored subjects are more likely to develop hypertension than the observed subjects and the opposite would be true for 1. In addition, sensitivity analyses based on assumptions that make the treatment effect just significant, , are also included. The results of sensitivity analysis are shown in Table 1. Figure 2 shows the cumulative incidence curves of hypertension for the 2 groups for different values for and and . The sensitivity analysis shows that the effect of candesartan varies by the assumptions made about the censored data and that the impact of the assumed censoring mechanism on the incidence curves increases over time. That reflects the fact that the cumulative number of the censored subjects that contribute to estimation of the incidence curves increases over time. The bold lines correspond to noniformative censoring, and c=0, and are the same as the PL estimates derived from a frequentist approach using Proc Lifetest in SAS.
Table 1.
Sensitivity analysis under different censoring mechanisms
| Placebo | Candesartan | p value | Z | Z | ||
| I48 (95% CR) | I48 (95% CR) | (48 months) | mean (SD) | p value | ||
| c = 0 | ||||||
| 1a | l0 = 1, l1 = 1 | 60.8 (55.1–66.4) | 50.9 (45.3–56.6) | 0.015 | 0.296 (0.049) | < 0.0001 |
| 2a | l0 = 1.2, l1 = 1 | 62.1 (56.3–67.7) | 50.9 (45.4–56.6) | 0.007 | 0.309 (0.050) | < 0.0001 |
| 3a | l0 = 1, l1 = 1.2 | 60.8 (55.1–64.4) | 52.2 (46.5–57.8) | 0.034 | 0.288 (0.049) | < 0.0001 |
| 4a | l0 = 1.2, l1 = 0.8 | 62.0 (56.3–67.7) | 49.6 (44.1–55.2) | 0.002 | 0.317 (0.049) | < 0.0001 |
| 5a | l0 = 0.8, l1 = 1.2 | 59.4 (53.8–65.0) | 52.2 (46.5–57.8) | 0.074 | 0.273 (0.048) | < 0.0001 |
| 6a | l0 = 1, 1.32 | 60.8 (55.2–66.4) | 52.8 (47.2–58.4) | 0.05 | 0.283 (0.049) | < 0.0001 |
| 7a | 1 | 58.8 (53.3–64.3) | 50.9 (45.4–56.6) | 0.05 | 0.275 (0.048) | < 0.0001 |
| c = 0.5 | ||||||
| 1b | l0 = 1, l1 = 1 | 60.7 (55.0–66.3) | 50.8 (45.2–56.5) | 0.016 | 0.295 (0.049) | < 0.0001 |
| 2b | l0 = 1.2, l1 = 1 | 61.9 (56.1–67.6) | 50.8 (45.2–56.5) | 0.007 | 0.308 (0.050) | < 0.0001 |
| 3b | l0 = 1, l1 = 1.2 | 60.7 (55.0–66.4) | 52.0 (46.3–57.7) | 0.034 | 0.288 (0.049) | < 0.0001 |
| 4b | l0 = 1.2, l1 = 0.8 | 61.9 (56.2–67.6) | 49.5 (44.0–55.1) | 0.002 | 0.316 (0.050) | < 0.0001 |
| 5b | l0 = 0.8, l1 = 1.2 | 59.3 (53.7–64.9) | 52.0 (46.3–57.7) | 0.017 | 0.273 (0.049) | < 0.0001 |
| 6b | l0 = 1, 1.32 | 60.7 (55.0–66.3) | 52.7 (46.9–58.3) | 0.05 | 0.283 (0.050) | < 0.0001 |
| 7b | = 1 | 58.8 (53.2–64.4) | 50.8 (45.2–56.6) | 0.05 | 0.275 (0.048) | < 0.0001 |
| c = 1.0 | ||||||
| 1c | l0 = 1, l1 = 1 | 60.5 (54.8–66.2) | 50.6 (44.9–56.3) | 0.016 | 0.294 (0.049) | < 0.0001 |
| 2c | l0 = 1.2, l1 = 1 | 61.7 (55.9–67.4) | 50.6 (45.0–56.3) | 0.008 | 0.307 (0.050) | < 0.0001 |
| 3c | l0 = 1, l1 = 1.2 | 60.5 (54.8–66.2) | 51.7 (46.0–57.4) | 0.033 | 0.287 (0.050) | < 0.0001 |
| 4c | l0 = 1.2, l1 = 0.8 | 61.7 (55.9–67.4) | 49.6 (44.1–55.2) | 0.002 | 0.317(0.049) | < 0.0001 |
| 5c | l0 = 0.8, l1 = 1.2 | 59.2 (53.5–64.8) | 51.7 (46.0–57.4) | 0.068 | 0.274 (0.048) | < 0.0001 |
| 6c | l0 = 1, 1.32 | 60.5 (54.7–66.2) | 52.3 (46.6–58.1) | 0.05 | 0.283 (0.050) | < 0.0001 |
| 7c | 1 | 58.6 (53.0–64.2) | 50.6 (44.9–56.3) | 0.05 | 0.274 (0.048) | < 0.0001 |
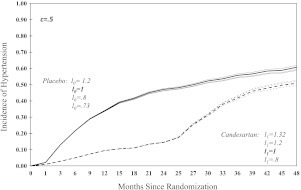
Fig. 2.
Sensitivity analysis for cumulative incidence of hypertension between placebo and candesartan groups over a 48-month period. Bold lines correspond to noninformative censoring, and .
The incidence of hypertension at 48 months ranges from 58.6% to 62.1% for the placebo group and from 49.6% to 52.8% for the candesartan group. The overall incidence of hypertension over time is significantly lower in candesartan group for all the assumed scenarios in Table 1. The incidence of hypertension at 48 months is also lower for the candesartan group under all assumed scenarios; but findings become borderline or nonsignificant for censoring mechanisms that increasingly favor the placebo group. For example, in scenario 6b, where the censored subjects compared with the observed subjects (in any interval) are on average 1.32 (95% credible interval ) times more likely to develop hypertension in the candesartan group but are just as likely (95% credible interval ) in the placebo group, then the group difference at 48 months is just significant. In scenario 5b, where the censored subjects compare with the observed subjects are on average 1.2 (95% credible interval ) times more likely to develop hypertension in the candesartan group, but 0.8 times as likely (95% credible interval ) in the placebo group, the effect of candesartan is nonsignificant at 48 months, . Under noninformative censoring mechanism the incidence of hypertension at 48 months was lower for the candesrtan group, at , compared with the placebo group, 60.8%. Finally, when comparing the overall incidence curves, the significance is strong regardless of the censoring mechanism. This is because large differences between curves occur early when the number of the censored subjects is small, and thus such differences are fairly robust to the assumptions on the censoring mechanism.
In summary, the differences in the incidence of hypertension between 2 groups over the entire study are robust for different censoring mechanisms. However, the group differences at 48 months are more sensitive to the censoring mechanism, varying from 12.4% under scenario 4a to 7.2% under scenario 5a. When the censoring mechanism is noninformative, a 2-year treatment with candesartan reduces the incidence of hypertension at 4 years by (95% credible interval ). In addition, the effect candesartan treatment is robust around a local region of the MAR mechanism. It would take differences on the odds of developing hypertension among dropouts favoring placebo (scenarios 6–7) for the effect of treatment to become borderline significant.
6. COMPARATIVE STUDIES
The proposed model can be directly applied to other studies with similar design. For illustration as well as comparison with other alternative approaches, we apply our method to the data analyzed by Scharfstein and others (2001) (SRER), which are relevant for this setting. SRER analyzed data from a randomized clinical trial focusing on the effect of 2 mg risperidone in treating schizophrenia. The endpoint outcome was defined as a 20% or more reduction in the Positive and Negative Symptom Scale relative to the baseline value. The subjects were followed up at 1, 2, 4, 6, and 8 weeks; data are shown in their Table 1 (for full details of the study refer to their paper). Following a selection model approach, SRER propose a model to perform sensitivity analysis under different censoring mechanisms by introducing censoring bias functions , for . They used intuitive parametric functions to reduce the number of the sensitivity parameters but, as the authors pointed out in their discussion, one can consider an infinite number of forms for , and therefore the results of the sensitivity analysis would be specific to the parametric functions . Specifically, they chose a parameterization where α is the log-odds ratio of dropping out between t and per unit increase in T for subjects who were at risk at t. As an alternative, we propose a full Bayesian approach within a PM setting using intuitive identifying parameters , for sensitivity analysis, which are assumed random following a log-normal distribution: , . There is no an exact relationship between our identifying parameters, l and c, and SRER sensitivity parameter α. However, our sensitivity parameters and theirs have an overall connection in terms of measuring the degree and the nature of the departure from a noninformative censoring mechanism.
Under noninformative censoring and that corresponds to α=0. When censoring is informative, corresponds to . That is, an indicates that compared with the observed subjects, the censored subjects are more likely to have an endpoint in t to and so are more likely to have a shorter event time T. Or equivalently, subjects with smaller T are more likely to be censored, which corresponds to a negative α. For small departures from a noninformative censoring mechanism, l is close to 1, which corresponds to α close to 0.
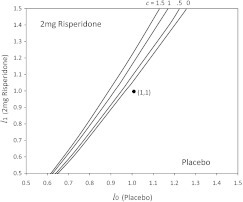
Next, we apply our method to the data analyzed by SRER. Our results of the sensitivity analysis are summarized in Figure 3, which similar to Figure 2(b) in SRER, showing a contour plot for . The region favoring risperidone is the upper left corner region of , . That is consistent with the corresponding region shown in SRER Figure 2(b) (the lower right corner region of , ). Under either approach, no significant difference appears at between treatment and placebo—assuming noninformative censoring. However, it requires only small departures from a noninformative censoring mechanism for the difference to become significant. For instance, treatment is better when , , and . That is, the censored subjects compared with the observed subjects (in any interval) are on average 1.07 times more likely to have an endpoint in the treatment group but are just as likely in the placebo group. Both methods show that small departures from noninformative censoring mechanism produce significant results.
Fig. 3.
Sensitivity analysis on the data from SRER.
The strength of our model lies in using a fully Bayesian approach with random sensitivity parameters, which are easy-to-use, and at the same time does not assume a parametric function on the censoring mechanism. Alternatively, one could expect that if a good parametric function for the censoring mechanism is known, the sensitivity analysis based on the correct specification of such function might well be more efficient.
7. CONCLUDING REMARKS
In this paper, we presented a Bayesian model for time-to-event data with informative censoring. We approached the missing data problem by using a PM model, which is identified by introducing an intuitive parameterization. The identifying parameters, , are defined for each group g as: the odds ratio of having an endpoint in , conditioned on being at risk at , between subjects from missing-data pattern r with the observed subjects in that interval (). Sensitivity analyses are performed using different prior distributions of . Even though the distribution of is unknown, it is possible for a subject matter expert to give it a range and then explore the sensitivity of statistical inferences over such a range. The new parameterization is a nonparametric approach for modeling the censoring mechanism; and therefore is robust to model misspecification that may result from parametric modeling. However, when the number of follow-up visits increases, so does the number of the sensitivity parameters under a nonparametric approach; and soon a fully unrestricted nonparametric approach becomes impractical. The robustness of modeling the correct dropout mechanism and reducing the number of the sensitivity parameters are competing requirements requiring compromise and adjustment according to the specific problem. We reduced the dimensionality of the sensitivity space by using a stochastic constraint to incorporate different levels of uncertainty. That is, we let follow a log-normal distribution with mean and variance . Then the set of the sensitivity parameters is reduced to and c. Our proposed parameterization is flexible for sensitivity analysis and contains noninformative censoring as a special case (, c=0). We assumed no knowledge on , and sensitivity analysis could be performed for values close to 1.
FUNDING
National Institute of Health (5P01HD039386-10).
Acknowledgments
We thank the Editor, Associate Editor, and a referee for careful review and insightful comments, which considerably improved this paper. Many thanks to Rhea Kish for her help with editing. Conflict of Interest: None declared.
Access to the WinBUGS code can be obtained from: nicola@umich.edu.
References
- Birmingham J, Rotnitzky A, Fitzmaurice GM. Pattern-mixture and selection models for analyzing longitudinal data with monotone missing patterns. Journal of the Royal Statistical Society, Series B. 2003;65:275–297. [Google Scholar]
- Daniels M, Hogan J. Reparameterizing the pattern mixture model for sensitivity analysis under informative dropout. Biometrics. 2000;56:1241–1248. doi: 10.1111/j.0006-341x.2000.01241.x. [DOI] [PubMed] [Google Scholar]
- Daniels M, Hogan J. Missing Data in Longitudinal Studies: Strategies for Bayesian Modeling and Sensitivity Analysis. Boca Raton, FL: Chapman & Hall; 2007. [Google Scholar]
- Demirtas H, Schafer JL. On the performance of random-coefficient pattern-mixture models for non-ignorable drop-out. Statistics in Medicine. 2003;22:2553–2575. doi: 10.1002/sim.1475. [DOI] [PubMed] [Google Scholar]
- Guo W, Ratcliffe SJ, Ten Have TT. A random pattern-mixture model for longitudinal data with dropouts. Journal of the American Statistical Association. 2004;99:929–937. [Google Scholar]
- Hjort NL. Nonparametric Bayes estimators based on beta processes in models for life history data. Annals of Statistists. 1990;18:1259–1294. [Google Scholar]
- Ibrahim JG, Chen M, Sinha D. Bayesian Survival Analysis. New York: Springer; 2001. [Google Scholar]
- Julius S, Kaciroti N, Nesbitt S, Egan BM, Michelson EL for the Trial of Preventing Hypertension (TROPHY) investigators. TROPHY study: outcomes based on the JNC 7 definition of hypertension. Journal of the American Society of Hypertension. 2008;2:39–43. doi: 10.1016/j.jash.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Julius S, Nesbitt S, Egan B, Weber MA, Michelson EL, Kaciroti N, Black HR, Grimm RH, Messerli FH, Oparil S, Schork MA for the Trial of Prevention Hypertension (TROPHY) Study Investigators. Feasibility of treating prehypertension with an angiotensin-receptor blocker. New England Journal of Medicine. 2006;354:1685–1697. doi: 10.1056/NEJMoa060838. [DOI] [PubMed] [Google Scholar]
- Kaciroti N, Raghunathan TE, Schork MA, Clark NM. A Bayesian model for longitudinal count data with non-ignorable dropout. Applied Statistics. 2008;57:521–534. doi: 10.1111/j.1467-9876.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaciroti N, Raghunathan TE, Schork MA, Clark NM, Gong M. A Bayesian approach for clustered longitudinal ordinal outcome with nonignorable missing data: evaluation of an asthma education program. Journal of the American Statistical Association. 2006;101:435–446. [Google Scholar]
- Kaciroti N, Schork MA, Raghunathan TE, Julius S. A Bayesian sensitivity model for intention-to-treat analysis on binary outcomes with dropouts. Statistics in Medicine. 2009;28:572–585. doi: 10.1002/sim.3494. [DOI] [PubMed] [Google Scholar]
- Kenward MG, Molenberghs G, Thijs H. Pattern-mixture models with proper time dependence. Biometrika. 2003;90:53–71. [Google Scholar]
- Little RJA. Pattern-mixture models for multivariate incomplete data. Journal of the American Statistical Association. 1993;88:125–134. [Google Scholar]
- Little RJA. Modeling the dropout mechanism in repeated measures studies. Journal of the American Statistical Association. 1995;90:1113–1121. [Google Scholar]
- Little RJA, Rubin DB. Statistical Analysis With Missing Data. 2nd edition. New York: John Wiley; 2002. [Google Scholar]
- Ma G, Troxel AB, Heitjan D. An index of local sensitivity to nonignorable drop-out in longitudinal modeling. Statistics in Medicine. 2005;24:2129–2150. doi: 10.1002/sim.2107. [DOI] [PubMed] [Google Scholar]
- Molenberghs G, Kenward MG. Incomplete Data in Clinical Studies. Chichester, UK: John Wiley; 2007. [Google Scholar]
- Molenberghs G, Michiels B, Kenward MG, Diggle PJ. Monotone missing data and pattern-mixture models. Statistica Neerlandica. 1998;52:153–161. [Google Scholar]
- Phadia EG, Susarla V. Nonparametric Bayesian estimation of a survival curve with dependent censoring mechanism. Annals of the Institute of Statistical Mathematics. 1983;35:389–400. [Google Scholar]
- Rotnitzky A, Scharfstein DO, Su T, Robins JM. Methods for conducting sensitivity analysis of trials with potentially nonignorable competing causes of censoring. Biometrics. 2001;57:103–113. doi: 10.1111/j.0006-341x.2001.00103.x. [DOI] [PubMed] [Google Scholar]
- Roy J, Daniels M. A general class of pattern-mixture models for nonignorable dropout with many possible dropout times. Biometrics. 2008;64:538–545. doi: 10.1111/j.1541-0420.2007.00884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfstein DO, Daniels MJ, Robins JM. Incorporating prior beliefs about selection bias into the analysis of randomized trials with missing outcomes. Biostatistics. 2003;4:495–512. doi: 10.1093/biostatistics/4.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfstein DO, Robins JM. Estimation of the failure time distribution in the presence of informative censoring. Biometrika. 2002;89:617–634. [Google Scholar]
- Scharfstein DO, Robins JM, Eddings W, Rotnitzky A. Inference in randomized studies with informative censoring and discrete time-to-event endpoints. Biometrics. 2001;57:404–413. doi: 10.1111/j.0006-341x.2001.00404.x. [DOI] [PubMed] [Google Scholar]
- Spiegelhalter DJ, Thomas A, Best NG, Lunn D. Sampling Based Approaches to Calculate Marginal Densities. Cambridge, UK: MRC Biostatistics Unit; 2003. WinBUGS User Manual: Version 1.4. [Google Scholar]
- Susarla V, Van Ryzin J. Nonparametric Bayesian estimation of survival curves from incomplete observations. Journal of the American Statistical Association. 1976;71:897–902. [Google Scholar]
- Tsai W. Estimation of survival curves from dependent censorship models via a generalized self-consistent property with nonparametric Bayesian estimation application. The Annals of Statistics. 1986;14:238–249. [Google Scholar]
- Wu MC, Carroll RJ. Estimation and comparison of changes in the presence of informative right censoring by modeling the censoring process. Biometrics. 1988;44:175–188. [Google Scholar]
- Zheng M, Klein JP. Estimates of marginal survival for dependent competing risks based on an assumed copula. Biometrika. 1995;82:127–138. [Google Scholar]