Abstract
The title compound, C15H12N2OS, exists as the thione tautomer in the solid state. The phenyl group is almost perpendicular [dihedral angle = 87.96 (5)°] to the fused ring system (r.m.s. deviation = 0.036 Å for 13 ring and exocyclic non-H atoms). In the crystal, centrosymmetric dimers, sustained by pairs of N—H⋯S hydrogen bonds, are connected into layers parallel to (-101) by C—H⋯O and C—H⋯S interactions.
Related literature
For recent studies on synthesis, drug discovery and crystal structures of quinazoline-4(3H)-one derivatives, see: El-Azab & El-Tahir (2012 ▶); El-Azab et al. (2011 ▶, 2010 ▶). For the antimicrobial activity of the title compound, see: Al-Omar et al. (2004 ▶). For the structures of related compounds, see: Bowman et al. (2007 ▶); Hashim et al. (2010 ▶).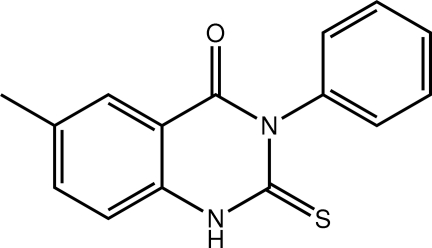
Experimental
Crystal data
C15H12N2OS
M r = 268.33
Monoclinic,
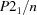
a = 12.7770 (3) Å
b = 5.1384 (1) Å
c = 19.0973 (4) Å
β = 91.814 (2)°
V = 1253.17 (5) Å3
Z = 4
Cu Kα radiation
μ = 2.23 mm−1
T = 100 K
0.35 × 0.15 × 0.05 mm
Data collection
Agilent SuperNova Dual diffractometer with an Atlas detector
Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011 ▶) T min = 0.967, T max = 0.998
4636 measured reflections
2576 independent reflections
2348 reflections with I > 2σ(I)
R int = 0.016
Refinement
R[F 2 > 2σ(F 2)] = 0.033
wR(F 2) = 0.098
S = 1.06
2576 reflections
177 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.25 e Å−3
Δρmin = −0.25 e Å−3
Data collection: CrysAlis PRO (Agilent, 2011 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812007301/qm2054sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812007301/qm2054Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812007301/qm2054Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H1n⋯S1i | 0.91 (2) | 2.49 (2) | 3.3662 (12) | 163.6 (17) |
| C3—H3⋯O1ii | 0.95 | 2.33 | 3.2522 (17) | 163 |
| C11—H11⋯S1iii | 0.95 | 2.86 | 3.7333 (16) | 154 |
| C15—H15⋯O1iv | 0.95 | 2.32 | 3.1988 (18) | 154 |
Symmetry codes: (i)  ; (ii)
; (ii) 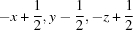 ; (iii)
; (iii) 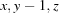 ; (iv)
; (iv) 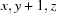 .
.
Acknowledgments
This work was supported by the Research Center of Pharmacy, King Saud University, Riyadh, Saudi Arabia. We also thank the Ministry of Higher Education (Malaysia) for funding structural studies through the High-Impact Research scheme (UM.C/HIR/MOHE/SC/12).
supplementary crystallographic information
Comment
Quinazoline-4(3H)-one derivatives are known for their various biological activities (El-Azab & El-Tahir, 2012; El-Azab et al. (2011, 2010). The title compound (I) has been investigated previously for its anti-microbial activity (Al-Omar et al., 2004). Herein, its crystal and molecular structure is described.
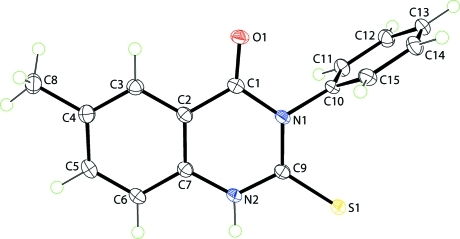
The key result of the crystal structure determination of (I), Fig. 1, is the confirmation that the compound exists as the thione tautomer in the solid-state. The non-hydrogen atoms comprising the fused ring system, including the exocyclic atoms, are co-planar with a r.m.s. deviation = 0.036 Å. The maximum deviations from the least-squares plane through these atoms are 0.038 (1) Å for the S1 atom and -0.085 (1) Å for N1, consistent with some pyramidal character for the latter atom. The phenyl group is almost perpendicular to the aforementioned plane: the dihedral angle = 87.96 (5)°.
The presence of the thione tautomer confirms the results of previous structure determinations on related compounds (Bowman et al., 2007; Hashim et al., 2010).
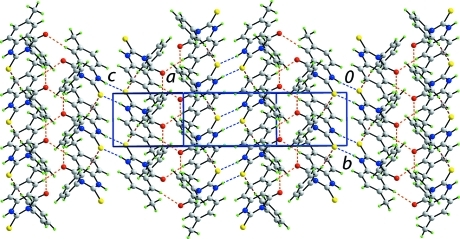
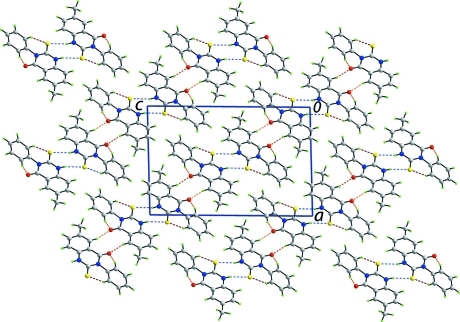
The key feature of the crystal packing is the formation of N—H···S hydrogen bonds between centrosymmetrically related molecules, Table 1. The dimeric aggregates thus formed are connected into layers parallel to (1 0 1) by C—H···O interactions, involving the bifurcated carbonyl-O atom, and C—H···S interactions, Fig. 2 and Table 1. Layers stack with no specific intermolecular interactions between them, Fig. 3.
Experimental
A mixture of 2-amino-5-methylbenzoic acid (1.51 g m, 10 mmol) and phenyl isothiocyanate (1.35 g m, 10 mmol) in absolute ethanol (30 ml) containing triethylamine (1.1 mg, 10 mmol) was refluxed for 3 h. The reaction mixture was allowed to cool, the solvent was removed under reduced pressure, and the solid obtained was dried and recrystallized from EtOH. Yield 90%; M.pt: 340–342 K; 1H NMR (500 MHz, DMSO-d6): δ 12.98 (s, 1H, exchangeable), 7.75 (s, 1H), 7.60 (d, 1H, J=8.0 Hz), 7.48 (t, 2H, J=7.0, 7.5 Hz), 7.41 (d, 1H, J=7.0 Hz), 7.36 (d, 1H, J=8.0 Hz), 7.26 (d, 2H, J=8.0 Hz), 2.37 (s, 3H). 13C NMR (DMSO-d6): δ 176.0, 160.2, 139.8, 138.1, 137.1, 134.4, 129.5, 129.3, 128.5, 127.2, 116.5, 116.2, 20.9.
Refinement
Carbon-bound H-atoms were placed in calculated positions [C—H = 0.95 to 0.98 Å, Uiso(H) = 1.2 to 1.5Ueq(C)] and were included in the refinement in the riding model approximation. The N—H atom was located in a difference Fourier map, and was refined with distance restraint of N—H = 0.88±0.01 Å; the Uiso value was refined.
Figures
Fig. 1.
The molecular structure of (I) showing the atom-labelling scheme and displacement ellipsoids at the 50% probability level.
Fig. 2.
A view of the supramolecular layer parallel to (1 0 1) in (I) mediated by N—H···S, C—H···O and C—H···S interactions, shown as blue, orange and brown dashed lines, respectively.
Fig. 3.
A view in projection down the b axis of the unit-cell contents of (I). The N—H···S, C—H···O and C—H···S interactions are shown as blue, orange and brown dashed lines, respectively.
Crystal data
| C15H12N2OS | F(000) = 560 |
| Mr = 268.33 | Dx = 1.422 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54184 Å |
| Hall symbol: -P 2yn | Cell parameters from 2386 reflections |
| a = 12.7770 (3) Å | θ = 3.5–76.4° |
| b = 5.1384 (1) Å | µ = 2.23 mm−1 |
| c = 19.0973 (4) Å | T = 100 K |
| β = 91.814 (2)° | Prism, colourless |
| V = 1253.17 (5) Å3 | 0.35 × 0.15 × 0.05 mm |
| Z = 4 |
Data collection
| Agilent SuperNova Dual diffractometer with an Atlas detector | 2576 independent reflections |
| Radiation source: SuperNova (Cu) X-ray Source | 2348 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.016 |
| Detector resolution: 10.4041 pixels mm-1 | θmax = 76.6°, θmin = 4.1° |
| ω scan | h = −15→16 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011) | k = −5→6 |
| Tmin = 0.967, Tmax = 0.998 | l = −14→24 |
| 4636 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.033 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.098 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0613P)2 + 0.3083P] where P = (Fo2 + 2Fc2)/3 |
| 2576 reflections | (Δ/σ)max = 0.001 |
| 177 parameters | Δρmax = 0.25 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.56616 (3) | 0.52832 (7) | 0.394981 (16) | 0.01717 (12) | |
| O1 | 0.35822 (8) | −0.1192 (2) | 0.25706 (5) | 0.0181 (2) | |
| N1 | 0.45653 (9) | 0.1596 (2) | 0.32532 (6) | 0.0149 (2) | |
| H1n | 0.4357 (15) | 0.305 (4) | 0.4827 (11) | 0.029 (5)* | |
| N2 | 0.42228 (9) | 0.2138 (2) | 0.44293 (6) | 0.0161 (2) | |
| C1 | 0.37571 (11) | −0.0266 (3) | 0.31493 (7) | 0.0150 (3) | |
| C2 | 0.31942 (10) | −0.0982 (3) | 0.37768 (7) | 0.0156 (3) | |
| C3 | 0.24261 (10) | −0.2920 (3) | 0.37432 (7) | 0.0173 (3) | |
| H3 | 0.2250 | −0.3722 | 0.3307 | 0.021* | |
| C4 | 0.19179 (11) | −0.3686 (3) | 0.43408 (7) | 0.0182 (3) | |
| C5 | 0.22073 (11) | −0.2484 (3) | 0.49797 (7) | 0.0195 (3) | |
| H5 | 0.1871 | −0.3001 | 0.5394 | 0.023* | |
| C6 | 0.29690 (11) | −0.0568 (3) | 0.50213 (7) | 0.0179 (3) | |
| H6 | 0.3156 | 0.0210 | 0.5459 | 0.021* | |
| C7 | 0.34592 (11) | 0.0206 (3) | 0.44129 (7) | 0.0156 (3) | |
| C9 | 0.47714 (10) | 0.2888 (3) | 0.38744 (7) | 0.0149 (3) | |
| C8 | 0.10873 (12) | −0.5770 (3) | 0.42997 (8) | 0.0229 (3) | |
| H8A | 0.1314 | −0.7176 | 0.3993 | 0.034* | |
| H8B | 0.0432 | −0.5024 | 0.4110 | 0.034* | |
| H8C | 0.0976 | −0.6465 | 0.4769 | 0.034* | |
| C10 | 0.52338 (10) | 0.2014 (3) | 0.26589 (7) | 0.0154 (3) | |
| C11 | 0.60838 (11) | 0.0364 (3) | 0.25902 (8) | 0.0188 (3) | |
| H11 | 0.6233 | −0.0942 | 0.2931 | 0.023* | |
| C12 | 0.67148 (12) | 0.0652 (3) | 0.20130 (8) | 0.0208 (3) | |
| H12 | 0.7303 | −0.0452 | 0.1959 | 0.025* | |
| C13 | 0.64818 (11) | 0.2554 (3) | 0.15178 (7) | 0.0208 (3) | |
| H13 | 0.6910 | 0.2746 | 0.1124 | 0.025* | |
| C14 | 0.56249 (12) | 0.4182 (3) | 0.15946 (7) | 0.0217 (3) | |
| H14 | 0.5471 | 0.5485 | 0.1254 | 0.026* | |
| C15 | 0.49894 (11) | 0.3912 (3) | 0.21705 (7) | 0.0193 (3) | |
| H15 | 0.4400 | 0.5013 | 0.2225 | 0.023* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0194 (2) | 0.01951 (19) | 0.01264 (19) | −0.00399 (12) | 0.00160 (13) | −0.00051 (12) |
| O1 | 0.0211 (5) | 0.0210 (5) | 0.0122 (5) | −0.0004 (4) | −0.0006 (4) | −0.0024 (4) |
| N1 | 0.0169 (5) | 0.0178 (6) | 0.0102 (5) | −0.0002 (4) | 0.0019 (4) | −0.0005 (4) |
| N2 | 0.0189 (6) | 0.0194 (6) | 0.0101 (5) | −0.0025 (5) | 0.0016 (4) | −0.0016 (5) |
| C1 | 0.0154 (6) | 0.0161 (6) | 0.0136 (6) | 0.0029 (5) | −0.0003 (5) | 0.0006 (5) |
| C2 | 0.0156 (6) | 0.0179 (6) | 0.0132 (6) | 0.0015 (5) | 0.0007 (5) | 0.0012 (5) |
| C3 | 0.0168 (6) | 0.0197 (7) | 0.0153 (6) | 0.0001 (5) | −0.0004 (5) | −0.0009 (5) |
| C4 | 0.0158 (6) | 0.0190 (7) | 0.0199 (7) | 0.0012 (5) | 0.0008 (5) | 0.0013 (5) |
| C5 | 0.0193 (6) | 0.0229 (7) | 0.0165 (6) | 0.0013 (5) | 0.0052 (5) | 0.0029 (6) |
| C6 | 0.0201 (7) | 0.0212 (7) | 0.0125 (6) | 0.0008 (5) | 0.0023 (5) | −0.0002 (5) |
| C7 | 0.0145 (6) | 0.0176 (6) | 0.0145 (6) | 0.0011 (5) | 0.0005 (5) | 0.0002 (5) |
| C9 | 0.0155 (6) | 0.0163 (6) | 0.0129 (6) | 0.0015 (5) | 0.0002 (5) | 0.0004 (5) |
| C8 | 0.0198 (7) | 0.0245 (7) | 0.0245 (7) | −0.0038 (6) | 0.0032 (6) | 0.0017 (6) |
| C10 | 0.0176 (6) | 0.0183 (6) | 0.0103 (6) | −0.0027 (5) | 0.0026 (5) | −0.0021 (5) |
| C11 | 0.0199 (7) | 0.0199 (7) | 0.0166 (7) | 0.0013 (5) | 0.0024 (5) | 0.0032 (5) |
| C12 | 0.0187 (7) | 0.0225 (7) | 0.0213 (7) | 0.0011 (6) | 0.0056 (6) | −0.0005 (6) |
| C13 | 0.0230 (7) | 0.0249 (7) | 0.0148 (6) | −0.0055 (6) | 0.0060 (5) | −0.0011 (6) |
| C14 | 0.0283 (8) | 0.0220 (7) | 0.0148 (7) | −0.0007 (6) | 0.0024 (6) | 0.0051 (6) |
| C15 | 0.0226 (7) | 0.0192 (7) | 0.0161 (6) | 0.0025 (6) | 0.0024 (5) | −0.0008 (6) |
Geometric parameters (Å, º)
| S1—C9 | 1.6788 (14) | C6—C7 | 1.3953 (19) |
| O1—C1 | 1.2175 (17) | C6—H6 | 0.9500 |
| N1—C9 | 1.3776 (17) | C8—H8A | 0.9800 |
| N1—C1 | 1.4171 (18) | C8—H8B | 0.9800 |
| N1—C10 | 1.4578 (16) | C8—H8C | 0.9800 |
| N2—C9 | 1.3454 (17) | C10—C15 | 1.379 (2) |
| N2—C7 | 1.3913 (18) | C10—C11 | 1.388 (2) |
| N2—H1n | 0.91 (2) | C11—C12 | 1.3940 (19) |
| C1—C2 | 1.4639 (18) | C11—H11 | 0.9500 |
| C2—C7 | 1.3918 (19) | C12—C13 | 1.386 (2) |
| C2—C3 | 1.398 (2) | C12—H12 | 0.9500 |
| C3—C4 | 1.3877 (19) | C13—C14 | 1.389 (2) |
| C3—H3 | 0.9500 | C13—H13 | 0.9500 |
| C4—C5 | 1.406 (2) | C14—C15 | 1.3945 (19) |
| C4—C8 | 1.508 (2) | C14—H14 | 0.9500 |
| C5—C6 | 1.385 (2) | C15—H15 | 0.9500 |
| C5—H5 | 0.9500 | ||
| C9—N1—C1 | 124.38 (11) | N2—C9—N1 | 116.73 (12) |
| C9—N1—C10 | 119.89 (11) | N2—C9—S1 | 120.73 (10) |
| C1—N1—C10 | 115.66 (11) | N1—C9—S1 | 122.54 (10) |
| C9—N2—C7 | 124.69 (12) | C4—C8—H8A | 109.5 |
| C9—N2—H1n | 115.0 (13) | C4—C8—H8B | 109.5 |
| C7—N2—H1n | 120.2 (13) | H8A—C8—H8B | 109.5 |
| O1—C1—N1 | 120.20 (12) | C4—C8—H8C | 109.5 |
| O1—C1—C2 | 124.34 (13) | H8A—C8—H8C | 109.5 |
| N1—C1—C2 | 115.45 (12) | H8B—C8—H8C | 109.5 |
| C7—C2—C3 | 120.25 (12) | C15—C10—C11 | 121.98 (12) |
| C7—C2—C1 | 119.46 (13) | C15—C10—N1 | 120.36 (12) |
| C3—C2—C1 | 120.24 (12) | C11—C10—N1 | 117.59 (12) |
| C4—C3—C2 | 120.66 (13) | C10—C11—C12 | 118.93 (13) |
| C4—C3—H3 | 119.7 | C10—C11—H11 | 120.5 |
| C2—C3—H3 | 119.7 | C12—C11—H11 | 120.5 |
| C3—C4—C5 | 118.17 (13) | C13—C12—C11 | 119.84 (14) |
| C3—C4—C8 | 120.35 (13) | C13—C12—H12 | 120.1 |
| C5—C4—C8 | 121.48 (13) | C11—C12—H12 | 120.1 |
| C6—C5—C4 | 121.79 (13) | C12—C13—C14 | 120.37 (13) |
| C6—C5—H5 | 119.1 | C12—C13—H13 | 119.8 |
| C4—C5—H5 | 119.1 | C14—C13—H13 | 119.8 |
| C5—C6—C7 | 119.22 (13) | C13—C14—C15 | 120.25 (13) |
| C5—C6—H6 | 120.4 | C13—C14—H14 | 119.9 |
| C7—C6—H6 | 120.4 | C15—C14—H14 | 119.9 |
| N2—C7—C2 | 118.93 (12) | C10—C15—C14 | 118.62 (13) |
| N2—C7—C6 | 121.17 (13) | C10—C15—H15 | 120.7 |
| C2—C7—C6 | 119.90 (13) | C14—C15—H15 | 120.7 |
| C9—N1—C1—O1 | 175.07 (13) | C5—C6—C7—N2 | 179.51 (13) |
| C10—N1—C1—O1 | −7.96 (18) | C5—C6—C7—C2 | −1.2 (2) |
| C9—N1—C1—C2 | −6.19 (19) | C7—N2—C9—N1 | −1.3 (2) |
| C10—N1—C1—C2 | 170.77 (11) | C7—N2—C9—S1 | 178.88 (10) |
| O1—C1—C2—C7 | −179.92 (13) | C1—N1—C9—N2 | 6.2 (2) |
| N1—C1—C2—C7 | 1.40 (19) | C10—N1—C9—N2 | −170.65 (12) |
| O1—C1—C2—C3 | 2.7 (2) | C1—N1—C9—S1 | −173.97 (10) |
| N1—C1—C2—C3 | −176.00 (12) | C10—N1—C9—S1 | 9.18 (18) |
| C7—C2—C3—C4 | 0.0 (2) | C9—N1—C10—C15 | −91.74 (16) |
| C1—C2—C3—C4 | 177.40 (13) | C1—N1—C10—C15 | 91.15 (16) |
| C2—C3—C4—C5 | −0.9 (2) | C9—N1—C10—C11 | 91.23 (16) |
| C2—C3—C4—C8 | 179.76 (13) | C1—N1—C10—C11 | −85.88 (15) |
| C3—C4—C5—C6 | 0.6 (2) | C15—C10—C11—C12 | 0.6 (2) |
| C8—C4—C5—C6 | −179.97 (13) | N1—C10—C11—C12 | 177.61 (13) |
| C4—C5—C6—C7 | 0.4 (2) | C10—C11—C12—C13 | −0.5 (2) |
| C9—N2—C7—C2 | −3.2 (2) | C11—C12—C13—C14 | 0.2 (2) |
| C9—N2—C7—C6 | 176.10 (13) | C12—C13—C14—C15 | −0.2 (2) |
| C3—C2—C7—N2 | −179.69 (12) | C11—C10—C15—C14 | −0.6 (2) |
| C1—C2—C7—N2 | 2.9 (2) | N1—C10—C15—C14 | −177.45 (13) |
| C3—C2—C7—C6 | 1.0 (2) | C13—C14—C15—C10 | 0.3 (2) |
| C1—C2—C7—C6 | −176.36 (12) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H1n···S1i | 0.91 (2) | 2.49 (2) | 3.3662 (12) | 163.6 (17) |
| C3—H3···O1ii | 0.95 | 2.33 | 3.2522 (17) | 163 |
| C11—H11···S1iii | 0.95 | 2.86 | 3.7333 (16) | 154 |
| C15—H15···O1iv | 0.95 | 2.32 | 3.1988 (18) | 154 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+1/2, y−1/2, −z+1/2; (iii) x, y−1, z; (iv) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: QM2054).
References
- Agilent (2011). CrysAlis PRO Agilent Technologies, Yarnton, Oxfordshire, England.
- Al-Omar, M. A., Abdel-Hamide, S. G., Al-Khamees, H. A. & El-Subbagh, H. I. (2004). Saudi Pharm. J. 12, 63–71.
- Bowman, W. R., Elsegood, M. R. J., Stein, T. & Weaver, G. W. (2007). Org. Biomol. Chem. 5, 103–113. [DOI] [PubMed]
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- El-Azab, A. S., Al-Omar, M. A., Abdel-Aziz, A. A.-M., Abdel-Aziz, N. I., El-Sayed, M. A.-A., Aleisa, A. M., Sayed-Ahmed, M. M. & Abdel-Hamide, S. G. (2010). Eur. J. Med. Chem. 45, 4188–4198. [DOI] [PubMed]
- El-Azab, A. S. & El-Tahir, K. H. (2012). Bioorg. Med. Chem. Lett. 22, 327–333.
- El-Azab, A. S., El-Tahir, K. H. & Attia, S. M. (2011). Monatsh. Chem. 142, 837–925.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Hashim, N. M., Osman, H., Rahim, A. A., Yeap, C. S. & Fun, H.-K. (2010). Acta Cryst. E66, o950. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812007301/qm2054sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812007301/qm2054Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812007301/qm2054Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report