Abstract
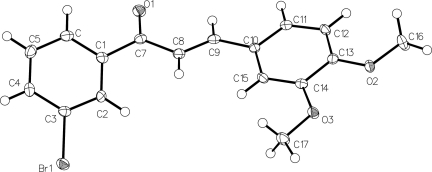
The molecular structure of the title compound, C17H15BrO3, consists of a bromophenyl and a 3,4-dimethoxyphenyl group linked through a prop-2-en-1-one spacer. The C=C double bond displays an E conformation, while the carbonyl group shows an S-cis conformation relative to the double bond.
Related literature
For the Suzuki reaction, see: Miyaura & Suzuki (1995 ▶); Bringmann et al. (2005 ▶). For bichalcone derivatives, see: Shetonde et al. (2010 ▶). For related structures, see: Escobar et al. (2008 ▶); Valdebenito et al. (2010 ▶); Chu et al. (2004 ▶); Radha Krishna et al. (2005 ▶); Wu et al. (2005 ▶).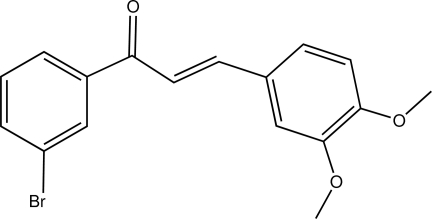
Experimental
Crystal data
C17H15BrO3
M r = 347.20
Monoclinic,
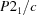
a = 12.7946 (5) Å
b = 3.9373 (1) Å
c = 29.8209 (10) Å
β = 109.219 (3)°
V = 1418.54 (8) Å3
Z = 4
Mo Kα radiation
μ = 2.91 mm−1
T = 120 K
0.2 × 0.12 × 0.08 mm
Data collection
Agilent Xcalibur Sapphire3 Gemini ultra diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011 ▶) T min = 0.802, T max = 1.000
12861 measured reflections
3429 independent reflections
2895 reflections with I > 2σ(I)
R int = 0.047
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.074
S = 1.10
3429 reflections
192 parameters
H-atom parameters constrained
Δρmax = 0.63 e Å−3
Δρmin = −0.41 e Å−3
Data collection: CrysAlis PRO (Agilent, 2011 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: OLEX2 (Dolomanov et al., 2009 ▶); software used to prepare material for publication: OLEX2.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812006836/fj2515sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812006836/fj2515Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812006836/fj2515Isup3.cdx
Supplementary material file. DOI: 10.1107/S1600536812006836/fj2515Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by FONDECYT through grant Nos. 1080147 and 3110066. MF thanks to Becaschile Programme (Chile) for support through a postdoctoral fellowship.
supplementary crystallographic information
Comment
From the synthetic point of view, bromochalcones are the choice precursors to accomplish the C—C bond formation, through the Suzuki reaction, one of the most popular and powerful methods for coupling aryl–aryl moieties (Miyaura & Suzuki, 1995; Bringmann et al., 2005), to produce symmetric or asymmetric biphenyls, this being the entry to bichalcones (Shetonde et al., 2010).
The molecular structure of the title compound displays two phenyl rings connected through the organic prop-2-en-1-one spacer. As shown in Fig. 1, one phenyl ring is substituted at positions 3 and 4 with methoxy groups, while the other is substituted at position 3' with one Br atom.
The dihedral angle between the two aromatic rings joined by the conjugated spacer is 26.59 (9)°. On the other hand, the spacer formed by C10—C9—C8—C7—O1—C1 can be considered as a plane with a RMSD of 0.029 Å. This feature is also observed in other chalcones (Escobar et al. 2008; Valdebenito et al., 2010; Chu et al. 2004; Radha Krishna et al. 2005; Wu et al. 2005).
Finally, both inter- and intramolecular hydrogen bonds are not observed in the crystalline packing of title compound.
Experimental
A mixture of 3-bromoacetophenone (0.5 g, 2,5 mmol) and 3,4-dimethoxibenzaldehyde (0.41 g, 2,5 mmol), were dissolved in Methanol (50 ml), and were treated with KOH (2 g, dissolved in 20 ml methanol). After 20 min 30 ml of water were added, and the title compound precipitated as a yellow solid. Then, it was filtered and recrystallized in ethanol to yield 1.27 g (73%) of a yellow solid. m.p.117–120°C. IR (KBr): ν = 1657 (CO), cm-1. 1H-NMR (400 MHz, CDCl3): d = 3.94 (3H, s, OCH3), 3.96 (3H, s, OCH3), 6.90 (1H, d, J = 8.3 Hz, H5), 7.16 (1H, m, H2), 7.24 (1H, d, J = 7.2 Hz., H6), 7.32 (1H, d, J = 15.6 Hz., Ha), 7.38 (1H, t, J = 7.8 Hz., H5), 7.70 (1H, d, J = 7.9 Hz., H4), 7.77 (1H, d, J = 15.6 Hz., Hb), 7.93 (1H, d, J = 7.7 Hz., H6), 8.13 (1H, m, H2). 13C-NMR (400 MHz, CDCl3): d = 56.1, 110.2, 111.2, 119.4, 122.9, 123.5, 127.0, 127.6, 130.2, 131.4, 135.4, 140.4, 146.0, 149.3, 151.8, 189.1. HRMS calc. for C17H15BrO3 346.02046; Found 346.019994.
Refinement
The H atoms positions were calculated after each cycle of refinement using a riding model with C—H distances in the range 0.95—0.98 Å and Uiso(H) = 1.2–1.5Ueq(C).
Figures
Fig. 1.
The molecular structure of title compound with full atom numbering scheme. Displacement ellipsoids are presented at 30% probability level and H atoms are shown as spheres.
Crystal data
| C17H15BrO3 | F(000) = 704 |
| Mr = 347.20 | Dx = 1.626 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.7107 Å |
| a = 12.7946 (5) Å | Cell parameters from 3587 reflections |
| b = 3.9373 (1) Å | θ = 2.6–29.0° |
| c = 29.8209 (10) Å | µ = 2.91 mm−1 |
| β = 109.219 (3)° | T = 120 K |
| V = 1418.54 (8) Å3 | Prism, colourless |
| Z = 4 | 0.2 × 0.12 × 0.08 mm |
Data collection
| Agilent Xcalibur Sapphire3 Gemini ultra diffractometer | 3429 independent reflections |
| Radiation source: Enhance (Mo) X-ray source | 2895 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.047 |
| Detector resolution: 16.1511 pixels mm-1 | θmax = 29.0°, θmin = 2.6° |
| ω scans | h = −17→16 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011) | k = −5→5 |
| Tmin = 0.802, Tmax = 1.000 | l = −40→40 |
| 12861 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.074 | H-atom parameters constrained |
| S = 1.10 | w = 1/[σ2(Fo2) + (0.019P)2 + 1.5139P] where P = (Fo2 + 2Fc2)/3 |
| 3429 reflections | (Δ/σ)max = 0.002 |
| 192 parameters | Δρmax = 0.63 e Å−3 |
| 0 restraints | Δρmin = −0.41 e Å−3 |
Special details
| Experimental. Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | −0.11568 (2) | 0.90741 (7) | 0.528213 (9) | 0.01853 (9) | |
| O1 | −0.25274 (14) | 0.3062 (5) | 0.31571 (6) | 0.0198 (4) | |
| O2 | 0.40922 (14) | 0.3072 (5) | 0.32327 (6) | 0.0173 (4) | |
| O3 | 0.37580 (14) | 0.5457 (5) | 0.39741 (6) | 0.0190 (4) | |
| C16 | 0.4322 (2) | 0.1612 (7) | 0.28340 (10) | 0.0205 (6) | |
| H16A | 0.3873 | 0.2751 | 0.2542 | 0.031* | |
| H16B | 0.4141 | −0.0814 | 0.2813 | 0.031* | |
| H16C | 0.5109 | 0.1902 | 0.2874 | 0.031* | |
| C9 | −0.0247 (2) | 0.2800 (7) | 0.32953 (9) | 0.0143 (5) | |
| H9 | −0.0759 | 0.1507 | 0.3052 | 0.017* | |
| C11 | 0.1119 (2) | 0.1273 (6) | 0.29162 (9) | 0.0143 (5) | |
| H11 | 0.0535 | 0.0184 | 0.2676 | 0.017* | |
| C12 | 0.2171 (2) | 0.1330 (6) | 0.28749 (9) | 0.0148 (5) | |
| H12 | 0.2298 | 0.0357 | 0.2606 | 0.018* | |
| C14 | 0.2834 (2) | 0.4212 (7) | 0.36346 (9) | 0.0143 (5) | |
| C1 | −0.2297 (2) | 0.6078 (7) | 0.38681 (9) | 0.0140 (5) | |
| C15 | 0.1780 (2) | 0.4234 (7) | 0.36627 (9) | 0.0144 (5) | |
| H15 | 0.1649 | 0.5243 | 0.3928 | 0.017* | |
| C5 | −0.3827 (2) | 0.8723 (7) | 0.40364 (10) | 0.0201 (6) | |
| H5 | −0.4579 | 0.9429 | 0.3932 | 0.024* | |
| C10 | 0.0897 (2) | 0.2760 (6) | 0.32972 (9) | 0.0130 (5) | |
| C3 | −0.2072 (2) | 0.8273 (6) | 0.46427 (9) | 0.0142 (5) | |
| C4 | −0.3163 (2) | 0.9344 (7) | 0.45004 (10) | 0.0178 (6) | |
| H4 | −0.3450 | 1.0479 | 0.4716 | 0.021* | |
| C8 | −0.0668 (2) | 0.4429 (7) | 0.35928 (9) | 0.0149 (5) | |
| H8 | −0.0187 | 0.5619 | 0.3858 | 0.018* | |
| C2 | −0.1622 (2) | 0.6663 (6) | 0.43363 (9) | 0.0135 (5) | |
| H2 | −0.0869 | 0.5968 | 0.4442 | 0.016* | |
| C13 | 0.3031 (2) | 0.2829 (6) | 0.32325 (9) | 0.0138 (5) | |
| C7 | −0.1871 (2) | 0.4390 (7) | 0.35127 (9) | 0.0143 (5) | |
| C17 | 0.3612 (2) | 0.6874 (7) | 0.43882 (9) | 0.0212 (6) | |
| H17A | 0.3096 | 0.8790 | 0.4298 | 0.032* | |
| H17B | 0.4327 | 0.7667 | 0.4604 | 0.032* | |
| H17C | 0.3313 | 0.5140 | 0.4548 | 0.032* | |
| C6 | −0.3401 (2) | 0.7083 (7) | 0.37247 (10) | 0.0175 (6) | |
| H6 | −0.3866 | 0.6638 | 0.3409 | 0.021* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.02037 (14) | 0.02143 (14) | 0.01387 (14) | −0.00093 (12) | 0.00574 (10) | −0.00270 (12) |
| O1 | 0.0164 (9) | 0.0267 (11) | 0.0153 (10) | −0.0041 (8) | 0.0037 (8) | −0.0030 (8) |
| O2 | 0.0138 (9) | 0.0241 (10) | 0.0154 (10) | 0.0002 (8) | 0.0068 (7) | −0.0033 (8) |
| O3 | 0.0141 (9) | 0.0302 (11) | 0.0126 (10) | −0.0035 (8) | 0.0042 (7) | −0.0081 (9) |
| C16 | 0.0222 (14) | 0.0244 (15) | 0.0198 (15) | 0.0009 (12) | 0.0138 (12) | −0.0032 (12) |
| C9 | 0.0151 (13) | 0.0143 (12) | 0.0115 (13) | −0.0014 (11) | 0.0017 (10) | 0.0032 (11) |
| C11 | 0.0151 (12) | 0.0144 (13) | 0.0115 (13) | 0.0002 (10) | 0.0018 (10) | 0.0005 (11) |
| C12 | 0.0198 (13) | 0.0146 (13) | 0.0115 (13) | 0.0031 (11) | 0.0073 (10) | −0.0011 (11) |
| C14 | 0.0173 (12) | 0.0132 (12) | 0.0113 (13) | −0.0004 (11) | 0.0030 (10) | −0.0002 (11) |
| C1 | 0.0150 (12) | 0.0128 (12) | 0.0148 (13) | −0.0034 (11) | 0.0058 (10) | 0.0026 (11) |
| C15 | 0.0190 (13) | 0.0143 (12) | 0.0111 (13) | 0.0019 (11) | 0.0065 (10) | −0.0001 (11) |
| C5 | 0.0137 (13) | 0.0235 (15) | 0.0237 (15) | 0.0011 (11) | 0.0071 (11) | 0.0050 (13) |
| C10 | 0.0150 (12) | 0.0115 (12) | 0.0126 (13) | 0.0025 (10) | 0.0048 (10) | 0.0031 (10) |
| C3 | 0.0156 (12) | 0.0133 (13) | 0.0131 (14) | −0.0031 (10) | 0.0039 (10) | 0.0019 (10) |
| C4 | 0.0168 (13) | 0.0176 (13) | 0.0217 (15) | 0.0010 (11) | 0.0102 (11) | −0.0001 (12) |
| C8 | 0.0146 (12) | 0.0164 (13) | 0.0128 (13) | −0.0009 (11) | 0.0033 (10) | 0.0017 (11) |
| C2 | 0.0104 (12) | 0.0140 (13) | 0.0174 (14) | −0.0024 (10) | 0.0064 (10) | 0.0030 (11) |
| C13 | 0.0141 (12) | 0.0135 (12) | 0.0141 (13) | 0.0016 (10) | 0.0052 (10) | 0.0031 (11) |
| C7 | 0.0155 (12) | 0.0124 (12) | 0.0141 (13) | −0.0005 (11) | 0.0036 (10) | 0.0036 (11) |
| C17 | 0.0201 (14) | 0.0285 (16) | 0.0147 (14) | −0.0036 (12) | 0.0054 (11) | −0.0065 (12) |
| C6 | 0.0159 (13) | 0.0203 (14) | 0.0141 (14) | −0.0026 (11) | 0.0019 (10) | 0.0025 (11) |
Geometric parameters (Å, º)
| Br1—C3 | 1.907 (3) | C1—C2 | 1.398 (3) |
| O1—C7 | 1.232 (3) | C1—C7 | 1.498 (4) |
| O2—C16 | 1.436 (3) | C1—C6 | 1.391 (4) |
| O2—C13 | 1.361 (3) | C15—H15 | 0.9500 |
| O3—C14 | 1.369 (3) | C15—C10 | 1.411 (3) |
| O3—C17 | 1.422 (3) | C5—H5 | 0.9500 |
| C16—H16A | 0.9800 | C5—C4 | 1.387 (4) |
| C16—H16B | 0.9800 | C5—C6 | 1.383 (4) |
| C16—H16C | 0.9800 | C3—C4 | 1.384 (4) |
| C9—H9 | 0.9500 | C3—C2 | 1.384 (4) |
| C9—C10 | 1.462 (3) | C4—H4 | 0.9500 |
| C9—C8 | 1.343 (4) | C8—H8 | 0.9500 |
| C11—H11 | 0.9500 | C8—C7 | 1.477 (3) |
| C11—C12 | 1.391 (3) | C2—H2 | 0.9500 |
| C11—C10 | 1.388 (3) | C17—H17A | 0.9800 |
| C12—H12 | 0.9500 | C17—H17B | 0.9800 |
| C12—C13 | 1.387 (3) | C17—H17C | 0.9800 |
| C14—C15 | 1.379 (3) | C6—H6 | 0.9500 |
| C14—C13 | 1.413 (4) | ||
| C13—O2—C16 | 116.6 (2) | C11—C10—C15 | 118.5 (2) |
| C14—O3—C17 | 117.0 (2) | C15—C10—C9 | 123.0 (2) |
| O2—C16—H16A | 109.5 | C4—C3—Br1 | 118.8 (2) |
| O2—C16—H16B | 109.5 | C2—C3—Br1 | 118.96 (19) |
| O2—C16—H16C | 109.5 | C2—C3—C4 | 122.3 (2) |
| H16A—C16—H16B | 109.5 | C5—C4—H4 | 120.7 |
| H16A—C16—H16C | 109.5 | C3—C4—C5 | 118.5 (2) |
| H16B—C16—H16C | 109.5 | C3—C4—H4 | 120.7 |
| C10—C9—H9 | 115.8 | C9—C8—H8 | 119.6 |
| C8—C9—H9 | 115.8 | C9—C8—C7 | 120.8 (2) |
| C8—C9—C10 | 128.5 (2) | C7—C8—H8 | 119.6 |
| C12—C11—H11 | 119.0 | C1—C2—H2 | 120.7 |
| C10—C11—H11 | 119.0 | C3—C2—C1 | 118.7 (2) |
| C10—C11—C12 | 122.0 (2) | C3—C2—H2 | 120.7 |
| C11—C12—H12 | 120.5 | O2—C13—C12 | 124.7 (2) |
| C13—C12—C11 | 119.0 (2) | O2—C13—C14 | 115.4 (2) |
| C13—C12—H12 | 120.5 | C12—C13—C14 | 119.9 (2) |
| O3—C14—C15 | 125.2 (2) | O1—C7—C1 | 119.5 (2) |
| O3—C14—C13 | 114.5 (2) | O1—C7—C8 | 121.5 (2) |
| C15—C14—C13 | 120.3 (2) | C8—C7—C1 | 118.9 (2) |
| C2—C1—C7 | 122.0 (2) | O3—C17—H17A | 109.5 |
| C6—C1—C2 | 119.4 (2) | O3—C17—H17B | 109.5 |
| C6—C1—C7 | 118.6 (2) | O3—C17—H17C | 109.5 |
| C14—C15—H15 | 119.9 | H17A—C17—H17B | 109.5 |
| C14—C15—C10 | 120.1 (2) | H17A—C17—H17C | 109.5 |
| C10—C15—H15 | 119.9 | H17B—C17—H17C | 109.5 |
| C4—C5—H5 | 119.8 | C1—C6—H6 | 119.6 |
| C6—C5—H5 | 119.8 | C5—C6—C1 | 120.7 (2) |
| C6—C5—C4 | 120.3 (2) | C5—C6—H6 | 119.6 |
| C11—C10—C9 | 118.4 (2) | ||
| Br1—C3—C4—C5 | −179.3 (2) | C10—C11—C12—C13 | 2.0 (4) |
| Br1—C3—C2—C1 | 179.90 (18) | C4—C5—C6—C1 | −1.0 (4) |
| O3—C14—C15—C10 | −177.6 (2) | C4—C3—C2—C1 | −0.5 (4) |
| O3—C14—C13—O2 | −2.7 (3) | C8—C9—C10—C11 | −171.3 (3) |
| O3—C14—C13—C12 | 176.7 (2) | C8—C9—C10—C15 | 7.1 (4) |
| C16—O2—C13—C12 | −0.9 (4) | C2—C1—C7—O1 | 159.4 (2) |
| C16—O2—C13—C14 | 178.5 (2) | C2—C1—C7—C8 | −22.0 (4) |
| C9—C8—C7—O1 | −4.7 (4) | C2—C1—C6—C5 | 1.6 (4) |
| C9—C8—C7—C1 | 176.7 (2) | C2—C3—C4—C5 | 1.1 (4) |
| C11—C12—C13—O2 | −179.5 (2) | C13—C14—C15—C10 | 2.3 (4) |
| C11—C12—C13—C14 | 1.1 (4) | C7—C1—C2—C3 | 179.2 (2) |
| C12—C11—C10—C9 | 175.6 (2) | C7—C1—C6—C5 | −178.5 (2) |
| C12—C11—C10—C15 | −2.9 (4) | C17—O3—C14—C15 | 0.2 (4) |
| C14—C15—C10—C9 | −177.7 (2) | C17—O3—C14—C13 | −179.8 (2) |
| C14—C15—C10—C11 | 0.7 (4) | C6—C1—C2—C3 | −0.8 (4) |
| C15—C14—C13—O2 | 177.3 (2) | C6—C1—C7—O1 | −20.6 (4) |
| C15—C14—C13—C12 | −3.3 (4) | C6—C1—C7—C8 | 158.1 (2) |
| C10—C9—C8—C7 | 175.1 (2) | C6—C5—C4—C3 | −0.3 (4) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FJ2515).
References
- Agilent (2011). CrysAlis PRO Agilent Technologies Ltd, Yarnton, Oxfordshire, England.
- Bringmann, G., Price Mortimer, A. J., Keller, P. A., Gresser, M. J., Garner, J. & Breuning, M. (2005). Angew. Chem. Int. Ed. 44, 5384–5427. [DOI] [PubMed]
- Chu, H.-W., Wu, H.-T. & Lee, Y.-J. (2004). Tetrahedron, 60, 2647–2655.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Escobar, C. A., Vega, A., Sicker, D. & Ibañez, A. (2008). Acta Cryst. E64, o1834. [DOI] [PMC free article] [PubMed]
- Miyaura, N. & Suzuki, A. (1995). Chem. Rev. 95, 2457–2483.
- Radha Krishna, J., Jagadeesh Kumar, N., Krishnaiah, M., Venkata Rao, C., Koteswara Rao, Y. & Puranik, V. G. (2005). Acta Cryst. E61, o1323–o1325.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shetonde, O. M., Wendimagegn, M., Merhatibeb, B., Kerstin, A.-M. & Berhanu, M. A. (2010). Bioorg. Med. Chem. 18, 2464–2473.
- Valdebenito, C., Garland, M. T., Fuentealba, M. & Escobar, C. A. (2010). Acta Cryst. E66, m838. [DOI] [PMC free article] [PubMed]
- Wu, H., Xu, Z. & Liang, Y.-M. (2005). Acta Cryst. E61, o1434–o1435.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812006836/fj2515sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812006836/fj2515Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812006836/fj2515Isup3.cdx
Supplementary material file. DOI: 10.1107/S1600536812006836/fj2515Isup4.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report