Abstract
Tumorigenesis is a clonal evolution process initiated from single cells within otherwise histologically normal tissue1. How single, sporadic mutant cells that have sustained oncogenic alterations evolve within tightly regulated tissue environment remains elusive. Here, we investigated the effects of inducing oncogene expression in single cells within organotypic mammary acini as a model to elucidate the processes by which oncogenic alterations initiate clonal progression from organized epithelial environments. We found that sporadic cells induced to overexpress oncogenes that specifically perturb cell-cycle checkpoints (HPV16-E7 and cyclinD1), deregulate c-Myc transcription, or activate AKT signaling, remain quiescent within growth-arrested acini. In contrast, single ErbB2-overexpressing cells initiate a cellular cascade involving cell translocation from the epithelial layer and luminal outgrowth characteristic of neoplastic progression in early-stage epithelial tumors. ErbB2-mediated luminal cell translocation is dependent on ERK and matrix metalloproteinase (MMP) activities, and genetic alterations that perturb local cell-matrix adhesion can drive cell translocation. We further provide evidence that luminal cell translocation may drive clonal selection by promoting either death or expansion of quiescent oncogene-expressing cells depending on whether preexisting alterations allow anchorage-independent survival and growth. Our data show that the initial outgrowth of single oncogene-expressing cells from organized epithelial structures is a highly regulated process, and we propose that a cell translocation mechanism allows sporadic mutant cells to evade suppressive microenvironments and provokes clonal selection for survival and proliferative expansion outside their native niches.
The outgrowth of sporadic mutant cells within tightly regulated cellular environments is fundamental to tumor evolution. However, oncogenic alterations are usually not sufficient to predict the behaviors and fates of sporadic cell variants2–7, in particular within complex cellular context such as tissues. Limitations in examining single-cell evolution within native tissues have precluded systematic analysis. Three-dimensional (3D) organotypic cultures recapitulate many characteristics of cell dynamics and organization found in tissues, while permitting complex manipulations and long-term monitoring at single-cell resolution. MCF10A, a non-transformed human mammary epithelial cell line, develop into polarized, growth-arrested acinar structures containing a hollow lumen when cultured on reconstituted basement membrane (Matrigel™) (Fig. 1a, b). By modeling induction of oncogenes in single cells within 3D acini, we explore how sporadic mutant cells evolve within organized epithelial environments.
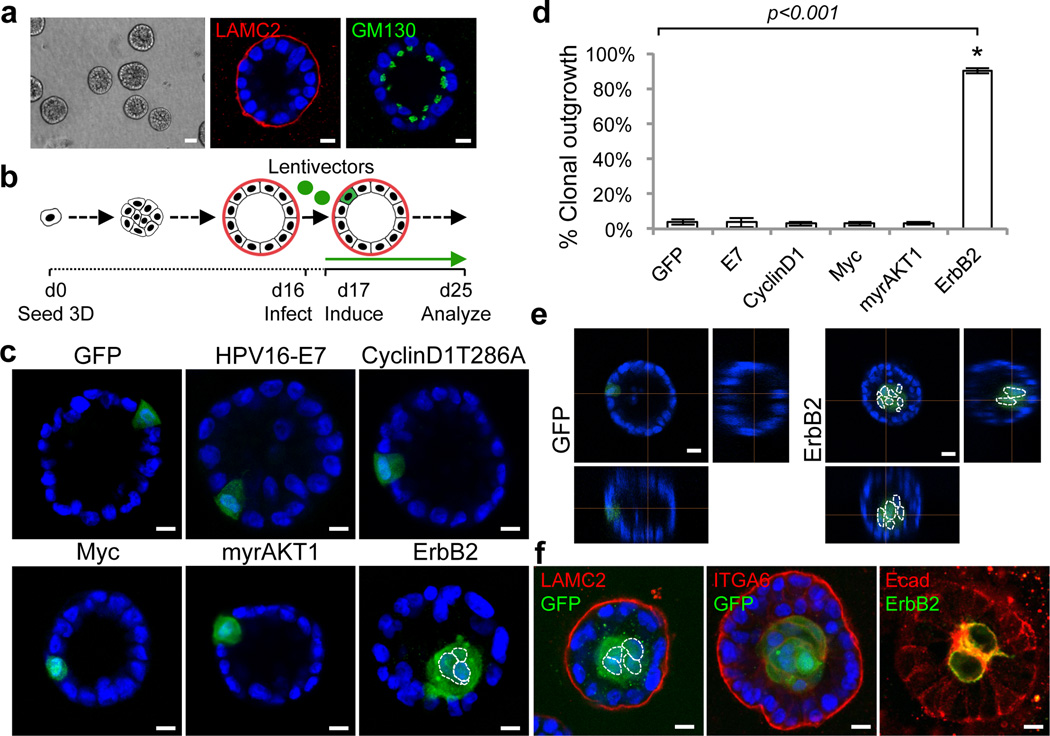
Figure 1. Single-cell induction of oncogenic alternation in mammary acinar culture.
a) Representative images of Day16 MCF10A acini. Left: Low magnification phase contrast image. Scale bar, 50mm. Middle and right: Representative acini immunostained with laminin-γ2 (LAMC2, red) and GM130 (green). Scale bars, 10µm. b) Schematic of single-cell lentiviral infection and doxycycline (Dox)-induction of oncogenes and reporters (green) in growth-arrested, polarized acini. c) Representative images and d) quantification of clonal expansion eight days after induction. e) Representative images of 3D confocal reconstructions of infected acini. f) Acini with expanded ErbB2-clones in lumen were immunostained for GFP (ErbB2- cells), laminin-γ2 (LAMC2), α6-integrin (ITGA6), E-cadherin (Ecad), or ErbB2. Nuclei were counterstained with DAPI (blue). Nuclei of GFP-expressing clones are outlined in dotted white lines in some cases to aid visualization. Raw data are shown in Supplementary Table 1. Error bars represent standard deviations of four experiments. Asterisks denote statistically significant change calculated using two-tailed t-tests. Scale bars, 10µm.
To induce sporadic single oncogene-expressing cells, growth-arrested (Day 16) MCF10A acini stably expressing rtTA (reverse tetracycline transactivator) were infected with low-dose lentiviral vectors driving tetracycline (Tet)-inducible bicistronic expression of oncogenes and fluorescent reporters (pLT-iG) to transduce <0.5% of cells (1 cell per ~10 acini) (Fig. 1b, Supplementary Fig. 2). Overexpressing c-Myc, a master transcription factor deregulated in many cancers, or myrAKT1, which constitutively activates AKT signaling, or perturbing cell-cycle checkpoints by overexpressing HPV16-E7 or Cyclin D1T286A (degradation-resistant CyclinD1) was not sufficient to drive clonal outgrowth. Transduced cells remained quiescent as single cells in the acinar epithelial layer, similar to GFP controls (Fig. 1c,d). MCF10As induced constitutively to express these oncogenes from Day1 of 3D cultures developed aberrant hyperproliferative structures (Supplementary Fig. 3), indicating the lack of clonal expansion from the single-cell context is not due to sub-threshold expression. In contrast, overexpressing ErbB2, a receptor tyrosine kinase amplified in 30% of breast tumors8, in sporadic cells within 3D acini effectively drove clonal outgrowth (90±2%, GFP-labeled cell clusters containing multiple nuclei) (Fig. 1c, d and Supplementary Fig. 4). Interestingly, these ErbB2 clones were confined in the lumen (Fig. 1e), resembling the histological feature of early-stage carcinoma-in-situ breast tumors9. Laminin-γ2, α6-integrin, and E-cadherin immunostaining indicated that gross acinar structures remained intact and luminal translocation is not associated with epithelial-mesenchymal transition (Fig. 1f). No invasive structures were observed (data not shown). Overexpressing ErbB2 in single cells within 3D acini derived from primary mouse mammary epithelial cells or a highly polarized ovarian cell line, MCAS, also led to luminal localization of transduced cells (Supplementary Fig. 5). The unique ability of ErbB2 to initiate clonal expansion and the striking pattern of luminal filling suggest that outgrowth of sporadic mutant cells from organized epithelial structures is tightly regulated.
Long-term (56–85 hours) time-lapse confocal microscopy indicated that single GFP control cells remained quiescent (5/5) within growth-arrested acini. In contrast, single ErbB2-overexpressing cells dissociated from the epithelial layer, exhibited elevated protrusive activity, and translocated into the lumen (6/7) (Fig. 2a, Supplementary Movies 1, 2 and Fig. 6). Blocking cell proliferation by aphidicolin did not block translocation (Fig. 2b–d), further indicating that translocation is independent of proliferation. Together, these data reveal an initial luminal cell translocation step in ErbB2-mediated clonal outgrowth.
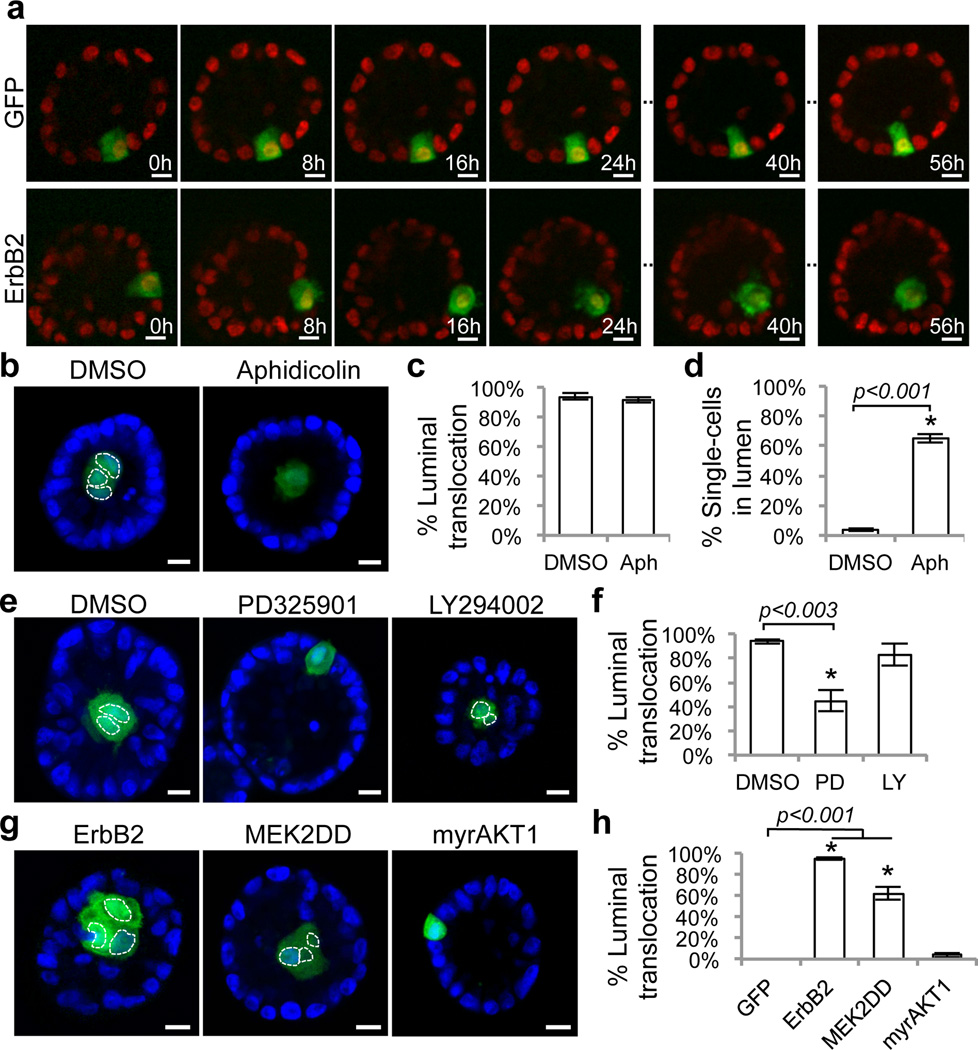
Figure 2. Mechanisms of luminal cell translocation.
a) Confocal sections from time series of acini (red nuclei) containing single ErbB2- or GFP-cell (green) captured starting from approximately 24 hours after induction (0h). b) Representative images and quantification of c) translocation and d) single ErbB2-cells in lumen of Day16 acini treated with aphidicolin (10µM) or DMSO for eight days. e) Representatives images and f) quantification of ErbB2-cells translocation in 3D acini treated with MEK inhibitor (PD325901, 1µM), PI3K/mTor inhibitor (LY294002, 20µM), or DMSO for eight days. g) Representative images and h) quantifications of luminal translocation of MEK2DD- or myrAKT1-cells from growth-arrested acini. Nuclei of GFP-expressing clones are outlined in dotted white lines to aid visualization. Raw data are shown in Supplementary Table 2. Error bars represent standard deviations of four experiments. Asterisks denote statistically significant change calculated using two-tailed t-tests. Scale bars, 10µm.
We next examined two major ErbB2 downstream pathways, mitogen-activated protein kinase (MAPK/ERK) and phosphatidylinositol-3-kinase (PI3K) in luminal translocation. MEK, but not PI3K/mTOR, inhibition greatly reduced ErbB2-mediated translocation (Fig. 2e,f). Moreover, constitutively active MEK (MEK2DD), but not AKT1 (myrAKT1), was sufficient to drive luminal cell translocation (Fig. 2g,h). We do not rule out possible ERK-independent ErbB2 downstream mechanisms, because perturbing ERK activity only partially inhibits or promotes ErbB2-mediated luminal translocation. Recent studies reported that Ras(V12) and v-Src transformed MDCK cells adjacent to normal neighbors are extruded from monolayer cultures by an ERK-dependent mechanism, although Raf-driven ERK activation is not sufficient to drive extrusion10,11. Another study showed that universal expression of activated Raf induces overall cell motility in MCF10A acini12. Our results suggest that this conserved role of ERK on cell motility is also critical for ErbB2-mediated single-cell luminal translocation in 3D.
We found that the Erk-regulated transcription factor Ets1 can drive luminal cell translocation (Fig. 3a–b). Ets1 transactivates proteinases including matrix-metalloproteinases (MMPs)13, which have been implicated in tissue remodeling. Interestingly, MMP inhibition significantly blocked ErbB2-, MEK2DD-, and Ets1-induced luminal translocation (Fig. 3d–e and Supplementary Fig. 7). Although which specific MMPs involved are unclear, these data indicate that MMP activity is important for cell translocation. We overexpressed MT1-MMP to examine whether MMP activity can promote translocation. MT1-MMP was chosen because its membrane localization and broad substrate specificity make it an attractive tool to modulate local MMP activity. Single-cell MT1-MMP overexpression in MCF10A acini was sufficient to drive luminal cell translocation. Notably, Ets1- and MT1-MMP-induced translocation did not drive clonal expansion (Fig. 3a–c) and was independent of Erk signaling (Supplementary Fig. 8). Together, these results identified specific proliferation-independent pathways that promote luminal cell translocation.
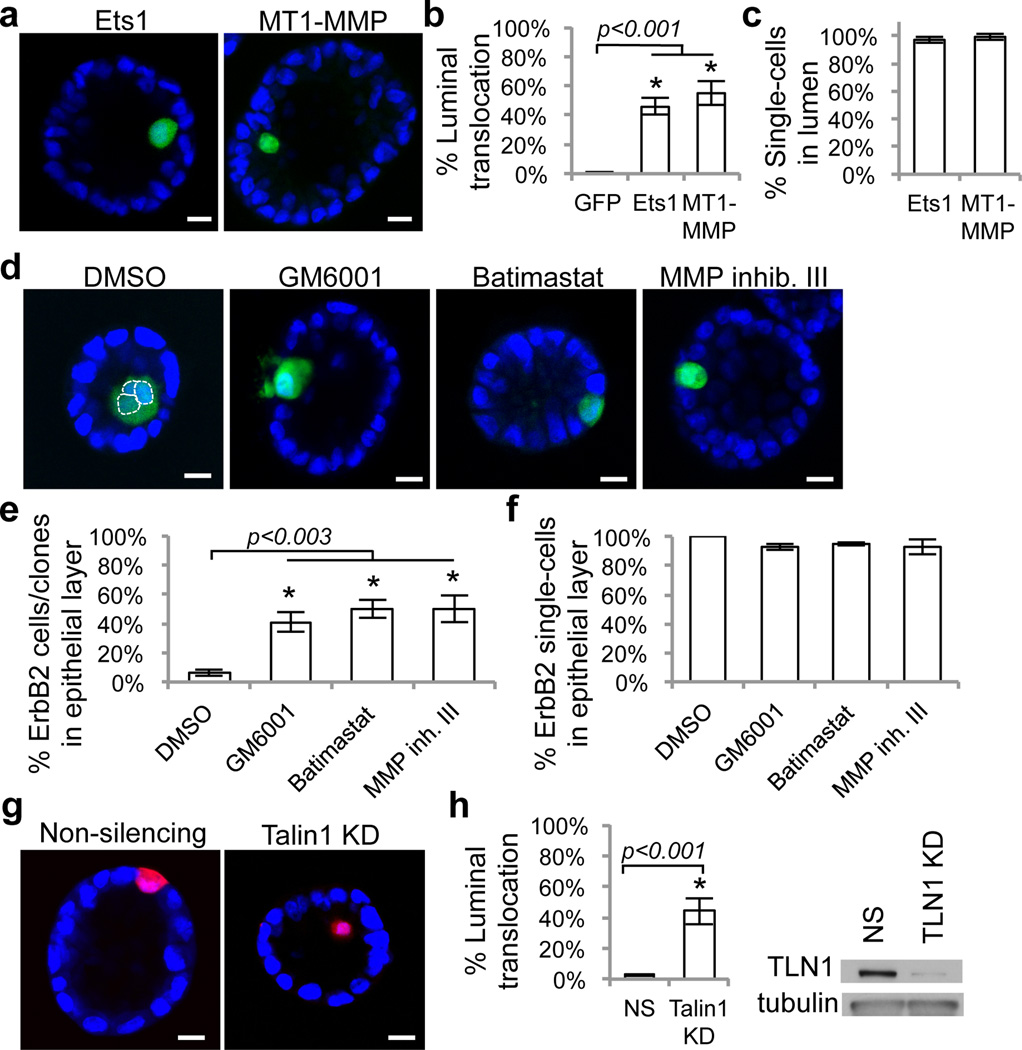
Figure 3. Luminal translocation and clonal outgrowth from suppressive epithelial environment.
a) Representative images, and quantification of b) luminal translocation and c) single-cell in lumen eight days after induction of Ets1 and MT1-MMP expression in 3D acini. d) Representative images of ErbB2 cells/clones and quantification of e) translocation and f) single-cells in epithelial layer of acini treated with GM6001, Batimastat, MMP inhibitor III, or DMSO for eight days. g) Representative images and h) quantification of translocation of talin-1 knockdown cells in 3D acini. Western blot indicates efficient talin-1 knockdown. Similar results were obtained by a different talin-1 knockdown construct (data not shown). Raw data are shown in Supplementary Table 3. Error bars represent standard deviations of three-four experiments. Asterisks denote statistically significant change calculated using two-tailed t-tests. Scale bars, 10µm.
We hypothesized that perturbation of local cell-matrix adhesion by MMP activities may trigger cell translocation. Consistent with this, MCF10As overexpressing ErbB2, but not Myc, myrAKT1 or GFP, showed impaired adhesion on Matrigel-coated plates (Supplementary Fig. 9). In addition, we observed compromised basement membrane adjacent to single ErbB2-, but not Myc-, myrAKT1-, or GFP-overexpressing cells in 3D acini (Supplementary Fig. 9). Moreover, weakening cell-matrix adhesion strength by knocking down talin-114, a protein that links integrins to actin filaments and localizes to the basal cell membrane in 3D acini (Supplementary Fig. 10), is sufficient to induce translocation (Fig. 3g,h).
Intriguingly, the ErbB2-cells that stayed in the epithelial layer due to MMP inhibition were unable to proliferate (Fig. 3f and Supplementary Fig. 11). MMP inhibition did not affect ErbB2-cells proliferation in monolayer cultures, and treatment of acini four days after single-cell ErbB2 induction, at which time most induced cells had already translocated, did not affect luminal outgrowth (Supplementary Fig. 12), suggesting that MMP activities are required specifically for the initial translocation step, but not proliferation.
Cell displacement has been proposed as a mechanism to remove aberrant cells from epithelia11,15–17. Our data predict that cell displacement by translocation may also facilitate outgrowth of sporadic mutant cells. Using MT1-MMP as a tool to drive cell translocation, and myrAKT1- and Myc-expressing cells as models, we examined the outcome of forced-translocation of quiescent oncogene-expressing cells. Single cells within MCF10A acini stably carrying Tet-inducible MT1-MMP-IRES-GFP (pLT-MT1-MMP-iGSP) or IRES-GFP (pLT-iGSP) vectors were infected with another lentiviral vector encoding Tet-inducible myrAKT1-IRES-mCherry (or Myc-IRES-mCherry) and constitutive rtTA expression (pLT-myrAKT1-iCSA or pLT-Myc-iCSA). Only those transduced single-cells contain the complete Tet-inducible components to drive doxycycline-dependent expression of myrAKT1 (or Myc), MT1-MMP, and the two fluorescent reporters (Supplementary Fig. 13). This combinatorial inducible approach overcomes size limitations for efficient virus packaging and allows flexible, multiplex genetic manipulations within single cells.
Forced-translocation of myrAKT1-mCherry/MT1-MMP-GFP cells, but not Myc-mCherry/MT1-MMP-GFP or mCherry/MT1-MMP-GFP cells, led to luminal expansion (Fig. 4a,b). myrAKT1-mCherry/MT1-MMP-GFP cells that failed to translocate remained as single-cells in the epithelial layer (99±2%), indicating that translocation, but not simply MT1-MMP and myrAKT1 co-expression, facilitates clonal outgrowth. Indeed, MT1-MMP did not increase colony formation of MCF10A/myrAKT1 cells in soft-agar (Supplementary Fig. 14).
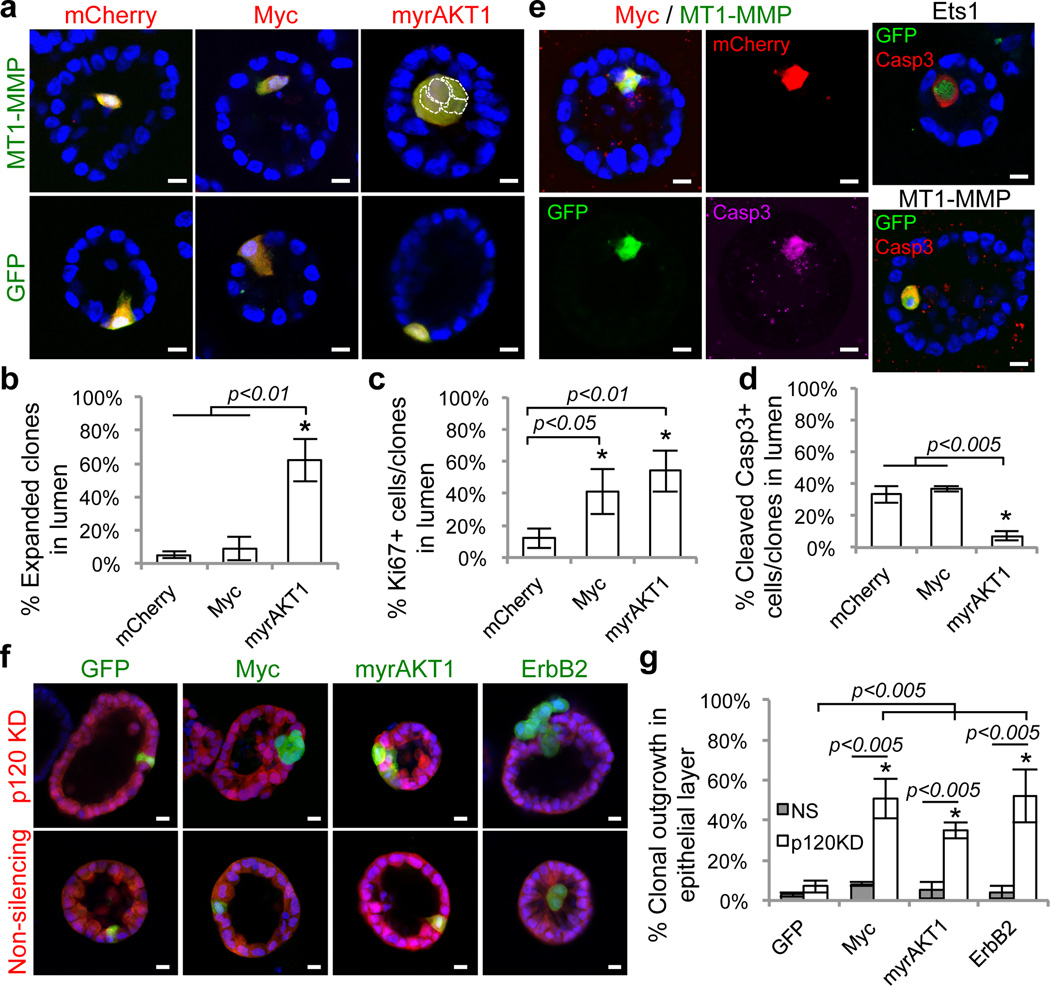
Figure 4. Cell translocation provokes clonal selection or outgrowth of quiescent mutant cells.
a–c) Single cells within Day16 MCF10A/pLT-MT1-MMP-iGSP or MCF10A/pLT-iGSP (IRES-GFP) acini were infected with pLT-Myc-iCSA, pLT-myrAKT1-iCSA or pLT-iCSA (IRES-mCherry) to inducibly drive oncogenes and reporters co-expression. b) Representative images and c) quantifications of Myc/mCherry, myrAKT1/mCherry, or mCherry cells/clones co-expressing MT1-MMP/GFP or GFP control (yellow) 12 days after doxycycline induction. Nuclei (DAPI, blue) of MT1-MMP/myrAKT1 clones are outlined with white dotted line. Quantification of translocated cells/clones for c) proliferation and d) apoptosis. e) Acini with Myc/MT1-MMP, Ets1-, and MT1-MMP-expressing cells were immunostained with antibody against cleaved Caspase-3 eight days after induction. f–g) Single cells within pre-formed, Day16 MCF10A/pTRIPZ-p120KD or MCF10A/pTRIPZ-NS (non-silencing) acini were infected with pLT-Myc-iG, pLT-myrAKT-iG, pLT-ErbB2-iG or pLT-iG. Acini were induced with doxycycline to drive expression of p120KD or NS shRNA in all cells (red) with co-expression of Myc, myrAKT1, ErB2, or GFP in single cells (green). Acini were induced with doxycycline for 48 hours to express p120KD or NS shRNA before infection with pLT-ErbB2-iG. f) Representative images and g) quantification of expanded clones in epithelial layer eight days after induction. Raw data are shown in Supplementary Table 4. Error bars represent standard deviations of four experiments. Asterisks denote statistically significant change calculated using two-tailed t-tests. Scale bars, 10µm.
Both translocated Myc- and myrAKT1-cells re-enter cell cycle (Fig. 4c), but only Myc-cells show increased apoptosis (Fig. 4d, e). These results suggest that the anti-apoptotic activity of myrAKT1 contributes to support clonal expansion in the matrix-deprived lumen. Consistent with these observations, 42±8% and 29±12% of translocated Ets1 and MT1-MMP single-cells, respectively, also display apoptosis (Fig. 4e). Although we could not directly trace these cells, apoptosis of single translocated Ets1-, MT1-MMP- or Myc-cells suggest that the clonal lineage would likely be eliminated.
This dichotomous fate of Myc- and myrAKT1-cells suggests that translocation is not sufficient for outgrowth, but may rather unleash cells from their suppressive epithelial environment. We tested whether perturbing the epithelial organization of 3D acini allows expansion of quiescent mutant cells. Knockdown of p120-catenin to destabilize epithelial cell-cell junctions18 in pre-formed acini greatly reduced β-catenin and E-cadherin staining at cell junctions without disrupting gross acinar structures (Supplementary Fig. 15). In contrast to the distinct outcomes upon forced-translocation, both Myc- and myrAKT1-cells, but not GFP cells, underwent clonal expansion upon p120-catenin knockdown (Fig. 4f,g). These data suggest that p120/cadherin junction-mediated epithelial organization is critical to suppress oncogene-induced proliferation in mature acini. Notably, single ErbB2-cells within pre-formed acini subjected to p120-catenin knockdown also proliferated but did not translocate (Fig. 4f,g), suggesting that intact epithelial organization also plays a role in supporting luminal cell translocation.
These findings highlight the suppressive influence of mature epithelial environment on sporadic mutant cells, and raise the question whether expression of oncogenes in neighboring cells would abrogate this proliferative suppression. Interestingly, induction of Myc or myrAKT1 in all cells of growth-arrested acini did not drive cell proliferation or disrupt acinar structure (Supplementary Fig. 16). This observation is consistent with previous finding that tamoxifen-induced Myc activation in growth-arrested MCF10A acini does not drive proliferation19. These results further demonstrate the strong suppressive control of organized epithelium, as oncogenes like Myc or myrAkt1 is not sufficient to abrogate this suppressive effect.
We have utilized 3D organotypic cultures to model the genetic and tissue architectural context in which sporadic oncogene-expressing cells arise in early stages of human breast tumorigenesis, and demonstrated that the initial outgrowth of these sporadic mutant cells within organized epithelial environments is highly regulated. We showed that while perturbing suppressive epithelial environment permits general expansion of quiescent mutant cells, a different process involving luminal cell translocation allows mutant cells to evade suppressive epithelial environment and drive selection for survival and expansion in the ECM-deprived lumen (Model, Supplementary Fig. 1). Our data also highlight the suppressive influence of organized epithelial structure on preneoplastic cell expansion.
Displacement of cell variants from epithelia10,11,15,17,20–22 and stem cell compartments16 are widely observed in diverse organ systems and species. Previous studies have shown that extrusion of apoptotic cells from epithelial monolayers involves force-dependent expulsion process from neighboring cells17. We show that luminal cell translocation in 3D is induced by ERK and MMP activities intrinsic to the translocating cells, and involves local perturbation of cell-matrix adhesion.
Cell migration from specialized niches and microenvironments underlies cell differentiation and tissue morphogenesis during development and regeneration23,24. Our data suggest that similar spatial cell translocation within tissue compartments may also play a role in tumorigenesis by provoking clonal selection. Oncogenic alterations have been observed in cells within otherwise histologically normal tissue of healthy individuals3,5, and disruption of tissue organization has been associated with tumor progression. Our findings raise the possibility that mechanisms, such as cell translocation or compromising tissue integrity, may initiate neoplastic progression from these dormant mutant cells.
Methods Summary
3D Matrigel culture and virus infection
Three-dimensional cultures of MCF-10A cells on basement membrane were set up in 8-well chamber slides (BD Biosciences), or coverglass bottom 8-chamber slides (MatTekII) as previously described25 with 4500–5000 cells in assay media (DMEM/F12 supplemented with 2% donor horse serum, 5ng/ml EGF, 10µg/ml insulin, 1ng/ml cholera toxin, 100 µg/ml hydrocortisone, 50U/ml penicillin, and 50µg/ml streptomycin, and 2% Matrigel™). Media were replaced every 4 days. On Day 16, 3D cultures were infected with the corresponding lentiviruses diluted in assay media without EGF for 6–8 hours at 37°C. Virus dosages were adjusted to infect less than 1 cell per 10 acini to achieve sporadic single-cell infection. The viruses were removed, and the chamber wells were rinsed with 500µl PBS and replaced with normal 3D assay media without Matrigel. Doxycycline was added at 1µg/ml on the next day along with drug treatment or vehicle control as indicated. Drugs and vehicles were replenished every two days, and the complete media were changed every 4 days. Acinar structures were analyzed eight days after induction with doxycycline or at extended times as indicated.
Supplementary Material
Acknowledgements
We thank Scott Valastyan, Taru Muranen and Wendy Lee for critical reading of the manuscript. We thank the members of the Brugge laboratory for comments and discussion, the Nikon Imaging Center at Harvard Medical School for providing imaging equipment and software, and the laboratory of Gaudenz Danuser for imaging software support. This work was supported by a grant from the NCI CA080111 (JSB) and an American Cancer Society postdoctoral fellowship (CTL).
Footnotes
Authors Contributions
C.T.L conceived the study, performed the experiments, analyzed the data, and drafted the manuscript. J.S.B. supervised the study and edited the manuscript.
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Dolberg DS, Bissell MJ. Inability of Rous sarcoma virus to cause sarcomas in the avian embryo. Nature. 1984;309:552–556. doi: 10.1038/309552a0. [DOI] [PubMed] [Google Scholar]
- 3.Holst CR, et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–1601. [PubMed] [Google Scholar]
- 4.Illmensee K, Mintz B. Totipotency and normal differentiation of single teratocarcinoma cells cloned by injection into blastocysts. Proc Natl Acad Sci U S A. 1976;73:549–553. doi: 10.1073/pnas.73.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonason AS, et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci U S A. 1996;93:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 7.Weaver VM, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slamon DJ, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 9.Hebner C, Weaver VM, Debnath J. Modeling morphogenesis and oncogenesis in three-dimensional breast epithelial cultures. Annu Rev Pathol. 2008;3:313–339. doi: 10.1146/annurev.pathmechdis.3.121806.151526. [DOI] [PubMed] [Google Scholar]
- 10.Kajita M, et al. Interaction with surrounding normal epithelial cells influences signalling pathways and behaviour of Src-transformed cells. J Cell Sci. 2010;123:171–180. doi: 10.1242/jcs.057976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan C, et al. Characterization of the interface between normal and transformed epithelial cells. Nat Cell Biol. 2009;11:460–467. doi: 10.1038/ncb1853. [DOI] [PubMed] [Google Scholar]
- 12.Pearson GW, Hunter T. Real-time imaging reveals that noninvasive mammary epithelial acini can contain motile cells. J Cell Biol. 2007;179:1555–1567. doi: 10.1083/jcb.200706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lincoln DW, 2nd, Bove K. The transcription factor Ets-1 in breast cancer. Front Biosci. 2005;10:506–511. doi: 10.2741/1546. [DOI] [PubMed] [Google Scholar]
- 14.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 15.Gibson MC, Perrimon N. Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science. 2005;307:1785–1789. doi: 10.1126/science.1104751. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Han Y, Xi R. Polycomb group genes Psc and Su(z)2 restrict follicle stem cell self-renewal and extrusion by controlling canonical and noncanonical Wnt signaling. Genes Dev. 2010;24:933–946. doi: 10.1101/gad.1901510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenblatt J, Raff MC, Cramer LP. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr Biol. 2001;11:1847–1857. doi: 10.1016/s0960-9822(01)00587-5. [DOI] [PubMed] [Google Scholar]
- 18.Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partanen JI, Nieminen AI, Makela TP, Klefstrom J. Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci U S A. 2007;104:14694–14699. doi: 10.1073/pnas.0704677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen J, Dahmann C. Extrusion of cells with inappropriate Dpp signaling from Drosophila wing disc epithelia. Science. 2005;307:1789–1790. doi: 10.1126/science.1104784. [DOI] [PubMed] [Google Scholar]
- 21.Bullen TF, et al. Characterization of epithelial cell shedding from human small intestine. Lab Invest. 2006;86:1052–1063. doi: 10.1038/labinvest.3700464. [DOI] [PubMed] [Google Scholar]
- 22.Marshall TW, Lloyd IE, Delalande JM, Nathke I, Rosenblatt J. The tumor suppressor adenomatous polyposis coli controls the direction a cell extrudes from an epithelium. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-05-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 24.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Debnath J, et al. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.