Abstract
Background
The D1275N SCN5A mutation has been associated with a range of unusual phenotypes including conduction disease and dilated cardiomyopathy (DCM) as well as atrial and ventricular tachyarrhythmias. However, when D1275N is studied in heterologous expression systems, most studies show near-normal sodium channel function. Thus, the relationship of the variant to the clinical phenotypes remains uncertain.
Methods and results
We identified D1275N in a patient with atrial flutter, atrial standstill, conduction disease, and sinus node dysfunction. There was no major difference in biophysical properties between wild-type and D1275N channels expressed in CHO or tsA201 cells in the absence or presence of β1 subunits. To determine D1275N function in vivo, the Scn5a locus was modified to knock out the mouse gene, and the full-length wild-type (H) or D1275N (DN) human SCN5A cDNAs were then inserted at the modified locus using recombinase mediated cassette exchange. Mice carrying the DN allele displayed slow conduction, heart block, atrial fibrillation, ventricular tachycardia, and a DCM phenotype, with no significant fibrosis or myocyte disarray on histological examination. The DN allele conferred gene-dose dependent increases in SCN5A mRNA abundance, but reduced sodium channel protein abundance and peak sodium current amplitudes (H/H, −41.0±2.9 pA/pF at −30mV; DN/H, 19.2±3.1 pA/pF, P<0.001 versus H/H; DN/DN, −9.3±1.1 pA/pF, P<0.001 versus H/H).
Conclusions
Although D1275N produces near normal currents in multiple heterologous expression experiments, our data establish this variant as a pathological mutation that generates conduction slowing, arrhythmias, and a DCM phenotype by reducing cardiac sodium current.
Keywords: genetics, ion channels, cardiomyopathy, electrophysiology
Introduction
Voltage-gated sodium channels play a critical role in the generation and propagation of the cardiac action potential, and mutations in SCN5A, the gene encoding the major pore-forming sodium channel α subunit in the heart (Nav 1.5), cause multiple inherited cardiac arrhythmia syndromes including long QT syndrome, the Brugada syndrome, isolated cardiac conduction disease, sinus node dysfunction, and atrial fibrillation.1–7 More recently, SCN5A mutations have been associated with dilated cardiomyopathy (DCM), and such DCM mutations have also been associated with a similar range of arrhythmias.8–15
The D1275N SCN5A mutation was initially reported in a Dutch family affected by atrial standstill, mild conduction disease, and atrial enlargement, but no ventricular structural abnormality; only subjects that carried a variant in the connexin 40 (Cx40) promoter displayed the clinical phenotype.16 Subsequently D1275N was implicated in a large family affected by DCM and various arrhythmias including sinus node dysfunction, atrial and ventricular tachyarrhythmias, and conduction disease.8, 9, 17 Most recently, the mutation was reported in a family with atrial tachyarrhythmias, conduction disease, and ventricular enlargement without impaired contractility.18
Heterologous expression systems are conventionally used to assess function of ion channel mutations.2, 19 Most studies (including our own data described below) that have compared wild-type and D1275N channels in heterologous expression systems have not shown major differences in the biophysical properties of the variant channel.16, 20 Thus, although the mutation has been reported as a cause of unusual phenotypes in a number of kindreds, its relationship to the clinical phenotypes remains uncertain.
To address this discrepancy, we have used recombinase-mediated cassette exchange21 to engineer mice expressing the mutant human channel (here termed DN); we compared the functional properties of these animals to those expressing wild-type human alleles (H), that we previously generated in an identical fashion.21 The data demonstrate that D1275N causes a severe defect in sodium channel function in vivo, consistent with the reported clinical phenotypes.
Methods
Study subjects
The proband and family members were screened for mutations in SCN5A by PCR amplification of coding regions and flanking intronic sequences, followed by direct sequencing of amplicons on an ABI PRISM 3730 DNA Sequence Detection System (Applied Biosystems, Foster City, CA). Informed consent was obtained for presentation of the kindred.
Animal model
All studies using animals were approved by the institutional animal care and use committees at Vanderbilt University, and performed in accordance with NIH guidelines. We have previously modified the Scn5a locus in mouse embryonic stem cells to enable the technique of recombinase mediated cassette exchange.21–23 In our initial studies, we inserted the full-length human SCN5A cDNA into the targeted locus.21 Mice homozygous for the exchanged allele (termed H/H) expressed only the human allele and had normal electrocardiograms (ECGs) and ventricular sodium current, supporting the hypothesis that expression of the exchanged allele was under control of endogenous Scn5a regulatory mechanisms.
For the present study, we used the same technique to generate DN mice, in which the exchanged construct was identical to that previously used for the H/H mice with the exception of (1) a c.3823G→A mutation resulting in p.D1275N and (2) insertion of a FLAG epitope between residues 153 and 154 of the extracellular linker S1-S2 in domain I; the FLAG insertion into S1-S2 linker has previously been found to have no effect on channel gating or cell surface expression.24, 25 We also generated FG mice bearing the wild-type SCN5A allele with the FLAG tag. Initial matings between mice heterozygous for engineered alleles resulted in H/H, DN/H, and FG/H mice, and these were then bred into the 129/Sv background. H/H, DN/H, and DN/DN mice were generated from DN/H × DN/H matings and H/H littermates were used as controls for all experiments. To genotype mice, genomic DNA was isolated from mouse tails, the target SCN5A PCR amplicon (c.3688 to c.4082) was incubated with Taq I (New England Biolabs, Ipswich, MA), and then was electrophoresed in agarose gels. Taq1 digests the fragment containing p.D1275 but does not digest that with p.N1275.
Surface electrocardiogram
ECGs were recorded during administration of isoflurane vapor titrated to maintain light anesthesia.26 Baseline ECG (leads I and II) was recorded for 15 minutes. Heart rate was measured as the average during a 30-second interval at baseline when a steady state was reached during anesthesia. For measurement of all other ECG parameters, 30 seconds of data in each lead were signal averaged using a custom-built LabVIEW program (National Instruments, Austin, TX) and the resultant waveform was analyzed using an electric caliper by an electrophysiologist blind to the genotype.27 The larger value from each lead was used. QRS duration was measured from the first deflection of the Q-wave (or R-wave when the Q-wave was absent) and the end of the S-wave defined as the point of minimum voltage in the terminal phase of the QRS complex. QT interval was measured from the beginning of the QRS complex to the end of the T-wave defined as the point where the T-wave merges with the isoelectric line. Heart rate-corrected QT interval (QTc) was calculated using a formula developed for mice: QTc = QT/(RR/100)1/2.28
Echocardiogram
Transthoracic echocardiograms were performed on resting conscious mice and analyzed by a sonographer blind to the genotype. Signals were acquired using a 15-MHz transducer (Sonos 5500 system, Agilent, Santa Clara, CA) at the Murine Cardiovascular Core, Vanderbilt University as previously described.29
Histology
Hearts were fixed overnight in 10% formalin, paraffin embedded, sectioned at 5 µm, and stained with Masson trichrome.
mRNA quantitation
Real-Time PCR was conducted using a 7900HT Real-Time Instrument (Applied Biosystems). mRNA was isolated from the left ventricles and cDNA was synthesized from 2 µg of the RNA using Transcriptor First Strand cDNA Synthesis Kit with random hexamer primers (Roche Applied Science, Indianapolis, IL) and used as template. To generate a standard curve for absolute quantification, genes of interest were subcloned into the pGEM-T vector (Clontech, Mountain View, CA). cDNA and 5 different dilutions of the vector with target DNA were prepared with pre-designed 6-carboxyfluorescein (FAM)-labeled fluorogenic TaqMan probe and primers (Applied Biosystems) for SCN5A (Hs00165693_m1) or β-actin (Mm00607939_S1) in triplicate in a same 94-well plate for real-time PCR amplification. Data were collected with instrument spectral compensation and analyzed using absolute quantification and a standard curve by SDS 2.2 software (Applied Biosystems). Each value was normalized to that for β-actin.
Western blotting
Protein was extracted from flash frozen hearts that were pulverized into powder and homogenized in a Dounce apparatus with 1×RIPA. Lysates were centrifuged at 10000 ×g for five minutes and protein content was analyzed using a bicinchoninic acid assay (Pierce Biochemicals, Rockford, IL). Forty to one hundred micrograms of protein from each cardiac sample was separated by running the sample on a NuPage 8% Tris-Acetate gel (Invitrogen, Carlsbad, CA). The protein was transferred to 0.2 µm nitrocellulose membranes (Amersham Biosciences, Sweden) which were blocked overnight in (TTBS) 0.05%Tween-20 Tris-buffed saline plus 5% non-fat dry milk at 4 °C, and then were incubated with antibodies targeting anti-Nav1.5 (pAb 1:200, Alomone Labs, Israel), or anti-calnexin (pAb 1:1000, Stressgen Bioreagents, Belgium) at room temperature for 2 hours. Membranes were washed three times with TTBS for 10 minutes each and incubated with secondary anti-mouse and anti-rabbit horseradish peroxidase-linked antibodies (Amersham Biosciences) in TTBS at room temperature for 1 hour. The blots were then washed four times for 10 minutes each in TTBS. We visualized antibody interactions using the ECL system (Amersham Biosciences).
Immunostaining/confocal microscopy
Unfixed hearts were frozen in Tissue Tek and sectioned at 6 µm. Sections were washed in 1× DPBS and then incubated in 1× DPBS containing 0.3% fish gelatin and 0.1% Triton (block) for one hour at 4°C. Sections were immunostained with antibodies targeting anti-Nav 1.5 (pAb 1:50, Alomone Labs) diluted in block solution overnight. Samples were then washed three times and incubated with Allexa 488 conjugated goat anti-mouse IgG (1:400, Invitrogen) secondary antibody for one hour at room temperature. Sections were then washed and coverslips were applied with Vectashield (Vector Labs, Burlingame, CA). Images were collected using a Zeiss LSM510 Meta confocal imaging system using 20×1.3 NA lens (pinhole equals one airy disc) with 2× zoom and analyzed using LSM 4.0 software.
Sodium current recordings
Sodium current was recorded using whole-cell voltage-clamp technique in single ventricular myocytes isolated by a modified collagenase/protease method or in Chinese hamster ovary (CHO) cells transiently expressing wild-type or D1275N SCN5A.21, 30, 31 The SCN5A DNA (NM_198056) was subcloned into the pBK-CMV vector (Stratagene, La Jolla, CA) and the mutation was prepared using the QuickChange II XL site-directed mutagenesis kit (Stratagene) followed by verification by resequencing. SCN5A DNA (1 µg) was transfected with the plasmid encoding the eGFP (pEGFP-IRES, Clontech) using Fugene6 (Roche Applied Science, Indianapolis, IN) in CHO cells. Cells were grown for 48 hours after transfection before study. Similar methods were used to study the biophysical properties of wild-type and D1275N sodium channels transfected with the sodium channel β1 subunit in human embryonic kidney cells (tsA201). Late sodium current was measured at the end of 200-ms test pulses to −20 mV from a holding potential of −120 mV (interpulse duration, 5 s).
The extracellular bath solution contained (in mmol/l): 145 NaCl, 4.0 KCl, 1.0 MgCl2, 1.8 CaCl2, 10 glucose, 10 HEPES, pH 7.4 (NaOH) for sodium current recording in CHO and tsA201 cells. Patch pipettes (~1.5 MΩ) contained (in mmol/l): 10 NaF, 110 CsF, 20 CsCl, 10 EGTA, and 10 HEPES, pH 7.4 (CsOH). To allow recording of sodium current in cardiomyocytes, the external Na+ concentration was lowered to 5 mM, electrodes with tip resistance <1 MΩ were used, and experiments were conducted at 18°C. Data acquisition was carried out using an Axopatch 200B patch-clamp amplifier and pCLAMP software (version 9.2, Molecular Devices, Sunnyvale, CA). Currents were filtered at 5 kHz and digitized using an analog-to-digital interface (Digidata 1322A, Molecular Devices). To minimize capacitive transients, capacitance and series resistance were adjusted to 70–85%. Details of the pulse protocols are presented schematically in the figures.
Action Potential Recordings
Action potentials from isolated mouse ventricular myocytes were elicited with injection of brief stimulus current (1–2 nA, 2–6 ms) at 5 Hz in current clamp mode (Axopatch 200A amplifier, Molecular Devices). The extracellular bath solution contained (in mmol/L): NaCl 140, KCl 5.4 , CaCl2 1.8, and MgCl2 1, HEPES 5, glucose 10, with a pH of 7.4 (adjusted by NaOH). Patch pipettes contained (in mmol/L): KCl 110, K2-ATP 5, MgCl2 1, BAPTA 0.1, HEPES 10, with a pH of 7.2 (adjusted by KOH). Microelectrodes of 3–5 MΩ were used. Data acquisition was carried out using an Axopatch 200B patch-clamp amplifier and pCLAMP. The action potential durations at 50% and 90% repolarization and the action potential amplitude were measured.
Data analysis
Results are presented as mean±SEM. The unpaired t-test was used for comparisons of electrophysiologic characteristics between D1275N and wild-type channels expressed in heterologous expression systems. ANOVA followed by a post hoc analysis with Bonferroni correction was used for all of comparisons among the genotypes of mice, except for the linear mixed-effects models with Bonferroni correction for comparisons of in vitro electrophysiologic characteristics of mice. All statistical analyses were performed with SPSS, version 12.0 (SPSS Inc, Chicago, IL). A value of two-tailed P <0.05 was considered statistically significant.
Results
Clinical case presentation
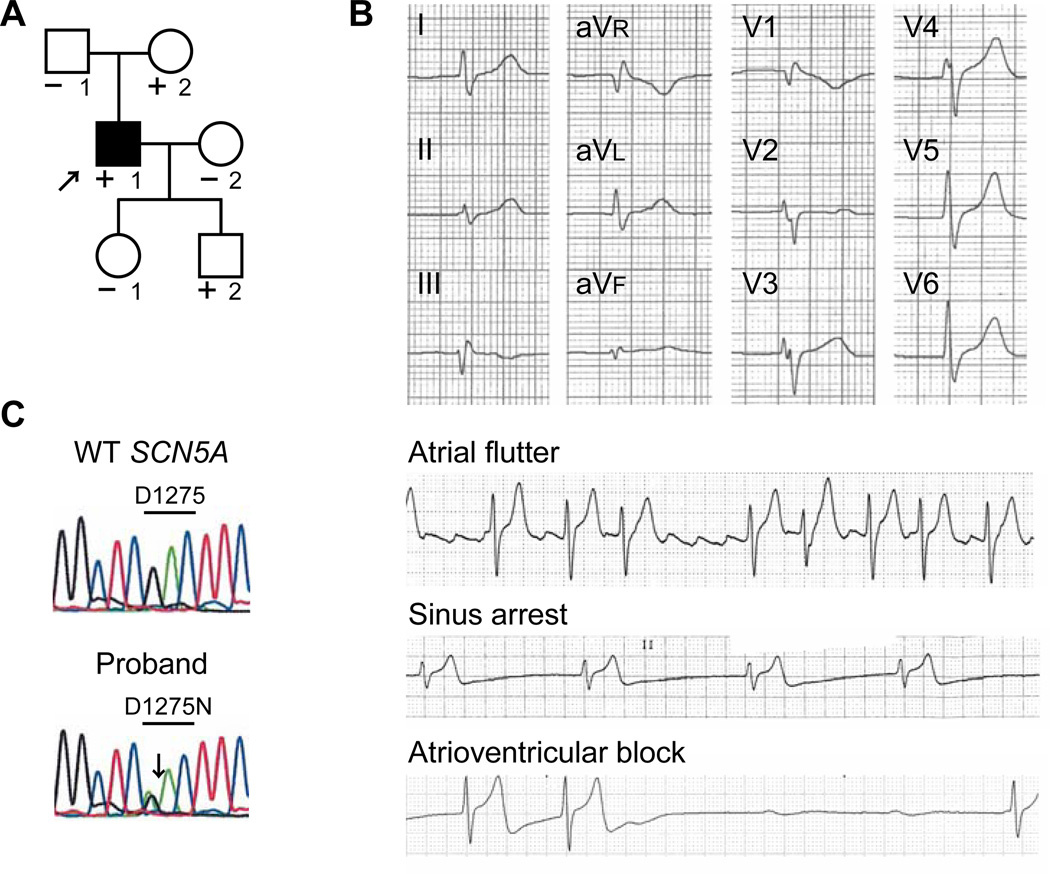
A 19-year-old Caucasian male (II-1) presented with recurrent exertional syncope (Figure 1A, arrow). Physical examination and echocardiography were normal, and his ECG demonstrated unusually slow atrial flutter that was conducted 1:1 to the ventricles with hypotension during exertion (Figure 1B). After catheter ablation of the cavotricuspid isthmus for atrial flutter, he had atrial standstill, prolonged QRS duration, sinus node dysfunction, high degree atrioventricular block, and normal QT interval. A cardioverter-defibrillator was implanted, and he has been asymptomatic for 10 years and echocardiography has been normal. We identified a missense mutation in SCN5A, c.3823G→A in exon 21 (Figure 1C), resulting in p.D1275N within a transmembrane domain of the protein (segment 3, domain III); the variant Cx40 associated with atrial standstill in the reported Dutch kindred was absent.16 Both his mother (I-2) and one son (III-2) share the mutation but have no clinical findings.
Figure 1.
D1275N SCN5A mutation in a patient with sinus node dysfunction, atrial flutter, and conduction disease. A, Pedigree. The proband is indicated by the arrow. Individuals carrying the mutation are indicated (+). Individuals tested negative for the mutation are indicated (−). A filled symbol indicates phenotype positive. B, Electrocardiogram and rhythm strips in the proband. C, Heterozygous single-nucleotide change in SCN5A (c.3823G→A) resulting in p.D1275N.
D1275N mice are viable and display gene-dose-dependent conduction slowing and arrhythmias
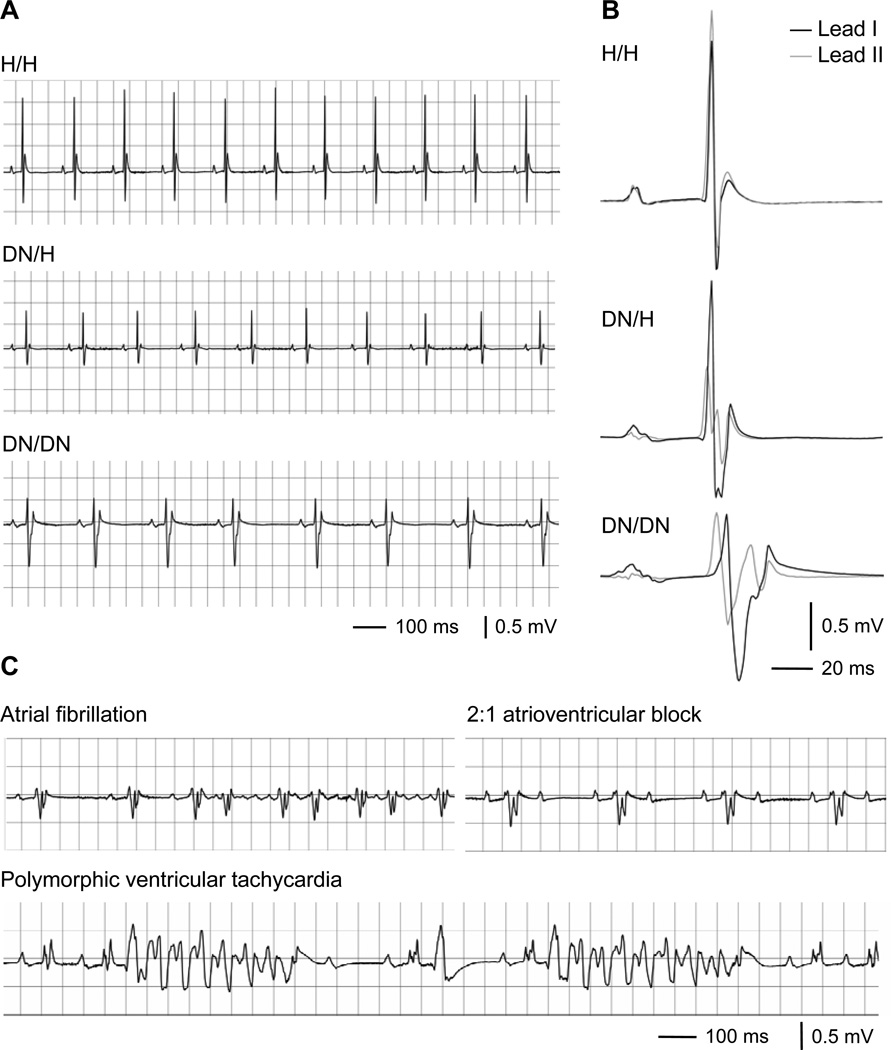
The distribution of pups from DN/H × DN/H matings was in Hardy-Weinberg equilibrium (52 H/H, 107 DN/H, 54 DN/DN). During a follow-up of 12 weeks, one DN/DN mouse died suddenly, but there were no deaths in DN/H or H/H mice. ECG recordings revealed that the DN allele caused abnormal phenotypes in a gene-dose-dependent fashion at 3 weeks (Figure 2 A and 2B, Table 1). The DN allele was associated with slow heart rate and slow cardiac conduction (prolongation of the P-wave duration, PR interval, and QRS duration) at 3 weeks and similar changes were observed at 12 weeks. In mice with ECGs recorded at both 3 and 12 weeks, the prolongation of the P-wave duration, PR interval, and QRS duration associated with the DN allele were progressive with age. In addition, spontaneous monomorphic and polymorphic ventricular tachycardia were observed in 7 out of 9 DN/DN mice during 15 minute recording periods under light anesthesia at 12 weeks, but no arrhythmia was observed in 18 DN/H or 10 H/H littermates studied under the same conditions (Figure 2C). Sinus node dysfunction (n = 3), atrioventricular block (≥second degree) (n = 4), and atrial fibrillation/tachycardia (n = 5) also occurred only in DN/DN mice, but not in DN/H or H/H littermates.
Figure 2.
Electrocardiography in mice. A, Representative ECG traces in leads I at 3 weeks. B, Representative signal-averaged ECG traces in leads I (black) and II (gray) at 3 weeks. See Table 1 for detailed results. C, Arrhythmias recorded in DN/DN mice at 12 weeks.
Table 1.
Electrocardiographic phenotype
| H/H | DN/H | DN/DN | |
|---|---|---|---|
| 3 weeks | n=11 | n=20 | n=9 |
| Heart rate, bpm | 388 ± 8 | 354 ± 8* | 335 ± 15* |
| P-wave duration, ms | 13.0 ± 0.6 | 17.0 ± 0.3* | 19.4 ± 0.5*† |
| PR interval, ms | 33.7 ± 0.7 | 35.6 ± 0.7 | 44.1 ± 0.9*† |
| QRS duration, ms | 9.8 ± 0.2 | 11.8 ± 0.3* | 22.3 ± 2.2*† |
| QT interval, ms | 48.5 ± 1.9 | 50.2 ± 1.1 | 67.6 ± 4.2*† |
| QTc interval, ms | 38.9 ± 1.6 | 38.5 ± 0.8 | 50.2 ± 2.5*† |
| 12 weeks | n=10 | n=18 | n=9 |
| Heart rate, bpm | 387 ± 8 | 368 ± 11 | 317 ± 18* |
| P-wave duration, ms | 14 ± 0.5 | 18.6 ± 0.3* | 27.1 ± 1.2*† |
| PR interval, ms | 37.8 ± 0.8 | 39.7 ± 0.7 | 57.1 ± 3.1*† |
| QRS duration, ms | 10.7 ± 0.4 | 12.9 ± 0.3 | 33.4 ± 2.7*† |
| QT interval, ms | 51.2 ± 0.6 | 54.8 ± 0.8 | 77.9 ± 3.7*† |
| QTc interval, ms | 41.1 ± 0.6 | 42.8 ± 0.9 | 55.9 ± 1.8*† |
| % ratio of 12 weeks to 3 weeks ‡ | n=9 | n=14 | n=6 |
| Heart rate | 101 ± 2 | 103 ± 4 | 119 ± 18 |
| P-wave duration | 104 ± 4 | 112 ± 5 | 143 ± 11*† |
| PR interval | 113 ± 3 | 115 ± 2 | 125 ± 5* |
| QRS duration | 111 ± 4 | 116 ± 4 | 154 ± 14*† |
| QT interval | 106 ± 5 | 114 ± 3 | 117 ± 11 |
| QTc interval | 106 ± 5 | 115 ± 3 | 123 ± 6 |
QTc = QT/(RR/100)1/2 (mouse-specific).
P<0.05 versus H/H.
P<0.05 versus DN/H.
For animals with measurements at both time points.
Reduced contractile function in DN mice
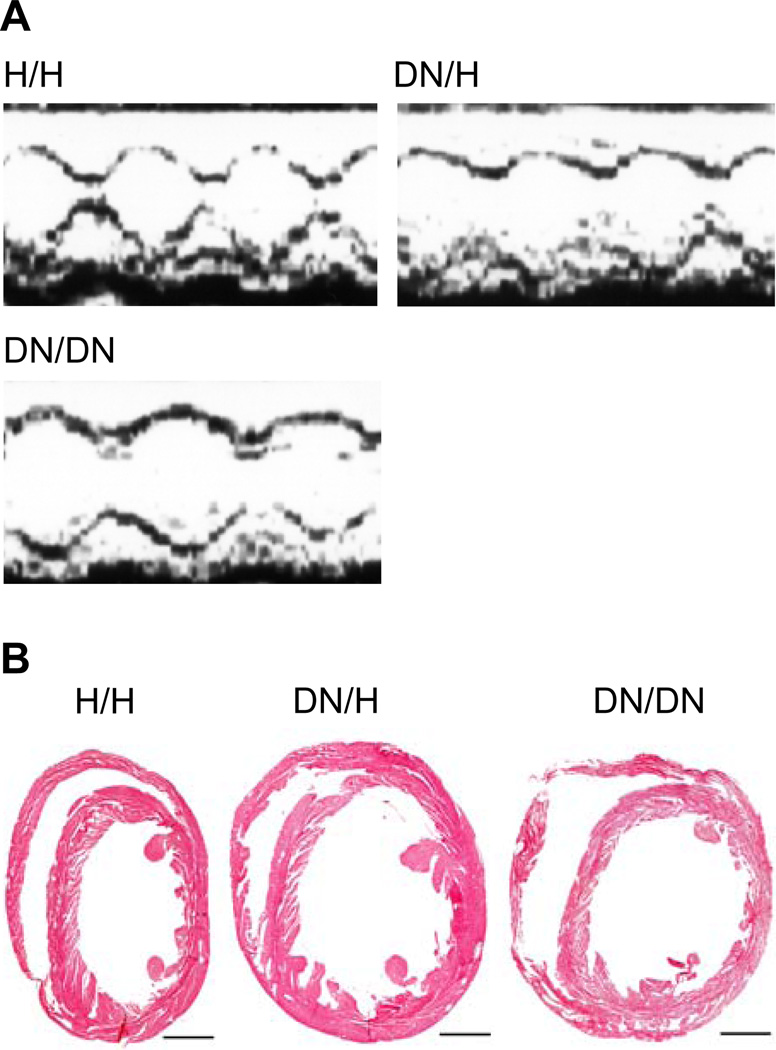
There was consistent and statistically significant end-diastolic and end-systolic left ventricular dilatation and calculated left ventricular fractional shortening reduction in a gene-dose-dependent fashion (Figure 3A, Table 2). Histological examination of mouse hearts revealed that the DN allele was associated with ventricular dilatation, but was not associated with significant fibrosis or myocyte disarray (Figure 3B). One possibility is that the FLAG tag incorporated into the DN allele contributes to the phenotypes in the DN animals. However, we found no difference in electrocardiographic and echocardiographic phenotypes between H/H and FG/FG animals, indicating that the FLAG tag does not contribute to the ventricular dysfunction or other phenotypes observed in DN animals.
Figure 3.
Dilated cardiomyopathy phenotype. A, Representative echocardiograms showing prominent increased end-systolic dimensions in DN/H and DN/DN mice at 12 weeks. See Table 2 for summary results. B, Masson’s trichrome staining in mice hearts. Scale bars indicate 1 mm.
Table 2.
Echocardiographic phenotype at 12 weeks
| H/H, n=9 | DN/H, n=19 | DN/DN, n=12 | |
|---|---|---|---|
| Septal wall, mm | 0.75 ± 0.02 | 0.72 ± 0.02 | 0.69 ± 0.03 |
| Posterior wall, mm | 0.51 ± 0.03 | 0.47 ± 0.01 | 0.51 ± 0.04 |
| Left ventricle, mm | |||
| End diastole | 3.01 ± 0.08 | 3.09 ± 0.08 | 3.33 ± 0.07* |
| End systole | 1.49 ± 0.09 | 1.76 ± 0.05* | 2.01 ± 0.05*† |
| Fractional shortening, % | 52.0 ± 1.6 | 43.1 ± 0.7* | 39.9 ± 0.7*† |
P <0.05 versus H/H.
P <0.05 versus DN/H.
Sodium current is reduced in DN myocytes
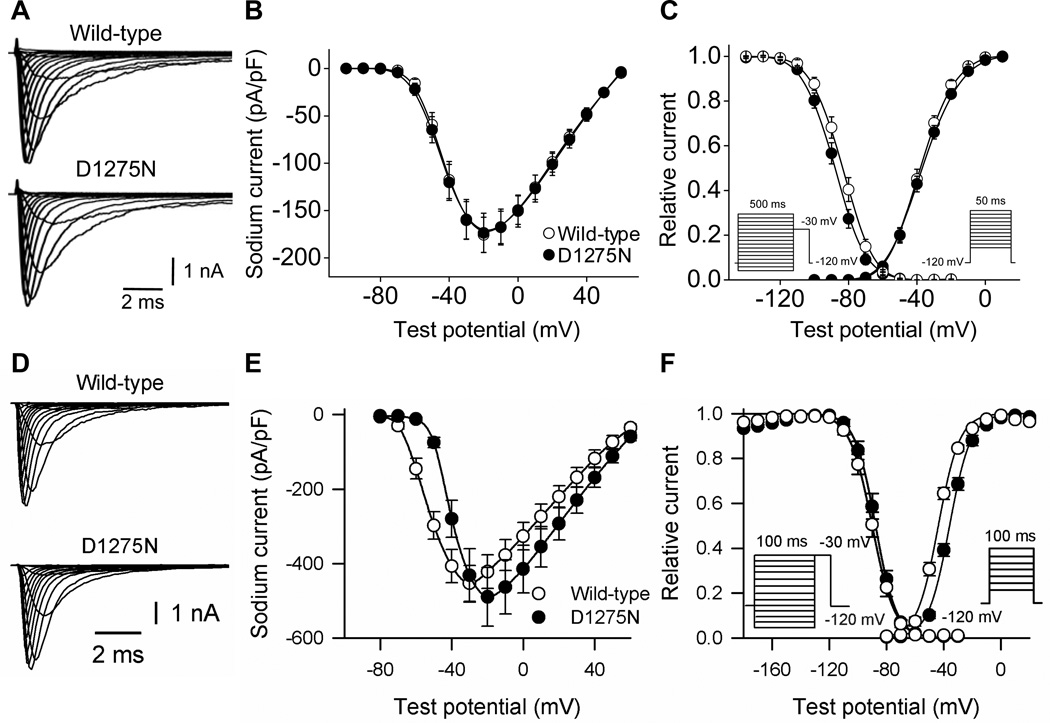
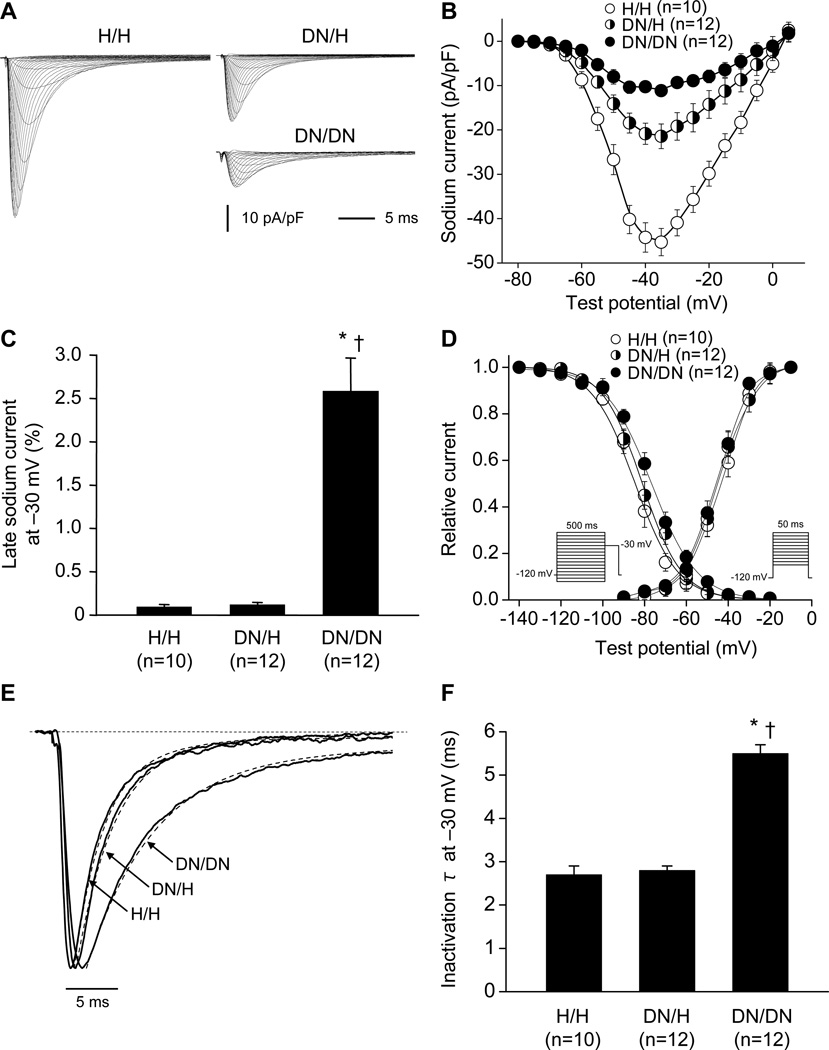
The manifest conduction slowing in DN mice is consistent with loss of sodium channel function. However, sodium current amplitudes and gating observed with heterologous expression of wild-type and D1275N channels in CHO cells were near-indistinguishable (Figure 4A, 4B and 4C; Table 3). In CHO cells, there was also no difference in the voltage dependence of activation and inactivation, or in the time course of inactivation. Similarly, only minor differences were observed between wild-type and D1275N channels coexpressed with β1 subunits in tsA201 cells: current amplitudes were near-identical, but there was a slight shift in the voltage dependence of activation and an increase in late sodium current (percent to peak current; wild-type, 0.22±0.05%, n=7; D1275N, 1.34±0.11%, n=8; P<0.001) (Figure 4D, 4E, and 4F; Table 3).
Figure 4.
Wild-type and D1275N sodium current in Chinese hamster ovary cells (A, B, C) and tsA201 cells (D, E, F). Wild-type or D1275N channels were coexpressed with β1 subunits in tsA201 cells. A and D, Representative current traces. See Table 3 for summary results. B and E, Current voltage relationships. C and F, Voltage dependence of activation and inactivation. The pulse protocols are shown in the inset.
Table 3.
Sodium channel gating in heterologous expression systems and ventricular cardiomyocytes
| Peak Current Density at −30 mV |
Voltage Dependence of Activation |
Voltage Dependence of Inactivation |
||||
|---|---|---|---|---|---|---|
| (pA/pF) | n | V1/2 (mV) | n | V1/2 (mV) | n | |
| CHO cells | ||||||
| Wild-type | −160 ± 20 | 25 | −35.4 ± 0.6 | 25 | −84.5 ± 1.0 | 24 |
| D1275N | −159 ± 21 | 28 | −34.7 ± 0.6 | 28 | −88.4 ± 0.8 | 27 |
| ts A201 cells | ||||||
| Wild-type | −454 ± 48 | 16 | −47.7 ± 1.1 | 16 | −89.4 ± 0.7 | 19 |
| D1275N | −432 ± 71 | 13 | −35.7 ± 1.1* | 13 | −88.0 ± 1.6 | 18 |
| Cardiomyocytes∥ | ||||||
| H/H | −40.9 ± 2.9 | 10 | −44.1 ± 1.0 | 10 | −84.1 ± 1.0 | 10 |
| DN/H | −19.2 ± 3.1† | 12 | −44.3 ± 1.4 | 12 | −81.2 ± 1.1 | 12 |
| DN/DN | −9.3 ± 1.1‡,§ | 12 | −45.6 ± 0.9 | 12 | −76.5 ± 0.8‡,§ | 12 |
Study conditions differ for heterologous expression systems and cardiomyocytes as described in Methods. N indicates the number of cells; CHO, Chinese hamster ovary.
P<0.001 versus wild-type.
P<0.05 versus H/H.
P<0.01 versus H/H.
P<0.01 versus DN/H.
Cardiomyocytes from three mice for each genotype.
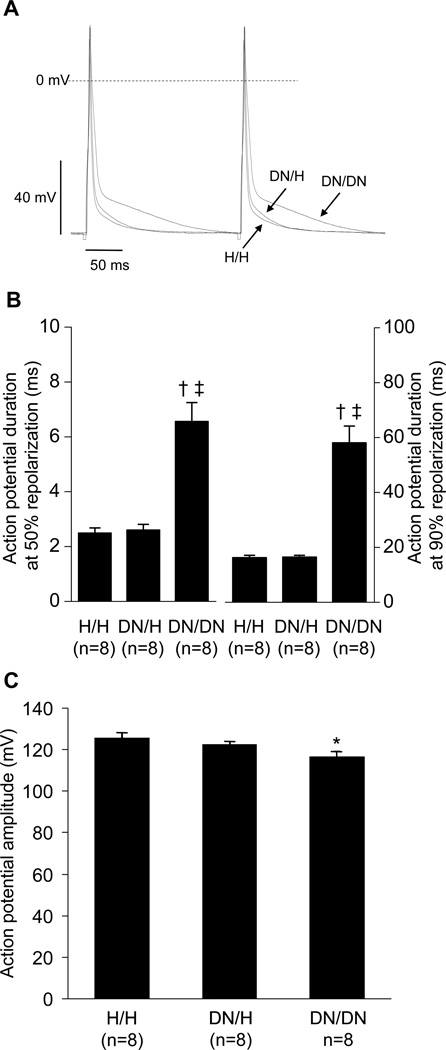
By contrast, in ventricular cardiomyocytes, peak sodium current amplitude was markedly reduced in DN/H and DN/DN mice compared to that observed in H/H littermates (Figure 5A and 5B, Table 3). In addition, late sodium current was increased in DN/DN mice compared to that in DN/H and H/H littermates (Figure 5C). We also found that sodium current in DN/DN myocytes displayed consistent changes in gating. The voltage dependence of inactivation was positively shifted in DN/DN mice compared to that in DN/H and H/H mice (Figure 5D). The time course of inactivation was slower in DN/DN mice compared to that in DN/H and H/H littermates (time constant at −30 mV; DN/DN, 5.5±0.2 ms; DN/H, 2.8±0.1 ms; H/H, 2.7±0.2 ms) (Figure 5E and 5F). There was no difference in the voltage-dependence of activation. The DN allele was associated with decreased action potential amplitude consistent with the decrease in peak sodium current, and with prolonged action potential duration, consistent with the increase in late current (Figure 6).
Figure 5.
Sodium current in male ventricular cardiomyocytes at 3 weeks showing altered sodium channel function by the DN allele. A, Representative current traces in H/H, DN/H, and DN/DN cells. See Table 3 for summary results. B, Current voltage relationships. C, Late sodium current at −30 mV. Late current amplitude was normalized to peak current amplitude. D, Voltage dependence of activation and inactivation. E and F, Inactivation time constant (τ) at −30 mV. *P<0.001 versus H/H. †P<0.001 versus DN/H. N indicates the number of cardiomyocytes from 3 mice.
Figure 6.
Action potential in male ventricular cardiomyocytes at 3 weeks. A, Representative action potential traces. B, Action potential duration at 50% and 90% repolarization. C, Action potential amplitude. *P<0.05 versus H/H. †P<0.01 versus H/H. ‡P<0.01 versus DN/H. N indicates the number of cardiomyocytes from 3 mice.
Sodium channel protein abundance is reduced in DN myocytes
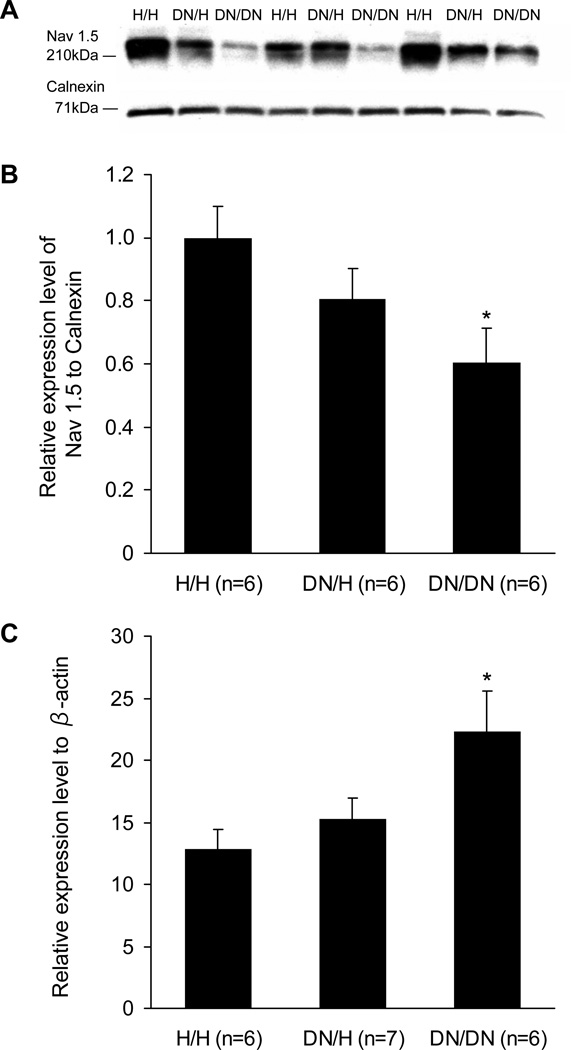
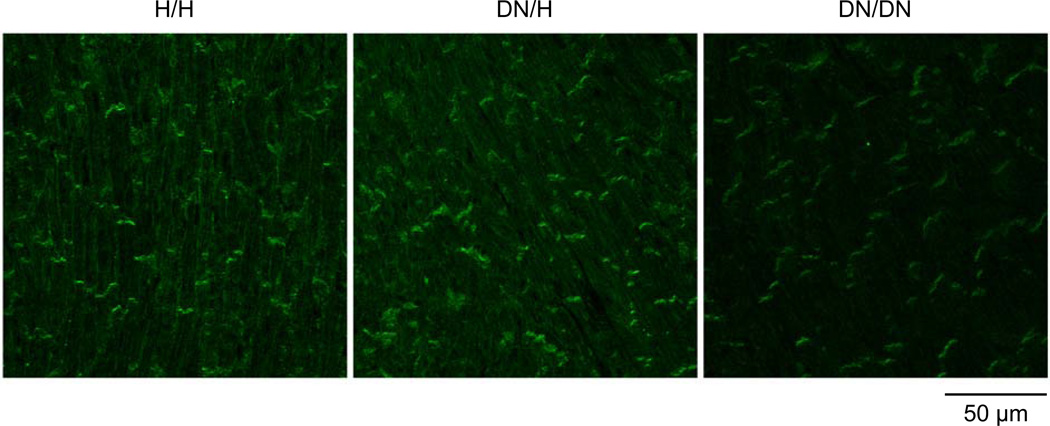
Western blotting showed reduction in sodium channel protein abundance associated with the DN allele and the changes were much more dramatic in DN/DN compared to DN/H hearts (Figure 7A and 7B). The abundance of the control calnexin protein was similar among H/H (reference, 100±6%), DN/H (103±4% of H/H), and DN/DN mice (100±5% of H/H) (P=NS for each). Although sodium current and sodium channel protein were reduced in DN/DN and DN/H mice compared to H/H littermates, real-time PCR showed that SCN5A transcript levels were elevated in mice with the DN allele (Figure 7C). Expression levels of β-actin transcripts were similar among H/H (reference, 100±1%), DN/H (100±1% of H/H), and DN/DN mice (102±5% of H/H) (P=NS for each). Immunostaining experiments were conducted in heart sections at 3 weeks (Figure 8). The DN allele was associated with reduced levels of cell surface expression. Notably, staining was obvious on the lateral myocyte aspects in H/H hearts but was near-absent in DN/DN hearts stained under identical conditions.
Figure 7.
Sodium channel expression levels at 3 weeks. A, Representative Western blots in ventricles. B, Sodium channel expression levels normalized to those of H/H. Calnexin was used as the loading control. C, Relative expression levels of SCN5A transcript normalized to those of β-actin in ventricle. *P<0.05 versus H/H.
Figure 8.
Immunostaining for sodium channel (Nav1.5) at 3 weeks. Heart sections from the ventricles were stained with anti Nav1.5 (green). Note the obvious lateral staining in the H/H heart and its absence in the DN/DN heart.
Discussion
The D1275N mutation has been associated with a range of sinus node dysfunction, conduction abnormalities, tachyarrhythmias, and contractile dysfunction.8, 9, 16–18 However, in previous studies, the evidence implicating D1275N as the causative mutation has been weak: for example, in the large Dutch kindred, the contribution of an additional connexin variant was invoked to explain why only a minority of subjects displayed a clinical phenotype, but that variant was absent in the proband reported here. In addition, D1275N does not generate major changes in sodium channel function in heterologous expression studies.8, 9, 16–18 Thus, despite the previous and the present clinical case reports, the formal possibility remained that D1275N does not actually contribute to the abnormal phenotypes. To address the role of this (and other) variants in mediating sodium channel-linked clinical phenotypes, we generated a series of mouse lines in which the murine cardiac sodium channel was ablated and human alleles were substituted in the murine Scn5a locus. The technique of recombinase mediated cassette exchange allowed us to place wild-type or mutant human sodium channel cDNAs in the murine cardiac sodium channel locus.21 We have previously reported that this approach eliminates expression of the murine channel and that sodium currents from unmodified wild-type murine ventricular myocytes and those expressing wild-type human SCN5A are indistinguishable, indicating that expression of the exchanged sequence is determined by endogenous sodium channel regulatory mechanisms.21
D1275N mice display sodium channel dysfunction
Sodium current amplitude was similar between D1275N and wild-type channels when expressed in heterologous expression systems in the present study, either in the absence or presence of β1 subunit.16 This is in agreement with most results previously reported, although one group has found that D1275N channels generate significantly less current than wild-type channels in tsA201 cells; the reason for this discrepancy is unknown.20 In our mouse model, D1275N was associated with decreased levels of sodium channel protein by Western analysis of total ventricular protein, decreased expression of sodium channels at the ventricular myocyte surface, and marked reduction of sodium current. In addition, we observed increased late current and altered voltage-dependence of channel inactivation. Thus, channel dysfunction conferred by D1275N becomes evident in the myocyte environment. The major change, reduction in peak sodium current, could represent decreased cell surface expression and/or altered gating of the channel protein. One possible explanation in either case is altered interactions with sodium channel partners, present in myocytes and absent in CHO and tsA201 cells.32, 33 There is precedent for such a hypothesis: the E1053K SCN5A mutation, which is associated with a loss of sodium channel function phenotype, has no effect on current density when studied in heterologous expression systems, but abolishes binding of the channel to ankyrin-G and reduces cell surface expression and sodium current in cultured cardiomyocytes.34 However, while E1053K does affect channel gating under heterologous expression,34 altered channel gating by D1275N was only found in the mice, but not in heterologous systems in our study. This is clearly not a general rule, since channel dysfunction observed with heterologous expression of other mutants (delKPQ1505-1507 and 1795insD) does recapitulate phenotypes observed clinically and in genetically modified mice.7, 35–37
Association of sodium channel with cardiomyopathy
In addition to arrhythmias, SCN5A mutations have been associated with cardiomyopathy.8–15 To date, 12 rare variants in SCN5A have been identified in cardiomyopathy and all of the variants have also been associated with arrhythmia phenotypes that result from loss of sodium channel function.8–15 In our mouse model, the loss of sodium channel function by D1275N is consistent with biophysical properties of other SCN5A mutations associated with DCM,10, 11, 38 and findings in clinical and experimental studies suggest that marked reduction of sodium current is critical for development of cardiomyopathy.13, 14, 39, 40 In prior studies, mice with 90% reduction of Scn5a expression level develop cardiac dysfunction,39 while heterozygous Scn5a knockdown mice (Scn5a+/−) display normal cardiac function.40 In our study, mice expressing D1275N, one of the initially reported SCN5A mutations in a cardiomyopathy kindred,8, 9 showed reduction of sodium current with disrupted channel gating and developed evident cardiomyopathy at 12 weeks. This is consistent with other reports describing that both R814Q occurring homozygously and the compound heterozygous occurrence of the W156X and R225W are associated with cardiomyopathy.13, 14 In these settings, the cardiomyopathy phenotype is generally absent in heterozygotes.13, 14
Among 12 rare variants in SCN5A associated with cardiomyopathy, 7 are located in transmembrane domains, and 6 of them including D1275N are predicted to change the electrical charge of substituted amino acids.8–15 These substitutions may lead changes in channel structure, resulting in altered channel gating and/or reduced channel expression levels directly or by disrupted interaction with sodium channel accessory proteins.
Although this study and previous work strongly imply that loss of sodium channel function has a critical role for development of cardiomyopathy,10, 41 the mechanisms remain controversial. The surface ECG tracings in DN mice (Figure 2) not only demonstrate gene-dose-dependent conduction slowing but suggest altered activation sequence (with ECG complex splintering); thus, electromechanical dyssynchrony, a well-recognized cause of cardiac contractile dysfunction,42 may be sufficient to explain the DCM phenotype. Another possibility raised by a recent report that suggests two pools of sodium channel protein in heart is that the mutant channel does not target to the appropriate subcellular domain to support normal cell-propagation.43 Among causative genes for DCM, cytoskeletal components such as syntrophins and dystrophins have been associated with SCN5A channel and disrupted interaction with such proteins may result in cardiomyopathy.44–46 Although SCN5A mutations have been associated with cardiac fibrosis,3, 40, 47 we did not observe fibrosis when the mice carrying the DN allele developed cardiac dysfunction. It has been reported that SCN5A-related DCM phenotype usually develops later (>10 years) than onset of arrhythmia phenotypes suggesting a possibility that DCM phenotype is secondarily mediated by arrhythmia.8–10, 48 However, in our study, the cardiomyopathy phenotype was evident relatively early, in the absence of sustained arrhythmia. Taken together, we propose that sodium channel dysfunction and electromechanical dyssynchrony represent the primary pathophysiology for DCM in this setting.
In conclusion, we found that the D1275N SCN5A mutation was associated with cardiomyopathy and multiple arrhythmias in vivo, in line with clinical findings in our and other studies.8, 9, 16–18 Although D1275N did not generate serious channel dysfunction when studied in heterologous expression systems, the mutation produced extensive channel dysfunction, notably marked reduction in peak current amplitude, and a cardiomyopathy phenotype in our mouse model. Further experiments along the lines outlined above will be required to elucidate the precise mechanisms for channel dysfunction and how this leads to the DCM phenotype. Defining the mechanisms underlying the disconnect between the results in heterologous expression systems and those in myocytes will contribute to further understanding of the variable phenotypes and penetrance of D1275N and other SCN5A mutations.
Acknowledgments
We thank Christiana Ingram, Justine Stassun, Laura Short, and Wei Zhang at Vanderbilt University for their assistance in performing or analyzing this work. We also acknowledge the expert performance of the staff of the Vanderbilt Transgenic Mouse/Embryonic Stem Cell Shared Resource for the blastocyst microinjections.
Sources of Funding
This work was supported by grants from the United States Public Health Service, Bethesda, MD (HL65962, HL49989, DK42502, and DK72473) and from the Fondation Leducq, Paris, France (Trans-Atlantic Network of Excellence: Preventing Sudden Cardiac Death, 05-CVD-01). The Murine Cardiovascular Core is supported in part by U24 DK59637, Cardiovascular Core, Mouse Metabolic Physiology Centers, Vanderbilt University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
References
- 1.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 2.Chen Q, Kirsch GE, Zhang D, Brugada R, Brugada J, Brugada P, Potenza D, Moya A, Borggrefe M, Breithardt G, Ortiz-Lopez R, Wang Z, Antzelevitch C, O'Brien RE, Schulze-Bahr E, Keating MT, Towbin JA, Wang Q. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 3.Schott JJ, Alshinawi C, Kyndt F, Probst V, Hoorntje TM, Hulsbeek M, Wilde AA, Escande D, Mannens MM, Le Marec H. Cardiac conduction defects associate with mutations in SCN5A. Nat Genet. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- 4.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, Rhodes TH, George AL., Jr Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellinor PT, Nam EG, Shea MA, Milan DJ, Ruskin JN, MacRae CA. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5:99–105. doi: 10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, George AL, Jr, Roden DM. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–1935. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bezzina C, Veldkamp MW, van Den Berg MP, Postma AV, Rook MB, Viersma JW, van Langen IM, Tan-Sindhunata G, Bink-Boelkens MT, van Der Hout AH, Mannens MM, Wilde AA. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 8.McNair WP, Ku L, Taylor MR, Fain PR, Dao D, Wolfel E, Mestroni L. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163–2167. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- 9.Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, Horton SC, Rodeheffer RJ, Anderson JL. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge J, Sun A, Paajanen V, Wang S, Su C, Yang Z, Li Y, Wang S, Jia J, Wang K, Zou Y, Gao L, Wang K, Fan Z. Molecular and Clinical Characterization of a Novel SCN5A Mutation Associated With Atrioventricular Block and Dilated Cardiomyopathy. Circ Arrhythmia Electrophysiol. 2008;1:83–92. doi: 10.1161/CIRCEP.107.750752. [DOI] [PubMed] [Google Scholar]
- 11.Shi R, Zhang Y, Yang C, Huang C, Zhou X, Qiang H, Grace AA, Huang CL, Ma A. The cardiac sodium channel mutation delQKP 1507–1509 is associated with the expanding phenotypic spectrum of LQT3, conduction disorder, dilated cardiomyopathy, and high incidence of youth sudden death. Europace. 2008;10:1329–1335. doi: 10.1093/europace/eun202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapplinger JD, Tester DJ, Alders M, Benito B, Berthet M, Brugada J, Brugada P, Fressart V, Guerchicoff A, Harris-Kerr C, Kamakura S, Kyndt F, Koopmann TT, Miyamoto Y, Pfeiffer R, Pollevick GD, Probst V, Zumhagen S, Vatta M, Towbin JA, Shimizu W, Schulze-Bahr E, Antzelevitch C, Salisbury BA, Guicheney P, Wilde AA, Brugada R, Schott JJ, Ackerman MJ. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bezzina CR, Rook MB, Groenewegen WA, Herfst LJ, van der Wal AC, Lam J, Jongsma HJ, Wilde AA, Mannens MM. Compound heterozygosity for mutations (W156X and R225W) in SCN5A associated with severe cardiac conduction disturbances and degenerative changes in the conduction system. Circ Res. 2003;92:159–168. doi: 10.1161/01.res.0000052672.97759.36. [DOI] [PubMed] [Google Scholar]
- 14.Frigo G, Rampazzo A, Bauce B, Pilichou K, Beffagna G, Danieli GA, Nava A, Martini B. Homozygous SCN5A mutation in Brugada syndrome with monomorphic ventricular tachycardia and structural heart abnormalities. Europace. 2007;9:391–397. doi: 10.1093/europace/eum053. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Chung MK, Martin D, Rozich R, Tchou PJ, Wang Q. SNP S1103Y in the cardiac sodium channel gene SCN5A is associated with cardiac arrhythmias and sudden death in a white family. J Med Genet. 2002;39:913–915. doi: 10.1136/jmg.39.12.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groenewegen WA, Firouzi M, Bezzina CR, Vliex S, van Langen IM, Sandkuijl L, Smits JP, Hulsbeek M, Rook MB, Jongsma HJ, Wilde AA. A cardiac sodium channel mutation cosegregates with a rare connexin40 genotype in familial atrial standstill. Circ Res. 2003;92:14–22. doi: 10.1161/01.res.0000050585.07097.d7. [DOI] [PubMed] [Google Scholar]
- 17.Greenlee PR, Anderson JL, Lutz JR, Lindsay AE, Hagan AD. Familial automaticity-conduction disorder with associated cardiomyopathy. West J Med. 1986;144:33–41. [PMC free article] [PubMed] [Google Scholar]
- 18.Laitinen-Forsblom PJ, Makynen P, Makynen H, Yli-Mayry S, Virtanen V, Kontula K, Aalto-Setala K. SCN5A mutation associated with cardiac conduction defect and atrial arrhythmias. J Cardiovasc Electrophysiol. 2006;17:480–485. doi: 10.1111/j.1540-8167.2006.00411.x. [DOI] [PubMed] [Google Scholar]
- 19.Bennett PB, Yazawa K, Makita N, George AL., Jr Molecular mechanism for an inherited cardiac arrhythmia. Nature. 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 20.Gui J, Wang T, Jones RP, Trump D, Zimmer T, Lei M. Multiple loss-of-function mechanisms contribute to SCN5A-related familial sick sinus syndrome. PLoS One. 2010;5:e10985. doi: 10.1371/journal.pone.0010985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu K, Hipkens S, Yang T, Abraham R, Zhang W, Chopra N, Knollmann B, Magnuson MA, Roden DM. Recombinase-mediated cassette exchange to rapidly and efficiently generate mice with human cardiac sodium channels. Genesis. 2006;44:556–564. doi: 10.1002/dvg.20247. [DOI] [PubMed] [Google Scholar]
- 22.Jones JR, Shelton KD, Magnuson MA. Strategies for the use of site-specific recombinases in genome engineering. Methods Mol Med. 2005;103:245–257. doi: 10.1385/1-59259-780-7:245. [DOI] [PubMed] [Google Scholar]
- 23.Lauth M, Spreafico F, Dethleffsen K, Meyer M. Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res. 2002;30:e115. doi: 10.1093/nar/gnf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye B, Valdivia CR, Ackerman MJ, Makielski JC. A common human SCN5A polymorphism modifies expression of an arrhythmia causing mutation. Physiol Genomics. 2003;12:187–193. doi: 10.1152/physiolgenomics.00117.2002. [DOI] [PubMed] [Google Scholar]
- 25.Liu K, Yang T, Viswanathan PC, Roden DM. New mechanism contributing to drug-induced arrhythmia: rescue of a misprocessed LQT3 mutant. Circulation. 2005;112:3239–3246. doi: 10.1161/CIRCULATIONAHA.105.564008. [DOI] [PubMed] [Google Scholar]
- 26.Knollmann BC, Chopra N, Hlaing T, Akin B, Yang T, Ettensohn K, Knollmann BE, Horton KD, Weissman NJ, Holinstat I, Zhang W, Roden DM, Jones LR, Franzini-Armstrong C, Pfeifer K. Casq2 deletion causes sarcoplasmic reticulum volume increase, premature Ca2+ release, and catecholaminergic polymorphic ventricular tachycardia. J Clin Invest. 2006;116:2510–2520. doi: 10.1172/JCI29128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casimiro MC, Knollmann BC, Ebert SN, Vary JC, Jr, Greene AE, Franz MR, Grinberg A, Huang SP, Pfeifer K. Targeted disruption of the Kcnq1 gene produces a mouse model of Jervell and Lange-Nielsen Syndrome. Proc Natl Acad Sci U S A. 2001;98:2526–2531. doi: 10.1073/pnas.041398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell GF, Jeron A, Koren G. Measurement of heart rate and Q-T interval in the conscious mouse. Am J Physiol. 1998;274(3 Pt 2):H747–H751. doi: 10.1152/ajpheart.1998.274.3.H747. [DOI] [PubMed] [Google Scholar]
- 29.Rottman JN, Ni G, Brown M. Echocardiographic evaluation of ventricular function in mice. Echocardiography. 2007;24:83–89. doi: 10.1111/j.1540-8175.2006.00356.x. [DOI] [PubMed] [Google Scholar]
- 30.Knollmann BC, Knollmann-Ritschel BE, Weissman NJ, Jones LR, Morad M. Remodelling of ionic currents in hypertrophied and failing hearts of transgenic mice overexpressing calsequestrin. J Physiol. 2000;525(Pt 2):483–498. doi: 10.1111/j.1469-7793.2000.t01-1-00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe H, Koopmann TT, Le Scouarnec S, Yang T, Ingram CR, Schott JJ, Demolombe S, Probst V, Anselme F, Escande D, Wiesfeld AC, Pfeufer A, Kaab S, Wichmann HE, Hasdemir C, Aizawa Y, Wilde AA, Roden DM, Bezzina CR. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest. 2008;118:2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abriel H. Roles and regulation of the cardiac sodium channel Na(v)1.5: Recent insights from experimental studies. Cardiovasc Res. 2007;76:381–389. doi: 10.1016/j.cardiores.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Meadows LS, Isom LL. Sodium channels as macromolecular complexes: implications for inherited arrhythmia syndromes. Cardiovasc Res. 2005;67:448–458. doi: 10.1016/j.cardiores.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Mohler PJ, Rivolta I, Napolitano C, LeMaillet G, Lambert S, Priori SG, Bennett V. Nav1.5 E1053K mutation causing Brugada syndrome blocks binding to ankyrin-G and expression of Nav1.5 on the surface of cardiomyocytes. Proc Natl Acad Sci U S A. 2004;101:17533–17538. doi: 10.1073/pnas.0403711101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumaine R, Wang Q, Keating MT, Hartmann HA, Schwartz PJ, Brown AM, Kirsch GE. Multiple mechanisms of Na+ channel--linked long-QT syndrome. Circ Res. 1996;78:916–924. doi: 10.1161/01.res.78.5.916. [DOI] [PubMed] [Google Scholar]
- 36.Nuyens D, Stengl M, Dugarmaa S, Rossenbacker T, Compernolle V, Rudy Y, Smits JF, Flameng W, Clancy CE, Moons L, Vos MA, Dewerchin M, Benndorf K, Collen D, Carmeliet E, Carmeliet P. Abrupt rate accelerations or premature beats cause life-threatening arrhythmias in mice with long-QT3 syndrome. Nat Med. 2001;7:1021–1027. doi: 10.1038/nm0901-1021. [DOI] [PubMed] [Google Scholar]
- 37.Remme CA, Verkerk AO, Nuyens D, van Ginneken AC, van Brunschot S, Belterman CN, Wilders R, van Roon MA, Tan HL, Wilde AA, Carmeliet P, de Bakker JM, Veldkamp MW, Bezzina CR. Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A-1795insD. Circulation. 2006;114:2584–2594. doi: 10.1161/CIRCULATIONAHA.106.653949. [DOI] [PubMed] [Google Scholar]
- 38.Keller DI, Acharfi S, Delacretaz E, Benammar N, Rotter M, Pfammatter JP, Fressart V, Guicheney P, Chahine M. A novel mutation in SCN5A, delQKP 1507–1509, causing long QT syndrome: role of Q1507 residue in sodium channel inactivation. J Mol Cell Cardiol. 2003;35:1513–1521. doi: 10.1016/j.yjmcc.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Hesse M, Kondo CS, Clark RB, Su L, Allen FL, Geary-Joo CT, Kunnathu S, Severson DL, Nygren A, Giles WR, Cross JC. Dilated cardiomyopathy is associated with reduced expression of the cardiac sodium channel Scn5a. Cardiovasc Res. 2007;75:498–509. doi: 10.1016/j.cardiores.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 40.Royer A, van Veen TA, Le Bouter S, Marionneau C, Griol-Charhbili V, Leoni AL, Steenman M, van Rijen HV, Demolombe S, Goddard CA, Richer C, Escoubet B, Jarry-Guichard T, Colledge WH, Gros D, de Bakker JM, Grace AA, Escande D, Charpentier F. Mouse model of SCN5A-linked hereditary Lenegre's disease: age-related conduction slowing and myocardial fibrosis. Circulation. 2005;111:1738–1746. doi: 10.1161/01.CIR.0000160853.19867.61. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen TP, Wang DW, Rhodes TH, George AL., Jr Divergent biophysical defects caused by mutant sodium channels in dilated cardiomyopathy with arrhythmia. Circ Res. 2008;102:364–371. doi: 10.1161/CIRCRESAHA.107.164673. [DOI] [PubMed] [Google Scholar]
- 42.Cazeau S, Leclercq C, Lavergne T, Walker S, Varma C, Linde C, Garrigue S, Kappenberger L, Haywood GA, Santini M, Bailleul C, Daubert JC. Effects of multisite biventricular pacing in patients with heart failure and intraventricular conduction delay. N Engl J Med. 2001;344:873–880. doi: 10.1056/NEJM200103223441202. [DOI] [PubMed] [Google Scholar]
- 43.Petitprez S, Zmoos AF, Ogrodnik J, Balse E, Raad N, El-Haou S, Albesa M, Bittihn P, Luther S, Lehnart SE, Hatem SN, Coulombe A, Abriel H. SAP97 and dystrophin macromolecular complexes determine two pools of cardiac sodium channels Nav1.5 in cardiomyocytes. Circ Res. 2011;108:294–304. doi: 10.1161/CIRCRESAHA.110.228312. [DOI] [PubMed] [Google Scholar]
- 44.Towbin JA, Bowles NE. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 45.Karkkainen S, Peuhkurinen K. Genetics of dilated cardiomyopathy. Ann Med. 2007;39:91–107. doi: 10.1080/07853890601145821. [DOI] [PubMed] [Google Scholar]
- 46.Gavillet B, Rougier JS, Domenighetti AA, Behar R, Boixel C, Ruchat P, Lehr HA, Pedrazzini T, Abriel H. Cardiac sodium channel Nav1.5 is regulated by a multiprotein complex composed of syntrophins and dystrophin. Circ Res. 2006;99:407–414. doi: 10.1161/01.RES.0000237466.13252.5e. [DOI] [PubMed] [Google Scholar]
- 47.Coronel R, Casini S, Koopmann TT, Wilms-Schopman FJ, Verkerk AO, de Groot JR, Bhuiyan Z, Bezzina CR, Veldkamp MW, Linnenbank AC, van der Wal AC, Tan HL, Brugada P, Wilde AA, de Bakker JM. Right ventricular fibrosis and conduction delay in a patient with clinical signs of Brugada syndrome: a combined electrophysiological, genetic, histopathologic, and computational study. Circulation. 2005;112:2769–2777. doi: 10.1161/CIRCULATIONAHA.105.532614. [DOI] [PubMed] [Google Scholar]
- 48.Shinbane JS, Wood MA, Jensen DN, Ellenbogen KA, Fitzpatrick AP, Scheinman MM. Tachycardia-induced cardiomyopathy: a review of animal models and clinical studies. J Am Coll Cardiol. 1997;29:709–715. doi: 10.1016/s0735-1097(96)00592-x. [DOI] [PubMed] [Google Scholar]