Abstract
TDP-43 is the major disease protein in ubiquitin-positive inclusions of amyotrophic lateral sclerosis and frontotemporal lobar degeneration (FTLD) characterized by TDP-43 pathology (FTLD-TDP). Accumulation of insoluble TDP-43 aggregates could impair normal TDP-43 functions and initiate disease progression. Thus, it is critical to define the signalling mechanisms regulating TDP-43 since this could open up new avenues for therapeutic interventions. Here, we have identified a redox-mediated signalling mechanism directly regulating TDP-43. Using in vitro and cell-based studies, we demonstrate that oxidative stress promotes TDP-43 cross-linking via cysteine oxidation and disulphide bond formation leading to decreased TDP-43 solubility. Biochemical analysis identified several cysteine residues located within and adjacent to the second RNA-recognition motif that contribute to both intra- and inter-molecular interactions, supporting TDP-43 as a target of redox signalling. Moreover, increased levels of cross-linked TDP-43 species are found in FTLD-TDP brains, indicating that aberrant TDP-43 cross-linking is a prominent pathological feature of this disease. Thus, TDP-43 is dynamically regulated by a redox regulatory switch that links oxidative stress to the modulation of TDP-43 and its downstream targets.
Keywords: cysteine, disulphide, oxidative, redox
Introduction
Trans-activation response (TAR) DNA-binding protein of 43 kDa (TDP-43) encoded by the TARDBP gene on chromosome 1 is a major component of τ-negative and ubiquitin-positive inclusions that characterize amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) linked to TDP-43 pathology (FTLD-TDP) (Neumann et al, 2006). Recent studies have identified TDP-43 aggregation and neuropathology in a wide spectrum of distinct neurodegenerative disorders collectively known as TDP-43 proteinopathies, supporting a central role for TDP-43 in neurodegenerative disease pathogenesis (Pesiridis et al, 2009; Lagier-Tourenne et al, 2010). Currently, >35 missense mutations in the TARDBP gene have been identified as being pathogenic for familial and sporadic ALS as well as in rare familial cases of ALS and FTLD-TDP (Lagier-Tourenne and Cleveland, 2009; Pesiridis et al, 2009). Moreover, TDP-43 pathologies are not limited to the brain and spinal cord, as TDP-43-positive cytosolic muscle aggregates have been identified in familial and sporadic inclusion body myositis (Salajegheh et al, 2009). These studies have sparked intense efforts to elucidate the physiological functions of TDP-43 and the molecular underpinning of TDP-43 proteinopathies.
TDP-43 is abundantly expressed in nearly all tissues and is highly conserved among mammals and invertebrates (Ayala et al, 2005). Structural studies have identified two RNA-recognition motifs, termed RRM1 and RRM2, capable of binding nucleic acids (Buratti and Baralle, 2001), and a glycine-rich C-terminal domain implicated in protein interactions. TDP-43 is expressed mainly in the nucleus and localizes prominently to discrete nuclear foci that partially overlap with gems and Cajal bodies (Wang et al, 2002), supporting a role for TDP-43 in RNA processing and splicing. Indeed, TDP-43 was shown to bind and stabilize human neurofilament mRNA (Volkening et al, 2009), promote exon skipping of the cystic fibrosis transmembrane conductance regulator (CFTR) (Buratti and Baralle, 2001; Buratti et al, 2001), facilitate exon 7 inclusion of the survival of motor neuron (SMN) 2 gene (Bose et al, 2008), and directly stabilize the mRNA encoding histone deacetylase 6 (HDAC6) (Fiesel et al, 2010). Unbiased global RNA sequencing approaches have recently identified TDP-43-binding sites in a large number of mRNAs including those that are involved in regulating synaptic function, RNA metabolism, neuronal development as well as neurodegeneration including FUS/TLS and TDP-43 itself (Polymenidou et al, 2011; Sephton et al, 2011; Tollervey et al, 2011). Further supporting a role in RNA processing came recently from studies showing that TDP-43 localizes to punctate neuronal granules and cytoplasmic stress granules (SGs) in primary neurons and cultured cells exposed to various forms of stress (Wang et al, 2008; Colombrita et al, 2009; Freibaum et al, 2010; Liu-Yesucevitz et al, 2010; Dewey et al, 2011; McDonald et al, 2011). Although the significance of TDP-43 re-localization is not yet clear, SGs represent cytoplasmic hubs regulating mRNA expression, processing, and sorting that may be critical for neuronal survival. However, despite these studies implicating TDP-43 in RNA regulation, any potential signalling mechanisms controlling TDP-43 function remains to be determined.
TDP-43 proteinopathies are characterized by cytoplasmic and/or nuclear inclusions containing hyper-phosphorylated, truncated, ubiquitinated, and aggregated TDP-43 protein (Neumann et al, 2006). Several studies have proposed TDP-43 loss-of-function as potential mechanisms of neurodegeneration. In both transgenic (Tg) mouse models and cell-based systems, expression of wild-type (WT) TDP-43 or mutant TDP-43 directed to the cytoplasm (i.e. TDP-43-ΔNLS) led to depletion of endogenous nuclear TDP-43 (Winton et al, 2008; Igaz et al, 2010; Tsai et al, 2010; Wils et al, 2010), and a similar loss of nuclear TDP-43 occurred in Tg mice expressing the ALS-associated A315T mutation (Wegorzewska et al, 2009). Moreover, since neuron loss was observed following nuclear clearance of endogenous TDP-43 in Tg mouse models, this suggests that nuclear depletion and loss of TDP-43 function is associated with neurodegeneration. Elucidating the mechanisms that control TDP-43 function could provide avenues for therapeutic intervention. Here, we identify a novel signalling mechanism in which oxidative stress and redox status rapidly and reversibly regulate TDP-43 solubility and nuclear function. We demonstrate that TDP-43 solubility is not dependent on canonical signalling transduction pathways, but instead is regulated via direct stress-induced cysteine oxidation and disulphide bond formation, resulting in the accumulation of insoluble cross-linked TDP-43 species. These results implicate redox signalling as a novel regulatory switch controlling TDP-43 and highlights stress-induced reactive oxygen species (ROS) as a potential factor in the pathogenesis of ALS, FTLD-TDP, and related TDP-43 proteinopathies.
Results
TDP-43 subcellular localization and solubility are regulated by oxidative stress
To determine if oxidative stress implicated in neurodegenerative diseases triggers alterations in TDP-43 subcellular localization and/or solubility, Cos7 cells were treated with various stressors including hydrogen peroxide (H2O2) and arsenite, which generate ROS through distinct mechanisms (Valko et al, 2005; Calabrese et al, 2010; Jomova et al, 2010). Consistent with the previous reports that TDP-43 localizes to SGs, H2O2 induced a rapid translocation of TDP-43 into cytosolic SGs, as determined by co-localization with the SG marker TIAR (Kedersha et al, 1999) (Figure 1Aa–c and B). In contrast, although arsenite treatment led to robust formation of TIAR-positive SGs, TDP-43 was not prominently detected within these SGs (Figure 1Ad–f and B), supporting observations that the majority of TDP-43 remains nuclear localized in response to arsenite treatment (Liu-Yesucevitz et al, 2010; Dewey et al, 2011). More detailed immunofluorescence analysis of arsenite-treated cells using a Triton X-100 extraction method revealed accumulation of insoluble nuclear TDP-43 aggregates (Supplementary Figure S1). Thus, TDP-43 preferentially accumulates within SGs in response to peroxide, while arsenite, heat shock, and cadmium caused TDP-43 accumulation within discrete nuclear aggregates (Supplementary Figure S2) that resemble those observed with RNA-binding deficient TDP-43 mutants (Ayala et al, 2008).
Figure 1.
Oxidative stress regulates TDP-43 subcellular localization and solubility. (A) Cos7 cells were exposed to either 1 mM H2O2 (a–c) or 0.25 mM sodium arsenite (d–f) for 1 h at 37°C in complete media. Cells were subsequently fixed and analysed by immunofluorescence microscopy using rabbit polyclonal TDP-43 and mouse monoclonal TIAR antibodies. Scale bar represents 50 μm. (B) SG localization of either TDP-43 or TIAR in response to H2O2 or arsenite was quantified using 10 fields per image (N=3 independent experiments) and represented as percent TIAR or TDP-43 localization to SGs. P-values were calculated by Student's t-test (***)=2.05 × 10−5 and (**)=5.1 × 10−4. Notably, TDP-43 showed preferential localization to H2O2, but not arsenite-induced SGs. (C) Cos7 cells were treated with the indicated oxidative stressors for 1 h and lysates were sequentially extracted with RIPA (R) and urea (U) buffers for analysis by immunoblotting using TDP-43 and GAPDH antibodies. (D) Arsenite-treated cells were washed twice in PBS and allowed a 2-h recovery in arsenite-free complete media followed by immunoblotting analysis as in (C). (E) Cells were pretreated overnight with 20 μg/ml cycloheximide (CHX), where indicated, prior to arsenite exposure and analysed similar to (C) above. Amyloid precursor protein (APP) was used as a positive control for cycloheximide treatment and was detected using an anti-APP-specific antibody (Karen). (F) Cells were treated with arsenite in the presence of 10 mM NAC, where indicated, for subsequent biochemical analysis similar to (C). Figure source data can be found in Supplementary data.
Sequential biochemical extraction of stressed cells showed a dramatic shift in TDP-43 solubility converting it from a soluble to an insoluble pool under all stress conditions analysed, with arsenite-treated cells showing the most profound change in solubility (Figure 1C). To determine if insoluble TDP-43 is reversed upon removal of the stress, arsenite-treated cells were allowed a 2-h recovery in full media, which rapidly restored the soluble TDP-43 pool (Figure 1D), suggesting that TDP-43 solubility is dynamically regulated by oxidative stress. Moreover, the restoration of soluble TDP-43 was not due to new protein synthesis, as cycloheximide treatment prior to arsenite addition yielded a similar solubility transition (Figure 1E, lanes 5–8) that was reversible upon stress recovery (Figure 1E, lanes 9–12). To evaluate whether stress-induced ROS are responsible for decreased TDP-43 solubility, cells were treated with the free radical scavenger N-acetyl-cysteine (NAC), which completely prevented the arsenite-mediated accumulation of insoluble TDP-43 (Figure 1F).
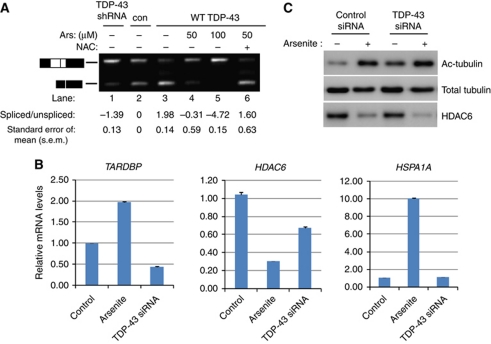
To determine whether decreased solubility compromised TDP-43 function (Buratti et al, 2001), we utilized a previously established cell-based CFTR splicing assay to assess if CFTR splicing is affected by arsenite treatment. QBI-293 cells were sequentially transfected with a CFTR hybrid minigene followed by either control, TDP-43 shRNA, or TDP-43 over-expressing plasmids, and CFTR exon 9 inclusion was monitored by RT–PCR (Figure 2A). As expected, TDP-43 shRNA led to accumulation of the exon 9 included transcript (Figure 2A, lanes 1 and 2), while WT TDP-43 over-expression inhibited exon 9 inclusion (Figure 2A, compare lanes 2 and 3). Strikingly, exposure to arsenite caused a dramatic increase in exon 9 inclusion, similar to knockdown using a TDP-43 shRNA (Figure 2A, lanes 3 and 5, see quantification below), reflecting a loss of TDP-43 function and reduced exon 9 splicing. Furthermore, reduced exon 9 splicing was completely reversed by treatment with NAC (Figure 2A, lane 6).
Figure 2.
Oxidative stress causes inhibition of TDP-43-mediated RNA regulatory functions. (A) Cells expressing vector control, TDP-43shRNA, or WT TDP-43 plasmids were co-transfected with a CFTR minigene and treated with arsenite and/or NAC where indicated. Spliced and unspliced CFTR transcripts were determined by RT–PCR using specific primers flanking exon 9 followed by gel electrophoresis. RT–PCR products were quantified using the Agilent 2100 Bioanalyzer and relative values of CFTR transcripts are represented as a log ratio of spliced/unspliced products. All samples are normalized to the vector control (lane 2) set to 0. Splicing analysis was performed in triplicate using three independent experiments (N=3), and error bars represent standard error of the mean (s.e.m.). (B) Control siRNA or TDP-43-specific siRNA expressing QBI-293 cells were exposed to 50 μM arsenite overnight, where indicated, to allow full redox-mediated inactivation of TDP-43, and subsequently total RNA was analysed by quantitative RT–PCR using transcript-specific primers for TARDBP, HDAC6, or HSPA1A, which encodes the Hsp70 protein as a control for arsenite exposure. Statistical analysis from N=3 independent replicates was performed using Student's t-test and P-values <2 × 10−4 for all control versus arsenite and control versus TDP-43 siRNA samples. (C) Control and TDP-43 siRNA samples prepared in parallel similar to (B) above were harvested for protein extraction and western analysis using acetylated tubulin, total tubulin, or HDAC6-specific antibodies. Figure source data can be found in Supplementary data.
We next evaluated the consequences of stress-induced inactivation of TDP-43 on several known TDP-43 target RNAs including the TARDBP transcript itself, the stability of which is regulated via a negative auto-regulatory mechanism (Ayala et al, 2011; Polymenidou et al, 2011), and the HDAC6 transcript, which was shown previously to be stabilized by TDP-43 (Fiesel et al, 2010; Kim et al, 2010). Consistent with the inhibition of TDP-43 function, arsenite treatment increased TARDBP mRNA levels and reduced HDAC6 mRNA levels, similar to that observed using a specific TDP-43 siRNA (Figure 2B). Furthermore, stress-induced inactivation of TDP-43 and depletion of HDAC6 correlated with increased levels of acetylated tubulin, a major substrate for HDAC6 (Hubbert et al, 2002) (Figure 2C). Thus, the insoluble accumulation of TDP-43 aggregates in response to oxidative stress led to loss of TDP-43 function as reflected by alterations in downstream RNA targets, further supporting ROS-mediated regulation of TDP-43 activity.
TDP-43 undergoes cysteine oxidation and disulphide bond formation in response to oxidative stress
Given the dramatic shift in TDP-43 solubility, we next determined whether ROS might activate stress-regulated signalling pathways and trigger downstream conformational changes, resulting in altered TDP-43 solubility. We performed studies using pharmacological inhibition or constitutive activation of several stress-regulated signalling cascades including p38, JNK, and ERK pathways and did not observe altered TDP-43 solubility (data not shown), suggesting an alternative signalling mechanism. Therefore, we hypothesized that oxidative damage might directly modify TDP-43 via cysteine oxidization. To investigate this possibility, full-length recombinant TDP-43 protein was exposed to H2O2, which, in contrast to arsenite, directly oxidizes proteins in vitro. The presence of reduced cysteine residues was determined by incubating TDP-43 with [14C]-iodoacetate, which irreversibly alkylates free thiol groups on cysteine residues. Indeed, untreated TDP-43 was efficiently labelled with [14C]-iodoacetate, but treatment with increasing concentrations of H2O2 blocked [14C]-iodoacetate-mediated alkylation, indicating cysteine oxidation upon exposure to increasing concentrations of H2O2 (Figure 3A).
Figure 3.
TDP-43 cysteine residues undergo oxidation and disulphide bond formation in vitro. (A) Purified recombinant TDP-43 protein was incubated with the indicated concentrations of H2O2 in the presence of [14C]-iodoacetate. Samples were analysed by gel electrophoresis, transferred to nitrocellulose membrane, and developed using a phosphor-imager. (B) Recombinant TDP-43 was treated with 1 mM H2O2 and analysed by non-reducing immunoblotting using a panel of antibodies detecting different TDP-43 epitopes: Ab-1, rabbit polyclonal anti-TDP-43 (Proteintech), Ab-2, polyclonal C-terminal 1038, Ab-3, monoclonal anti-TDP-43 (Proteintech), and Ab-4, polyclonal N-terminal 1065 antibodies. (C) Recombinant TDP-43 treated with H2O2 was subsequently treated with the reducing agent DTT where indicated. All proteins were subsequently cross-linked as a final step using EGS according to the procedures detailed in the Materials and methods section and analysed by immunoblotting using anti-TDP-43 1038. Figure source data can be found in Supplementary data
Given that oxidized cysteines can readily form disulphide bonds, we asked if H2O2 induced TDP-43 disulphide cross-linking in vitro. A dramatic accumulation of high molecular weight (HMW) TDP-43 immunoreactive bands was observed following H2O2 treatment using a panel of TDP-43 antibodies (Figure 3B, see HMW species). HMW disulphide bond formation was also observed using the sensitive cross-linking agent ethylene glycol disuccinate (EGS) (Figure 3C). Notably, in addition to HMW TDP-43 species, peroxide treatment led to a significant mobility shift in monomeric TDP-43, consistent with intra-molecular cysteine interactions (Figure 3C, lane 2). To further confirm the formation of intra- and inter-molecular interactions, oxidized TDP-43 was subsequently treated with the reducing agent dithiothreitol (DTT), which resulted in the complete collapse of all TDP-43 species into a distinct 43 kDa protein band (Figure 3C, compare lanes 2 and 4).
We next determined if TDP-43 disulphide cross-linking also occurs in cultured cells. To accomplish this, QBI-293 cells transiently expressing WT TDP-43 were acutely treated with arsenite and analysed by immunoblotting using a TDP-43 antibody (Figure 4A). Analysis of cell lysates specifically under non-reducing conditions revealed a smear of HMW TDP-43 immunoreactive bands with a prominent ∼120 kDa disulphide species (Figure 4A, compare –DTT versus +DTT, lanes 4 and 8). Using naive untransfected Cos7 cells, endogenous TDP-43 readily formed HMW species in response to several ROS generators including arsenite and cadmium and these responses were specific since they were completely inhibited by treatment with NAC (Figure 4B). In addition to HMW species, a less pronounced, but readily detectable, intra-molecular mobility shift was observed with endogenous monomeric TDP-43 in response to arsenite treatment (Figure 4C). Finally, we confirmed TDP-43 disulphide bond formation using cultured hippocampal neurons, which showed robust accumulation of insoluble HMW species following exposure to arsenite (Figure 4D, see HMW TDP-43 species in darker exposure and compare −DTT with +DTT lanes). These results reveal that TDP-43 is highly susceptible to stress-induced cysteine oxidation, resulting in conformational changes consistent with both intra- and inter-molecular cross-links.
Figure 4.
TDP-43 disulphide bond formation occurs in cultured cells exposed to oxidative stress. (A) QBI-293 cells transfected with a WT TDP-43 expression plasmid were treated with 0.25 mM arsenite for 1 h where indicated. Cell lysates were sequentially extracted and analysed under reducing and non-reducing conditions followed by immunoblotting with anti-TDP-43 1038. Bracketed bands represent HMW TDP-43 species as indicated. (B) Cos7 cell lysates from naive untransfected cells treated with either 0.25 mM arsenite or 10 μM cadmium in the presence of 10 mM NAC, where indicated, were analysed by non-reducing western blotting using anti-TDP-43 1038 or anti-GAPDH antibodies. (C) Urea extracted fractions from arsenite-treated QBI-293 or Cos7 cells were immunoblotted in the presence or absence of DTT and gel mobility of endogenous monomeric TDP-43 was evaluated using anti-TDP-43 1038. (D) Hippocampal neurons isolated from WT C57/BL6 mice were cultured for 10 days prior to treatment with 0.25 mM arsenite for 2 h. Neurons were sequentially extracted similar to (A) and soluble/insoluble lysates were immunoblotted using anti-TDP-43 1038. The asterisk (*) indicates a non-specific cross-reactive band detected with anti-TDP-43 1038 in mouse cells. The bracketed bands highlight HMW TDP-43 species detected under non-reducing conditions. Figure source data can be found in Supplementary data
Conserved TDP-43 cysteine residues within and surrounding RRM2 are required for disulphide bond formation
To map cysteine residues that mediate WT TDP-43 cross-linking, mass spectrometry (nanoLC/nanospray/MS/MS) was used to identify disulphide-linked TDP-43 peptides generated after treatment with oxidative stress. Peroxide-treated recombinant TDP-43 protein, including both HMW and monomeric species (Figure 5A, TDP-43ox protein bands identified with bracket and arrows), were excised from a non-reducing gel and subjected to mass spectrometry analysis. The peptide WCDCKLPNSK (mass 1190.52) was identified in both monomeric and HMW TDP-43 species, in which Cys173 and Cys175 formed intra-molecular disulphide bonds, as indicated by the following: (1) specific de-hydrogenation of the indicated cysteine residues within the peptide, (2) impaired peptide fragmentation peaks surrounding the disulphide bond, and (3) impaired ability to alkylate these modified cysteine residues (see Figure 5C and Supplementary Figure S3). Mass spectrometry analysis of all samples in the presence of DTT did not detect disulphide-bonded peptides. To confirm these results using a cell-based approach, over-expressed TDP-43 was immunoprecipitated from insoluble fractions of peroxide or arsenite-treated cells, and the TDP-43 protein bands were analysed by gel electrophoresis and Coomassie staining (Figure 5B). TDP-43 immunoprecipitates from insoluble fractions were excised for mass spectrometry analysis (Figure 5B, lanes 2 and 3). Similar to the in vitro analysis, the identical Cys173/Cys175 cross-linked peptide, WCDCKLPNSK, was observed in either peroxide or arsenite-treated cells, further confirming the presence of this intra-molecular interaction in cells. Thus, an intra-molecular Cys173/Cys175 interaction contributes, in part, to the fully cross-linked TDP-43 species generated in response to oxidative stress.
Figure 5.
Oxidative stress induces TDP-43 intra-molecular cysteine interactions as detected by mass spectrometry. (A) Untreated or H2O2 treated (10 mM) recombinant WT TDP-43 were processed in triplicate using non-reducing SDS–PAGE and Coomassie blue staining. HMW multimeric and monomeric TDP-43 species were gel excised for analysis by nanoLC/nanospray/MS/MS. (B) TDP-43 was immunoprecipitated from either soluble or insoluble fractions of arsenite or peroxide-treated cells as described in the Materials and methods section. Immunoprecipitated proteins were analysed by gel electrophoresis and Coomassie staining, and monomeric TDP-43 was gel excised for analysis by nanoLC/nanospray/MS/MS. All data were acquired using a specialized software (Mass Matrix, Case Western University) to detect disulphide bonds. (C) A heat map diagram was generated using Mass Matrix software, which highlights the intra-molecular disulphide-bonded species (C173–C175) identified by mass spectrometry from gel excised TDP-43 protein bands isolated in (A, B). TDP-43 cysteine residues are displayed on x- and y-axes and increasing shades of red indicate significance of disulphide bonding between the indicated residues. Note, only Cys173 and Cys175 undergo highly significant disulphide bond formation. The peptide spectrum and mass peak values corresponding to the disulphide-bonded peptide, WCDCKLPNSK, are shown in Supplementary Figure S3. Figure source data can be found in Supplementary data.
We were unable to determine the contribution of potential disulphide bonds present within RRM2, as only ∼50% TDP-43 protein sequence coverage was obtained by mass spectrometry. However, TDP-43 sequence alignment from several species revealed significant conservation among a cluster of four cysteine residues (Cys173, 175, 198, 244), suggesting that additional cysteines (i.e. Cys198, 244) might also contribute to disulphide formation (Figure 6A). To test this hypothesis, we generated recombinant cysteine-deficient mutants, TDP-2CS (C173/175S) and TDP-4CS (C173/175/198/244S), and these proteins were analysed by immunoblotting. WT and 2CS proteins readily formed HMW cross-linked species in response to peroxide treatment (Figure 6B, see lanes 4 and 5 as well as lanes 7 and 8), which were reduced with DTT treatment even following 10 mM peroxide treatment (Figure 6B, lanes 10 and 11). However, the formation of peroxide-induced HMW species was almost completely impaired in the absence of all four cysteine residues (Figure 6B, compare WT and 2CS to 4CS in lanes 6 and 9), indicating that Cys173, 175, 198, and 244 are likely critical for TDP-43 cross-linking. In addition to the fact that the 4CS mutant abrogated formation of HMW TDP-43 species, the gel mobility of monomeric 4CS was more compact in the absence of reducing agents compared with the more diffusely migrating WT or 2CS protein bands (Figure 6C, lane 3), suggesting impaired formation of intra-molecular disulphides. To confirm that these cysteines undergo disulphide cross-linking, WT, C173/C175S, C198/244S, or 4CS mutant TDP-43 proteins were over-expressed in QBI-293 cells followed by exposure to arsenite. Insoluble fractions enriched in TDP-43 disulphide cross-linked species were analysed under reducing or non-reducing conditions (Figure 6D). Cells expressing WT, C173/175S, or C198/244S double mutants formed prominent HMW bands, which were dramatically reduced in 4CS mutant expressing cells lacking all four cysteines (Figure 6D, compare HMW TDP-43 bands in lanes 1, 3, 5 versus lane 7). Normalization of monomeric TDP-43 protein levels was achieved using two-fold more insoluble extracts from 4CS expressing cells compared with WT extracts (Figure 6E), and further confirmed a significant reduction in disulphide bond formation in the absence of these critical cysteine residues.
Figure 6.
Conserved TDP-43 cysteine residues within and surrounding RRM2 are required for stress-induced disulphide bond formation. (A) A partial TDP-43 sequence alignment from several species is shown. Matched amino-acid residues from the indicated species are shaded in red, conserved cysteine residues are shown in black boxes, and the single non-conserved cysteine residue is boxed in green. (B) Purified recombinant WT, 2CS (C173/175S), or 4CS (C173/175/198/244S) proteins were treated with 1–10 mM H2O2, and subsequently with DTT where indicated, and analysed by non-reducing immunoblotting. The 4CS mutant shows abrogated formation of HMW TDP-43 species in response to H2O2 treatment compared with WT or TDP-2CS proteins. (C) WT, 2CS, or 4CS proteins were similarly immunoblotted for analysis of monomeric TDP-43 gel mobility. Under non-reducing conditions, monomeric WT and 2CS proteins migrated as diffuse TDP-43 species, while the 4CS mutant migrated as a compact monomeric band (lanes 1–3), more similar to the reduced WT protein (lane 4). (D) QBI-293 cells expressing WT TDP-43, the cysteine mutants C173/175S, C198/244S, or the 4CS mutant were treated with arsenite and insoluble fractions were processed in parallel under either reducing or non-reducing conditions followed by immunoblotting with anti-myc (9E10) detecting over-expressed TDP-43 proteins. A darker exposure revealed a significant decrease in HMW TDP-43 bands in the absence of all four cysteine residues. (E) Insoluble lysates from WT and 4CS cell extracts prepared as described in (D) above were analysed by non-reducing immunoblotting using two-fold more 4CS (2.5–10% total insoluble extract) compared with WT (1.2–5% total insoluble extract) to more accurately normalize the monomeric protein input and evaluate formation of HMW TDP-43 species. Figure source data can be found in Supplementary data.
Pathological TDP-43 aggregates or TDP-43 genetic mutations are associated with increased disulphide bond formation
Since oxidative stress increases TDP-43 cross-linking and enhances aggregation, we asked if this might be a potential mechanism responsible for the formation of prominent inclusion bodies observed in postmortem FTLD-TDP brains (Figure 7A, see TDP-43 pathology in FTLD-TDP). Sequential biochemical extractions of frontal cortex from control and FTLD-TDP subjects were performed with extraction buffers of increasing strength supplemented with iodoacetamide to prevent non-specific oxidation during tissue processing. Different TDP-43 species observed in the presence and absence of DTT were assessed by immunoblotting of sarkosyl and urea extractable fractions (Neumann et al, 2006). Control brains showed mild accumulation of TDP-43 disulphide species ranging from ∼90 to 130 kDa in both sarkosyl and urea fractions as detected with multiple TDP-43 antibodies (Figure 7B–D, lanes 2 and 4). Strikingly, analysis of frontal cortex extracts of FTLD-TDP brains showed a dramatic accumulation of additional cross-linked HMW TDP-43 species ranging from ∼90 to 300 kDa, and a distinct urea extractable ∼250 kDa TDP-43 species that was not prominently detected in control subjects (Figure 7B and C, lanes 6 and 8). Significantly, the majority of HMW species in sarkosyl and urea fractions were reduced in the presence of DTT, suggesting that they are cross-linked by disulphide bond formation (Figure 7B and C, lanes 5 and 7). However, we also observed DTT resistant TDP-43 HMW protein bands, which we attribute to the presence of more heavily cross-linked and fibrillar species of TDP-43 that represent aggregated C-terminal TDP-43 fragments (Figure 7D, lanes 5 and 7). Taken together, our results provide evidence that a range of cross-linked TDP-43 species are present physiologically in normal human brain, but these are far more prominent in pathological FTLD-TDP brain tissue containing TDP-43 aggregates.
Figure 7.
TDP-43 disulphide bond formation occurs in normal and pathological FTLD-TDP brain. (A) Cortical brain sections from control or FTLD-TDP subjects were analysed by immunohistochemistry (IHC) using polyclonal TDP-43 1039, which robustly detects TDP-43 aggregates. Inset represents a × 60 magnification of a characteristic perinuclear TDP-43 inclusion in FTLD-TDP. Scale bar represents 50 μm. (B–D) Sequential biochemical extractions of frontal cortex from control or FTLD-TDP subjects was performed and sarkosyl (B) and urea (C, D) fractions were analysed by immunoblotting in parallel under either reducing (lanes 1, 3, 5, and 7) or non-reducing (lanes 2, 4, 6, and 8) conditions using a commercially available polyclonal anti-TDP-43 (B, C) or polyclonal anti-TDP-43 2089 (D), raised against a C-terminal TDP-43 epitope that prominently detects aggregated and ubiquitinated TDP-43 C-terminal fragments. Note, HMW TDP-43 disulphide species bracketed by solid black lines are readily detectable in control subjects (∼90–130 kDa), and more prominently observed in FTLD-TDP subjects (∼90–300 kDa). The reduction of these bands in the presence of DTT was readily observed (B, C), but partially impaired by fibrillar C-terminal aggregated species (D). Figure source data can be found in Supplementary data.
Genetic studies have identified ∼35 missense mutations in the TARDBP gene that are pathogenic for familial and sporadic ALS. However, the functional consequences of these mutations on TDP-43 function or any pathogenic mechanisms associated with mutant TDP-43 have not been determined. Interestingly, two of these mutations, G348C and S379C, introduce an additional cysteine residue that could enhance TDP-43 cross-linking via generation of additional disulphide bonds. To examine this possibility in vitro, recombinant WT TDP-43 or a panel of ALS mutant TDP-43 proteins were oxidized and analysed by Coomassie staining and immunoblotting. Interestingly, peroxide-induced oxidation of G348C and S379C mutants generated a distinct TDP-43 banding pattern characterized by a prominent ∼90 kDa species, a putative TDP-43 dimer, that was absent in WT as well as G294A and R361S mutant samples, as detected by Coomassie staining (Figure 8A) and anti-TDP-43 antibodies (Figure 8B, see bands marked with asterisks). Moreover, a similar ∼90 kDa species was detected in cultured cells transfected with G348C and S379C cysteine-generating mutants (Figure 8C, see TDP-43 band marked with asterisk) but not in those expressing WT TDP-43 and non-cysteine-generating A90V, G294A, and R361S mutants, although expression of all TDP-43 constructs showed the expected ∼120–250 kDa HMW species upon arsenite treatment (Figure 8C, and see quantification of ∼90 versus ∼120 kDa band). Thus, introducing cysteine residues as genetic mutations in the TARDBP gene resulted in the generation of distinct, abnormally cross-linked TDP-43 species that could represent a potential pathogenic mechanism associated with a subset of familial ALS patients.
Figure 8.
Cysteine-generating ALS mutations produce abnormal disulphide cross-linking in response to oxidative stress. (A) Recombinant purified WT and ALS mutant TDP-43 proteins were left untreated or treated with 10 mM H2O2 for 10 min at 4°C and disulphide banding patterns were analysed by Coomassie staining. The asterisk highlights the ∼90 kDa TDP-43 species specifically produced by the cysteine-generating G348C and S379C mutants. (B) Recombinant TDP-43 proteins were similarly exposed to 1–10 mM H2O2 and analysed by non-reducing immunoblotting using anti-TDP-43 1038. WT, G294A, and R361S proteins showed a typical accumulation of >120 kDa HMW species, while the cysteine-generating mutants showed a unique smeared banding pattern characterized by an ∼90 kDa protein band, as indicated with the asterisk. (C) Cells transfected with WT TDP-43 or a panel of ALS mutations (G348C, S379C, A90V, G294A, or R361S) were treated with 100 μM arsenite for 1 h, sequentially extracted as detailed in the Materials and methods, and insoluble fractions were analysed by non-reducing immunoblotting. Protein bands were visualized using anti-myc 9E10 to detect over-expressed myc-tagged TDP-43 proteins migrating as both monomeric and HMW TDP-43 species. The ∼90 kDa TDP-43 immunoreactive band indicated with the asterisk (*) was detected in G348C and S379C mutants but not control or non-cysteine-generating mutants. Quantification of the 90 and 120 kDa disulphide cross-linked bands is depicted below and was determined by band intensity signals (Multi Guage v2.3) that were estimated as percent disulphide species/total TDP-43 monomer, as assessed on identical image exposures. Figure source data can be found in Supplementary data.
Discussion
Genetic, biochemical, and neuropathological evidence strongly implicate TDP-43 in the pathogenesis of ALS, FTLD-TDP, and related TDP-43 proteinopathies. However, the signalling mechanisms that regulate normal physiological or pathological TDP-43 functions are not well understood. Here, we provide evidence that oxidative stress-mediated accumulation of ROS is a critical factor linked to TDP-43 aggregation, solubility, and nuclear activity. Indeed, we demonstrate that a novel redox-based mechanism directly regulates TDP-43, in which conserved cysteine residues undergo oxidation and disulphide cross-linking, resulting in altered conformation and impaired nuclear function. Thus, our data suggest that cysteine-dependent redox modifications represent a rapid and reversible regulatory switch to modulate TDP-43 functions in response to fluctuating oxidative conditions in neurons.
Interestingly, treatment with multiple oxidative stressors including H2O2, arsenite, cadmium, and heat shock all resulted in a similar shift in TDP-43 solubility from a soluble to an insoluble pool (Figure 1C), which partially accumulated as HMW cross-linked TDP-43 species despite the fact that these stressors likely induce oxidative stress through different mechanisms. For example, following treatment with H2O2, TDP-43 translocated from the nucleus to cytoplasmic SGs, but treatment with arsenite showed reduced translocation and instead aggregated within the nucleus, suggesting that stress-type differentially regulates TDP-43 solubility and functions (Figure 1A). However, since both H2O2 and arsenite treatment resulted in the formation of similar C173–C175 cross-linked and insoluble species of TDP-43 (Figure 5), these data support redox regulation of TDP-43 as a dominant response to general oxidant stress, which could underlie the aggregation and/or altered solubility previously observed in response to a variety of cellular stressors including lipids, heavy metals, proteasome inhibition, and membrane stress (Caragounis et al, 2010; Zhang et al, 2010; Dewey et al, 2011).
Stress-induced cysteine oxidation may represent an upstream event that triggers subsequent TDP-43 post-translational modifications that emerge with further disease progression, suggesting a potential two-step mechanism of TDP-43 pathogenesis. For example, disulphide bonds could structurally alter TDP-43 conformation thereby allowing increased accessibility of putative TDP-43 kinases and proteases. However, cells exposed to oxidative stress did not generate hallmark pathological signatures including phosphorylation and C-terminal fragmentation, suggesting that these modifications occur later during subsequent disease progression. Given the proximity of the four identified cysteine residues to the RRM2 domain, cysteine cross-linking of TDP-43 could abrogate binding to RNA targets, an effect that would functionally mimic RNA-binding deficient TDP-43 mutants (Buratti and Baralle, 2001; Ayala et al, 2008). Supporting this possibility, stress-induced disulphide cross-linking impaired TDP-43 splicing function (Figure 2A) and led to accumulation as nuclear aggregates similar to RNA-binding deficient TDP-43 mutants (Supplementary Figures S1 and S2). Residue mapping using a previously described crystal structure of TDP-43 (Kuo et al, 2009) suggests that several of these cysteines are exposed and readily accessible to ROS-mediated oxidization. Importantly, however, redox-regulated TDP-43 is reversible since stress recovery reversed the solubility of TDP-43 from an insoluble to soluble fraction (Figure 1D). The conversion of insoluble TDP-43 back to a soluble form within hours occurred at the protein level (Figure 1E) and agrees with the previously reported half-life of WT TDP-43 in cultured cells of ∼12 h (Ling et al, 2010).
TDP-43 was highly susceptible to cysteine oxidation, resulting in both intra- and inter-molecular interactions (Figures 3 and 4), although the exact pairing and contribution of individual cysteine residues is not yet known. Indeed, a wide range of disulphide cross-linked species (from ∼90 to 300 kDa) were generated in response to stress in vitro as well as in cultured cells and in diseased human brain containing TDP-43 aggregates (Figure 7). These data suggest that cysteine modifications are a critical determinant of TDP-43 protein conformation and function in vivo. Although we cannot rule out the possible involvement of N-terminal cysteine residues (Cys39, 50), our mass spectrometry and mutational analysis support Cys173, 175, 198, and 244 as the major redox-regulated cysteine residues (Figure 6), potentially generating a complex, heterogeneous mixture of cross-linked TDP-43 species with altered gel mobility properties. Supporting this observation, mutation of all four residues resulted in reduced disulphide bond formation. Although we do not fully understand the unique solubility properties of TDP-43, the exquisite sensitivity of TDP-43 to cysteine modifications suggests that disulphide bond formation could have a major structural impact on TDP-43, which is supported by functional inactivation of TDP-43 in response to ROS (Figure 2). The increasing number of post-translational modifications regulating TDP-43 (e.g. phosphorylation, ubiquitination, C-terminal cleavage) suggests that complex regulatory mechanisms exist to maintain and/or fine-tune TDP-43 solubility and function. Future studies could shed light on any cross-talk between these regulatory modifications and potential synergism in promoting TDP-43 pathogenesis in the diseased brain.
TDP-43 disulphide cross-links were readily detected in control brain tissue with several TDP-43 antibodies (Figure 7), indicating that dynamic TDP-43 redox regulation occurs constitutively in the normal human brain. The future characterization of TDP-43 targets could identify a subset of redox-regulated RNAs that are critical for maintaining proper neuronal homeostasis. Such a redox-based mechanism could link ROS generated by normal metabolism to the physiological regulation of RNAs, providing a rapid response to globally regulate protein expression. Several studies have previously shown that RNA-binding proteins are subject to physiological redox regulation. SMN complex activity, which is deficient in spinal muscular atrophy, is essential for the biogenesis of small nuclear ribonucleoproteins, and is negatively regulated by SMN cysteine oxidation (Wan et al, 2008). Cellular redox status similarly regulates iron regulatory protein 2 (IRP2), in which two cysteines predicted to lie within the RNA-binding cleft undergo oxidation, causing inhibition of IRP2–RNA interactions (Zumbrennen et al, 2009). Finally, Hu-antigen R (HuR) homodimerization may have a functional significance in redox modulation of HuR activity in response to oxidative stress (Benoit et al, 2010), further implicating redox regulation as a general physiological mechanism regulating mRNA expression.
Misfolded mutant Cu, Zn-superoxide dismutase (SOD1) protein found in familial ALS patients with SOD1 mutations forms aberrant cysteine cross-links, which accumulate as insoluble SOD1 aggregates (Tiwari and Hayward, 2003). Strikingly, we observed a similar phenomenon for WT TDP-43 under mild oxidative conditions and a more prominent accumulation of distinct disulphide cross-linked species with the ALS-associated mutants G348C and S379C, which introduce an additional cysteine residue (Figure 8). Although the exact role of disulphide species in either TDP-43 or SOD1 aggregation is currently unclear, it has been proposed that SOD1 disulphides could have a subtle modulatory, rather than causal effect on SOD1 aggregation (Karch and Borchelt, 2008). The similarities in redox sensitivity between TDP-43 and SOD1 proteins suggest a common underlying mechanism in which ROS promotes cross-linking of similarly aggregation-prone proteins. Indeed, neurodegenerative diseases have been characterized by accumulation of ROS and oxidative damage in regions including the motor system of ALS spinal cord (Agar and Durham, 2003). Although the full repertoire of TDP-43 nuclear targets awaits further investigation, the elucidation of redox-regulated TDP-43 provides a molecular framework to better understand TDP-43 as a critical stress-responsive RNA-associated factor. Moreover, these studies suggest that antioxidant-based therapies could have neuroprotective effects by increasing TDP-43 solubility and protein function in individuals with ALS, FTLD-TDP, and related TDP-43 proteinopathies.
Materials and methods
Plasmids and cell culture
Human TDP-43 was cloned into pCDNA5/TO vector (Invitrogen) and site-directed mutagenesis (Quikchange kit; Stratagene, La Jolla, CA) was used to create all Cys → Ser or Cys → Ala (data not shown) mutations at residues C173, C175, C198, C244 in the combinations indicated. Plasmids were transfected into QBI-293 cells using Fugene-6 (Roche) as per the manufacturer's protocols. QBI-293 and Cos7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, and 1% L-glutamate. TDP-43 siRNA (Qiagen) sequence was as follows: AACACTACAATTGATATCAAA and was transfected using RNAi Max reagent (Invitrogen) following the manufacturer's protocols. Cells were exposed to oxidative stressors in the following manner: 1 mM H2O2 for 1 h, 10 mM cadmium sulphate for 4 h, 0.1–0.25 mM sodium arsenite for 1 h (western blotting) or heat shock at 42°C for 1 h. Primary neuronal cultures were prepared from C57BL/6 mouse brains (Charles River, Wilmington, MA) following the NIH Guide for the Care and Use of Experimental Animals and were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Dissociated hippocampal neurons were plated onto poly-D-lysine-coated plates and treated with 0.25 mM sodium arsenite, where indicated, prior to lysis and protein extraction.
Immunocytochemistry and immunohistochemistry
Control and FTLD-TDP human brain sections were processed and analysed as previously described (Neumann et al, 2006). Cultured cells were fixed in 4% paraformaldehyde (PFA) for 10 min, rinsed 3 × in phosphate-buffered saline (PBS), and permeabilized with 0.2% Triton X-100 (Sigma) in PBS for 10 min. Cells were then blocked in 5% milk for 1 h and subsequently incubated with primary antibodies of interest overnight at 4°C following the standard immunofluorescence techniques. For Triton X-100 extraction, cells were left untreated or treated with 0.25 mM arsenite for 1 h followed by extraction with 1% Triton X-100 for 10 min where indicated, washed 3 × with PBS, and subsequently fixed with 4% PFA as described above. Triton extraction removed the majority of nuclear proteins, as assessed by immunostaining analysis of several unrelated RNA-binding proteins. Primary antibodies used for immunofluorescence were as follows: polyclonal anti-TDP-43 (Proteintech) 1:500, mouse anti-TIAR (BD Biosciences) 1:1000, polyclonal anti-myc (Sigma). Double labelling was performed using Alexa Fluor 488- and 594-conjugated secondary antibodies (Molecular Probes, Eugene, OR), and coverslipped with Vectashield mounting medium (Vector Laboratories). Digital images were obtained using an Olympus BX 51 microscope (Tokyo, Japan) equipped with bright-field and fluorescence light sources using a ProgRes C14 digital camera (Jenoptik AG, Jena, Germany) and Adobe Photoshop, version 9.0 (Adobe Systems, San Jose, CA) or digital camera DP71 (Olympus) and DP manager (Olympus).
Biochemical procedures
Fractionation of the human brain tissue was performed as previously reported (Neumann et al, 2006), with the notable addition of 50 mM iodoacetamide to all extraction buffers to prevent non-specific oxidation of proteins during processing. The human brain samples were analysed by reducing and non-reducing immunoblotting using commercial polyclonal anti-TDP-43 (Proteintech) or C-terminal anti-TDP-43 antibodies (polyclonal 2089). Fractionation of all cell lysates was performed by sequential extraction using buffers of increasing strength. Cells from six-well culture dishes were scraped into 300 ml RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40, 5 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS) containing 1 mM phenylmethylsulphonyl fluoride, a mixture of protease inhibitors (1 mg/ml pepstatin, leuptin, N-p-Tosyl-l-phenylalanine chloromethyl ketone, Nα-Tosyl-l-lysine chloromethyl ketone hydrochloride, trypsin inhibitor; Sigma), and a mixture of phosphatase inhibitors (2 mm imidazole, 1 mm NaF, 1 mm sodium orthovanadate; Sigma). Samples were sonicated and centrifuged at 100 000g for 30 min at 4°C, and then re-extracted in RIPA buffer to ensure complete removal of soluble proteins. Resultant insoluble pellets were extracted in 100 ml urea buffer (7 M urea, 2 M Thiourea, 4% CHAPS, 30 mM Tris pH 8.5), sonicated and centrifuged at 100 000g for 30 min at room temperature. Soluble and insoluble fractions were analysed by either reducing (+DTT) or non-reducing (−DTT) western blotting using the indicated antibodies to detect TDP-43 protein. Antibodies for western analysis were as follows: rabbit polyclonal anti-TDP-43 (Proteintech) 1:1000, mouse monoclonal antibody (mAb) anti-TDP-43 (Proteintech) 1:1000, rabbit polyclonal anti-TDP-43 C-terminal 1038 1:10,000, rabbit polyclonal anti-TDP-43 N-terminal 1065 1:10 000, anti-GAPDH (6C5 mouse mAb; Advanced ImmunoChemical Inc.) 1:3000, anti-acetylated tubulin (Sigma) 1:1000, anti-α-tubulin (Sigma) 1:1000, anti-HDAC6 (H-300; Santa Cruz) 1:1000.
Splicing analysis
TDP-43 functional activity was assessed using a CFTR splicing assay. Either control (pcDNA-5TO vector), TDP-43 over-expression constructs, or TDP-43 shRNA construct (Origene) were transiently transfected into QBI-293 cells using Lipofectamine 2000 reagent (Invitrogen) following the standard manufacturer's protocols. After 48 h, a hybrid minigene construct TG(13)T(5) (a generous gift from Dr F Baralle, International Centre for Genetic Engineering and Biotechnology, Trieste, Italy), which monitors CFTR exon 9 splicing was transiently transfected into the same cells. Twelve hours after CFTR minigene transfections, cells were treated with 50 mM arsenite and 10 mM NAC, where indicated, for an additional 12 h prior to cell harvest. CFTR exon 9 inclusion versus exclusion in the presence of the indicated TDP-43 plasmids was then evaluated by RT–PCR from isolated total RNA prepared from cells 72 h after transfection of TDP-43 constructs and 24 h after transfection of the TG(13)T(5) CFTR minigene reporter construct. The primers used were Bra2, TAGGATCCGGTCACCAGGAAGTTGGTTAAATCA; a2–3, CAACTTCAAGCTCCTAAGCCACTGC. PCR conditions were as follows: 95°C for 10 min (hot start), followed by 30 cycles of denaturing at 95°C for 30 s, annealing at 57°C for 30 s, and elongation at 72°C for 60 s. The PCR products were visualized on a 1.5% agarose gel as shown. Exon 9 splice products were quantified using the Agilent 2100 Bioanalyzer on a DNA 1000 chip and values were calculated as log ratios of spliced/unspliced products.
Recombinant TDP-43 methods
TDP-43 recombinant proteins (WT, C173/175S, C173/175/198/244S, and panel of mutant forms of TDP-43 found in ALS patients with TARDP mutations) were purified from BL21 cells using the pCOLD vector expression system (Takara Bio Inc.). After protein induction, all TDP-43 proteins were present predominantly in insoluble fractions of bacterial lysates. To generate soluble purified TDP-43 protein, bacterial pellets were washed extensively 5 × in RIPA buffer to remove soluble proteins, and insoluble pellets containing TDP-43 proteins were subsequently washed repeatedly in 0.1% SDS, 0.1 M MOPS pH 7.5 and TDP-43 solubility was monitored until total protein was solubilized. Re-solubilized TDP-43 proteins were estimated to be ∼95% pure based on visualization by Coomassie staining. Protein stocks were maintained at high concentration (10 mg/ml) in 0.1% SDS, 0.1 M MOPS pH 7.5 to prevent loss of TDP-43 solubility and diluted a minimum of 1000-fold in PBS for subsequent experiments. For cross-linking experiments, TDP-43 proteins were diluted into PBS to a final concentration of 10 ng/ml and H2O2 treated (1–10 mM, 15°C) for 10 min. Where indicated, EGS (ethylene glycol bis(succinimidyl succinate)) (Sigma) was added to a final concentration of 50 mM, followed by incubation at 22°C for 15 min. After quenching of the cross-linking reactions with excess glycine, samples were resolved by electrophoresis and analysed by immunoblotting with polyclonal anti-TDP-43 1038 antibody. For [14C]-iodoacetate labelling, WT TDP-43 protein was treated with H2O2 for 10 min at 15°C and then incubated with [14C]-iodoacetic acid (final concentration of 2.4 mM, 7.9 mCi/mmol; Perkin-Elmer) for 30 min. The carboxymethylation reaction was quenched by a 5-min incubation in the presence of 100 mM DTT, and the samples were resolved by electrophoresis, electrotransfer to nitrocellulose membrane, and PhosphorImaging using STORM software.
In vitro and cell-based mass spectrometry analysis
For in vitro mass spectrometry analysis, 1 μg recombinant WT TDP-43 was left untreated or exposed to 1 mM peroxide for 10 min at 15°C. Samples were analysed by non-reducing SDS–PAGE electrophoresis and visualized by Coomassie staining. Monomeric and HMW multimeric TDP-43 were gel excised and analysed by nanoLC/nanospray/MS/MS using the University of Pennsylvania proteomics core facility. Data were acquired using Xcalibur software (Thermo Scientific) and analysed using Mascot software (Matrix Science). Mass Matrix software (Case Western University) was used to confirm disulphide-bonded peptides, and to generate statistical significance and heat map data. For cell-based analysis, 10 cm dishes of QBI-293 cells were transiently transfected with WT TDP-43, and 48 h later treated with 0.25 mM arsenite for 1 h. Soluble (RIPA) and insoluble (Urea) lysates were prepared and total TDP-43 was immunoprecipitated from these fractions using anti-TDP-43 clone 171 complexed to protein A/G beads (Sigma). Urea solubilized fractions were diluted 10-fold in Triton buffer (1% Triton X-100, 40 mM Tris pH 8.0) prior to immunoprecipitation reaction. Immunoprecipitated samples were analysed by reducing or non-reducing SDS–PAGE electrophoresis and Coomassie staining. Gel excised TDP-43 monomer and HMW protein bands were similarly analysed by nanoLC/nanospray/MS/MS in the absence of reducing agents.
Quantitative RT–PCR
Cells were exposed to 50 μM arsenite overnight, where indicated, to allow full inactivation of TDP-43 for subsequent analysis of TARDBP, HDAC6, and HSP70 mRNA levels. Total RNA was prepared from cell lysates using RNA easy (Qiagen) and cDNA synthesis kits (Superscript III; Invitrogen). QRT–PCR was performed using the Applied Biosystems 7900HT Fast Real-Time PCR system. PCR was carried out using either Sybr Green or the Taqman Gene Expression System (Applied Biosystems). Q-PCR conditions used were 95°C for 10 min, followed by 40 cycles of denaturing at 95°C for 15 s, and annealing/extension at 60°C for 1 min, in a 20-μl reaction volume. All primers for QRT–PCR were ordered from Applied Biosystems (Taqman Gene Expression Assay System) and assessed for comparable efficiency using a standard dilution curve. Primer information is as follows: Cyclophilin A (PPIA) Hs99999904_m1 (Applied Biosystems), β-actin (BACT) 4352935E (Applied Biosystems), and TDP-43 Hs00540114_s1. β-Actin and cyclophilin A were used as reference genes. Other primers used were as follows: HDAC6 forward: CGCACAGGGCTGGTCTATG, HDAC6 reverse: TGGTGGCTGTCCCACAAGTT, HSPa1a (HSP70) forward: GGTGCTGATCCAGGTGTACGA, HSPa1a reverse: GCGCCCCAACAGATTGTT.
Supplementary Material
Acknowledgments
We thank Dr Linda Kwong for critical reading of this manuscript. We thank Dr Lionel Igaz for technical assistance and insightful discussions and Dr Chao-Xing Yuan for assistance with mass spectrometry analysis. The mass spectrometry analysis was provided by Proteomics Core Facility, University of Pennsylvania, supported by Grant P30CA016520 (Abramson Cancer Center), and by Grant ES013508–04 (CEET). This study was supported by grants from the National Institutes of Health (AG17586, AG32953), the Association for Frontotemporal Degeneration (TJC), the Koller Foundation for ALS Research, the David S and Emily Scott Pottruck ALS Program, as well as a gift from the Podolin family. VMYL is the John H Ware III Chair in Alzheimer's Disease Research, and JQT is the William Maul Measey-Truman G Schnabel, Jr Chair of Geriatric Medicine and Gerontology.
Author contributions: TJC performed all in vitro studies, cell-based experiments, human IHC and tissue extraction, and mass spectrometry analysis. AHW assisted with human tissue extraction and immunoblotting. TU performed the CFTR splicing assay, quantification of splicing changes, and assisted with quantitative RT–PCR analysis. VMYL supervised and designed the experiments. TJC, JQT, and VMYL were involved in the writing of this study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agar J, Durham H (2003) Relevance of oxidative injury in the pathogenesis of motor neuron diseases. Amyotroph Lateral Scler Other Motor Neuron Disord 4: 232–242 [DOI] [PubMed] [Google Scholar]
- Ayala YM, De Conti L, Avendaño-Vázquez SE, Dhir A, Romano M, D'Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, Baralle FE (2011) TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J 30: 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE (2005) Human Drosophila and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol 348: 575–588 [DOI] [PubMed] [Google Scholar]
- Ayala YM, Zago P, D'Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE (2008) Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci 121: 3778–3785 [DOI] [PubMed] [Google Scholar]
- Benoit RM, Meisner NC, Kallen J, Graff P, Hemmig R, Cèbe R, Ostermeier C, Widmer H, Auer M (2010) The x-ray crystal structure of the first RNA recognition motif and site-directed mutagenesis suggest a possible HuR redox sensing mechanism. J Mol Biol 397: 1231–1244 [DOI] [PubMed] [Google Scholar]
- Bose JK, Wang IF, Hung L, Tarn WY, Shen CK (2008) TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem 283: 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343 [DOI] [PubMed] [Google Scholar]
- Buratti E, Dörk T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20: 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese V, Cornelius C, Mancuso C, Lentile R, Stella AM, Butterfield DA (2010) Redox homeostasis and cellular stress response in aging and neurodegeneration. Methods Mol Biol 610: 285–308 [DOI] [PubMed] [Google Scholar]
- Caragounis A, Price KA, Soon CP, Filiz G, Masters CL, Li QX, Crouch PJ, White AR (2010) Zinc induces depletion and aggregation of endogenous TDP-43. Free Radic Biol Med 48: 1152–1161 [DOI] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, Ratti A (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem 111: 1051–1061 [DOI] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P 3rd, Good SK, Johnson BA, Herz J, Yu G (2011) TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol 31: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiesel FC, Voigt A, Weber SS, Van den Haute C, Waldenmaier A, Görner K, Walter M, Anderson ML, Kern JV, Rasse TM, Schmidt T, Springer W, Kirchner R, Bonin M, Neumann M, Baekelandt V, Alunni-Fabbroni M, Schulz JB, Kahle PJ (2010) Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J 29: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Chitta RK, High AA, Taylor JP (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res 9: 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458 [DOI] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, Malunda J, Xu Y, Winton MJ, Trojanowski JQ, Lee VM (2010) Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest 121: 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomova K, Vondrakova D, Lawson M, Valko M (2010) Metals, oxidative stress and neurodegenerative disorders. Mol Cell Biochem 345: 91–104 [DOI] [PubMed] [Google Scholar]
- Karch CM, Borchelt DR (2008) A limited role for disulfide cross-linking in the aggregation of mutant SOD1 linked to familial amyotrophic lateral sclerosis. J Biol Chem 283: 13528–13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Shanware NP, Bowler MJ, Tibbetts RS (2010) Amyotrophic lateral sclerosis-associated proteins TDP-43 and FUS/TLS function in a common biochemical complex to co-regulate HDAC6 mRNA. J Biol Chem 285: 34097–34105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. (2009) Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res 37: 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW (2009) Rethinking ALS: the FUS about TDP-43. Cell 136: 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 19: R46–R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, Cleveland DW (2010) ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci USA 107: 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B (2010) Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 5: e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W, Rouleau GA, Vande Velde C (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet 20: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133 [DOI] [PubMed] [Google Scholar]
- Pesiridis GS, Lee VM, Trojanowski JQ (2009) Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet 18: R156–R162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14: 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salajegheh M, Pinkus JL, Taylor JP, Amato AA, Nazareno R, Baloh RH, Greenberg SA (2009) Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve 40: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J, Moore MJ, Yu G (2011) Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem 286: 1204–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, Hayward LJ (2003) Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J Biol Chem 278: 5984–5992 [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14: 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KJ, Yang CH, Fang YH, Cho KH, Chien WL, Wang WT, Wu TW, Lin CP, Fu WM, Shen CK (2010) Elevated expression of TDP-43 in the forebrain of mice is sufficient to cause neurological and pathological phenotypes mimicking FTLD-U. J Exp Med 207: 1661–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT (2005) Metals, toxicity and oxidative stress. Curr Med Chem 12: 1161–1208 [DOI] [PubMed] [Google Scholar]
- Volkening K, Leystra-Lantz C, Yang W, Jaffee H, Strong MJ (2009) Tar DNA binding protein of 43 kDa (TDP-43), 14–3–3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res 1305: 168–182 [DOI] [PubMed] [Google Scholar]
- Wan L, Ottinger E, Cho S, Dreyfuss G (2008) Inactivation of the SMN complex by oxidative stress. Mol Cell 31: 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, Reddy NM, Shen CK (2002) Higher order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci USA 99: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, Wu LS, Chang HY, Shen CK (2008) TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem 105: 797–806 [DOI] [PubMed] [Google Scholar]
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH (2009) TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 106: 18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, Cuijt I, Smits V, Ceuterick-de Groote C, Van Broeckhoven C, Kumar-Singh S (2010) TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 107: 3858–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM (2008) Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem 283: 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Tanji K, Yoshida H, Hayakari M, Shibata T, Mori F, Uchida K, Wakabayashi K (2010) Alteration of biochemical and pathological properties of TDP-43 protein by a lipid mediator, 15-deoxy-Delta(12,14)-prostaglandin J(2). Exp Neurol 222: 296–303 [DOI] [PubMed] [Google Scholar]
- Zumbrennen KB, Wallander ML, Romney SJ, Leibold EA (2009) Cysteine oxidation regulates the RNA-binding activity of iron regulatory protein 2. Mol Cell Biol 29: 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.