Abstract
The minimal RNA synthesis machinery of non-segmented negative-strand RNA viruses comprises a genomic RNA encased within a nucleocapsid protein (N-RNA), and associated with the RNA-dependent RNA polymerase (RdRP). The RdRP is contained within a viral large (L) protein, which associates with N-RNA through a phosphoprotein (P). Here, we define that vesicular stomatitis virus L initiates synthesis via a de-novo mechanism that does not require N or P, but depends on a high concentration of the first two nucleotides and specific template requirements. Purified L copies a template devoid of N, and P stimulates L initiation and processivity. Full processivity of the polymerase requires the template-associated N protein. This work provides new mechanistic insights into the workings of a minimal RNA synthesis machine shared by a broad group of important human, animal and plant pathogens, and defines a mechanism by which specific inhibitors of RNA synthesis function.
Keywords: de-novo RNA synthesis initiation, negative-strand RNA viruses, ribavirin, RNA-dependent RNA polymerases, vesicular stomatitis virus
Introduction
RNA-dependent RNA polymerases (RdRPs) catalyse replication and transcription of RNA virus genomes and as such are central to their replicative cycle. RdRPs, however, lack obvious proofreading capacity with the result that their error rates are three orders of magnitude higher than DNA-dependent polymerases. Maintaining the integrity of the viral genome during exponential genome replication demands that RdRPs correctly initiate synthesis. There are two distinct mechanisms by which RdRPs initiate (Paul et al, 1998; Kao et al, 2001). In one mechanism, the RdRP initiates synthesis de-novo by forming a phosphodiester bond between the initiation nucleoside triphosphate (NTPi) and a second nucleoside triphosphate (NTP). Examples of RdRPs that utilize this mechanism are those of hepatitis C virus (HCV) and rotavirus (Kao et al, 1999, 2001; Chen and Patton, 2000; Zhong et al, 2000). The second mechanism requires a nucleic acid or protein primer to initiate (Paul et al, 1998). The polymerase of influenza A virus, for example, either cleaves host cell pre-mRNA cap structures to serve as a primer for transcription (Plotch et al, 1981), or initiates de-novo during RNA replication. Evidence has also accumulated that a dinucleotide pppApG primer is synthesized internally on the template to prime replication (Deng et al, 2006). Similarly, such prime and realign mechanisms exist for members of the families Arenaviridae and Bunyaviridae (Garcin et al, 1995; Kranzusch et al, 2010).
Atomic structures of RdRPs from positive-strand RNA viruses and dsRNA viruses have shown that they share a common overall fold and that there are similarities in the mechanism of catalysis (for a review, see Bressanelli et al, 1999; Butcher et al, 2001; Tao et al, 2002; O’Farrell et al, 2003; Salgado et al, 2004; van Dijk et al, 2004 and Ferrer-Orta et al, 2006). RdRPs contain two specific channels: (i) a well-defined ‘template tunnel’ into which the template binds and (ii) a small positively charged tunnel involved in the interaction with the NTPi and the incoming NTP during elongation. Most RdRPs therefore require specific interaction with both the 3′ end of the template and the NTPi to correctly initiate RNA synthesis, and high concentrations of the NTPi usually stimulate de-novo initiation (Lohmann et al, 1999; Nomaguchi et al, 2003).
The polymerases of non-segmented negative-strand (NNS) RNA viruses are widely thought to initiate by a de-novo mechanism. Direct tests of this and a characterization of the mechanism are, however, lacking. The RNA synthesis machinery of the NNS RNA viruses is a ribonucleoprotein (RNP) complex, our understanding of which has been largely shaped by studies of vesicular stomatitis virus (VSV). In the RNP complex, the genomic RNA is completely coated by the viral nucleoprotein (N) to form an N-RNA template for transcription and replication by the polymerase complex. Atomic structures of the N-RNA complex of VSV and other NNS RNA viruses reveal that the RNA is sequestered between the N- and C-terminal lobes of the viral nucleocapsid protein (Albertini et al, 2006; Green et al, 2006). Those structures suggest that the RNA must transiently dissociate from N to allow polymerase access to the bases. How this dissociation occurs, and how polymerase loads onto the RNA to initiate synthesis are uncertain. It is also unclear whether the template-associated N plays a direct role in mediating viral RNA synthesis. The enzymatic subunit of the polymerase is contained within a 241-kDa large protein (L) which also possesses a GDP:polyribonucleotidyltransferase that caps the mRNA, and a dual specificity mRNA cap methyltransferase (Sleat and Banerjee, 1993; Li et al, 2005, 2006, 2008; Ogino and Banerjee, 2007). Although L contains all the enzymatic activities for RNA synthesis, it requires a 29-kDa phosphoprotein (P) to engage the N-RNA template. Engagement of the N-RNA is mediated by a C-terminal domain of P (PCTD) which binds between adjacent N-monomers on the template (Green and Luo, 2009). Atomic structures of L are currently unavailable, although a recent electronic microscopic (EM) analysis reveals that VSV L contains a core ring-like domain to which the RdRP maps, and an appendage involved in mRNA capping (Rahmeh et al, 2010). On complex formation with P, L undergoes a conformational rearrangement that is readily visible by EM. Whether such rearrangements themselves are required for engagement and or copying of the N-RNA template is unknown.
Existing systems to study RNA synthesis in-vitro for NNS RNA viruses are limited by the necessity to purify the N-RNA template from viral particles or from cells. As a consequence, only templates that are replication competent can be examined, and this greatly hampers our understanding of the mechanism by which the polymerase initiates. In the present study, we examine the mechanism of initiation of RNA synthesis for VSV, by establishing a simple in-vitro system. We show that purified recombinant L uses naked RNA without the need for either N or P. VSV L preferentially initiates RNA synthesis de-novo at the 3′ terminus of the Leader (Le) RNA. This initiation depends upon a specific requirement for both the NTPi and the template, and is further enhanced by formation of the first phosphodiester bond. By comparing the activity of L, with the L–P complex we define that P plays a separate role in facilitating L initiation independent of its need to engage N protein. We also demonstrate that P acts directly on L to enhance processivity and that the template-associated N protein is required for the full processivity of the L–P complex. This new in-vitro system represents a powerful method for understanding how inhibitors of RNA synthesis function, and we test this directly with ribavirin triphosphate (RTP). The simplicity of this system should readily translate to the study of any NNS RNA virus polymerase.
Results
A minimal in-vitro RNA synthesis assay for VSV
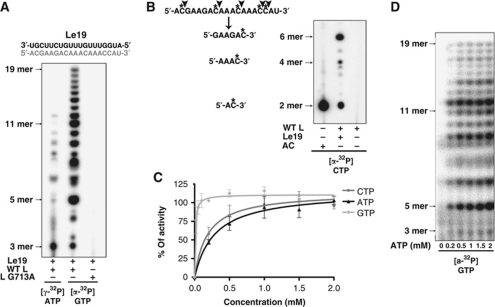
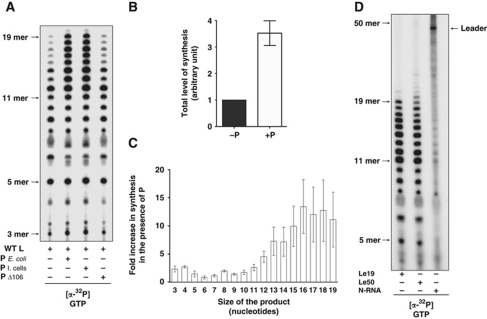
The RNA synthesis machinery of NNS RNA viruses comprises an encapsidated template (N-RNA) and a polymerase complex (L–P). We hypothesized that N and P play largely structural roles and thus we sought to develop a simple in-vitro RNA synthesis assay, using only the enzyme and substrate components: L and naked RNA. The 3′ end of the Le region contains essential signals for RNA synthesis initiation. Therefore, we chemically synthesized RNA corresponding to the first 19 nt of the genome (Le19) and used this as template for RNA synthesis in-vitro. In the absence of either N or P, L initiates RNA synthesis on the Le19 RNA (Figure 1A). The products of RNA synthesis ranged in size from 3 to 19 nt suggesting that L was not fully processive on naked RNA. Reaction products were readily visualized following 30 min of incubation and steadily increased for up to 2–3 h with no apparent differences in the mobilities of the accumulated products (Supplementary Figure S1). To verify that the RNA products depend upon a functional RdRP, we employed a catalytically inactive L G713A (Sleat and Banerjee, 1993). As expected, reactions reconstituted with L G713A were defective for RNA synthesis (Figure 1A). The products of RNA synthesis represent de-novo initiation as they were labelled by incorporation of [γ32P]-ATP (Figure 1A). To examine whether the products correspond to initiation at position 1 of the genome and were template dependent, reactions were performed in the presence of [α32P]-CTP and digested with RNase A, an endoribonuclease that cleaves 3′ of C and U residues. RNase digestion of transcripts that initiate at position 1 should yield up to five labelled products containing a single labelled (*) nucleotide AC*, GAAGAC*, AAAC*, AAAC*, and C*, corresponding to positions 1–2, 3–8, 9–12, 13–16, and position 17 of the template, respectively. RNase A digestion yielded a 2-nt RNA that co-migrates with the AC marker, a 6-nt RNA corresponding to positions 3–8, and lesser quantities of a 4-nt RNA corresponding to AAAC template by positions 9–12 and 13–16 (Figure 1B, lanes 1 and 2). No products were observed in the absence of template (Figure 1B, lane 3), indicating that the polymerase cannot synthesize short primers in the absence of template. Collectively, these data demonstrate that L initiates RNA synthesis by a de-novo mechanism, predominantly at the 3′ end of the template. These results also show that RNA synthesis does not require P or the template-associated N protein, however, the appearance of RNA <19 nt demonstrates that the L is not fully processive on naked RNA (Figure 1A).
Figure 1.
Development and optimization of an in-vitro RNA synthesis assay for VSV L. (A) De-novo RNA synthesis by L on an Le19 RNA template. Sequence of Le19 (black) and its expected synthesis product (grey) are indicated on top. Activity assays were set up with WT L or a catalytically inactive G713A mutant as described in Materials and methods, using 0.2 μM of Le19, 0.2 μM of protein and either [α32P]-GTP or [γ32P]-ATP. Reactions were quenched by the addition of EDTA/formamide and analysed on a 20% polyacrylamide/7 M urea gel. De-novo initiation product sizes are indicated on the left. (B) RNase A digestion of the synthesized products. The full-length sequence of the expected synthesis product is shown, stars indicate radiolabelled nucleotides and arrows RNase A cleavage sites (left). Expected RNase A cleavage products are indicated below. RNase A cleavage products were resolved on a 20% polyacrylamide/7 M urea gel (right). An AC RNA was labelled with T4 PNK, using [γ32P]-ATP, as a control. (C) Effect of increasing concentrations of ATP, CTP, and GTP on RNA synthesis. Reactions were performed as above with [α32P]-GTP and variable concentration of ATP or CTP. For titration of GTP, products were labelled with [α32P]-CTP. The total amount of synthesis was quantified by summing the band intensities using a PhosphorImager. The activity curves are derived from three independent experiments. Error bars represent the standard deviation from the mean of independent experiments. (D) Effect of increasing ATP concentrations on RNA synthesis. Reactions were performed as above in the presence of [α32P]-GTP and the indicated concentration of ATP. De-novo initiation product sizes are indicated on the left.
High concentrations of the first two NTPs stimulate initiation
To characterize the mechanism of de-novo initiation, we varied the concentration of the NTPs in the reaction. For VSV (Testa and Banerjee, 1979), RNA synthesis on the N-RNA template is stimulated by high concentrations of ATP, which is also the initiating nucleotide. Varying the concentration of ATP from 0 to 2 mM, we find that initiation on naked RNA is stimulated at >500 μM of ATP as well as similarly high concentrations of the second nucleotide, CTP (Figure 1B and C). By contrast, only low concentrations of the third nucleotide GTP >20 μM were required for efficient initiation, and initiation was insensitive to the presence of UTP (Figure 1C). Collectively, these results support a model in which ATP and CTP play a role for initiation, likely by their interaction within the L active site for formation of a correct initiation complex.
Template requirements for initiation
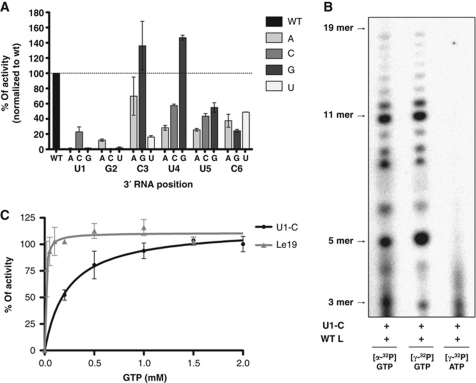
Crystallographic structures of RdRPs that initiate de-novo in complex with their RNA template reveal an interaction between the 3′ end of the template and the RdRP catalytic site as critical for formation of the initiation complex (Butcher et al, 2001; Tao et al, 2002; O’Farrell et al, 2003; Salgado et al, 2004). Recent work on the NNS RNA virus, respiratory syncytial virus (RSV) shows, however, that initiation can occur even with mutation or deletion of the 3′ terminal nucleotide (Noton et al, 2010). We therefore tested the importance of the 3′ terminal nucleotides in control of initiation and found that substitutions at any of the first 6 nt of the RNA affect initiation (Figure 2A). Positions 1 and 2 were most sensitive to substitution, as U1A, U1G, G2C, and G2U ablate the ability of L to initiate RNA synthesis, and U1C and G2A permit only low levels of initiation (Figure 2A). Substitution of U1C directs initiation at ∼20% of wild-type levels; therefore, we examined whether L could also initiate with GTP, instead of ATP. Such a purine preference for de-novo initiation has been documented for other RdRPs (Kao et al, 2001) that is reflective of a common evolutionary ancestry of these polymerases. Reactions carried out in the presence of [γ32P]-ATP or [γ32P]-GTP demonstrate that, on the U1C template, L initiates with GTP but not with ATP (Figure 2B). These results thus demonstrate that the de-novo initiation is templated and occurs at the 3′ end of the genome. Initiation on the U1C template was also stimulated by increasing the concentration of GTP, in a similar manner to the effect of ATP concentration on the wild-type template (Figures 1C and 2C). Collectively, these results demonstrate that the first two positions of the template, 3′-UG-5′, are crucial for initiation by L, with positions 3–6 influencing initiation. The results also support a specific interaction with the first two positions of the RNA to form an initiation complex. The initiation site of VSV L is therefore a major determinant of the template specificity.
Figure 2.
Mapping the requirements for initiation by L. (A) A saturation mutagenesis of the first six positions of Le19 was tested for de-novo RNA synthesis. Total RNA synthesis was quantified, normalized to levels of RNA synthesis on WT Le19 and graphed. Error bars represent the standard deviation from the mean of independent experiments. (B) De-novo RNA synthesis by L on U1C template with either [α32P]-GTP, [γ32P]-GTP or [γ32P]-ATP. (C) Effect of increasing GTP concentrations on RNA synthesis on Le19 and U1C template. Reactions were performed as described above and graphed as in Figure 1C. Error bars represent the standard deviation from the mean of independent experiments.
The minimal template requirements for initiation
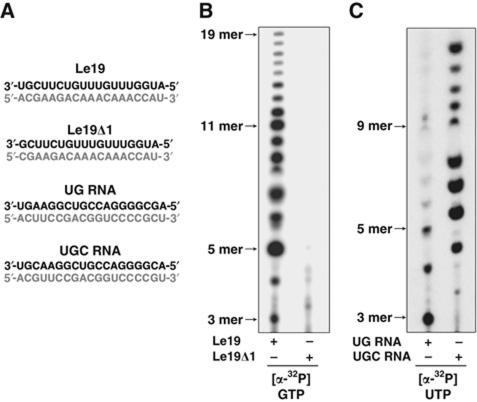
The above experiments demonstrate that VSV L has specific template requirements to initiate RNA synthesis. To further delineate the minimal template requirements for VSV L, we next tested whether L can initiate on a template lacking the first U. Deletion of position 1 (Le19Δ1) ablates RNA synthesis (Figure 3A and B), further underscoring that the 3′ terminal U is essential for the de-novo initiation mechanism. We next compared the ability of L to use random synthetic RNA onto which the 3′ terminal sequence UG or UGC was appended (Figure 3A). In both cases, L initiates RNA synthesis, but the 3′-UGC template directs higher levels of RNA synthesis than the 3′-UG (Figure 3C). These results demonstrate that the 3′-UG is sufficient to direct initiation of RNA synthesis, but synthesis is favoured by 3′-UGC. The elongation properties of L differ on the two templates, with 3′-UGC directing the synthesis of longer products. This result suggests that while 3′-UG is a minimal initiation site for L, the presence of a C residue at the third position stabilizes the initiation complex.
Figure 3.
The minimal requirements for initiation. (A) A Le19 template deleted at the first position (Le19Δ1) or random RNA templates starting with the two (UG RNA) or three (UGC RNA) nucleotide of Le were designed. Sequences of different templates (black), and their expected synthesis product (grey) are shown. (B) Comparison of de-novo RNA synthesis by L on Le19 and Le19Δ1. (C) De-novo RNA synthesis by VSV L on UG RNA or UGC RNA templates.
Internal initiation is not favoured
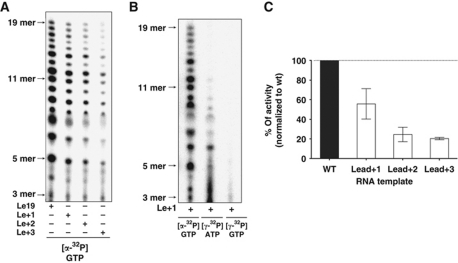
RdRPs can use two different mechanisms to initiate RNA synthesis de-novo: 3′ terminal initiation or internal initiation. The results described above show that VSV L initiates at the 3′ end of the RNA. To further test whether L can also initiate internally, we appended additional non-viral sequence onto the 3′ end of the RNA template. Addition of one (Le+1), two (Le+2), or three (Le+3) nucleotide to the 3′ end was tolerated for initiation (Figure 4A). The products of RNA synthesis were identical for each template, demonstrating that the nucleotides incorporated during synthesis are the same. Moreover, as the longest RNA product did not extend beyond 19 nt these results strongly suggest that L initiates internally on the first U of the template. In support of this, reactions performed in the presence of [γ32P]-ATP or [γ32P]-GTP demonstrate that ATP is the initiating nucleotide (Figure 4B). The efficiency of initiation was, however, progressively diminished as the first U was moved further from the 3′ end of the template (Figure 4C). Collectively, these results show that L initiates internally at the position of the first Le nt, but the efficiency of initiation is progressively reduced as the initiating nucleotide is displaced further from the 3′ end of the RNA.
Figure 4.
VSV L can initiate internally. (A) De-novo RNA synthesis by VSV L on Le19 and Le19 with addition of one (Le+1), two (Le+2), or three (Le+3) nucleotides at the 3′ end. (B) De-novo RNA synthesis by VSV L on Le+1 with [α32P]-GTP, [γ32P]-GTP, or [γ32P]-ATP. (C) Total amount of product synthesized in (A) was quantified with a PhosphorImager, normalized, and graphed. Error bars represent the standard deviation from the mean of independent experiments.
P enhances RNA synthesis initiation and processivity, and the template-associated N protein is required for full processivity
P is the essential cofactor that bridges the interaction between L and N-RNA, and is also generally thought to serve as a processivity factor for RNA synthesis. Evidence that P directly influences the activities of L in RNA synthesis, beyond its critical role in bringing L to the template is however lacking. To investigate the effect of P on RNA synthesis initiation by L, we compared initiation of the L–P complex with initiation by L (Figure 5A). Quantitative analysis of the products of the reaction reveals that the L–P complex produces four-fold more RNA than L alone, whereas a P mutant lacking the L binding domain is unable to enhance RNA synthesis (Figure 5A and B). This result demonstrates that P plays more than a structural role in recruiting L to the N-RNA template, and directly enhances the ability of L to initiate RNA synthesis. Furthermore, the L–P complex appears significantly more processive than L alone, since we observe an increase in the abundance of the larger products (Figure 5C). Quantitative analysis of each product reveals that P increases the abundance of products <12-nt by 4-fold and >12 nt by 12-fold.
Figure 5.
P stimulates L initiation and processivity, and template-associated N is required for full processivity. (A) De-novo RNA synthesis by VSV L on Le19 with or without P. P protein was produced in E. coli or Sf21 cells. A deletion mutant P Δ106 lacks the ability to bind L. (B) Total amount of synthesis was quantified, normalized, and graphed. (C) Individual reaction products of different sizes were quantified and the ratio of the products generated with P over those without P was determined. Error bars represent the standard deviation from the mean of independent experiments. (D) RNA synthesis by L–P on naked 19 (Le19) and 50 (Le50) nucleotides long RNAs, and the encapsidated RNA (N-RNA) templates.
To determine the role of the template-associated N protein in polymerase processivity, we compared RNA synthesis by L–P on naked and encapsidated templates. As observed for the 19-nt template, the L–P complex is not fully processive on a naked RNA template (Le50) corresponding to the first 50 nt of the genome with few products visualized beyond 21 nt (Figure 5D). By contrast, the L-P complex is fully processive on the viral N-RNA template producing the 47-nt leader RNA from the first 50 nt of the genome with only minor amounts of prematurely terminated products (Figure 5D). Collectively, these results show that P enhances both the initiation and processivity of L, and the template-associated N protein is required for full processivity during RNA synthesis.
Inhibition of RNA synthesis by RTP
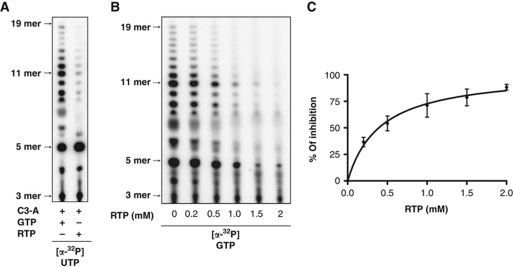
Ribavirin, a guanosine analogue, is a broad-spectrum antiviral whose mechanism of action has been hotly debated, and its impact on replication of NNS RNA viruses is unclear (Graci and Cameron, 2006; Leyssen et al, 2008). Among the cellular targets of ribavirin is inosine monophosphate dehydrogenase (IMPDH), inhibition of which results in a suppression of the intracellular pools of GTP. However, ribavirin is also converted into the triphosphate (RTP) where it can act to directly inhibit viral polymerase and can also be incorporated into genomic RNA by some viral RdRPs resulting in error catastrophe. To examine the mechanism of action of RTP on an NNS RNA viral RdRP, we used our in-vitro assay and a C3A template (Figure 2A), which supported efficient initiation while allowing us to clearly evaluate the incorporation of RTP at the sixth position of the RNA. Our results demonstrate that RTP is incorporated in the RNA resulting in the accumulation of 5 nt transcripts, however, elongation beyond that length was dramatically reduced (Figure 6A). These results suggest that RTP exerts a dual effect both by inhibiting RdRP activity, and may also be incorporated at low levels into the product RNA, which could lead to error catastrophe. RNA synthesis reactions supplemented with increasing concentrations of RTP showed a progressive reduction in products with an IC50 of ∼400 μM (Figure 6B and C). This provides direct evidence that RTP has antiviral activity against the VSV polymerase, and emphasizes that the assay developed in this study presents a powerful method to screen polymerase inhibitors.
Figure 6.
Inhibition of RNA synthesis initiation. (A) De-novo RNA synthesis by VSV L on C3-A with [α32P]-UTP, and either GTP or RTP. (B) Inhibitory effect of increasing RTP concentrations on RNA synthesis by VSV L. (C) Total amount of products was quantified and the percentage of VSV L RNA synthesis inhibition by the different RTP concentrations was plotted. Error bars represent the standard deviation from the mean of independent experiments.
Discussion
The principal breakthrough provided in this study is the demonstration that the minimal functional unit for RNA synthesis of NNS RNA viruses comprises a genomic RNA and L protein with N and P playing regulatory roles to ensure authentic copying. This achievement permitted us, for the first time, to define the mechanism by which L initiates RNA synthesis, and also to define the role of P and the template-associated N protein in RNA synthesis. Our work shows that the VSV polymerase uses a template-dependent de-novo initiation mechanism, and that initiation at the 3′ end of the template is strongly favoured over internal initiation. This work additionally defines that P serves as a processivity factor, but is insufficient for full polymerase processivity. Full processivity of the P–L complex additionally requires the template-associated N protein. We exploited this new in-vitro system to show that RTP, a broad-spectrum antiviral agent, directly blocks the VSV polymerase. This work has implications for the evolution of NNS RNA viruses, as well as the development of inhibitors that target polymerase function in this important group of human, animal, and plant pathogens.
The minimal machinery for RNA synthesis in NNS RNA viruses
Early biochemical experiments established that the minimal RNA synthesis machinery of NNS RNA viruses comprises the N encased genomic RNA associated with the viral polymerase, an L–P complex (Emerson and Yu, 1975; Mellon and Emerson, 1978). The atomic structure of N-RNA complexes from VSV and rabies virus provided evidence that the RNA must somehow be dissociated from N for copying by the polymerase (Albertini et al, 2006; Green et al, 2006). The co-crystal structure of the PCTD of VSV with the N-RNA complex led to a model in which P brings L to the RNA template by binding directly between N molecules, and this interaction is perhaps also required to keep L associated with the N-RNA during copying (Green and Luo, 2009). By now providing the first direct evidence that L can actually use RNA in the absence of the N and P, we have defined the minimal RNA synthesis components as L and RNA. We conclude that while N and P play important roles in viral RNA synthesis they are not essential for template recognition or nucleotide polymerization by L. Our study thus provides support for the model that N is displaced from the template during polymerase copying. Since the L–P complex itself is not fully processive on naked RNA, our findings further demonstrate that the template-associated N represents an important processivity factor for the RdRP.
Template structures of N-RNA complexes from NNS and segmented negative-strand (SNS) RNA viruses reveal that in each case the RNA is sequestered between two lobes of N (Ruigrok et al, 2011). Electron microscopic analysis and nuclease sensitivity of purified N-RNA templates demonstrate the template of SNS viruses is more loosely organized than the tightly shielded template of NNS viruses. Previous work with purified polymerase complexes from SNS viruses has revealed that those polymerases can initiate on naked nucleic acid (Kranzusch et al, 2010). Here, our observations with VSV L and naked nucleic acid extend this paradigm and reveal that likely all negative-strand RNA viruses polymerase complexes are capable of RNA synthesis in the absence of N. RNA synthesis initiation must therefore begins with a shared mechanism of remodelling of the N-RNA template and direct exposure of the viral nucleic acid to the polymerase machinery.
Although in the present study, we show that VSV L can use naked RNA, we do not think that substantial stretches of RNA are exposed during RNA synthesis. Rather we favour the view that a small number of N molecules are displaced locally during copying of the template by L. Such a view is consistent with the fact that the template RNA of VSV is resistant to nuclease digestion even during transcription (Green et al, 2000; Iseni et al, 2000). Our recent electron microscopic characterization of VSV L protein provides a rough estimate of the dimensions of a single L molecule (Rahmeh et al, 2010). Three-dimensional reconstruction from EM images of L showed that the core ring-like domain to which the RdRP was localized is ≈50 Å in thickness. Although speculative, if L copies the N-RNA template as a monomer, and if the RNA passes through the ring, a maximum of 2–3 molecules of N protein would be displaced during transit by any single L protein molecule. Each N protein molecule of VSV binds 9 bases of RNA, suggesting that 18–27 bases could be transiently dissociated from N by a single molecule of L. Perhaps, such a transient displacement of N from the template could be concurrent with an alteration of the processivity of L either by direct N–L interactions or perhaps through dynamic interactions mediated by P.
Prior to this study, other work examined whether the VSV polymerase can use RNA lacking N protein (Moyer et al, 1991). In those earlier studies, the RNA templates examined were substantially longer than that which we have utilized here, including templates obtained from DI particles and full-length genomic RNA (Pattnaik et al, 1992). We strongly suspect that this reflects the fact that such large RNA molecules readily adopt a range of structures and that those will impede the ability of the polymerase to copy the naked template. Because we demonstrate that the L–P complex is not fully processive in the absence of the template-associated N protein, such full-length unencapsidated templates would not serve to permit the synthesis of full-length products. Our work therefore also suggests that one function of the template-associated N may be to keep the RNA relatively unstructured so that it can be efficiently copied by L. As such, the polymerase would only transiently displace a few molecules of N at a time, to limit the potential for structure formation within the template. It is noteworthy that L lacks any obvious helicase motifs, and seems to be unable to utilize N-RNA as template without the need for supplemental P protein.
In the context of the RNP complex, P is an essential polymerase cofactor for RNA synthesis. It has been demonstrated that P has a structural role involving physical interaction with N and L (Green and Luo, 2009; Rahmeh et al, 2010). Previous work suggested that P serves as a processivity factor for L (Emerson and Yu, 1975), but given its essential role in bringing L to the N-RNA template it was not clear precisely whether the function of P was limited to that role (Green and Luo, 2009). Using naked RNA, we show that in the presence of P, L generates four-fold more products. Moreover, P enhances both RNA synthesis initiation and processivity. This suggests that P, in addition to allowing L access to the RNA during transcription and replication, has direct functional effects on the enzymatic properties of L to enhance RNA synthesis. These results provide the first direct demonstration that P is a processivity factor for L independent of its key role in engagement of the template-associated N protein. Precisely, how this functions is unclear, however, our EM analysis revealed substantial conformational rearrangements of L on complex formation with P. Perhaps, such structural rearrangements lead to a more processive L protein.
Previous work has also shown that phosphorylation of P is essential for transcription and virus growth in vivo (Barik and Banerjee, 1992a, 1992b; Das and Pattnaik, 2004). In the present study, we employed P protein expressed and purified from either Escherichia coli or Spodoptera fugiperda 21 cells which are presumed to differ in their phosphorylation status. Both sources of P served to promote initiation and processivity of L (Figure 5A), suggesting that early steps in catalysis of RNA synthesis do not require P protein phosphorylation.
A de-novo mechanism for RNA synthesis initiation in NNS RNA viruses
A second major contribution of this work is to define the mechanism by which the VSV polymerase initiates. Initiation depends upon high concentrations of the first two nucleotides and their incorporation is strictly template dependent. Other RdRPs that initiate de-novo are known to have a higher Km for NTPi than the other NTPs (Testa and Banerjee, 1979; Gaal et al, 1997; Lohmann et al, 1999; Laurila et al, 2002; Nomaguchi et al, 2003), and the NTPi specifically binds the active site to be positioned correctly prior to the first nucleotidyl transfer (Bressanelli et al, 1999; Butcher et al, 2001; Tao et al, 2002; O’Farrell et al, 2003; Salgado et al, 2004). Our results show that VSV L needs specific interaction of both the NTPi (ATP) and the incoming NTP (CTP) to generate a stable initiation complex. This suggests that VSV L initiation complex may require a double base pair with the RNA template to be in a correct conformation and/or sufficient stability to start synthesis.
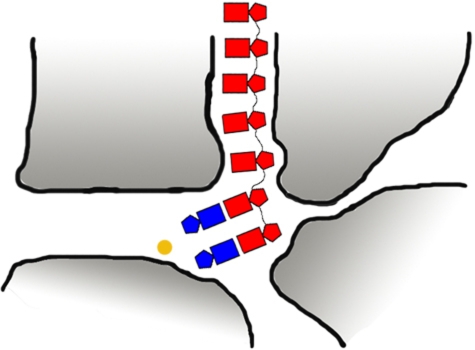
All RdRPs also contain a template channel, which regulates initiation complex formation. The first two positions of Le may interact specifically within the template tunnel and form one of the essential elements of the initiation complex. Although the third position of Le is not crucial for initiation, this position seems determinant in stabilization of the complex and enhances the efficiency of the initiation. We therefore propose that all three positions form important interactions in a template tunnel in L, with specific interactions with the incoming ATP and CTP (Figure 7). Nucleotides beyond this position (4–6) likely play a role in affecting polymerase/promoter affinity (Figure 2A).
Figure 7.
Model of VSV L de-novo initiation complex. The 3′ end of Le (red) gains access to the active site of L through a template tunnel and is stabilized by the interaction between the active site and its first 2 nt. The active site recognizes specifically the NTPi (ATP) and the second NTP (CTP) (blue) incorporated during initiation to create a double base pair between ATP, CTP and the UG nt at the 3′ end of Le, resulting in a stable initiation complex. RNA synthesis is initiated with formation of the first phosphodiester mediated by the divalent ion (yellow).
Recent studies on RSV show that its polymerase initiates even on templates in which the 3′ terminal nucleotide is mutated or deleted. Remarkably, the products of such initiation contain the authentic wild-type sequence (Noton et al, 2010). This led to the suggestion that the RSV L may be preloaded with the authentic initiating nucleotide. This interesting finding raises the question of whether the initiation mechanism used by VSV and RSV L are distinct, since we find for VSV that the position 1 efficiently templates the initiating nucleotide. One possible explanation for this difference is that the initiating nucleotides are indeed differently selected by the two polymerases. VSV utilizes a strictly templated mechanism whereas the template interactions for RSV are dictated by internal positions with the terminal nucleotide (ATP) being incorporated irrespective of template sequence. It would be of significant interest to reconstitute the same type of system as described here to study RNA synthesis initiation in RSV to test whether the initiation mechanism is distinct. Notably, we do not find evidence of non-templated initiation by the VSV L protein (Figure 1B) or evidence consistent with a preloading of polymerase with a non-templated nucleotide (Figures 2 and 4).
RdRPs can initiate synthesis de-novo at the 3′ terminus or in some cases internally on the template. Although we present evidence that L can initiate internally, this type of initiation is much less efficient than that at the 3′ terminus. This suggests that the active site of VSV L is sufficiently flexible to allow the 3′ end of the template to overshoot its position and still permit authentic initiation. Our findings that internal initiation is less favoured over 3′ initiation is consistent with prior work in which RNA synthesis was reconstituted on N-RNA templates with purified polymerase. In the presence of ATP and CTP, a dinucleotide product was generated that corresponds to initiation from the 3′ end of the genome and not a tetranucleotide product corresponding to internal initiation at a gene-start element (Emerson, 1982). Although evidence has accumulated that is consistent with the idea that RNA synthesis can also initiate internally at the first gene-start in infected cells, that study also found that in-vitro initiation was 3′ dependent (Whelan and Wertz, 2002). Our current system has not yet succeeded in recapitulating a fully processive enzyme on a naked RNA template (Figure 5D) and so we have been unable to study the process of sequential transcription. Our evidence is however incompatible with significant internal entry and initiation of polymerase, at least in-vitro.
For the SNS RNA viruses, a prime and realign mechanism for initiation is supported (Bouloy et al, 1990; Garcin and Kolakofsky, 1992; Kranzusch et al, 2010). In this model, the polymerase initiates internally on the template RNA to generate a dinucleotide primer, which subsequently realigns with the 3′ end of the template to promote elongation. Such a mechanism does not appear to operate in the case of VSV, since deletion of the terminal nucleotide completely blocks initiation and dinucleotide and trinucleotide products generated during in-vitro transcription do not chase into fully elongated products.
Determination of the mechanism by which inhibitors block RNA synthesis
The in-vitro assay developed here can be used to screen inhibitors specifically directed against the polymerase. As proof of concept, we show that RTP inhibits RNA synthesis by VSV L, through both inhibition of initiation and elongation. Importantly, the IC50 for RTP in this assay is relatively high and it may be difficult to reach such concentrations in vivo. For the positive-strand RNA virus, poliovirus (Crotty et al, 2000), as well as the ambisense RNA virus, LCMV (Moreno et al, 2011), evidence has accumulated that the polymerase can incorporate RTP in the genome, which leads to mis-incorporation of nucleotides during template copying and the inhibition of virus replication by ‘error catastrophe’. Such a mechanism may also operate to inhibit VSV and other NNS RNA viruses and would not be readily apparent in our assay, which reports on transcription of a short template. Ribavirin itself is also potent inhibitor of IMPDH, which likely also contributes to the reported antiviral activity against NNS RNA viruses (Graci and Cameron, 2006).
The approach described here to study VSV RNA synthesis should be readily adaptable to the study of other NNS RNA viruses obviating the need to purify the N-RNA template from infectious virus particles. The NNS RNA viruses contain some of the most significant human, animal, and plant pathogens extant; therefore, adaptation of this system will likely provide a powerful tool to screen for inhibitors of their polymerases.
Materials and methods
Protein expression and purification
Recombinant L and P were expressed as described in Rahmeh et al (2010). Briefly, 6 × His-tagged L was expressed in Spodoptera fugiperda 21 (Sf21) cells and affinity purified with Ni-NTA agarose (Qiagen) followed by ion exchange chromatography. In all, 10 × His-tagged P and P Δ106 were expressed in Escherichia coli BL21 (DE3) cells or Sf21 cells then affinity purified with Ni-NTA agarose (Qiagen) followed by gel filtration (Superdex 200 HR 10/30, GE Healthcare).
Template production
Naked RNA templates were chemically synthesized and purified (Integrated DNA Technologies). Genomic N-RNA templates were prepared as previously described (Rahmeh et al, 2010).
In-vitro VSV L polymerase assay
Standard polymerase assays were carried out using 0.2 μM of template with 0.2 μM of VSV L in a reaction mixture containing 20 mM Tris-base, pH 8, 50 mM NaCl, 2 mM DTT and 0.5% (vol/vol) Triton X-100, 6 mM MgCl2, 200 μM UTP, 1.5 mM ATP, 1.5 mM CTP and 165 nM of [α32P]-GTP (3000 Ci/mmol). Reactions were incubated at 30°C for 3 h, and stopped by the addition of EDTA/formamide. Reactions products were resolved using denaturing polyacrylamide gel electrophoresis (20% polyacrylamide, 7 M urea) in TBE buffer, and analysed by autoradiography. The sizes of the products were determined by comparison with a 19-nt marker RNA labelled by T4 polynucleotide kinase (PNK) (New England Biolabs) using [γ32P]-ATP (3000 Ci/mmol). Assays studying the effects of NTP concentration were carried out using the indicated concentrations. For monitoring the effect of varying GTP concentration, reactions were supplemented with 165 nM of [α32P]-ATP (3000 Ci/mmol). To determine the first nucleotide incorporated during initiation, standard reactions were carried out with 165 nM [γ32P]-GTP (3000 Ci/mmol) or [γ32P]-ATP (3000 Ci/mmol). On random RNA templates, AC RNA and ACG RNA, 200 μM of GTP, 1.5 mM of each, ATP and CTP and 165 nM of [α32P]-UTP (3000 Ci/mmol) were used. Internal initiation reactions were performed using 3′-AUGCUUCUGUUUGUUUGGUA-5′ (Le+1), 3′-GAUGCUUCUGUUUGUUUGGUA-5′ (Le+2), or 3′-GAAUGCUUCUGUUUGUUUGGUA-5′ (Le+3) as a template. The effect of P was examined by the addition of 0.2 μM of P to the standard reaction. The standard polymerase assay was supplemented with 250 μM GTP and 0.2 μM of P when N-RNA was used as a template. The effect of RTP was evaluated on the C3A template with 165 nM of [α32P]-UTP (3000 Ci/mmol), 1.5 mM of each, ATP and CTP and 200 μM of either GTP or RTP. Assays studying the effects of RTP concentrations were carried out by adding the indicating concentrations of RTP in the standard polymerase assay. All radioisotopes were purchased from Perkin-Elmer.
RNase A analysis of the products
Standard polymerase assays were carried out with 200 μM GTP and 165 nM of [α32P]-CTP (3000 Ci/mmol). Reactions were incubated at 30°C for 3 h, following by 5 min at 75°C and cooled down on ice. In all, 5 μM of RNase A was added to the reactions and incubated at 30°C for 1 h. RNA products were then purified by phenol/chloroform extraction and digested by five units of Antarctic Phosphatase (AP) (New England Biolabs), at 37°C for 1 h. Products were then incubated at 65°C for 10 min and phosphorylated at their 5′ end by T4 polynucleotide kinase (New England Biolabs) and ATP. A 2-nt long AC RNA was chemically synthesized, purified (Ambion) and labelled by T4 polynucleotide kinase using [γ32P]-ATP (3000 Ci/mmol). All reactions were stopped by the addition of EDTA/formamide and resolved on a 20% polyacrylamide/7 M urea gel, and analysed by autoradiography.
Supplementary Material
Acknowledgments
We acknowledge Philip J Kranzusch for critical discussion and review of the manuscript. This study was supported by NIH grants AI059371 and AI057159 to SPJW. SPJW is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award, and the Genzyme Innovators in Biomedical Research Award.
Author contributions: BM and SPJW designed experiments and analysed data; BM and AAR performed research; BM and SPJW wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW (2006) Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313: 360–363 [DOI] [PubMed] [Google Scholar]
- Barik S, Banerjee AK (1992a) Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein P. Proc Natl Acad Sci USA 89: 6570–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S, Banerjee AK (1992b) Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J Virol 66: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloy M, Pardigon N, Vialat P, Gerbaud S, Girard M (1990) Characterization of the 5′ and 3′ ends of viral messenger RNAs isolated from BHK21 cells infected with Germiston virus (Bunyavirus). Virology 175: 50–58 [DOI] [PubMed] [Google Scholar]
- Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA 96: 13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI (2001) A mechanism for initiating RNA-dependent RNA polymerization. Nature 410: 235–240 [DOI] [PubMed] [Google Scholar]
- Chen D, Patton JT (2000) De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA 6: 1455–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE (2000) The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med 6: 1375–1379 [DOI] [PubMed] [Google Scholar]
- Das SC, Pattnaik AK (2004) Phosphorylation of vesicular stomatitis virus phosphoprotein P is indispensable for virus growth. J Virol 78: 6420–6430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Vreede FT, Brownlee GG (2006) Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J Virol 80: 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson SU (1982) Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell 31 (3 Pt 2): 635–642 [DOI] [PubMed] [Google Scholar]
- Emerson SU, Yu Y (1975) Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol 15: 1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N (2006) A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol 16: 27–34 [DOI] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL Jr, Gourse RL (1997) Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278: 2092–2097 [DOI] [PubMed] [Google Scholar]
- Garcin D, Kolakofsky D (1992) Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol 66: 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D (1995) The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol 69: 5754–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graci JD, Cameron CE (2006) Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol 16: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Luo M (2009) Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci USA 106: 11713–11718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Macpherson S, Qiu S, Lebowitz J, Wertz GW, Luo M (2000) Study of the assembly of vesicular stomatitis virus N protein: role of the P protein. J Virol 74: 9515–9524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Zhang X, Wertz GW, Luo M (2006) Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313: 357–360 [DOI] [PubMed] [Google Scholar]
- Iseni F, Baudin F, Blondel D, Ruigrok RW (2000) Structure of the RNA inside the vesicular stomatitis virus nucleocapsid. RNA 6: 270–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CC, Del Vecchio AM, Zhong W (1999) De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253: 1–7 [DOI] [PubMed] [Google Scholar]
- Kao CC, Singh P, Ecker DJ (2001) De novo initiation of viral RNA-dependent RNA synthesis. Virology 287: 251–260 [DOI] [PubMed] [Google Scholar]
- Kranzusch PJ, Schenk AD, Rahmeh AA, Radoshitzky SR, Bavari S, Walz T, Whelan SP (2010) Assembly of a functional Machupo virus polymerase complex. Proc Natl Acad Sci USA 107: 20069–20074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurila MR, Makeyev EV, Bamford DH (2002) Bacteriophage phi 6 RNA-dependent RNA polymerase: molecular details of initiating nucleic acid synthesis without primer. J Biol Chem 277: 17117–17124 [DOI] [PubMed] [Google Scholar]
- Leyssen P, De Clercq E, Neyts J (2008) Molecular strategies to inhibit the replication of RNA viruses. Antiviral Res 78: 9–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Fontaine-Rodriguez EC, Whelan SP (2005) Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol 79: 13373–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Rahmeh A, Morelli M, Whelan SP (2008) A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol 82: 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang JT, Whelan SP (2006) A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci USA 103: 8493–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V, Overton H, Bartenschlager R (1999) Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J Biol Chem 274: 10807–10815 [DOI] [PubMed] [Google Scholar]
- Mellon MG, Emerson SU (1978) Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J Virol 27: 560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno H, Gallego I, Sevilla N, de la Torre JC, Domingo E, Martin V (2011) Ribavirin can be mutagenic for arenaviruses. J Virol 85: 7246–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SA, Smallwood-Kentro S, Haddad A, Prevec L (1991) Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J Virol 65: 2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomaguchi M, Ackermann M, Yon C, You S, Padmanabhan R (2003) De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: nucleotide, primer, and template parameters. J Virol 77: 8831–8842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton SL, Cowton VM, Zack CR, McGivern DR, Fearns R (2010) Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion. Proc Natl Acad Sci USA 107: 10226–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell D, Trowbridge R, Rowlands D, Jager J (2003) Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J Mol Biol 326: 1025–1035 [DOI] [PubMed] [Google Scholar]
- Ogino T, Banerjee AK (2007) Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell 25: 85–97 [DOI] [PubMed] [Google Scholar]
- Pattnaik AK, Ball LA, LeGrone AW, Wertz GW (1992) Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 69: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Paul AV, van Boom JH, Filippov D, Wimmer E (1998) Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393: 280–284 [DOI] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM (1981) A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23: 847–858 [DOI] [PubMed] [Google Scholar]
- Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SP (2010) Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA 107: 20075–20080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok RW, Crepin T, Kolakofsky D (2011) Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol 14: 504–510 [DOI] [PubMed] [Google Scholar]
- Salgado PS, Makeyev EV, Butcher SJ, Bamford DH, Stuart DI, Grimes JM (2004) The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure 12: 307–316 [DOI] [PubMed] [Google Scholar]
- Sleat DE, Banerjee AK (1993) Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol 67: 1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Farsetta DL, Nibert ML, Harrison SC (2002) RNA synthesis in a cage—structural studies of reovirus polymerase lambda3. Cell 111: 733–745 [DOI] [PubMed] [Google Scholar]
- Testa D, Banerjee AK (1979) Initiation of RNA synthesis in vitro by vesicular stomatitis virus. Role of ATP. J Biol Chem 254: 2053–2058 [PubMed] [Google Scholar]
- van Dijk AA, Makeyev EV, Bamford DH (2004) Initiation of viral RNA-dependent RNA polymerization. J Gen Virol 85 (Pt 5): 1077–1093 [DOI] [PubMed] [Google Scholar]
- Whelan SP, Wertz GW (2002) Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome. Proc Natl Acad Sci USA 99: 9178–9183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Uss AS, Ferrari E, Lau JY, Hong Z (2000) De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol 74: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.