Abstract
Appropriate number of neurons and glial cells is generated from neural stem cells (NSCs) by the regulation of cell cycle exit and subsequent differentiation. Although the regulatory mechanism remains obscure, Id (inhibitor of differentiation) proteins are known to contribute critically to NSC proliferation by controlling cell cycle. Here, we report that a transcriptional factor, RP58, negatively regulates all four Id genes (Id1–Id4) in developing cerebral cortex. Consistently, Rp58 knockout (KO) mice demonstrated enhanced astrogenesis accompanied with an excess of NSCs. These phenotypes were mimicked by the overexpression of all Id genes in wild-type cortical progenitors. Furthermore, Rp58 KO phenotypes were rescued by the knockdown of all Id genes in mutant cortical progenitors but not by the knockdown of each single Id gene. Finally, we determined p57 as an effector gene of RP58-Id-mediated cell fate control. These findings establish RP58 as a novel key regulator that controls the self-renewal and differentiation of NSCs and restriction of astrogenesis by repressing all Id genes during corticogenesis.
Keywords: cell cycle control, cortical development, cyclin-dependent kinase inhibitor, fate of NSCs, p57
Introduction
In the vertebrate central nervous system (CNS), different subtypes of neurons and glial cells are produced from neural stem cells (NSCs) at a precisely regulated order and timing, which can be replicated in the culture of NSCs in vitro (Qian et al, 2000; Shen et al, 2006; Guillemot, 2007; Miller and Gauthier, 2007). The enhancement of astrogenesis followed by neurogenesis is regulated by intrinsic and extrinsic cellular mechanisms working in harmony. Abnormal astrogenesis may be one of the causative factors that induce epilepsy, learning disabilities, and mental retardation (Sosunov et al, 2008; Napolioni et al, 2009). Tuberous sclerosis complex is a multisystem genetic disorder that involves enhanced astrogenesis including gliosis and brain tumour (Sosunov et al, 2008; Napolioni et al, 2009; Ess, 2010).
Among intrinsic regulators, the 4 Id genes (Id1, Id2, Id3, and Id4) are known to serve an important role in astrogenesis. Id overexpression inhibits neuronal differentiation while promoting cell proliferation and astrogenesis in vivo and in vitro (Cai et al, 2000; Jung et al, 2010). Several candidate molecules involved in the upregulation of Id genes in neuroepithelial cell culture or neural cell lines have been described, including bone morphogenic protein 2, fibroblast growth factor 2 (FGF2), and nerve growth factor (Nagata and Todokoro, 1994; Nakashima et al, 2001; Passiatore et al, 2011), which may underlie Id gene upregulation at the time of neural-lineage cell proliferation. However, the mechanism of the timely repression of Id genes upon NSCs exiting the cell cycle is poorly understood.
Downstream targets of Id proteins have also been reported. Several lines of evidence have implicated the involvement of cyclin-dependent kinase inhibitors, consisting of the Cip/Kip family (p21/Cip1, p27/Kip1, and p57/Kip2) and INK4 family (p16/INK4a, p15/INK4b, p18/INK4c, and p19/INK4d) in Id gene-mediated signalling (Cánepa et al, 2007; Joseph and Hermanson, 2010). Id proteins can inhibit transcription of p21, p57, and p16, which depend on the sequestration of type I and type II bHLH transcription factor or Ets domain proteins by Id proteins (Sun et al, 1991; Langlands et al, 1997; Prabhu et al, 1997; Ohtani et al, 2001; Zheng et al, 2004; Rothschild et al, 2006).
We have previously identified RP58, a sequence-specific transcriptional repressor, sharing homology with a number of transcriptional regulators at the amino terminus, termed the POZ domain (Aoki et al, 1998), and provided evidence for its essential role in normal brain development (Okado et al, 2009). In the developing mouse brain, RP58 is strongly expressed in the cerebral cortex and hippocampus, and its expression gradually increases in parallel with neuronal maturation (Ohtaka-Maruyama et al, 2007). Rp58 knockout (KO) mice demonstrated interruption of the cell cycle exit of progenitor cells, leading to increased mitotic cell populations such as radial glial progenitors and intermediate progenitors. Consistent with these observations, RP58 expression is lost in some human-derived brain tumour cell lines. Exogenous RP58 expression in both medulloblastoma and glioblastoma reduced their proliferation and increased cell death in vivo and in vitro (Tatard et al, 2010). Thus, the previous observations imply a strong association between RP58 and NSC cell-cycle regulation, but the molecular mechanisms connecting them have remained unknown.
In the present study, we observed strongly upregulated mRNA expression of Id1–Id4 and excess astrogenesis in the cortex of Rp58 KO mice and identified all Id genes as direct targets of RP58. Moreover, the excess number of progenitors and astrocytes in Rp58 KO cortex was rescued by either the downregulation of Id genes or p57 overexpression in vivo. These results suggest that RP58 is a novel regulator that promotes cell cycle exit from the germinal zone through the transcriptional repression of all Id genes, leading to p57 upregulation.
Results
Rp58 deletion causes increased progenitors and enhanced astrogenesis
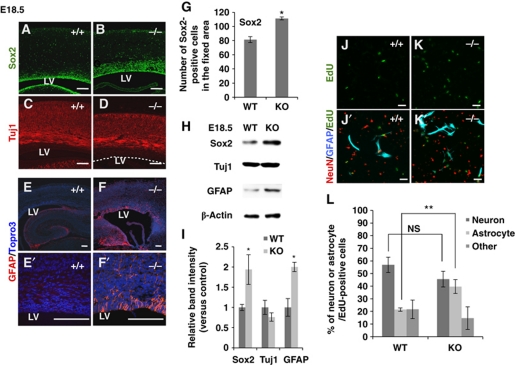
To investigate the function of RP58 in mammalian CNS development, we previously generated Rp58 KO mice (Okado et al, 2009). RP58 deletion in the developing cortex led to an enlarged Sox2-positive progenitor pool (Figure 1A, B and G). Similarly, the cells expressing cyclin-E, a marker of cell cycle re-entry, were increased in Rp58 KO mouse cortex at E18.5, while few cyclin-E-positive cells were observed in wild-type (WT) E18.5 cortex (Supplementary Figure S1). Nevertheless, no substantial difference in the expression level of an early neuronal marker (Tuj1) was observed between Rp58 KO and control cortex (Figure 1C and D).
Figure 1.
RP58 depletion causes increased astrogenesis both in vivo and in vitro. (A–D) Representative images of coronal sections from E18.5 WT and mutant mice stained for Sox2 (A, B) and Tuj1 (C, D). Scale bars: 0.1 mm. LV, lateral ventricle. The dotted line in (D) indicates the edge of the neocortical VZ (n=3). (E–F′) Representative images of caudal coronal sections from E18.5 WT and mutant mice stained for GFAP. (E′, F′) Show magnified areas in (E, F), respectively. Nuclei were stained with Topro3 (E–F′). Scale bars: 0.1 mm. (G) The number of Sox2-positive cells within a fixed area was counted. (H) Immunoblotting for Sox2, Tuj1, GFAP, and β-actin in cortical homogenates from E18.5 WT and mutant mice. β-Actin was used as an internal control. (I) Quantification of protein expression levels in (H) using Fiji software available online (http://pacific.mpi-cbg.de/wiki/index.php/Fiji) (t-test: *P<0.05, **P<0.01). Error bars indicate the s.d. (n=3). (J–L) Cerebral cortical cells prepared from E16.5 WT or mutant mice were cultured in EdU-containing medium for 12 h and then incubated for 5 days. (J–K′) Cells were stained with antibodies against GFAP (blue) and NeuN (red). Cells incorporating EdU were detected using the alkyl-azide reaction (green). Scale bars: 50 μm. (L) Percentage of neurons, astrocytes, and other cell types within the EdU-labelled progenitor cell population in (J′, K′) (t-test: **P<0.01). Error bars indicate the s.d. (n=7: WT=4, mutant=3).
We then addressed whether the increased number of progenitors influences the subsequent generation of astrocytes. A previous study reported few GFAP-positive astrocytes in the mouse cerebral cortex at around P0 (Pereira et al, 2010), although hippocampal GFAP expression was already detected at E17.5 (Favaro et al, 2009). Intriguingly, GFAP immunostaining in E18.5 KO cortex revealed many GFAP-positive astrocytes (Figure 1E–F′). The increase in astrocytes (at the same rostral-caudal level) was prominent in the caudal but not rostral cortex (Supplementary Figure S2). In E16.5 cortex, no GFAP-expressing astrocytes were detected in either WT or mutant mice (Supplementary Figure S3A–B′). Double immunostaining with GFAP and the astrocyte marker S100β was performed to confirm that the GFAP-positive cells were astrocytes rather than radial glia (Raponi et al, 2007) (Supplementary Figure S4). Immunoblot analysis of cerebral cortex lysates demonstrated a marked increase in Sox2 and GFAP levels in mutant mice at E18.5, whereas Tuj1 levels were unchanged (Figure 1H and I).
Since Rp58 KO mice died soon after birth, postnatal analysis of astrogenesis in vivo was impossible. Cells, including NSCs from the E16.5 cerebral cortex, were labelled with EdU (a thymidine analogue) in the culture medium for 12 h and incubated for a further 5 days. Fluorescence labelling was then performed for EdU, GFAP, and NeuN, a neuronal marker, to confirm whether the increased progenitors observed in the mutant mice could differentiate into GFAP-positive astrocytes. Approximately 40% of mutant cells and 20% of WT cells did differentiate into astrocytes. No significant difference in neuronal differentiation was observed between mutant and WT cells (Figure 1J–L). Thus, RP58 deletion leads to enhanced astrogenesis during the late neurogenic period in the cerebral cortex in vivo and excess generation of astrocytes from progenitors in vitro.
All four Id genes are upregulated in the cortex of Rp58 KO mice
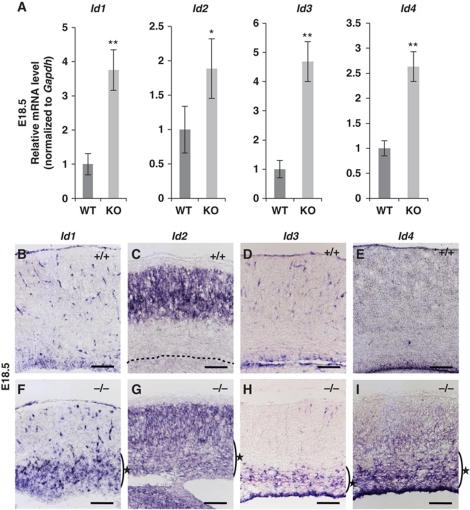
To identify downstream target genes repressed by RP58 in the developing cortex, DNA microarray analysis was performed using mRNA from E16.5 mutant and WT mouse cortex. The obtained data were analysed by MAPP pathway analysis, which indicated that the genes showing significantly altered expression in Rp58 KO compared with WT were involved in six pathways (Supplementary Tables SI and SII). First, we focussed on the pathways associated with cell-cycle progression and astrogenesis. Second, because RP58 acts as a transcriptional repressor (Aoki et al, 1998; Fuks et al, 2001; Takahashi et al, 2008), we noted the genes showing increased expression in the mutant cortex, with particular reference to those that also contained the RP58-binding consensus sequence in human genomic loci corresponding to those in the mouse (Supplementary Figure S5). As shown in Supplementary Tables SII and SIII, Id1and Id3 mRNAs were significantly increased in the mutant cortex compared with the control. To confirm the DNA microarray analysis results, real-time PCR analyses were conducted using E18.5 cortex. At this stage, all four tested Id mRNAs were upregulated in the mutant cerebral cortex (Figure 2A).
Figure 2.
Rp58 KO mice showed aberrant expression of Id mRNA. (A) Real-time PCR analysis of all four Id mRNAs in WT and mutant cerebral cortices at E18.5. Gapdh mRNA expression was used for normalization (*P<0.05, **P<0.01; t-test). Data represent the mean±s.d. (n=3). (B–I) Representative in situ hybridization images showing the four Id mRNAs in coronal brain sections from E18.5 WT (B–E) and mutant mice (F–I). All four Id mRNAs were ectopically expressed around the mutant SVZ (F–I) (indicated with star) (n=3). The dotted line in (C) indicates the edge of the VZ. Scale bars: 250 μm.
In situ hybridization was then performed to detect the regions where Id mRNAs were upregulated in the mutant (Figure 2B–I). In general, Id1 and Id3 mRNAs are expressed in proliferating neural progenitor cells in the cortex throughout embryonic CNS development, while Id2 and Id4 mRNAs are observed in the same cell types at the early stages of neurogenesis and continue to be expressed in post-mitotic neurons (Jen et al, 1996, 1997; Tzeng and de Vellis, 1998; Tzeng, 2003). In Rp58 KO brains, all four Id mRNAs were ectopically expressed in the ventricular zone (VZ)/subventricular zone (SVZ) (Figure 2F–I, stars). Although Id2 was expressed throughout the mutant cortex, its increase in mutant mice could not be detected clearly by DNA microarray analysis because Id2 was also strongly expressed in the control cortical plate (Figure 2C). In addition, we used immunohistochemistry to confirm that the expression of Id proteins was increased in the mutant SVZ compared with that in WT SVZ (Supplementary Figure S6Ac–H′c). In the WT cortex, explicit expression of each Id was observed only in the VZ (Supplementary Figure S6A′c–H′c). In the rostral cortex, few differences in the Id expression patterns were observed between WT and KO mice. Mice of both genotypes showed expression of Id1 and Id4 in VZ/SVZ, Id2 in the total cortical area, and Id3 alone in the VZ (Supplementary Figure S6Ar–Hr).
To determine whether the increase in Id mRNAs in the Rp58 KO cortex was not only due to an increased number of progenitors but also to the loss of direct RP58-dependent regulation, we estimated Id mRNA expression at the single cell level in WT- and mutant-derived neurospheres by normalizing it to that of Gapdh. The mRNA level of Pax6, a progenitor cell marker, was used as a negative control (Supplementary Figure S7A and B). Quantitative real-time PCR analysis revealed higher levels of all four Id mRNAs in Rp58 KO neurospheres compared with the control. The expression level of Pax6 mRNA was unchanged, suggesting that each Id mRNA increased in the individual cells. Taken together, these results indicate that the expression levels of all Id genes are increased in Rp58 KO cortex compared with control, particularly due to overexpression in the mutant mouse proliferative zone.
RP58 directly represses the transcription of all Id family members
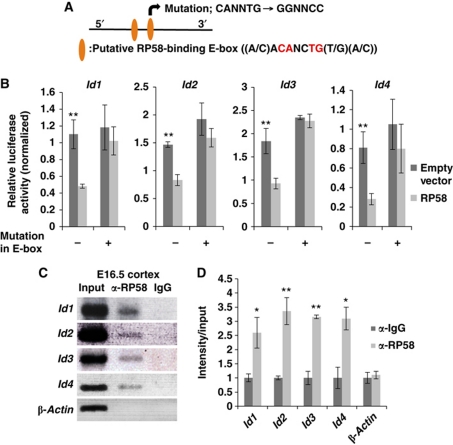
Next, to determine whether Id gene expression was directly downregulated by RP58, luciferase reporter assays and ChIP analysis were performed (Figure 3). Genomic regions around putative RP58-binding sites for each Id gene (Supplementary Figure S5) were cloned from the E18.5 WT mouse genome and inserted upstream of a luciferase gene in a reporter plasmid, and the impact of RP58 expression on Id gene expression was analysed. The same analysis was also performed using the mutated binding site (Figure 3A and B). The activity of the luciferase reporter gene fused to putative RP58-binding sites was reduced following RP58 expression in Cos7 cells. The activity of each mutated Id reporter gene did not significantly differ from WT, indicating that this sequence is important for the RP58-mediated reporter gene repression (Figure 3B). In addition, in vivo ChIP analysis of E16.5 cortex using an anti-RP58 antibody (Takahashi et al, 2008) demonstrated that RP58 bound to the chromatin of putative RP58-binding sites at each Id gene (Figure 3C and D). These results suggest that RP58 directly inhibits the transcriptional activity of Id1–Id4 genes by binding to their regulatory sequences.
Figure 3.
RP58 directly bound to the regulatory regions of Id genes and repressed their activity. (A) Schematic shows two E-boxes (CANNTG) with specific sequences bound by RP58. The mutation was introduced into the latter E-box. Besides the typical E-boxes (CANNTG), there is only one pair of Rp58-binding E-boxes (ACANCTG) situated in close proximity to the id-encoding genomic sequences. (B) Luciferase assay representing the effects of RP58 on the putative RP58-binding region of each Id. Cos7 cells were transiently transfected with 0.2 μg pGL4P-Id1, -Id2, -Id3, or -Id4 regulatory region-luc as a reporter. The luciferase activity of cells co-transfected with Rp58 expression vector or pCDNA3.1 (+) empty vector (0.2 μg) was measured. The mean Photinus pyralis firefly activities were normalized to the mean activities of Renilla luciferase vector (0.1 μg) (t-test: **P<0.01). Data were represented as mean values±s.d. (n=3). (C) ChIP analysis of the putative RP58-binding region of each Id gene. Mouse E16.5 homogenized cerebral cortex was immunoprecipitated with an antibody specific for RP58 or rabbit-IgG as a negative control. As a positive control, 0.2% total input chromatin DNA was used, and the amplification products of the β-actin promoter region were used as a negative control. (D) The agarose gel image presented in (C) was quantified using Fiji software available online (http://pacific.mpi-cbg.de/wiki/index.php/Fiji) (t-test: *P<0.05, **P<0.01). Data were represented as mean±s.d. (n=3).
RP58 attenuates NSC proliferation and the downregulation of the four Id genes in neurospheres
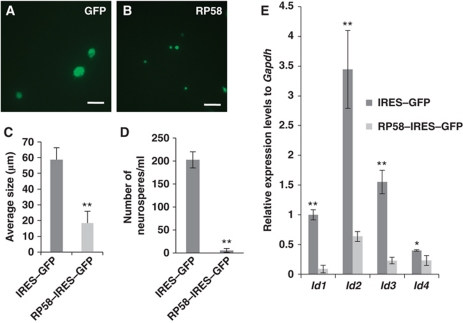
The effect of ectopic RP58 expression on NSC proliferation was then examined. Isolated E14.5 cortical NSCs were infected with either RP58–IRES–GFP (Lv–RP58) or control IRES–GFP (Lv–GFP) lentivirus and cultured under nonadherent conditions for 7 days with EGF mitogen. Under these conditions, a single NSC could grow to a neurosphere by repeated proliferation (Figure 4A–C). RP58-infected neurospheres (15.23±2.67 μm) demonstrated an approximately two-fold decrease in diameter compared with control (55.42±1.85 μm) (Figure 4C). The dissociated primary neurospheres were then replated to analyse their self-renewal competency. Few secondary neurospheres were formed from the Lv–Rp58-infected neurospheres (5.67±4.04), whereas control Lv–GFP-infected cells formed ∼200 secondary neurospheres (202.33±17.5; Figure 4D). The same trend was observed when the experiments were performed in FGF2- or EGF/FGF2-supplemented medium (Supplementary Figure S8). In addition, Id mRNA expression levels were quantified in cultures of Lv–Rp58- or Lv–GFP-infected cells in the presence of EGF. Cultures infected with Lv–RP58 demonstrated lower expression levels of Id mRNAs than those infected with control Lv–GFP (Figure 4E). Lv–Rp58-infected neurospheres lost their competency for self-renewal, which was partially rescued by the co-induction of each individual Id, or all four Ids, along with RP58 (Supplementary Figure S9). By contrast, quaternary-passaged mutant neurospheres were much larger than the WT neurospheres, as shown by a shift to the right in the size distribution plots (Supplementary Figure S10). These data suggest that RP58 attenuates NSC proliferation, which is accompanied by downregulation of all four Id mRNAs.
Figure 4.
Ectopic expression of RP58 in mouse NSC-derived neurospheres reduced their size and Id1–Id4 mRNA expression levels. (A, B) Representative images of neurospheres transduced with either IRES–GFP lentivirus (Lv–GFP) (A) or RP58–IRES–GFP lentivirus (Lv–RP58) (B). The neurospheres were harvested after 7 days from E14.5. Scale bars: 100 μm. (C) Quantitative analysis of the diameter of neurospheres in (A, B) (t-test: **P<0.01). Error bars indicate s.d. (n=40). (D) The number of secondary generated neurospheres was counted after replating the dissociated primary neurospheres at a density of 0.5 × 105 cells/ml. Three independent experiments were performed (t-test: **P<0.01). Error bars indicate the s.d. (E) Real-time PCR analysis of Id1–Id4 mRNAs in Lv–RP58 or Lv–GFP-infected neurospheres. Gapdh mRNA expression was used for normalization (t-test: *P<0.05, **P<0.01). Data were represented as mean±s.d. (n=4).
Increase in progenitors and astrocytes induced by Id gene overexpression mimics the phenotype observed in Rp58 KO mice
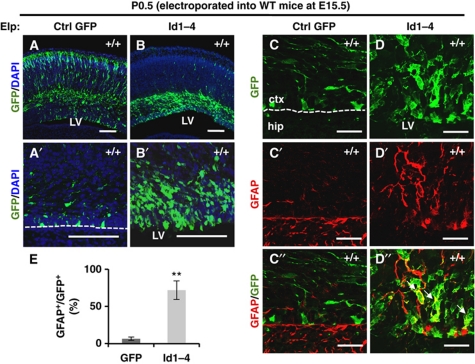
We next investigated the issue of whether increased and ectopic expression of Id genes could accelerate astrogenesis. Id1–Id4 and control Gfp were overexpressed in E15.5 WT cortex using in utero electroporation techniques, and then the embryos were sacrificed at P0.5 (Figure 5). Id1–Id4 genes were transfected together into the WT cerebral cortex because all Id mRNAs were upregulated in Rp58 KO cortex. Many Id-overexpressing cells accumulated from the VZ to the intermediate zone. In contrast, almost all control GFP-electroporated cells migrated to the upper layers of the cortex (Figure 5A–B′). Tissue was then immunostained for GFAP and Ki67, a cell proliferation marker (Figure 5C–D″; Supplementary Figure S11A–C). Many GFP-positive Id-expressing cells colocalized with GFAP in the VZ/SVZ (71.9±12.5%) compared with control (6.6±2.5%; Figure 5E). The Ki67-positive fraction also increased among Id-electroporated cells, comprising 51.8±8.6% compared with 13.9±3.7% of cells in WT littermates (Supplementary Figure S11). Taken together, these results demonstrate that high expression levels of Id1–Id4 during late neurogenesis induce excess astrocytes and proliferating cells similar to those observed in Rp58 KO mice.
Figure 5.
Id1–Id4 overexpression mimicked the Rp58 KO phenotype in the promotion of astrogenesis. (A–D″) Representative images showing coronal sections of WT brains electroporated at E15.5 with Gfp expression vector (A, A′, C–C″) or co-electroporated with Gfp, Id1, Id2, Id3, and Id4 expression vectors (B, B′, D–D″) fixed at P0.5 and stained with anti-GFP (green) and Topro3 (blue, nuclear stain) (A–B′), or anti-GFP (green, C, D, C″, D″) and anti-GFAP (red) (C′–D″). Scale bars: 100 μm (A–B′), 25 μm (C–D″). The dotted line in (A′, C) indicates the edge of the VZ. The arrows in (D″) indicate the double-stained cells. (E) Quantification of the ratio of GFAP-positive cells to GFP-positive cells in (C″, D″) (t-test: **P<0.01). Data were represented as mean±s.d. (n=4).
Transfection with either Id shRNAs or p57 rescues the number of progenitors and astrocytes
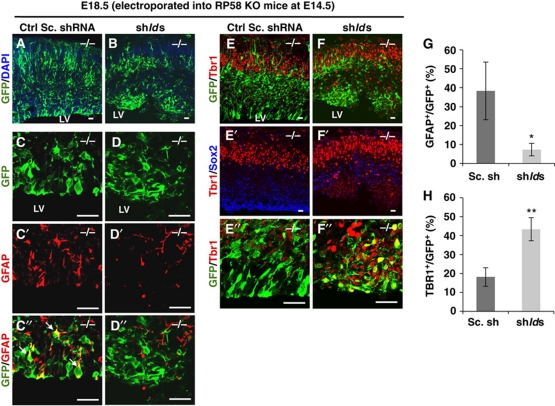
To confirm the hypothesis that Rp58 KO phenotypes are induced by failure of RP58-mediated repression of Id mRNA expression, we examined whether the phenotypes are rescued by Id knockdown in the Rp58 KO mouse cerebral cortex (Figure 6). Downregulation of id1–4 by shRNAs was confirmed by RT–PCR (Supplementary Figure S12). Fewer Id1–Id4 knockdown cells were colocalized with GFAP compared with the control scramble shRNA-expressing cells in KO mice (shIds, 7.3±3.3% versus control shRNA, 38.3±15.2%; Figure 6C–D″ and G). Moreover, Id knockdown rescued the increase of proliferating cells compared with control GFP-expressing mutant cortex (shIds, 9.1±4.3% versus control shRNA, 48.7±13.5%; Supplementary Figure S13). In contrast, Id downregulation resulted in the promotion of neuronal differentiation around the VZ (shIds, 43.3±6.1% versus control shRNA, 18.2±4.9%; Figure 6E–F″ and H), demonstrating that Id knockdown in Rp58 KO cortex rescued progenitors from cell cycle exit defects. However, the cells that exited from the cell cycle failed to migrate to the upper layers of the cortex compared with the WT (Supplementary Figure S14).
Figure 6.
Id1–Id4 knockdown rescued the precocious astrogenesis defects and promoted neuronal differentiation in Rp58 KO mice. (A–F″) Representative images of coronal sections of Rp58 KO cortices electroporated at E14.5 with control scramble (Sc) shRNA (A, C–C″, E–E″) or co-electroporated with Sc shRNA and shId1, shId2, shId3, or shId4 (B, D–D″, F–F″) and fixed at E18.5 and stained with anti-GFP (green, A, B, C, D, C″, D″, E, F, E″, F″) and anti-GFAP (red, C′, D′, C″, D″), anti-Sox2 (blue, E′, F′), and anti-Tbr1 (red, E, F, E″, F″). Scale bars: 25 μm. (G, H) Quantification of the ratio of GFAP-positive cells to GFP-positive cells in (C″, D″, G), and the ratio of Tbr1-positive cells to GFP-positive cells in (E″, F″, H) (t-test: *P<0.05, **P<0.01). Error bars indicate s.d. (n=3). The arrows in (C″) indicate the double-stained cells.
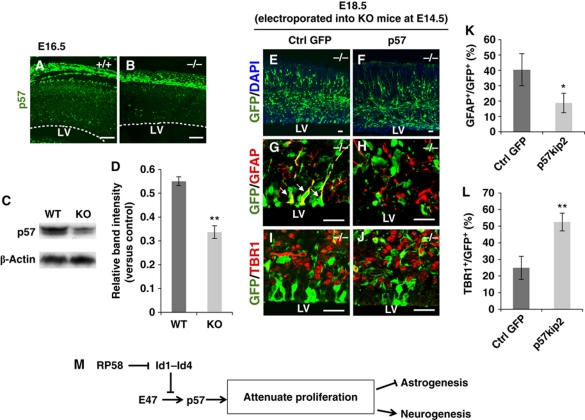
Finally, we focussed on the downstream targets of Id proteins. Referring to the microarray analysis data generated using E16.5 WT and Rp58 KO cortex (Supplementary Table SIII), the expression level of p57 decreased in Rp58 KO cortex compared with WT. Note that there was no significant difference in the expression level of the other Cip/Kip family genes (p21 and p27) (a fold change of 1.5 were used as criteria) (Supplementary Table SIII). The results of immunoblotting and real-time PCR revealed the same tendency indicated by microarray analysis (Figure 7A–D). In the developing cortex, p57 has been identified as a functional target of E protein (E47), and Id2 has been shown to prevent p57 expression by entrapping E47 (Rothschild et al, 2006). Members of the Cip/Kip family are well-known positive regulators of progenitors exiting the cell cycle in various tissues (Joseph and Hermanson, 2010).
Figure 7.
p57 overexpression rescued the precocious astrogenesis defects and promoted cell cycle exit in Rp58 KO mice. (A, B) Representative images showing p57 in coronal sections from E18.5 WT and mutant mice. Scale bars: 100 μm. LV, lateral ventricle. The dotted line in (A, B) indicates the edge of the neocortical VZ (n=3). (C) Immunoblotting for p57 and β-actin protein in cortical homogenates of E18.5 WT and mutant mice. β-Actin was used as an internal control for immunoblotting. (D) Quantification of protein expression levels demonstrated in (C) using Fiji software (t-test: **P<0.01). Error bars indicate s.d. (n=3). (E–J) Rp58 KO cortex electroporated at E14.5 with control GFP expression vector (E, G, I) and co-electroporated with p57 expression vector (F, H, J) were fixed at E18.5 and coronal sections were stained with anti-GFP (green, E–J), anti-GFAP (red, G, H), and anti-Tbr1 (red, I, J). Scale bars: 25 μm. (K, L) Quantification of the ratio of GFAP-positive cells to GFP-positive cells in (G, H), and the ratio of Tbr1-positive cells to GFP-positive cells in (I, J) (t-test: *P<0.05, **P<0.01). Error bars indicate s.d. (n=3). The arrows in (G) indicate the double-stained cells. (M) Summary scheme illustrating the RP58-mediated fate determination of NSCs in the developing cortex.
We then examined whether the phenotypes are rescued after compensation of p57 expression in the Rp58 KO cerebral cortex using in utero electroporation (Figure 7E–L; Supplementary Figure S15A–C). Fewer p57-overexpressing cells were colocalized with GFAP compared with control GFP-expressing cells in KO mice (p57, 18.79±6.38% versus control GFP, 40.36±10.41%; Figure 7G, H, and K) and TBR1-positive differentiating neurons were increased in p57 electroporated cortex compared with control (p57, 52.46±5.32% versus control GFP, 24.97±6.98%; Figure 7I, J, and L). Although shIds electroporation caused ectopic neuronal differentiation in Rp58 KO mouse, such phenotype was not observed after p57 electroporation. Moreover, p57 overexpression rescued the increase of proliferating cells compared with control GFP-expressing KO cortex (p57, 24.29±10.26% versus control GFP, 45.78±3.67%; Supplementary Figure S15A–C). Conversely, to determine the effect of astrocyte differentiation observed in the WT cortex on p57 functional inhibition, we used the C-terminal portion of p57 (Cterp57, a Cip/Kip family member, which lacks the cyclin/CDK binding/inhibitory domains and contains unique proline and acidic domains). It is reported that the inhibition of p57 function by Cterp57 enhances cell proliferation (Tury et al, 2011); therefore, we asked whether inhibiting the function of p57 affects astrocyte differentiation. In utero electroporation was performed at E15.5, followed by immunostaining of the cortices with GFAP at P9.5. Upon electroporation with a control plasmid, most cells produced differentiated neurons that migrated into the cortical plate over the next few days; however, large numbers of GFAP+/GFP+ cells were observed in Cterp57 electroporated cortices (100.0±0.0% control and 82.74±5.37% Cterp57-induced cells; Supplementary Figure S16). These results further support our conclusion that the decreased expression of p57 in the Rp58 KO cortex results in increased numbers of progenitors and astrocytes. Taken together, the results suggest that either repressed expression of all four Id genes or compensation for p57 expression was able to rescue the increased number of progenitors and enhanced astrogenesis observed in Rp58 KO mice, and that the functional inhibition of p57 mimics this mutant phenotype, suggesting that RP58-mediated p57 expression through the repression of Id mRNA expression is the key mechanism by which RP58 regulates the population of cortical neurons and astrocytes.
Discussion
RP58 mediates cell cycle control of NSCs
Two primary parameters define the output of NSCs: (1) how fast progenitor cells divide and (2) the balance between cell cycle re-entry and the exit of progenitors (Dehay and Kennedy, 2007). It was observed that Id1–Id4 mRNAs and Id1–Id4 proteins were upregulated in the cortex of Rp58 KO mice (Figure 2; Supplementary Figure S6), and the excess progenitors in E16.5 Rp58 KO cortex exhibited biased differentiation towards astrocytes relative to neurons compared with control (Figure 1J–L). Id proteins are known to act by antagonizing the activity of bHLH transcription factors (NeuroD, Myogenin and E proteins), retinoblastoma (Rb), and Ets domain proteins, which results in the inhibition of cell cycle exit and enhanced cell cycle re-entry (Iavarone et al, 1994; Prabhu et al, 1997; Yates et al, 1999; Ohtani et al, 2001; Trabosh et al, 2009). In the developing cortex, Id2 prevents p57 expression by antagonizing E protein activity (Rothschild et al, 2006). Since Id1, 3, and 4 interact with E proteins (Sun et al, 1991; Langlands et al, 1997; Pagliuca et al, 2000), all Id members are expected to prevent the expression of p57 in the cortex by dimerizing with E proteins. Rp58 deletion caused elevated expression of all Id genes, which may directly lead to the reduction of p57 expression. In the primary culture of cortical progenitors derived from p57 KO mice, the ratio of progenitors to total cells was increased (Tury et al, 2011), similar to the present observations in Rp58 KO (Figure 1A, B, G and H). Moreover, additional rounds of the cell cycle have been observed in the developing mouse retina and spinal cord with the loss of p57 (Dyer and Cepko, 2000; Gui et al, 2007). Similar abnormalities in cell proliferation were also observed in the Rp58 KO mouse (Supplementary Figure S1).
Progenitors isolated from the Rp58 KO cortex at E18.5 produced neurospheres twice as large as those produced from WT cortex under undifferentiated conditions (Supplementary Figure S17). Therefore, in Rp58 KO mice, progenitors of the late phase of neurogenesis may maintain proliferative competency as strong as those present in the early phase of neurogenesis. In general, as corticogenesis proceeds, the cell cycle of NSCs is delayed depending on the length of the G1-phase (Takahashi et al, 1995), and a cell cycle delay favours the pathway for neuronal differentiation (Lukaszewicz et al, 2002; Canzoniere et al, 2004; Nguyen et al, 2006; Katsimpardi et al, 2008). The length of the G1-phase changes the length of cytokine exposure time, which in turn regulates cell cycle re-entry or neuronal differentiation (Burdon et al, 2002; Kioussi et al, 2002; Baek et al, 2003; Oliver et al, 2003; Fluckiger et al, 2006; Pilaz et al, 2009). p57 is intimately involved in cell cycle delay by preventing cells from entering G1- to S-phase (Besson et al, 2008; Neganova and Lako, 2008; Pateras et al, 2009). The lack of p57 in Rp58 KO mice may cause progenitors to have a high proliferating potential in late neurogenesis, leading to difficulty in neuronal differentiation.
Collectively, increased Id expression resulting from the absence of RP58 may lead to reduced p57 expression during late neurogenesis, which in turn impairs progenitors from exiting the cell cycle as neurons. Missing the appropriate timing for neuronal development may switch the competency of progenitors from neural to astroglial, possibly resulting in the decreased ratio of neurons to astrocytes observed in Rp58 KO cortex (Figure 7M).
RP58 is a prominent negative regulator of all four Id family members in the nervous system
The present study has confirmed a causal relationship between RP58 and Id-dependent regulation of astrogenesis via in vivo experiments. First, the phenotype of Rp58 KO mice, excess progenitors and astrogenesis, could be reversed by the downregulation of Id mRNAs with in utero electroporation (Figure 6C–D″ and G; Supplementary Figure S13). Second, the overexpression of Id genes in NSCs of the WT cortex led to cell-autonomous abnormalities similar to those observed in Rp58 KO embryos (Figure 5C–D″ and E; Supplementary Figure S11).
Some reports have indicated that Id overexpression promotes cell proliferation and astrogenesis in vivo and in vitro (Cai et al, 2000; Jung et al, 2010). These abnormal phenotypes were also observed in Rp58 KO cortex. In the Rp58 KO cortex, NSCs and astrocytes were increased (Figure 1A, B, and E–F′), and we have previously reported defects in NSC cell cycle exit (Okado et al, 2009). In addition, analyses of Id KO mice have revealed that Id1 and Id3 are required to maintain the timing of neuronal differentiation during brain development, while Id4 regulates lateral expansion of the proliferating zone in the developing cortex and hippocampus (Lyden et al, 1999; Tzeng, 2003; Yun et al, 2004). Rp58 KO and Id KO mice have opposite phenotypes regarding the number of NSCs and their exit from the cell cycle (Lyden et al, 1999; Tzeng, 2003; Kyuson et al, 2004; Okado et al, 2009). This evidence supports our finding that all Id genes are targets for transcriptional repression by RP58 (Figures 3 and 4E).
The expression patterns of each Id gene are known to partially overlap in the CNS and they may demonstrate a certain level of functional redundancy. In fact, Id1, Id2, or Id3 single-KO mice are viable and demonstrate no reported defects in cephalic structure (Yan et al, 1997; Pan et al, 1999; Yokota et al, 1999). Id4 KO mice show only mild defects such as small brain size, as well as Id1, Id2, and Id3 mRNA upregulation in the telencephalon (Yun et al, 2004; Bedford et al, 2005). In fact, we found that cortices electroporated with the single Id showed similar phenotypes to those electroporated with all four Ids. Quantitative analysis of Ki67+/GFP+ progenitors (13.9±3.7%) and GFAP+/GFP+ astrocytes (7.4±1.6%) in the control cortices revealed the distribution of electroporated cells. Embryos electroporated with a single Id vector showed significantly more Ki67+/GFP+ progenitors (Id1: 49.0±6.5%, Id2: 46.3±9.4%, Id3: 51.8±8.6%, Id4: 45.9±3.6%) and GFAP+/GFP+ astrocytes (Id1: 48.3±2.0%, Id2: 41.8±3.3%, Id3: 47.0±7.5%, Id4: 41.4±10.0%) (Supplementary Figure S18). Conversely, the excess progenitors and astrocytes in the Rp58 KO mouse cortex could not be rescued by single Id knockdown (Supplementary Figure S19). These findings suggested that the four Id genes show functional redundancy in terms of increased numbers of astrocytes and proliferating cells. The enlarged astrocytes were more clearly visible in the KO caudal cortex than in the rostral cortex. This is probably because the Id expression pattern in the rostral region in KO mice was not altered to the same extent as that in the WT (Supplementary Figure S6). This suggests that a stronger Rp58 repression mechanism for Ids operates in the caudal cortex than in the rostral cortex. The present study revealed that all Id genes were repressed by RP58 (Figures 3 and 4E), indicating that RP58 plays a crucial role in repressing Id genes without mutual compensation to allow NSCs to exit the cell cycle and generate post-mitotic neurons.
Recently, the ubiquitin proteasome pathway was reported to degrade Id proteins. Id1, Id2, and Id4 proteins have a recognition site for anaphase-promoting complex (APC), which has E3 ubiquitin ligase activity, and Id2 was shown to be a substrate of APC (Lasorella et al, 2006). COP9 signalosome-mediated ubiquitination degrades Id1 and Id3 (Berse et al, 2004). Post-translational regulation through these ubiquitin proteasome pathways is possibly an important mechanism for the repression of Id proteins in the cell cycle exit of NSCs. In the Rp58 KO cortex, interruption of Id expression in NSCs rescued the phenotype of defective cell cycle exit. This finding indicates that the timely transcriptional repression of Id genes by RP58 also play a crucial role in NSC cell cycle exit. Moreover, RP58-mediated transcriptional repression of Id2 and Id3 was also reported as important for skeletal myogenesis (Yokoyama et al, 2009). Therefore, we propose that, in addition to post-transcriptional regulation, the transcriptional regulation of Id genes by RP58 may be a crucial mechanism for cell differentiation from various types of progenitor cells.
RP58 and neuronal differentiation
In the early phase of corticogenesis, a large progenitor pool produces the appropriate number of neurons by NSC proliferation and subsequent differentiation, while in the late phase, an increase of neuron-generating divisions compensates for the reduced size of the progenitor pool (Polleux et al, 1997; Lukaszewicz et al, 2005). Little difference was observed between Tuj1 expression levels in Rp58 KO and control cortex (Figure 1C, D, H, and I). This observation may seem counterintuitive due to the cell cycle exit defects of NSCs in Rp58 KO mice. The increased number of NSCs in Rp58 KO mice may compensate for their reduced ability to exit the cell cycle to limit the impairment of neuronal production.
Incidentally, another characteristic phenotype that we observed for Rp58 KO mice is neuronal migration defects of developing cortex (Supplementary Figure S14). However, this phenotype was not rescued by Id downregulation or p57 overexpression (Figures 6B and 7F; Supplementary Figure S14), suggesting that RP58 controls NSC differentiation and neuronal migration in different molecular mechanisms. We are currently investigating RP58 targeting of neuronal migration-related genes, which could underlie migration defects in Rp58 KO mice.
RP58 and brain cancer
RP58 expression was reported to show downregulation in brain tumour cell lines and, moreover, RP58 transduction into human glioma drastically decreased its proliferation and promoted apoptosis (Tatard et al, 2010). However, the relationship between RP58 downregulation and brain tumour induction is not understood. In the present study, RP58 transduction into neurospheres derived from E14.5 NSCs was found to result in reduced cell proliferation (Figure 4A–D), and the excess progenitors in the Rp58 KO mouse cortex could be rescued either by Id knockdown (Figure 6; Supplementary Figure S13) or p57 compensation (Figure 7E–L; Supplementary Figure S15). Some groups have reported that the expression levels of Id genes and p57 are also related to brain tumours. Various combinations of Id members are upregulated in human glioma and human cell lines derived from tumours of the nervous system, and they are frequently associated with enhanced malignancy and aggressive clinical behaviour (Lasorella et al, 2001; Fong et al, 2004; Perk et al, 2005; Jeon et al, 2008; Kuzontkoski et al, 2010; Zeng et al, 2010). Targeting Id dimerization to other proteins in human neuroblastoma cell lines suppresses tumourigenic properties including cell invasion (Ciarapica et al, 2009). Decreased expression of p57 is observed in glioblastoma multiforme, and induction of p57 into glioma inhibits malignant transformation (Tsugu et al, 2000; Sakai et al, 2004). The findings of the present study raise the hypothesis that abnormal Rp58 downregulation causes Id upregulation and p57 downregulation, ultimately resulting in the development of brain tumours. However, progenitors derived from the Rp58 KO cortex were unable to form colonies in soft agar, suggesting that Rp58 single KO is insufficient to initiate cancer (Supplementary Figure S20). Increased understanding of RP58 will contribute to the development of cancer therapies and diagnostic criteria.
Materials and methods
Animals
Rp58 KO mice were generated as previously described (Okado et al, 2009). All experimental protocols were approved by the Animal Care and Use Committee of the Tokyo Metropolitan Institute for Neuroscience.
Plasmid construction
Each Id gene coding sequence was cloned from E14.5 mouse cerebral cortex into the pBlueScript II SK (−) vector for in situ hybridization, the PCDNA3.1 (+) vector for luciferase assay, and the pCAG vector (a kind gift of Dr Y Kawaguchi) for in utero electroporation. See Supplementary Table SI for the cloned Id accession numbers. For the downregulation of Id genes, the following shRNA expression vectors were purchased and used for in utero electroporation and lentivirus infection: clone ID V3LHS_397678 (shId1), RHS4430_98841701 (shId2), V2LMM_217187 (shId3), and V3LHS_373602 (shId4) (Funakoshi).
Immunohistochemistry
Embryos were fixed with 4% paraformaldehyde (PFA) at 4°C overnight. The brains were then cryoprotected in 30% sucrose at 4°C overnight. Serial coronal sections (25 μm) were cut with a cryostat. Immunostaining was performed with the following primary antibodies diluted in PBS including 10% Block Ace (DS Pharma Biomedical): anti-GFP (Abcam, 1:500), anti-GFAP (Dako, 1:500), anti-NeuN (Chemicon, 1:200), anti-Sox2 (Santa Cruz, 1:200), anti-Tbr1 (Abcam, 1:500), anti-Ki67 (Novocastra, 1:200), anti-Tuj1 (Neuromics, 1:200), anti-cyclin-E (Santa Cruz, 1:200), anti-p57 (Abcam, 1:100), anti-Id1 (Santa Cruz, 1:500), anti-Id2 (a gift from Dr Mori and Dr Yokota, 1:500), anti-Id3 (Santa Cruz, 1:500), anti-Id4 (Santa Cruz, 1:500), and anti-S100β (Dako, 1:10 000). The antigens in tissues were reactivated by heating in 10 mM citrate buffer (adjusted to pH 6.0) using a microwave or autoclave. Sections were permeabilized with 0.2% Triton X-100 and 25% Block Ace (DS Pharma Biomedical) in PBS, incubated overnight with primary antibodies, washed three times with PBS-0.05% Tween-20, and then incubated for 2 h with fluorochrome-conjugated secondary antibodies and washed twice more. Anti-IgG antibodies conjugated to biotin (Vector, 1:200) followed by an avidin–biotin complex (Vector) or anti-IgG antibodies conjugated to HRP (Amersham, 1:200) were used as secondary antibodies to detect anti-Cyclin-E, S100β, Id1, Id2, Id3, and Id4, and the TSAplus Fluorescence System (Perkin-Elmer) was used to detect HRP activity. Finally, sections were treated with DAPI (Nacalai Tesque) or TOPRO3 (Molecular Probes) for nuclear staining, mounted in Permaflow (Thermo Scientific) and observed using a BX51WI Fluoview laser-scanning microscope (Olympus).
GFP-, GFAP-, Tbr1-, or Ki67-positive cell counts were done in a fixed area (from the VZ to the SVZ). The TSA or TSA Plus Fluorescence System (Perkin-Elmer) was adapted for the double staining of GFAP and S100β using rabbit polyclonal antibodies according to the method of Friocourt et al (2008). Briefly, sections were first incubated with diluted anti-S100β antibody (1:10 000) followed by incubation with or without rabbit anti-GFAP antibody (1:200).
Immunoblotting
Mouse E18.5 embryonic cerebral cortex extracts were homogenized in lysis buffer containing 20 mM Tris (pH 7.5), 0.1% SDS, 1% Triton X-100, 1% deoxycholate and protease inhibitor cocktail (Roche Diagnostics). The protein content in total cell lysates was quantified with the DC protein assay kit (Bio-Rad) and 30 μg total protein per lane was run in SDS–PAGE gels and transferred to PVDF membranes (Millipore). The proteins were probed with anti-GFAP (Dako, 1:1000), anti-Sox2 (Santa Cruz, 1:500), anti-Tuj1 (Neuromics, 1:500), and anti-β-actin antibodies (Sigma, 1:1000). Blots were processed by chemiluminescence using standard protocols (Amersham).
Neurosphere formation and differentiation
Cortex from E14.5 mice was digested enzymatically with 10 U/ml papain-EDTA for 30 min at 32°C, followed quickly by DNAse treatment. The tissue was then washed twice with complete medium, consisting of mouse NeuroCult NSC basal medium plus mouse NeuroCult NSC proliferation supplements (Stem Cell Technologies) with 20 ng/ml EGF, 10 ng/ml FGF2, or both EGF and FGF2, and 1 μl/ml penicillin/streptomycin (P/S; Sigma; P4333), and then mechanically triturated in complete medium. The cells were plated at a density of 2 × 105/ml cells per 3.5 cm dish (BD Falcon) and incubated for 7 days in a humidified 5% CO2 atmosphere. The primary neurospheres were then counted and collected for differentiation. Results of the neurosphere counts were expressed as mean±s.d. Some neurospheres were fixed in 4% PFA in PBS for 30 min at room temperature and their diameters were measured using an IX70 fluorescence microscope (Olympus). Approximately 10 neurospheres were deposited on poly-L-lysine-coated coverslips in individual wells of 24-well culture plates (Sumiron) containing differentiation medium (DMEM/F12 containing B27 supplement (GIBCO) and 1 μl/ml P/S). The neurospheres were allowed to differentiate for 3 days in humidified 5% CO2. The differentiated neurospheres were then fixed as above. After washing with PBS, they were stained.
Primary culture and EdU labelling
Cortex from E16.5 mice was digested enzymatically with 10 U/ml papain-EDTA for 30 min at 32°C, followed quickly by DNAse treatment. The tissue was then washed twice with medium (DMEM containing 10% FBS, 1 μl/ml P/S, and 5 mM Gln) and mechanically triturated in the medium. The cells were plated at a density of 0.4 × 106 cells per well in polyethyleneimine-coated 24-well plates. For the labelling of progenitor cells, cells were incubated with 10 μM EdU for 12 h before fixation. Detection of incorporated EdU was performed using the click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen) according to the manufacturer's protocol.
Microarray analysis
Sample RNAs were obtained from E16.5 Rp58 KO mouse cerebral cortex or control cortex. Two independent total RNA samples from each mouse strain were mixed and purified using the RNeasy Mini Kit (Qiagen). Oligonucleotide microarray analysis was performed using Panorama Micro Array gene expression chips, each containing ∼22 000 probe sets (Sigma-Aldrich) according to the manufacturer's instructions. The array data were analysed using GeneSpring GX 11.0 (Agilent Technologies). Downloaded log base 2 data were converted to linear values and uploaded to GeneSpring GX 11.0. The arrays and genes were normalized to the median values of the chips and genes. GenMAPP and MAPPfinder were then used to generate and validate molecular signalling pathways significantly enriched in the gene expression profiles (Dahlquist et al, 2002; Doniger et al, 2003). The z-score, a standardized difference score, was calculated using the expected value and the standard deviation of the number of genes meeting the criterion on a MAPP, taking into account the MAPP sizes. The criteria used for the selected MAPP pathways were as follows: Z-score ⩾0 and Permute P⩽0.05. The microarray data were deposited in GEO under the accession number GSE34327.
RNA isolation and quantitative real-time PCR
Total RNA was extracted with Trizol (Invitrogen) and the RNeasy Mini Kit (Qiagen) and then reverse-transcribed using ReverTraAce reverse transcriptase (Toyobo). cDNAs were used for quantitative real-time PCR, which was performed using the SYBER Green PCR Master Mix (Toyobo). All procedures were performed according to the manufacturers' instructions (Invitrogen, Toyobo). Gapdh expression served as a control for mRNA expression. Primer sequences are available upon request.
In situ hybridization
RNA in situ hybridization was performed according to the method of Sugitani et al (2002). Briefly, probes corresponding to the full-length mouse Id1–Id4 sequences were amplified by RT–PCR and subcloned into pBlueScript II SK (−) vector (Stratagene), followed by synthesis of anti-sense riboprobes using T7 polymerase (Stratagene). Sections were hybridized overnight at 58°C with 400 ng/ml of digoxigenin–dNTP-labelled probes (Roche Diagnostics) incubated with an alkaline phosphatase-conjugated anti-digoxigenin antibody (1:500 dilution) and developed with the BCPI/NTB substrate (Roche Diagnostics). Sections were mounted with CC/Mount (Diagnostic Bio Systems) and Entellan New (Merck) mounting media, and then observed with an Axiophoto2/Axio Cam microscope and camera (Carl Zeiss).
Luciferase assay
Cos7 cells were transfected with a luciferase reporter gene fused to a fragment of Id1 silencer (nucleotides +1189–1400; Id1-luc), Id2 silencer (nucleotides −2576 to −2966; Id2-luc), Id3 silencer (nucleotides −3318 to −3028; Id3-luc) and Id4 silencer (nucleotides +23 271–23 551; Id4-luc) using Lipofectamine Plus reagent (Invitrogen), as described by the manufacturer. An effector gene construct (pCDNA 3.1 (+) inserted with Rp58 or empty vector) and a plasmid containing a sea pansy luciferase expression construct (pRL-CMV; Promega) were co-transfected with the reporter constructs described above for normalization. After 24 h, cells were solubilized and luciferase activity was measured as recommended for the Pikkagene dual luciferase assay system (Toyo Ink). Lumit LB 9501 (Berthold) was used for quantification.
Chromatin immunoprecipitation assay
Cortex from E16.5 WT mice was digested enzymatically with 10 U/ml papain-EDTA for 30 min at 32°C and then washed three times with PBS. According to the manufacturer's instructions (Cosmo Bio; http://www.cosmobio.co.jp/support/technology/chip_assay_--/index.asp), the cells were fixed in 1% PFA for 10 min at room temperature. Cells were sonicated in SDS lysis buffer on ice using an ultrasonic processor (XL; Astrason) for five cycles of 30 s on, 1 min off, and the generation of genomic DNA fragments <1 kbp was confirmed. Immunoprecipitation with 10 μg rabbit anti-RP58 antibody and control rabbit anti-IgG antibodies was carried out overnight at 4°C. About 5% of the immunoprecipitated DNA was used for RT–PCR. Primer sequences used for ChIP assays are available upon request.
Lentivirus generation and infection
The Rp58 full-length cDNA (Ohtaka-Maruyama et al, 2007) was subcloned into the pCL36–C1L–CMp–IRES–GFP lentivirus expression vector (kind gift of Dr Hanawa; Hanawa et al, 2009). ShId expression vectors were purchased from Thermo (see Plasmid construction). HEK 293T cells were transfected with vector containing the insert or the empty vector along with Packaging Mix (pCAG-kGP4.1R, pCAG4-RTR2 and pCAGGS-VSV-G vectors) (kind gift of Dr Hanawa) for 40 h (with medium replaced after 6 h). Virus particles pelleted by ultra-centrifugation (5800 g, Beckman SW 28 rotor, 16 h, 4 °C). Viruses were then suspended in Hanks Balanced Salt Solution (HBSS) and stored at −80°C until use. For infection into primary or neurosphere cultures, 1 × 104 TU/ml virus was added. Samples were collected 4 days after infection.
In utero electroporation
In utero electroporation was performed as previously described (Tabata and Nakajima, 2001). Briefly, expression vectors were electroporated into the telencephalic vesicle. Vector concentration was 3 μg/μl in HBSS with 0.01% Fast green as a tracer and mixed in a 1:3 ratio when co-electroporated with pCAG–GFP. Pregnant mice at E14.5 or E15.5 were anaesthetized with 25 ml/kg pentobarbital. A square electroporator (CUY21SC, Nepa Gene) was used to deliver four cycles of 33 V for 50 ms at 95 ms intervals per embryo.
Colony formation assay
In all, 2.5 × 104 cells were suspended in 4 ml of 0.3% top agar containing DMEM plus 10% FBS and poured onto 3 ml of bottom agar (0.5% agar in DMEM containing 10% FBS) in a six-well plate. After 14 days incubation at 37°C, colonies with >0.05 mm diameter were scored. The cultures were plated in triplicate.
Statistical tests
Statistical analysis was performed using the standard t-test (two-sample assuming equal variance).
Supplementary Material
Acknowledgments
We thank Dr Hideki Hanawa (Heisei Tateishi Hospital) for critical suggestions regarding the preparation of recombinant lentivirus and providing materials for lentivirus preparation. We acknowledge St Jude Children's Research Hospital (Dr Arthur Nienhuis) and the George Washington University for accept to use the plasmid for lentivirus production. We thank Dr Kentaro Mori and Dr Yoshifumi Yokota (Fukui University) for kindly providing the anti-Id2 antibody. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, the Strategic Research Program for Brain Sciences by the Ministry of Education and a Grant-in-Aid for Scientific Research on Innovative Areas ‘Neural Diversity and Neocortical Organization’ from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Author contributions: SH, YH, and HO designed the study. SH, MK, SO, and HO wrote the manuscript. SH performed the experiments and data analysis. AM generated the viruses used in the study. COM designed the in utero electroporation analysis and prepared the samples for microarray analysis. HO performed the immunostaining analysis. COM, MK, SO, YH, and HO critically read the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aoki K, Meng G, Suzuki K, Takashi T, Kameoka Y, Nakahara K, Ishida R, Kasai M (1998) RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J Biol Chem 273: 26698–26704 [DOI] [PubMed] [Google Scholar]
- Baek SH, Kioussi C, Briata P, Wang D, Nguyen HD, Ohgi KA, Glass CK, Wynshaw-Boris A, Rose DW, Rosenfeld MG (2003) Regulated subset of G1 growth-control genes in response to derepression by the Wnt pathway. Proc Natl Acad Sci USA 100: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Walker R, Kondo T, van Crüchten I, King ER, Sablitzky F (2005) Id4 is required for the correct timing of neural differentiation. Dev Biol 280: 386–395 [DOI] [PubMed] [Google Scholar]
- Berse M, Bounphengb M, Huanga X, Christyc B, Pollmanna C, Dubie W (2004) Ubiquitin-dependent degradation of Id1 and Id3 is mediated by the COP9 signalosome. J Mol Biol 343: 361–370 [DOI] [PubMed] [Google Scholar]
- Besson A, Dowdy SF, Roberts JM (2008) CDK inhibitors: cell cycle regulators and beyond. Dev Cell 14: 159–169 [DOI] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P (2002) Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol 12: 432–438 [DOI] [PubMed] [Google Scholar]
- Cai L, Morrow EM, Cepko CL (2000) Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development 127: 3021–3030 [DOI] [PubMed] [Google Scholar]
- Cánepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, Ogara MF (2007) INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life 59: 419–426 [DOI] [PubMed] [Google Scholar]
- Canzoniere D, Farioli-Vecchioli S, Conti F, Ciotti MT, Tata AM, Augusti-Tocco G, Mattei E, Lakshmana MK, Krizhanovsky V, Reeves SA, Giovannoni R, Castano F, Servadio A, Ben-Arie N, Tirone F (2004) Dual control of neurogenesis by PC3 through cell cycle inhibition and induction of Math1. J Neurosci 24: 3355–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarapica R, Annibalid D, Raimondi L, Savino M, Nasi S, Rota R (2009) Targeting Id protein interactions by an engineered HLH domain induces human neuroblastoma cell differentiation. Oncogene 28: 1881–1891 [DOI] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR (2002) GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet 31: 19–20 [DOI] [PubMed] [Google Scholar]
- Dehay C, Kennedy H (2007) Cell-cycle control and cortical development. Nature Rev Neuro 8: 438–450 [DOI] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR (2003) MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol 4: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL (2000) p57 (Kip2) regulates progenitor cell proliferation and amacrine interneuron development in the mouse retina. Development 127: 3593–3605 [DOI] [PubMed] [Google Scholar]
- Ess KC (2010) Tuberous sclerosis complex: a brave new world? Curr Opin Neurol 23: 189–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V (2009) Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci 12: 1248–1256 [DOI] [PubMed] [Google Scholar]
- Friocourt G, Kanatani S, Tabata H, Yozu M, Takahashi T, Antypa M, Raguénès O, Chelly J, Férec C, Nakajima K, Parnavelas JG (2008) Cell-autonomous roles of ARX in cell proliferation and neuronal migration during corticogenesis. J Neurosci 28: 5794–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckiger AC, Marcy G, Marchand M, Négre D, Cosset FL, Mitalipov S, Wolf D, Savatier P, Dehay C (2006) Cell cycle features of primate embryonic stem cells. Stem Cells 24: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S, Debs RJ, Desprez PY (2004) Id genes and proteins as promising targets in cancer therapy. Trends Mol Med 10: 387–392 [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T (2001) Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J 20: 2536–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui H, Li S, Matise MP (2007) A cell-autonomous requirement for Cip/Kip cyclin-kinase inhibitors in regulating neuronal cell cycle exit but not differentiation in the developing spinal cord. Dev Biol 301: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F (2007) Cell fate specification in the mammalian telencephalon. Prog Neurobiol 83: 37–52 [DOI] [PubMed] [Google Scholar]
- Hanawa H, Yamamoto M, Zhao H, Shimada T, Persons DA (2009) Optimized lentiviral vector design improves titer and transgene expression of vectors containing the chicken beta-globin locus HS4 insulator element. Mol Ther 17: 667–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA (1994) The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev 8: 1270–1284 [DOI] [PubMed] [Google Scholar]
- Jen Y, Manova K, Benezra R (1996) Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Dev Dyn 207: 235–252 [DOI] [PubMed] [Google Scholar]
- Jen Y, Manova K, Benezra R (1997) Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn 208: 92–106 [DOI] [PubMed] [Google Scholar]
- Jeon HM, Jin X, Lee JS (2008) Inhibitor of differentiation 4 drives brain tumor-initiating cell genesis through cyclin E and notch signaling. Genes Dev 22: 2028–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Hermanson O (2010) Molecular control of brain size: regulators of neural stem cell life, death and beyond. Exp Cell Res 316: 1415–1421 [DOI] [PubMed] [Google Scholar]
- Jung S, Park RH, Kim S, Jeon YJ, Ham DS, Jung MY, Kim SS, Lee YD, Park CH, Suh-Kim H (2010) Id proteins facilitate self-renewal and proliferation of neural stem cells. Stem Cells Dev 19: 831–841 [DOI] [PubMed] [Google Scholar]
- Katsimpardi L, Gaitanou M, Malnou CE, Lledo PM, Charneau P, Matsas R, Thomaidou D (2008) BM88/Cend1 expression levels are critical for proliferation and differentiation of subventricular zone-derived neural precursor cells. Stem Cells 26: 1796–1807 [DOI] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG (2002) Identification of a Wnt/Dvl/β-Catenin → Pitx2 pathway mediating cell-type specific proliferation during development. Cell 111: 673–685 [DOI] [PubMed] [Google Scholar]
- Kuzontkoski PM, Mulligan-Kehoe MJ, Harris BT, Israel MA (2010) Inhibitor of DNA binding-4 promotes angiogenesis and growth of glioblastoma multiforme by elevating matrix GLA levels. Oncogene 29: 3793–3802 [DOI] [PubMed] [Google Scholar]
- Langlands K, Yin X, Anand G, Prochownik EV (1997) Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J Biol Chem 272: 19785–19793 [DOI] [PubMed] [Google Scholar]
- Lasorella A, Stegmüller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A (2006) Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature 442: 471–474 [DOI] [PubMed] [Google Scholar]
- Lasorella A, Uo T, Iavarone A (2001) Id proteins at the cross-road of development and cancer. Oncogene 20: 8326–8333 [DOI] [PubMed] [Google Scholar]
- Lukaszewicz A, Savatier P, Cortay V, Giroud P, Huissoud C, Berland M, Kennedy H, Dehay C (2005) G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron 47: 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Savatier P, Cortay V, Kennedy H, Dehay C (2002) Contrasting effects of basic fibroblast growth factor and neurotrophin 3 on cell cycle kinetics of mouse cortical stem cells. J Neurosci 22: 6610–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R (1999) Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401: 670–677 [DOI] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS (2007) Timing is everything: making neurons versus glia in the developing cortex. Neuron 54: 357–369 [DOI] [PubMed] [Google Scholar]
- Nagata Y, Todokoro K (1994) Activation of helix-loop-helix proteins Id1, Id2 and Id3 during neural differentiation. Biochem Biophys Res Commun 199: 1355–1362 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takizawa T, Ochiai W, Yanagisawa M, Hisatsune T, Nakafuku M, Miyazono K, Kishimoto T, Kageyama R, Taga T (2001) BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc Natl Acad Sci USA 98: 5868–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolioni V, Moavero R, Curatolo P (2009) Recent advances in neurobiology of tuberous sclerosis complex. Brain Dev 31: 104113. [DOI] [PubMed] [Google Scholar]
- Neganova I, Lako M (2008) G1 to S phase cell cycle transition in somatic and embryonic stem cells. J Anat 213: 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, Philpott A, Roberts JM, Guillemot F (2006) p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev 20: 1511–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaka-Maruyama C, Miwa A, Kawano H, Kasai M, Okado H (2007) Spatial and temporal expression of RP58, a novel zinc finger transcriptional repressor, in mouse brain. J Comp Neurol 502: 1098–1108 [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067–1070 [DOI] [PubMed] [Google Scholar]
- Okado H, Ohtaka-Maruyama C, Sugitani Y, Fukuda Y, Ishida R, Hirai S, Miwa A, Takahashi A, Aoki K, Mochida K, Suzuki O, Honda T, Nakajima K, Ogawa M, Terashima T, Matsuda J, Kawano H, Kasai M (2009) The transcriptional repressor RP58 is crucial for cell-division patterning and neuronal survival in the developing cortex. Dev Biol 331: 140–151 [DOI] [PubMed] [Google Scholar]
- Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, Wickramasinghe R, Scott MP, Wechsler-Reya RJ (2003) Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci USA 100: 7331–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca A, Gallo P, De Luca P, Lania L (2000) Class A helix-loop-helix proteins are positive regulators of several cyclin-dependent kinase inhibitors' promoter activity and negatively affect cell growth. Cancer Res 60: 1376–1382 [PubMed] [Google Scholar]
- Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y (1999) Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol 19: 5969–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passiatore G, Gentilella A, Rom S, Pacifici M, Bergonzini V, Peruzzi F (2011) Induction of Id-1 by FGF-2 involves activity of EGR-1 and sensitizes neuroblastoma cells to cell death. J Cell Physiol 7: 1763–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateras IS, Apostolopoulou K, Niforou K, Kotsinas A, Gorgoulis VG (2009) p57KIP2: “Kip”ing the cell under control. Mol Cancer Res 27: 1902–1919 [DOI] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ (2010) Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA 107: 15957–15962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R (2005) Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer 5: 603–614 [DOI] [PubMed] [Google Scholar]
- Pilaz LJ, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, Kennedy H, Dehay C (2009) Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci USA 106: 21924–21929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Dehay C, Moraillon B, Kennedy H (1997) Regulation of neuroblast cell-cycle kinetics plays a crucial role in the generation of unique features of neocortical areas. J Neurosci 17: 7763–7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S, Ignatova A, Park ST, Sun XH (1997) Regulation of expression of the cyclin dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol 17: 5888–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S (2000) Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28: 69–80 [DOI] [PubMed] [Google Scholar]
- Raponi E, Agenes F, Delphin C, Assard N, Baudier J, Legraverend C, Deloulme JC (2007) S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia 55: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild G, Zhao X, Iavarone A, Lasorella A (2006) E proteins and Id2 converge on p57Kip2 to regulate cell cycle in neural cells. Mol Cell Biol 26: 4351–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Peraud A, Mainprize T, Nakayama J, Tsugu A, Hongo K, Kobayashi S, Rutka JT (2004) Inducible expression of p57KIP2 inhibits glioma cell motility and invasion. J Neurooncol 68: 217–223 [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S (2006) The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci 9: 743–751 [DOI] [PubMed] [Google Scholar]
- Sosunov AA, Wu X, Weiner HL, Mikell CB, Goodman RR, Crino PD, McKhann GM II (2008) Tuberous sclerosis: a primary pathology of astrocytes? Epilepsia 49: 53–62 [DOI] [PubMed] [Google Scholar]
- Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T (2002) Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev 16: 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D (1991) Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 11: 5603–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K (2001) Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience 103: 865–872 [DOI] [PubMed] [Google Scholar]
- Takahashi A, Hirai S, Ohtaka-Maruyama C, Miwa A, Hata Y, Okabe S, Okado H (2008) Co-localization of a novel transcriptional repressor simiRP58 with RP58. Biochem Biophys Res Commun 368: 637–642 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS Jr (1995) The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci 15: 6046–6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatard VM, Xiang C, Biegel JA, Dahmane N (2010) ZNF238 is expressed in postmitotic brain cells and inhibits brain tumor growth. Cancer Res 70: 1236–1246 [DOI] [PubMed] [Google Scholar]
- Trabosh VA, Divito KA, Aguda BD, Simbulan-Rosenthal CM, Rosenthal DS (2009) Sequestration of E12/E47 and suppression of p27KIP1 play a role in Id2-induced proliferation and tumorigenesis. Carcinogenesis 30: 1252–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugu A, Sakai K, Dirks PB, Jung S, Weksberg R, Fei YL, Mondal S, Ivanchuk S, Ackerley C, Hamel PA, Rutka JT (2000) Expression of p57(KIP2) potently blocks the growth of human astrocytomas and induces cell senescence. Am J Pathol 157: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tury A, Mairet-Coello G, Dicicco-Bloom E (2011) The cyclin-dependent kinase inhibitor p57Kip2 regulates cell cycle exit, differentiation, and migration of embryonic cerebral cortical precursors. Cereb Cortex 21: 1840–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SF (2003) Inhibitors of DNA binding in neural cell proliferation and differentiation. Neurochem Res 28: 45–52 [DOI] [PubMed] [Google Scholar]
- Tzeng SF, de Vellis J (1998) Id1, Id2, and Id3 gene expression in neural cells during development. Glia 24: 372–381 [DOI] [PubMed] [Google Scholar]
- Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y (1997) High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol 17: 7317–7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates PR, Atherton GT, Deed RW, Norton JD, Sharrocks AD (1999) Id helix-loop-helix proteins inhibit nucleoprotein complex formation by the TCF ETS-domain transcription factors. EMBO J 18: 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P (1999) Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397: 702–706 [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Ito Y, Ueno-Kudoh H, Shimizu H, Uchibe K, Albini S, Mitsuoka K, Miyaki S, Kiso M, Nagai A, Hikata T, Osada T, Fukuda N, Yamashita S, Harada D, Mezzano V, Kasai M, Puri PL, Hayashizaki Y, Okado H et al. (2009) A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor RP58. Dev Cell 17: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Mantani A, Garel S, Rubenstein J, Israel MA (2004) Id4 regulates neural progenitor proliferation and differentiation in vivo. Development 131: 5441–5448 [DOI] [PubMed] [Google Scholar]
- Zeng W, Rushing EJ, Hartmann DP, Azumi N (2010) Increased inhibitor of differentiation 4 (Id4) expression in glioblastoma: a tissue microarray study. J Cancer 1: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Wang H, Xue L, Zhang Z, Tong T (2004) Regulation of cellular senescence and p16INK4a expression by Id1 and E47 proteins in human diploId fibroblast. J Biol Chem 279: 31524–31532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.