Abstract
Mutations in BRCA2 confer an increased risk of cancer development, at least in part because the BRCA2 protein is required for the maintenance of genomic integrity. Here, we use proteomic profiling to identify APRIN (PDS5B), a cohesion-associated protein, as a BRCA2-associated protein. After exposure of cells to hydroxyurea or aphidicolin, APRIN and other cohesin components associate with BRCA2 in early S-phase. We demonstrate that APRIN expression is required for the normal response to DNA-damaging agents, the nuclear localisation of RAD51 and BRCA2 and efficient homologous recombination. The clinical significance of these findings is indicated by the observation that the BRCA2/APRIN interaction is compromised by BRCA2 missense variants of previously unknown significance and that APRIN expression levels are associated with histological grade in breast cancer and the outcome of breast cancer patients treated with DNA-damaging chemotherapy.
Keywords: BRCA2, breast cancer, DNA repair
Introduction
Inheritance of a single mutated allele of BRCA2 strongly predisposes to breast cancer and a number of other malignancies (Wooster and Weber, 2003). BRCA2 encodes a very large protein (3418 amino acids (aa)) that is likely to have multiple cellular functions but is particularly important for the repair of DNA double-strand breaks (DSBs) by the process of homologous recombination (HR). HR is a sequence-conservative process that involves the identification of a homologous DNA sequence to that present at the DSB, and the utilisation of this sequence as a template for repair. As part of this process, BRCA2 sequesters the DNA recombinase RAD51, mobilises it to the site of damage and then facilitates the formation of helical RAD51-single-stranded (ss) DNA nucleoprotein filaments either side of the DSB. These nucleoprotein filaments invade double-stranded (ds) DNA, usually the sister chromatid, that has homology to the site of DNA damage. Following strand invasion, DNA synthesis is instigated using the homologous sequence as a template. This ultimately leads to the restoration of the original sequence at the damaged site. However, in the absence of functional BRCA2, cells use alternative, more error-prone forms of DNA repair, with the inevitable consequence that the genome becomes peppered with chromosomal rearrangements and breaks. This genetic instability is thought to foster the development of malignancy (Gudmundsdottir and Ashworth, 2006).
In addition to RAD51, BRCA2 has also been shown to interact with a number of other proteins that control HR including PALB2 (Xia et al, 2006), FANCG (Hussain et al, 2003), FANCD2 (Hussain et al, 2004), BRCA1 (Chen et al, 1998) and DSS1 (Marston et al, 1999). In a similar fashion to BRCA2 deficiency, mutations in BRCA2-binding proteins can also result in compromised HR efficiency and sensitisation to DNA damage. Notably, biallelic mutations in the BRCA2-interacting proteins PALB2, FANCD2 and FANCG (and also biallelic mutations in BRCA2 itself) cause Fanconi anaemia (FA), a disease characterised by cellular sensitivity to DNA cross-linking agents (Moldovan and D’Andrea, 2009). The interplay between other FA susceptibility genes and BRCA2 is currently unclear, although it has been demonstrated that FANCD2 and BRCA2 associate in response to damage and co-localise at stalled replication forks (Hussain et al, 2004).
Orthologues of BRCA2 have also been identified in lower organisms. Bioinformatic analyses identified a candidate BRCA2 orthologue in Drosophila melanogaster, CG30169/dmBrca2, (Lo et al, 2003) and subsequent analysis suggests that dmBrca2 shares many of the characteristics of its human orthologue despite being a much smaller and simpler protein in structure (Brough et al, 2008). The mechanism of DSB repair and the essential components of this process are also well conserved between mammals and Drosophila (Rong and Golic, 2003). Here, we exploited dmBRCA2 to identify additional BRCA2-interacting proteins and in doing so identify APRIN as a novel determinant of RAD51 localisation, HR and the clinical response to chemotherapy.
Results
Identification and validation of APRIN as a BRCA2-interacting protein
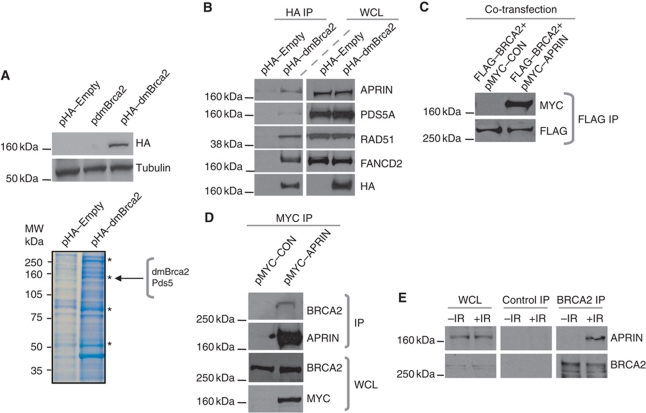
We reasoned that a rapid approach to identifying novel BRCA2-interacting proteins was to exploit the ease by which the relatively small dmBrca2 protein could be manipulated. To identify novel interactions, we expressed haemaglutanin (HA)-epitope-tagged dmBrca2 in Drosophila embryonic Kc cells and identified dmBrca2-interacting proteins by using anti-HA immunoprecipitation (IP) from total cell lysates, followed by gel electrophoresis and mass spectrometric (MS) analysis (Figure 1A). As expected, this approach identified three peptides with 100% identity to regions of the dmBrca2 protein (Supplementary Figure S1A). In addition, we also identified 11 peptide sequences with 100% identity to fragments of the Drosophila protein Pds5 (CG17509) (Supplementary Figure S1B and C), the likely orthologue of the yeast Pds5 protein (Celniker et al, 2002) and the human proteins PDS5A and APRIN/PDS5B (Hartman et al, 2000; Losada et al, 2005). Sequence alignment analysis (using clustalw software, http://www.ebi.ac.uk/Tools/msa/clustalw2/) indicated that PDS5A and PDS5B were not only very similar to each other (65% amino acid sequence identity) but also similar to the Drosophila Pds5 orthologue (35% amino acid sequence identity between PDS5A and Pds5; Supplementary Figure S1D) (Chenna et al, 2003), suggesting that there may be functional conservation between the different PDS5 species. Yeast Pds5 is a cohesin-associated protein that is involved in the maintenance of sister chromatid cohesion (Peters et al, 2008), but the role of the mammalian orthologue in this process is less clear (Losada et al, 2005; Zhang et al, 2007).
Figure 1.
BRCA2 interacts with APRIN. (A) (Top panel) Western blot analysis of whole cell lysates (WCL) from Drosophila Kc cells transfected with pHA–Empty, pdmBrca2 or pHA–dmBrca2 expression constructs. Immunoblots were probed with an HA-epitope tag-specific antibody or a β-tubulin antibody as shown, indicating expression of the HA–dmBrca2 fusion protein. (Bottom panel) Coomassie-stained polyacrylamide gel containing anti-HA immunoprecipitated material from Drosophila Kc cells transfected with either pHA–Empty or pHA–dmBrca2 expression constructs. Position of dmBrca2 and Pds5-containing bands removed for Q-TOF mass spectrometry analysis are shown. Asterisks indicate additional bands excised for mass spectrometry analysis. MW, molecular weight. (B) Western blot analysis of anti-HA immunoprecipitates or WCL from human 293T cells transiently expressing either pHA–Empty or pHA–dmBrca2 constructs. Blots were probed with anti-human APRIN, PDS5A, RAD51, FANCD2 and HA antibodies as shown. (C) Western blot analysis of anti-FLAG immunoprecipitates from human 293T cells transiently expressing FLAG-epitope-tagged human BRCA2 as well as MYC-epitope-tagged human APRIN or a control (CON) construct. Blots were probed with anti-FLAG or anti-MYC antibodies as shown, suggesting a human BRCA2/APRIN interaction. (D) Western blot analysis of anti-MYC immunoprecipitates or WCL from human 293T cells transiently expressing MYC-epitope-tagged APRIN or a control (CON) construct. Blots were probed with anti-human BRCA2, anti-human APRIN or anti-MYC antibodies as shown, suggesting a human BRCA2/APRIN interaction. (E) Western blot analysis of anti-human BRCA2 immunoprecipitates or WCL from untransfected human 293T cells exposed to 10 Gy of IR. Lysates were collected 2 h following treatment and blots were probed with anti-human BRCA2 or anti-human APRIN antibodies as shown, suggesting an endogenous human BRCA2/APRIN interaction following damage. Figure source data can be found in Supplementary data.
To extend the observations made using MS, we used IP and immunoblot analysis. In human 293T cells, we observed interactions between HA-epitope-tagged dmBRCA2 and endogenous human APRIN, PDS5A, RAD51 and FANCD2 (Figure 1B). The interaction between dmBrca2 and human APRIN was far stronger than the dmBRCA2–human PDS5A interaction. In addition, APRIN dysfunction has also been linked to cancer (Chen et al, 2007) and therefore our subsequent studies focused upon the APRIN/BRCA2 interaction. We went on to observe interactions between epitope-tagged human BRCA2 and epitope-tagged human APRIN (Figure 1C) and between epitope-tagged human APRIN and endogenous human BRCA2 (Figure 1D). Demonstrating an interaction between endogenous human APRIN and endogenous human BRCA2 initially proved problematic but we noted that this interaction was enhanced could be observed following exposure of cells to ionising radiation (IR) (Figure 1E), suggesting that this interaction could play a role in the response to such a cellular insult.
APRIN binds the BRC1 repeat region of BRCA2
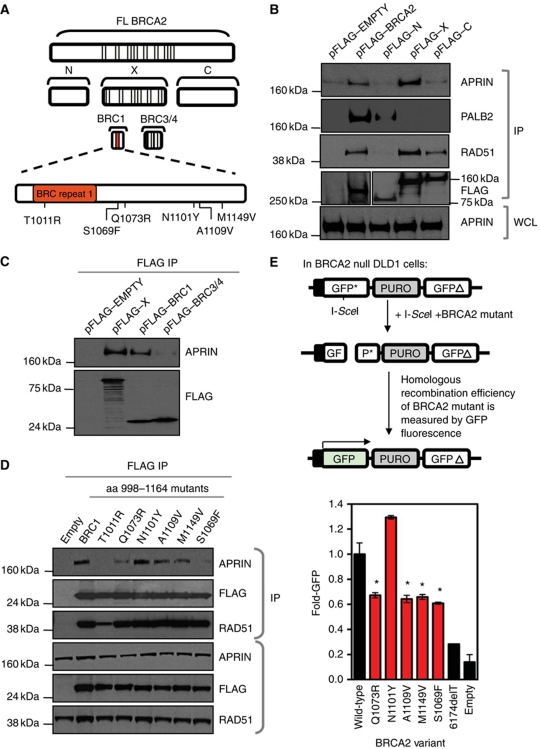
To gain insight into the nature of the APRIN–BRCA2 interaction, we expressed fragments of the BRCA2 protein and defined the domains of BRCA2 that interact with APRIN (Figure 2A). Using 293T cells transiently expressing FLAG-tagged fragments of BRCA2, we demonstrated that a BRCA2 fragment encompassing aa 786–1909 (fragment ‘X’ in Figure 2A) co-immunoprecipitated with APRIN, whereas BRCA2 fragments representing N-terminal (aa 1–627) or C-terminal (aa 2126–3418) fragments did not (Figure 2B). The APRIN-binding fragment of BRCA2 (aa 786–1909) contains BRC repeats 1–6 (Figure 2A). Using additional BRCA2 fragments encompassing repeats BRC1 or BRC3/4 (Figure 2A), we demonstrated that a 166-aa fragment (998–1164) including BRC1 bound APRIN, whereas the BRC3/4 fragment did not (Figure 2C). In addition to these co-IP experiments, we validated the interaction of BRCA2 998–1164 with APRIN using a mammalian two-hybrid system. This analysis also confirmed a lack of association between APRIN and the N- and C-portions of BRCA2 (Supplementary Figure S2A).
Figure 2.
APRIN interacts with the BRCA2 BRC1 repeat. (A) Schematic diagram of FLAG-tagged fragments of human BRCA2 used in the experiments described in (B–E). FL BRCA2=aa 1–3418; fragment N=aa 1–627; fragment X=aa 786–1909; fragment C=aa 2126–3418; fragment BRC1=aa 998–1164; fragment BRC3/4=aa 1386–1560. In addition, the position of six missense mutations within the BRC1 BRCA2 fragment (aa 998–1164) that were investigated in (D, E) are shown. Not to scale. (B) Western blot analysis of whole cell lysates (WCL) and anti-FLAG immunoprecipitates from 293T cells transiently expressing BRCA2 fragments from pFLAG–EMPTY, pFLAG–BRCA2 (FL), pFLAG–N, pFLAG–X and pFLAG–C constructs. Blots were probed with anti-APRIN and the control antibodies anti-PALB2, anti-RAD51 and anti-FLAG, as shown. This demonstrates an interaction between APRIN and fragment X of BRCA2 (aa 786–1909), as well as FL BRCA2. Confirmation of this association was also seen using a mammalian two-hybrid method (Supplementary Figure S2A). Validation of appropriate fragment behaviour was controlled by detection of PALB2 binding to the N fragment of BRCA2 only and RAD51 binding to both the X- and C-terminus. (C) Western blot analysis of anti-FLAG immunoprecipitates from 293T cells transiently expressing pFLAG–EMPTY, pFLAG–X, pFLAG–BRC1 or pFLAG–BRC3/4. Blots were probed with anti-APRIN or anti-FLAG antibodies as shown, demonstrating an interaction between APRIN and the BRC1 fragment of BRCA2 (aa 998–1164). (D) Western blot analysis of anti-FLAG immunoprecipitates and WCL from 293T cells transiently expressing six of the FLAG-tagged mutated fragments of BRCA2 (aa 998–1164) assessed in Supplementary Figure S2B and illustrated in (A). Blots were probed with anti-APRIN, anti-FLAG or anti-RAD51 antibodies as shown. A number of BRCA2 variants abrogate the BRCA2/APRIN interaction. (E) BRCA2 species containing the indicated missense variants were expressed in BRCA2−/− DLD1 cells along with DR-GFP and ISCE1 reporter constructs. After 48 h, GFP-positive cells were quantified by flow cytometry. Error bars represent standard errors of the mean. *P<0.05 (Student's t test) compared to wild type.
To assess the potential significance of these data, we examined the Breast Cancer Information Core (BIC) database to identify BRCA2 sequence polymorphisms that mapped to the 166 aa APRIN-binding fragment. The BIC database (http://research.nhgri.nih.gov/bic/) encompasses germline BRCA2 sequence information obtained principally from breast cancer patients but also from women potentially at an increased risk of the disease. Within the aa 998–1164 region, 37 missense BRCA2 variants with unknown functional significance have been identified. We assessed the effect of 15 variants on the BRCA2/APRIN interaction. To do this, we used site-directed mutagenesis to introduce each mutation into constructs expressing the 998–1164 aa BRCA2 fragment and used these in a mammalian two-hybrid analysis, where each variant BRCA2 fragment represented the ‘bait’ and a full-length (FL) APRIN cDNA expression construct encoded the ‘prey’. This analysis demonstrated that a number of the BRCA2 variants exhibited a compromised interaction with APRIN (Supplementary Figure S2B). To validate these observations, we selected the six BRCA2 variants that demonstrated the most reduced association with APRIN for further validation (T1011R, Q1073R, N1101Y, A1011V. M1149V and S1069F; Figure 2A). We performed immunoblot analysis on material that co-immunoprecipitated with each of the six FLAG-epitope-tagged BRCA2 variant fragments in human cells. This validation experiment (Figure 2D) indicated that while all six variants were expressed, two BRCA2 variants, T1011R and S1069F, almost completely ablated the BRCA2/APRIN interaction, while Q1073R and M1149V had less significant effects. Since the DNA recombinase, RAD51, also binds to the BRC repeat sequences, we assessed whether these same variants modulated the interaction between BRCA2 and RAD51. As expected, we observed decreased RAD51-binding capacity in the T1011R variant located within the BRC1 repeat but did not observe a similar reduction in RAD51 binding for any of the other BRCA2 variants that we had shown to modulate APRIN binding (Figure 2D). Taken together, these observations suggested that the S1069F, Q1073, A1109V and M1149V BRCA2 variants altered the BRCA2–APRIN interaction independently of the BRC1–RAD51 interaction.
To examine these effects further, we introduced five of the BRCA2 variants into a BRCA2 cDNA sequence (PIR1) that encodes a minimal BRCA2 protein isoform that is capable of mediating HR and which serves as a useful tool for manipulating BRCA2 for functional studies (Edwards et al, 2008) (Supplementary Figure S2C). We introduced each of the BRCA2 variants into PIR1 cDNA expression constructs and transfected these into BRCA2-deficient DLD1 cells (Hucl et al, 2008) harbouring a synthetic HR reporter substrate DR-GFP (Green Fluorescent Protein) that contains an inducible DSB (Pierce et al, 1999) (Supplementary Figure S2D). We then estimated the ability of BRCA2 variants to restore HR in BRCA2-deficient cells by measuring GFP fluorescence generated by the HR-repaired DR-GFP construct. As expected, the non-manipulated PIR1 cDNA construct restored HR in BRCA2 null DLD1 cells, matching previous observations (Edwards et al, 2008). Contrastingly, BRCA2 cDNAs with either Q1073R, A1109V, M1149V or S1069F variants did not rescue HR in DR-GFP (Figure 2E). We did note, however, that there was a lack of a direct correlation between the level of interaction impairment (Figure 2D) and the impairment of HR (Figure 2E). For example, the BRCA2 variants S1069F, A1109V and M1149V impaired the BRCA2–APRIN interaction to varying levels but had similar effects on HR efficiency. However, it is possible that a biological threshold effect occurs in this case, such that both complete ablation or a modest impairment of the BRCA2–APRIN interaction have similar effects on HR efficiency. As an additional control, the BRCA2 variant N1101Y, that did not abrogate the BRCA2–APRIN interaction (Figure 2D), did restore HR in BRCA2 null cells (Figure 2E). Taken together, these results suggested that BRCA2 missense variants present in the BIC database that modulate the BRCA2–APRIN interaction also impair HR.
APRIN is required for efficient DNA repair
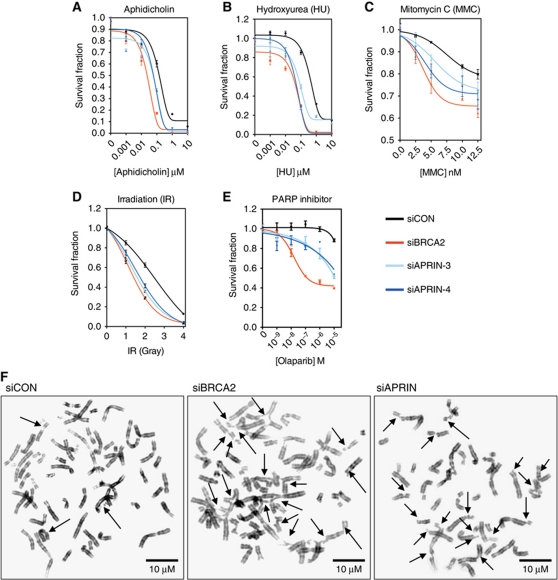
Given the DNA damage-dependent interaction of APRIN with BRCA2 (Figure 1E), we assessed whether APRIN played a non-redundant role in the cellular sensitivity to agents that cause DNA damage. APRIN silencing by RNA interference (Supplementary Figure S3A) sensitised human kidney 293T cells to the polymerase α inhibitor, aphidicholin (Figure 3A), the DNA synthesis inhibitor, hydroxyurea (HU; Figure 3B), and the interstrand cross-linking reagent, mitomycin C (MMC; Figure 3C) as measured by short-term viability assays. Using Hela cells, which are more adherent than 293T cells, we also used colony formation assays, to demonstrate that APRIN silencing sensitised cells to IR (Figure 3D) as well as the PARP inhibitor, olaparib (Figure 3E). Similar results for IR and MMC sensitivity were also seen in breast epithelial cancer T47D cells and non-tumour-derived breast epithelial MCF10A cells (Supplementary Figure S3B, C and Supplementary Table SI). In addition, APRIN silencing caused genomic instability in response to MMC treatment, as demonstrated by an increased prevalence of chromosomal aberrations (Figure 3F and summarised in Table I). These DNA-damaging reagents cause the formation of DNA lesions such as interstrand cross-links that inhibit replication forks and cause fork collapse and DNA DSBs.
Figure 3.
APRIN silencing increases genomic instability and heightens sensitivity to DNA-damaging agents. (A–C) Survival curves of 293T cells transfected with siRNA targeting APRIN and then treated with either (A) aphidicholin or (B) HU or (C) MMC for 5 days. (D, E) Survival curves generated from colony formation assays on adherent Hela cells transfected with siRNA targeting APRIN then exposed to either (D) IR or (E) PARP inhibitor. For (A–E), two different siRNA species (siAPRIN-3 and siAPRIN-4) were used in separate experiments. Non-targeting control siRNA (siCON) and siRNA targeting BRCA2 were used as negative and positive controls, respectively. Error bars represent standard errors of the mean from three replicate experiments. (F) Metaphase chromosome images generated from 293T cells treated with control, BRCA2 or APRIN siRNA after MMC exposure. The prevalence of specific chromosomal aberrations between the different siRNA treatments was compared using a two-tailed heteroscedastic t-test (n=50) and are summarised in Table I. Scale bar represents 10 μM.
Table 1. Unrepaired chromosomal aberrations in cells transfected with siRNA against BRCA2 and APRIN and treated with MMC.
| siCONa | siRNA BRCA2a | siRNA APRINa | P (CON versus BRCA2)b | P (CON versus APRIN)b | P (BRCA2 versus APRIN)b | |
|---|---|---|---|---|---|---|
| Chromatid gaps and breaks | 1 (0–12) | 4 (0–33) | 3 (0–17) | 0.000174 | 0.000759 | 0.414088 |
| Chromosomal gaps and breaks; acentric fragments | 0 (0–3) | 0 (0–7) | 0 (0–6) | 0.19571 | 0.272194 | 0.815242 |
| Chromatid or chromosome gaps and breaks; acentric fragments | 1.5 (0–12) | 4.5 (0–33) | 4 (0–17) | 0.000108 | 0.000383 | 0.4027 |
| Triradial | 0 (0–2) | 0 (0–3) | 0 (0–2) | 0.13393 | 0.244443 | 0.593049 |
| Quadriradial | 0 (0–1) | 0 (0–2) | 0 (0–6) | 0.00274 | 0.094461 | 0.897594 |
| Di or tricentric | 0 (0–0) | 0 (0–1) | 0 (0–1) | 0.322223 | 0.159386 | 0.562501 |
| Complex chromosomal aberrations | 0 (0–2) | 0 (0–6) | 0 (0–6) | 0.292147 | 0.269018 | 1 |
| Total | 2 (0–14) | 5.5 (0–39) | 4 (0–18) | 0.000116 | 0.000267 | 0.501974 |
| aMedian number of unrepaired aberrations (range). | ||||||
| bHeteroscedastic two-tailed t-test. | ||||||
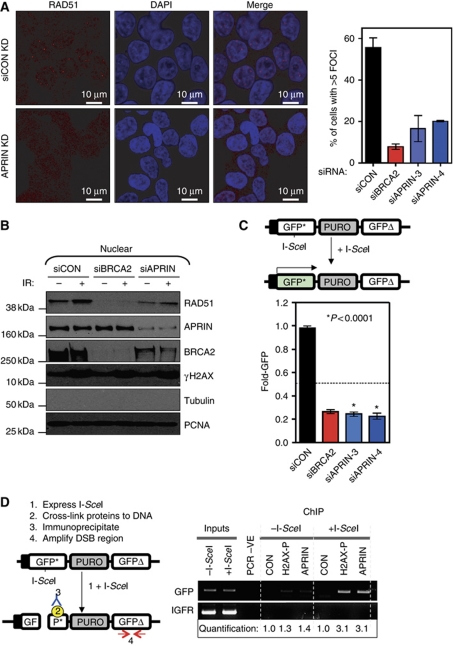
Previously, it has been demonstrated that BRCA2 forms S-phase nuclear foci that co-localise with other repair molecules following DNA damage (Wong et al, 1997). Using a MYC-tagged APRIN expression construct, we found that APRIN also forms nuclear foci that occasionally co-localise with RAD51 (Supplementary Figure S4A) and BRCA2 (Supplementary Figure S4B). Notably the APRIN–RAD51 and APRIN–BRCA2 co-localisation events were less frequent than BRCA2–RAD51 co-localised foci and this could be due to APRIN being localised elsewhere in the cell. Nevertheless, we were confident that the APRIN immunostaining was specific as the frequency of APRIN–BRCA2 foci was reduced following transfection of either APRIN or BRCA2 siRNA (data not shown).
The formation of nuclear foci involving RAD51 and BRCA2 most likely represents one of the critical events in the faithful repair of DNA damage by HR (Ammazzalorso et al, 2010), namely the BRCA2-coordinated loading of RAD51 onto DNA (Forget and Kowalczykowski, 2010). Silencing of APRIN caused a significant reduction in IR-induced RAD51 (Figure 4A) and BRCA2 (Supplementary Figure S4C and D) nuclear foci, supporting the hypothesis that APRIN modulates HR. Consistent with these observations, we also noticed that APRIN silencing led to reduced nuclear RAD51 and BRCA2 localisation after DNA damage, as estimated by western (Figure 4B; Supplementary Figure S4E). As RAD51 and BRCA2 foci primarily occur in S phase, we also assessed whether the reduction in RAD51/BRCA2 foci caused by APRIN silencing could be secondary to a change in the cell cycle. APRIN silencing did not obviously alter the cell cycle profile of cells (Supplementary Figure S4F), suggesting that limiting the extent of S phase was an unlikely explanation for the reduction in nuclear foci.
Figure 4.
APRIN is involved in DNA repair. (A) (Left panel) Representative confocal microscopy images from 293T cells transfected with non-targeting control siRNA or siRNA targeting APRIN. Forty-eight hours after transfection, cells were exposed to 10 Gy IR and then fixed and stained using a nuclear dye (DAPI, blue in image) and an anti-RAD51 antibody. Nuclear IR-induced RAD51 foci are shown in red. The scale bar represents 10 μM. (Right panel) Graphical representation of this data is shown. Cells were treated with control (siCON), BRCA2 or APRIN siRNA (siBRCA2 and siAPRIN, respectively), as shown. The proportion of cells containing five or more nuclear RAD51 foci after IR was estimated from three independent experiments (n>100 for each experiment). Error bars for each individual experiment represent standard errors of the mean. (B) Western blot analysis of nuclear proteins from 293T cells treated as in (A). Cells were transfected and irradiated as in (A). Following cytosolic/nuclear extraction, the nuclear fraction was analysed by western blot using antibodies detecting RAD51, APRIN, BRCA2, γH2AX, β-tubulin and PCNA. PCNA (nuclear specific) and β-tubulin (cytosolic) expression confirmed successful fractionation. See Supplementary Figure S3D for analysis of the cytosolic fraction. (C) (Top panel) Schematic of the pDR-GFP HR assay. The DR-GFP recombination substrate encompasses (from left to right); an hCMV enhancer/chicken β-actin promoter (black box); a modified GFP* (a GFP gene containing an integrated I-SceI endonuclease restriction site that leads to a premature stop codon); a puromycin drug selectable marker (PURO); and a second modified GFP-coding sequence (GFPΔ), which harbours 5′ and 3′ truncations. Neither GFP* nor GFPΔ are functional GFP ORFs. Expression of I-SceI in cells carrying a DR-GFP reporter induces a DSB in GFP*. Repair of this DSB by HR/gene conversion uses the GFPΔ gene as a template, and results in the removal of the termination codons from GFP*, reconstitution of a functional GFP ORF and GFP-mediated fluorescence. Non-conservative forms of repair do not reconstitute the GFP ORF and do not lead to GFP expression. (Bottom panel) Twenty-four hours following transfection with the indicated siRNA, Hela cells harbouring a single-copy genomic integration of the DR-GFP reporter, with or without I-SceI. GFP expression was estimated by FACS analysis and is represented in the graph. Error bars represent standard errors of the mean from three separate experiments. (D) (Left panel) Schematic of the ChIP–PCR assay used, which is based upon the HR reporter construct described in (C). 293T cells harbouring a genomic HR reporter were transfected with an I-SceI expression construct that caused a DSB within the GFP* gene. Twenty-four hours after transfection, chromatin-bound proteins were cross-linked to DNA and cells lysed. After sonication of the genomic DNA, IP was performed using an anti-APRIN antibody. To ascertain the presence of DNA flanking, the DSB site in immunoprecipitates, PCR amplifying a GFP sequence was used. Positive control (anti-γH2AX) and negative control (anti-FLAG) IPs were used to validate the assay. A region of the IGFR gene was also amplified to demonstrate specificity of γH2AX and APRIN to DSB regions rather than genomic DNA as a whole. (Right panel) 293T cells harbouring the pDR-GFP reporter construct were transfected with the I-SceI expression construct and 24 h later, ChIP–PCR was performed as described above. In the PCR-negative control, DNA was substituted with water in the PCR reaction. This analysis indicated that after transfection of the I-SceI expression construct, the amount of DSB-flanking DNA that co-immunoprecipitated with γH2AX was elevated, compared with levels in cells not expressing I-SceI, thus validating the assay. In addition, the amount of DSB-flanking DNA that co-immunoprecipitated with APRIN when cells were transfected with the I-SceI expression construct was also elevated, suggesting that APRIN is present at or near the site of DSBs. Quantification of the PCR bands are shown, normalised to the control ChIP without I-SceI treatment.
To directly measure the effect of APRIN deficiency upon HR, we assayed HR-mediated DNA repair using Hela cells harbouring a stable single-copy integration of the DR-GFP reporter system (Pierce et al, 1999; Slabicki et al, 2010). Silencing of APRIN caused a reduction in HR repair similar in scale to that caused by silencing of BRCA2 (Figure 4C). Since APRIN appeared to modulate HR capacity, we investigated whether APRIN was localised at the site of a DSB. To assess this, we designed a chromatin IP (ChIP) assay based upon the HR reporter described in Figure 4C and similar to that previously described by Potts and colleagues (Potts et al, 2006). Cells harbouring a genomic DR-GFP reporter were transfected with an I-SceI expression construct that caused a DSB within the reporter construct. Twenty-four hours after transfection, chromatin-bound proteins were cross-linked to DNA and then the cells were lysed. After sonication of the genomic DNA, we performed IP using antibodies specific for either APRIN or a phosphorylated form of the histone H2AX (γH2AX) as a control. H2AX around the site of DSBs is phosphorylated as part of the normal DNA damage response (Bonner et al, 2008). To assess whether either of these two proteins were associated with the reporter DNA bearing a DSB, we purified DNA from immunoprecipitates and performed PCR using primers specific for regions flanking the DSB caused by I-SceI expression. After transfection of the I-SceI expression construct, the amount of DR-GFP DNA that co-immunoprecipitated with γH2AX was elevated when compared to levels in cells not expressing I-SceI, thus validating the assay (Figure 4D). We also observed that the amount of DR-GFP DNA that co-immunoprecipitated with APRIN when cells were transfected with the I-SceI expression construct was also elevated (Figure 4D), suggesting that APRIN is present at or near the site of DSBs. Taken together with the effects of APRIN silencing upon RAD51 foci formation, DNA damage sensitivity and HR capacity, the localisation of APRIN at the site of a DSB implicated APRIN as a critical HR/DSB repair factor.
Further dissection of the BRCA2/APRIN interaction
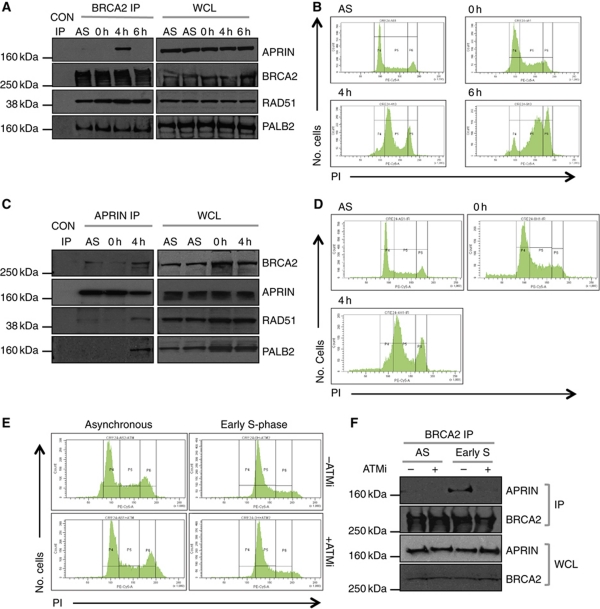
To further characterise the nature of the BRCA2/APRIN interaction, we assessed its temporal modulation in response to agents that stall replication forks, cause fork collapse/DSB formation and induce HR. In the first instance we used aphidicolin, a replicative DNA polymerase inhibitor (Rothkamm et al, 2003; Van et al, 2010). We exposed 293T cells to aphidicolin for 18 h and then analysed the BRCA2/APRIN interaction following release from aphidicolin blockade. Immunoprecipitation of endogenous BRCA2, followed by western blot analysis of G1/S synchronised cells indicated that the BRCA2/APRIN interaction was greatest at 4 h after release from aphidicolin-induced arrest (Figure 5A), a time point at which the majority (68%) of cells were in the first half of S-phase (Figure 5B). The BRCA2/APRIN interaction was not readily observed in asynchronous (AS) cells, in cells predominantly at G1/S (0 h), or when the majority of cells were in late S/G2 (6 h) (Figure 5A and B). A similar pattern was also observed in the breast cell lines, T47D and MCF10A (Supplementary Figure S5A–D) and was observed whether anti-BRCA2 or anti-APRIN IPs were used (Figure 5C and D). We also observed a very similar pattern of BRCA2/APRIN interaction after release from HU exposure (Supplementary Figure S5E and F). Recent models suggest that chronic HU exposure causes replication fork collapse, followed by new replication origin firing (Petermann et al, 2010). The firing of new replication origins eventually generates a DNA template that is used in long-tract HR to repair the collapsed replication fork (Petermann et al, 2010). It is possible that the delay in BRCA2/APRIN interaction after release from HU block could represent the time taken for the firing of replication origins and the generation of new DNA prior to HR repair.
Figure 5.
The BRCA2/APRIN interaction is cell-cycle dependent. (A) Western blot analysis of BRCA2 immunoprecipitated material or whole cell lysates (WCL) from AS or synchronous 293T cultures. For synchronous cultures, cells were arrested at the G1/S checkpoint by aphidicolin treatment (18 h) and then released into the cell cycle by the removal of aphidicolin (t=0 h). Samples were taken for IP at 0, 4 and 6 h after release as shown. Western blots were probed with antibodies detecting human endogenous APRIN, BRCA2, RAD51 or PALB2 as shown. (B) PI cell-cycle FACS profiles of cells treated in (A). (C) Western blot analysis of APRIN immunoprecipitated material or WCL from AS 293T cultures or 293T cells treated with aphidicolin as in (A). Samples were taken for IP at 0 and 4 h after release as shown. Western blots were probed with antibodies detecting human endogenous BRCA2, APRIN, RAD51 or PALB2 as shown. (D) PI cell-cycle FACS profiles of cells treated in (C). (E) PI cell-cycle FACS profiles of 293T cells treated with the ATM catalytic inhibitor KU0055933. Samples were collected for analysis in early S-phase. Cell-cycle arrest and synchronisation was achieved using the aphidicolin method (see (A)). (F) Western blot analysis of BRCA2 immunoprecipitates and WCL from cells in (E). Blots were probed with anti-APRIN or BRCA2 antibodies as shown, suggesting that the APRIN/BRCA2 interaction is abrogated by inhibition of ATM.
In the light of these observations, we assessed whether other BRCA2 interactions mirrored the temporal nature of the BRCA2/APRIN interaction. We assessed BRCA2 interactions with the BRCA2 partner protein PALB2 (Xia et al, 2006) and the DNA recombinase RAD51 (Wong et al, 1997). Using IP and western blot analysis of aphidicolin-treated cells, we found that the BRCA2/PALB2 and BRCA2/RAD51 interactions were relatively consistent following release from aphidicolin blockade (Figure 5A). We also assessed whether APRIN co-immunoprecipitated with these other BRCA2-binding proteins. RAD51 and PALB2 both co-immunoprecipitated with APRIN (Figure 5C); notably, the APRIN/BRCA2, APRIN/RAD51 and APRIN/PALB2 interactions were only observable 4 h after release into S-phase (Figure 5C and D).
When taken together with the observations that APRIN silencing modulates the response to various DNA-damaging agents (Figure 3), RAD51 foci formation (Figure 4A) and the repair of a synthetic HR substrate (Figure 4C), the increase in the BRCA2/APRIN interaction following chronic aphidicolin or HU exposure is consistent with the hypothesis that the BRCA2/APRIN interaction is associated with the repair of DSBs by HR following replication fork collapse. The HR process is, in part, orchestrated by the protein kinase ATM (Zhou et al, 2000; Bakkenist and Kastan, 2004), which phosphorylates a series of protein substrates in response to damage and cellular stress. In fact, APRIN has already been identified as a potential ATM/ATR target (Matsuoka et al, 2007). To investigate whether ATM signalling might be required for the BRCA2/APRIN association, we treated cells with the small molecule ATM inhibitor (ATMi), KU0055933 (Hickson et al, 2004). Inhibition of ATM abrogated the BRCA2/APRIN association in early S-phase (Figure 5E and F), suggesting that this interaction was governed by ATM catalytic activity, and implicates APRIN as part of the cellular response to DNA damage.
APRIN and BRCA2 interact with replication and cohesion factors
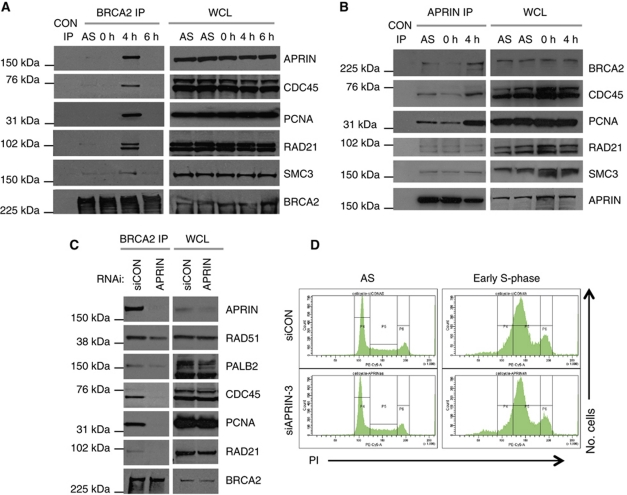
BRCA2 is thought to be associated with DSB repair at stalled replication forks (Hussain et al, 2004; Thompson and Hinz, 2009). Furthermore, the parasite Trypanosoma brucei orthologue of BRCA2 interacts with the pre-replication complex protein CDC45 (Oyola et al, 2009). We investigated whether the early S-phase APRIN/BRCA2 association coincided with binding of BRCA2 to components of the replication fork machinery. Using western blot analysis of BRCA2 immunoprecipitated proteins from human 293T cells, we first confirmed the interaction between human BRCA2 and human CDC45 (Figure 6A). As for the BRCA2/APRIN interaction, the BRCA2/CDC45 interaction was restricted to early S-phase. Similarly, an interaction between BRCA2 and the DNA replication/fork progression protein PCNA was also restricted to early S-phase (Figure 6A). We then assessed whether APRIN also interacted with these same replication proteins; APRIN/CDC45 and APRIN/PCNA interactions were also identified from APRIN immunoprecipitates, predominantly at early S-phase (Figure 6B).
Figure 6.
BRCA2 and APRIN associate with cohesion proteins and replication complex. (A) Western blot analysis of BRCA2 immunoprecipitated material or whole cell lysates (WCL) from AS or synchronous 293T cultures. For synchronous cultures, cells were arrested at the G1/S checkpoint by aphidicolin treatment (18 h) and then released into the cell cycle by the removal of aphidicolin (t=0 h). Samples were taken for IP at 0, 4 and 6 h after release as shown. Western blots were probed with antibodies detecting human endogenous APRIN, CDC45, PCNA, RAD21, SMC3 and BRCA2 as shown. (B) Western blot analysis of APRIN immunoprecipitated material or WCL from AS or synchronous 293T cultures as in (A). Samples were taken for IP at 0 and 4 h after release as shown. Western blots were probed with antibodies detecting human endogenous BRCA2, CDC45, PCNA, RAD21, SMC3 and APRIN as shown. (C) Western blot analysis of BRCA2 immunoprecipitated material or WCL from synchronised, early S-phase 293T cultures as in (A). Cells were transfected with siRNA as shown, 48 h prior to aphidicolin treatment and samples taken for analysis at 4 h after release from aphidicolin treatment. Western blots were probed with antibodies as shown. See also Supplementary Table SI. (D) PI FACS profiles of 293T cells treated with siRNA silencing APRIN. Cells were transfected with siRNA 48 h prior to aphidicolin treatment. FACS analysis was performed 4 h after release from aphidicolin treatment. Figure source data can be found in Supplementary data.
APRIN, and its lower organism orthologues, have previously been shown to interact with elements of the cohesin complex (Losada et al, 2005; Lengronne et al, 2006). This complex includes SMC3 and RAD21 and is involved in the tethering of sister chromatids after DNA synthesis (Lengronne et al, 2006). Having confirmed that APRIN associates with both SMC3 and RAD21 in 293T cells (Figure 6B), we investigated whether BRCA2 also interacted with these cohesin-related factors. We found that BRCA2 interacted with RAD21 in early S-phase. We also identified a weaker but reproducible association between BRCA2 and SMC3 (Figure 6A).
We next assessed the possibility that APRIN acts as a determinant of the ability of BRCA2 to bind other interacting partners in early S-phase. Using RNA interference to silence the expression of APRIN, we demonstrated that APRIN was required for BRCA2's interaction with replication and cohesin components (e.g., CDC45, PCNA and RAD21) (Figure 6C and D). Furthermore, although the effects were more modest, silencing of APRIN also reduced the binding of RAD51 and PALB2 to BRCA2 (Figure 6C; Supplementary Table SII), in part corroborating the reduction in nuclear RAD51 localisation and foci formation observed using APRIN siRNA (Figure 4A and B). Collectively, these data suggest that APRIN sits at a nodal point in terms of the interaction between BRCA2 and elements of the cohesin, replication and repair machinery.
Clinical relevance of APRIN in breast cancer
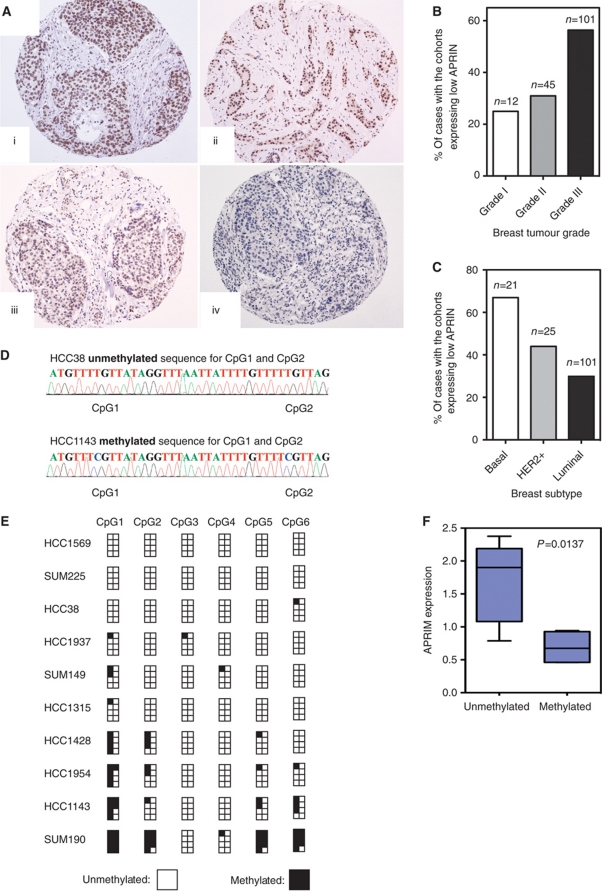
To date, a number of preliminary observations have suggested associations between APRIN expression levels and cancer (Beckmann et al, 1996; Chen et al, 1998; Edwards et al, 1998; Harada et al, 2001; Zhang et al, 2008). To investigate this in more detail, we examined the expression of APRIN in a panel of 160 invasive breast tumours using an anti-APRIN antibody optimised for immunohistochemistry (IHC). Tumours were derived from patients with no obvious family history of cancer. After IHC staining, each section was scored using a semi-quantitative scoring system based upon a combination of staining intensity and the proportion of IHC-positive cells in each tumour (see Materials and methods). The median expression of APRIN in the whole cohort was defined as a score equal to 8 and we considered tumours with a score of <8 having reduced APRIN expression (Figure 7A). The frequency of IHC scores for the breast tumour panel is summarised in Supplementary Figure S6A. From this analysis, we found that 46.9% (75/160) of breast tumours had reduced APRIN expression and six of these tumour samples appeared to express no detectable APRIN. We then cross-compared APRIN expression with histological grade in the 160 breast cancer biopsies. The frequency of low APRIN expression correlated with histological grade (P=0.005). In general, the expression of APRIN tended to be lower in grade III tumours, where 56.4% (57/101) of samples exhibited low-level expression. In grade I tumours, the frequency of low-level APRIN expression was 25% (3/12) and 31% (14/45) in grade II tumours (Figure 7B). We also compared APRIN levels with expression of the oestrogen receptor (ER) in breast tumours. Low APRIN expression correlated with the ER-negative phenotype (P=0.019), and was observed in 64.7% (22/34) of ER-negative cases. This correlation concurred with the frequency of APRIN expression in breast cancer molecular subtypes as defined by a validated immunohistochemical surrogate (Nielsen et al, 2004); low APRIN expression was most frequent in the tumours with a basal-like/triple-negative phenotype (67% (14/21)), although this did not reach statistical significance (P<0.1), less frequent in HER2-amplified tumours (44% (11/25)) and least frequent in tumours with a luminal phenotype (30% (45/105)) (Figure 7C). A summary of the correlations between APRIN expression and other clinicopathological markers is summarised in Supplementary Table SIII. We also assessed the correlation between APRIN transcript levels and breast cancer subtype. Analysis of microarray data (http://www.ebi.ac.uk/microarray-as/ae/ accession number: E-TABM-543) from a separate cohort of 55 sporadic tumours confirmed that reduced APRIN expression (P=0.009) in tumours correlated with the basal-like phenotype based on a validated immunohistochemical surrogate (Nielsen et al, 2004), when compared with HER2-positive and luminal subtypes (Supplementary Figure S6B).
Figure 7.
APRIN expression correlates with response to chemotherapy. (A) Representative micrographs of APRIN expression in invasive breast cancers; (i, ii) illustrate normal levels of APRIN expression with (i) generating a score of 9 and (ii) a score of 8; (iii, iv) show reduced levels of APRIN expression with (iii) generating a score of 6 and (iv) a score of 2. (i–iv) (haematoxylin/DAB × 100). For distribution of scores see Supplementary Figure S5A. (B) Correlation between high grade (grade III) and low APRIN expression (two-tailed t-test: P<0.005). (C) Low APRIN expression also correlates with basal breast cancer phenotype (two-tailed t-test: P<0.05). (D) The chromatograms illustrate the sequence of unmethylated CpG1 and CpG2 seen in all HCC38 clones (top) and the methylated sequence of CpG1 and CpG2 seen in most HCC1143 clones (bottom). (E) A graphical representation of the proportion of methylated CpGs detected for each of the 10 basal cell lines. (F) A box and whisker diagram illustrating that detectable APRIN mRNA levels are significantly lower in the APRIN methylated lines, when compared with unmethylated lines (P=0.0137, Student's t-test) as assessed by RT–PCR.
Remarkably, APRIN is located directly adjacent to human BRCA2 on chromosome 13 (Supplementary Figure S6C). This is a region often subject to genomic loss in breast and ovarian cancers. The close proximity of APRIN to BRCA2 raises the possibility that these genes are lost together following a gross genomic deletion at 13q13. However, low expression of APRIN in both the 160 invasive and 55 sporadic tumours did not correlate with genomic loss as assessed by array CGH (data not shown and Supplementary Figure S6D (http://www.rock.icr.ac.uk/ Project_ID=118)). To investigate this observation, we compared BRCA2 mRNA levels in the panel of invasive breast tumour samples with the corresponding APRIN IHC expression scores. This analysis revealed no correlation (P=0.183) between APRIN and BRCA2 expression levels, suggesting that these two genes are perhaps independently regulated in these tumours (Supplementary Figure S6E). Since variations in APRIN expression in breast tumours did not correlate with copy number, one possible mechanism of lowered levels of APRIN could be epigenetic silencing of the APRIN promoter.
Since variations in APRIN expression in breast tumours did not correlate with APRIN copy number, we investigated whether APRIN had undergone epigenetic silencing by promoter methylation, a common mechanism of transcriptional control. While we were unable to examine APRIN methylation in the same tumour panel described above, we did assess APRIN methylation in basal-like breast cancer cell lines. MethPrimer was used to design primer pairs that spanned the two CpG islands of APRIN and methylation-specific PCRs (MSPs) were performed on 20 basal-like breast cancer cell lines. Only CpG island 1, which contains six CpGs, was identified as a methylated region in some of the cell types. The position of the primer pair used (MSPF and MSPR) is depicted in Supplementary Figure S6F and the MSPs using these primer pairs are shown in Supplementary Figure S6G. To confirm that these MSPs represent methylated CpGs, we bisulphite sequenced 10 of the basal-like cell lines. The small cluster of CpGs were amplified by PCR using primer pairs BSF and BSR (Supplementary Figure S6F) and the products were TOPO-cloned. Eight individual colonies were sequenced using BSR (Figure 7D and E) and the proportion of clones with methylation-protected CpGs were calculated and are depicted in a methylation map illustrated in Figure 7E and Supplementary Figure S6G. When quantitative RT–PCR (qRT–PCR) was performed for APRIN mRNA transcript, we noted that the methylation status correlated with mRNA expression (P=0.0137) (Figure 7F), suggesting that promoter methylation may be a mechanism for APRIN silencing in breast tumours.
Our dissection of APRIN function suggested that loss of APRIN expression could sensitise tumour cells to agents that cause DNA damage (Figure 3; Supplementary Figure S3B and C). To investigate the clinical significance of these findings, we compared APRIN expression in breast tumours with the response to anthracycline, a drug which is thought to elicit a therapeutic effect by causing DNA damage. In ER-negative tumours, low APRIN expression correlated with a statistically significant longer disease-free survival (DFS) in patients treated with adjuvant anthracycline-based chemotherapy (whole cohort P=0.126, ER-negative P=0.0027, ER-positive P=0.576; Supplementary Figure S7Ai, Bi and Ci). Similar associations with outcome were seen for low APRIN expressing ER-negative patients when examined for metastasis-free survival (MFS) (Supplementary Figure S7Aii, Bii and Cii) and breast cancer specific survival (BCSS) (Supplementary Figure S7Aiii, Biii and Ciii). A multivariate survival Cox Hazard model analysis demonstrated that reduced APRIN expression was independently associated with outcome in this cohort of patients treated with adjuvant anthracycline-based chemotherapy (Supplementary Table SIV). Cumulatively, these results suggest that low APRIN expression correlates with outcome in patients following chemotherapy in particular subtypes of breast cancer. Since the intercalating agent, anthracycline generates DNA damage, this correlation is possibly a consequence of the role of APRIN in HR.
Discussion
We have identified APRIN as a protein that interacts with the breast and ovarian cancer tumour suppressor BRCA2. APRIN has been previously characterised as a cohesin-associated protein and has been shown to be dysregulated in cancer (Beckmann et al, 1996; Edwards et al, 1998; Harada et al, 2001; Zhang et al, 2008). We show here that APRIN modulates HR, is present at DSBs and influences the response to DNA-damaging agents. Importantly, analysis of the BRCA2/APRIN interaction suggests that BRCA2 variants with unassigned functional significance modulate the BRCA2/APRIN interaction. APRIN and BRCA2 likely sit within a network of proteins with known functions in DNA replication, repair and cohesion. APRIN appears to play a major role in regulating the interaction between BRCA2 and proteins such as CDC45, PCNA and RAD21 as well as having an influence on the association between BRCA2 and the key regulators of HR, RAD51 and PALB2. Finally, the clinical significance of our observations is underscored by the association of APRIN expression levels with pathological grade in breast cancer and clinical response to DNA-damaging chemotherapy.
Our data provide the first evidence of a direct linkage between elements of the cohesin complex and the tumour suppressor BRCA2. Previous reports have implicated elements of the cohesin complex and associated proteins in DNA repair. For example, the absence of a fully functional cohesin complex is essential for DSB repair after DNA replication in budding yeast (Sjogren and Nasmyth, 2001) and in human cells elements of the cohesin complex are recruited to DSBs and are essential for competent HR (Potts et al, 2006). A possible model is that cohesin enables HR by holding sister chromatids together when DSBs occur. This localisation of sister chromatids should allow strand invasion and DNA exchange between sister chromatids to take place, thus enabling the repair process (Sjogren and Strom, 2010). However, it should be noted that this model of DNA damage-induced cohesion is primarily based upon studies in budding yeast, although the formation of a DSB has been shown to increase sister chromatid proximity in chicken DT40 cells (Dodson and Morrison, 2009). Nevertheless, our findings support a model whereby cohesin-related proteins such as APRIN are crucial to DNA repair processes that involve sister chromatids, such as HR. Given the pathogenic implications of dysregulated HR, it also seems reasonable that some level of co-ordination and localisation between damage-induced sister chromatid cohesion and the HR machinery could exist. It is possible that the interactions between BRCA2, APRIN and other elements of the replication and cohesion machinery may underlie this link. Indeed, we have shown that APRIN is required for the interaction of BRCA2 with a number of components of the HR and cohesin complexes (Figure 6C), perhaps suggesting that APRIN serves as the interface between the ostensibly distinct HR and cohesion activities.
One of the remarkable features of the APRIN gene is its close proximity to BRCA2 on chromosome 13. Loss of heterozygosity (LOH) at 13q is more frequently found in higher-grade ovarian tumours rather than in lower-grade ovarian cancers (Dodson et al, 1993). LOH distal to BRCA2 is common and this is a region that also contains the RB1 locus. However, silencing of the RB1 gene has only been seen in a small proportion of ovarian tumours, supporting the hypothesis that an additional tumour suppressor gene exists, distal to BRCA2 (Liu et al, 1994). In fact, a strong association between genomic loss 250 kb distal to BRCA2 and reduced survival in breast cancer patients has been observed (Hansen et al, 2002). APRIN is positioned 180–380 kb distal to BRCA2 and LOH at the intragenic APRIN microsatellite marker, D13S171 (Geck et al, 2001), has linked this protein to cancers of the prostate (Edwards et al, 1998) and breast (Beckmann et al, 1996). More recently, LOH analysis demonstrated allelic loss in 72% of oesophageal squamous cell carcinomas, which significantly correlated with higher pathological grade (Harada et al, 2001; Zhang et al, 2008). Similarly, reduced expression of APRIN in primary breast tumour samples has also been reported (Chen et al, 2007). APRIN is also known as the androgen-induced proliferative shutoff protein, AS3, and has been associated with the risk of prostate cancer (Geck et al, 1997).
We demonstrate here that a significant proportion of breast tumours express low APRIN levels as assessed by IHC. We also demonstrate methylation at a number of CpGs located within the APRIN promoter in breast cancer cell lines, which suggests a potential mechanism for the reduced APRIN expression detected in the tumour samples. The presence of low APRIN expression statistically correlated with high histological grade and ER-negative disease. Importantly, examination of APRIN expression in tumours from patients subsequently treated with a DNA-damaging chemotherapy uncovered a correlation between low tumour APRIN expression and a favourable response to therapy. Given the demonstration that APRIN can modulate HR and APRIN is a BRCA2-binding protein, we hypothesise that this involvement with the DNA repair apparatus explains the clinical correlate. However, we cannot exclude the possibility that other functions of APRIN, such as in steroid hormone receptor signalling (Geck et al, 2000), also contribute to the overall response to therapy. Analysis of various tumour subtypes detected an increased survival correlation for low APRIN expression in both basal and HER2-amplified cancers after treatment with anthracycline-based chemotherapy (data not shown). Given that the cohort analysed in this study was retrospectively accrued, that all patients received the adjuvant systemic therapy and that an association with survival was observed in ER-negative diseases only, the associations with outcome reported here should be considered as hypothesis generating rather than definitive. However, based on our results, further studies testing APRIN as a potential biomarker of response to DNA-damaging agents in ER-negative breast cancers are warranted.
In summary, we have shown that APRIN plays a key role in DNA repair and genomic integrity and this, in turn, may lead to low APRIN expressing tumours, demonstrating increased response to DNA-damaging chemotherapy.
Materials and methods
Cell culture, transfections and drug treatments
Drosophila Kc embryonic cell lines (Echalier and Ohanessian, 1969; Yanagawa et al, 1998) were cultured at 25°C in Schneider's Drosophila medium (Invitrogen), supplemented with 10% fetal bovine serum (FBS) (PAA) and 1% penicillin–streptomycin. Cells were plated 24 h prior to transfection of plasmid DNA with the Effectene transfection reagent (Qiagen). Human 293T, Hela and T47D cells were cultured and maintained at 37°C (5% CO2) in Dulbecco's modified Eagle's medium (Sigma, Poole, UK) supplemented with FBS (PAA), 1% L-glutamine and penicillin–streptomycin (Gibco). MCF10A cells were grown in DMEM/F12 media supplemented with 5% horse serum, EGF (20 μg/ml), hydrocortisone (0.5 μg/ml), CholeraToxin (0.1 μg/ml) and insulin (10 μg/ml). BRCA2−/− DLD1 cells (Hucl et al, 2008) were grown in McCoys media supplemented with FBS (PAA), 1% L-glutamine and penicillin–streptomycin (Gibco). Lipofectamine 2000 was used for transfection of 293T cells with plasmid DNA by following the manufacture's instructions. RNAi transfection was carried out using RNAiMAX (Invitrogen) in six-well format following the manufacturer's instructions. APRIN, BRCA2, RAD21 and control siRNA was purchased from Dharmacon and their catalogue numbers are summarised in Supplementary Table SV. Treatment with aphidicolin (Sigma; 2 μg/ml) and HU (Sigma; 3 mM) was performed for 18 h followed by replacement with fresh media or collection for particular assay. ATMi (KU0055933) was used at a concentration of 1 μM. Damage sensitivity assays (Irradiation and MMC (Sigma)) were performed 48 h post-RNAi treatment in 96-well format and at the stated doses. Survival was assessed 5 days following treatment using cell-titre glow reagent.
Plasmids
The HA–dmBRCA2 cassette was cloned from the Drosophila expression vector, pAC5.1-HA–dmBrca2 (Brough et al, 2008), into pcDNA3.1(+) to generate pHA–dmBrca2. Human BRCA2 expression constructs, pFLAG–BRCA2, pFLAG–N, pFLAG–X, pFLAG–C, pFLAG–IV, pFLAG–BRC1 and pFLAG–BRC3/4 have previously been described (Marston et al, 1999; Edwards et al, 2008). The open reading frames (ORF) from these constructs were also fused to a DNA-binding domain by cloning them into the matchmaker mammalian two-hybrid vector, pM (Clontech). The APRIN ORF was amplified by PCR (using primers APRINF/R; see Supplementary Table SVI) and TOPO-clone into pCR4-TOPO (Invitrogen). Once sequence verified, the ORF was then fused to a MYC-epitope tag by cloning into pCMV-MYC (Clontech) to generate pMYC–APRIN. The APRIN ORF was also fused to the activation domain of pVP16 (Clontech) from the matchmaker mammalian two-hybrid system to generate pVP16-APRIN. The control mammalian two-hybrid construct pVP16-DSS1 has previously been described (Marston et al, 1999). Site-directed mutagenesis (Stratagene) was used to generate the BRC1 and PIR1 (Edwards et al, 2008) missense mutations identified on the BIC website (http://research.nhgri.nih.gov/bic/) using the oligonucleotides shown in Supplementary Table SVI.
IP and western blot analysis
Whole cell protein lysates were extracted from Kc, 293T, T47D and MCF10A cells by lysis with NP250 buffer (20 mM Tris pH 7.6, 1 mM EDTA, 0.5%. NP40, 250 mM NaCl); to generate 1–5 mg for interaction studies or 20 mg for MS analysis. Following a 20-min incubation at 4°C, lysates were sonicated to ensure that the nuclear proteins were solubilised. IPs were performed by incubating protein G beads (Sigma) bound by the anti-BRCA2 (2 μg (ab-1) Calbiochem) or anti-APRIN (2 μg, Novus-100-755) antibody or anti-HA, -MYC or -FLAG-conjugated agarose (Sigma), with precleared lysate at 4°C overnight. Beads were then washed 10 times in 1 ml cold NP420 buffer (Tris pH 7.6, 1 mM EDTA, 0.5% NP40, 420 mM NaCl) using Poly Prep chromatography columns (Bio-Rad). Elution of immunoprecipitated proteins from the beads was performed by passing 750 μl of elution buffer (0.2 M Glycine, 500 mM NaCl, 1 mM EDTA) through the bead-slurry containing column. VivaSpin-500 columns (Sigma) were used to increase concentration of eluted protein prior to loading onto gel. Western blots were performed using Novex precast TA or Bis-Tris gels (Invitrogen) as described previously (Farmer et al, 2005). Primary antibodies were immunoblotted overnight and those used include anti-HA (1/1000; Roche-11867423001), anti-MYC (1/1000; abcam-9106), anti-FLAG (1/1000; Sigma), anti-APRIN (1/1000; Novus-100-755), anti-BRCA2 (1/200; Calbiochem-OP95), anti-RAD51 (1/200; Santa Cruz-8349), anti-PALB2 (1/500; Bethyl Labs-A301-247A), anti-actin (1/2000; Santa Cruz-1616) and anti-α-tubulin (1/2000; Sigma). Anti-rabbit and anti-mouse secondary antibodies were incubated with the blot (1:5000) for 1 h at room temperature followed by exposure using the enhanced chemiluminescent detection kit (Amersham Pharmacia Biotech). Cytoplasmic/nuclear fractionation of cellular protein was performed using a proteome extraction kit according to the manufacturer's instructions (Calbiochem).
MS analysis
Protein separated on a Novex precast Bis-Tris gels were stained with coomassie blue by following the manufacturer's instructions (Invitrogen). Visible protein bands were excised using sterile conditions and the proteins contained within these bands were digested with trypsin. Tryptic peptides were then identified by MS analysis using a Q-TOF II tandem mass spectrometer (Micromass, UK), coupled to a CapLC liquid chromatography system (Waters, USA) carried out at the MRC Proteomics Department (Hammersmith Hospital, London). Following MS analysis, the sequences of tryptic peptides were analysed using Mascot software (Matrix Science).
Immunofluorescence
Immunofluorescence analysis was performed as described previously (Farmer et al, 2005). Briefly, 24 h following RNAi/plasmid transfection (if appropriate) 250 000 293T cells were seeded onto polylysene coverslips in a six well. After a further 24 h, the relevant plates were X-irradiated with 10 grays (Gy) and fixed with 4% PFA after 4 h. Cells were permeabilised with 0.2% triton for 30 min followed by a 3-h treatment with DNase (Roche) at 37°C. The coverslips were incubated with the primary antibody anti-RAD51 (1/1000; Santa Cruz-8349), MYC (1/1000; Santa Cruz-56633), BRCA2 (1/200; Santa Cruz-28235) or γH2AX (1/1000; Millipore-05-636) overnight at 4°C. After washing, the primary antibodies were visualised with Alexa Flour-555 and the nuclei with DAPI (Molecular Probes). Foci were captured using a Leica TCS-SP2 confocal microscope.
HR assay
Forty-eight hours following siRNA/plasmid transfection, 293T and BRCA2−/− DLD1 (Hucl et al, 2008) cells were transfected with pDR-GFP and either the I-SceI-expressing vector, pcBASce or an empty vector control. The number of GFP-positive cells was analysed 72 h after transfection by counting 50 000 cells by flow cytometry.
Fluorescence-activated cell analysis
Following various treatments (RNAi or aphidicolin), 293T and Hela cells were fixed in 70% ice-cold ethanol (in PBS) and stained with 4% propidium iodide (PI) and 10% RNase A in PBS for cell-cycle analysis. The sample readout was performed on the FACS LSR II (Becton Dickinson, USA), and the data were analysed using CellQuest Pro (Becton Dickinson, USA). For GFP fluorescence, cells were trypsinised and resuspended in 4°C PBS prior to FACS analysis.
ChIP assay
293T cells stably harbouring pDR-GFP were transfected with the I-SceI expression construct, pcBASce or an empty vector control. After 48 h, the cells were lysed and a ChIP assay was performed using the ChIP Assay Kit according to the manufacturer's instructions (17-295, Upstate, UK). Antibodies used for IP were anti-HA (Roche), anti-γH2A.X (Cell Signaling) and anti-APRIN (Novus-100-755). PCR primers were designed to amplify the GFP region (using GFPF/GFPR; see Supplementary Table SV). Control PCR primers flanked an amplicon of similar size in the IGFR gene.
Metaphase spreads
293T siCON, siBRCA2 and siAPRIN cells were treated with 0.04 μg/ml MMC (Sigma-Aldrich) for 48 h before fixation. KaryoMAX Colcemid 0.05 μg/ml (GibcoBRL) was added to the cultures at 4 h prior to fixation. Metaphase spreads were prepared as described previously (Gudmundsdottir and Ashworth, 2004). Chromosomes were stained with DAPI and images were captured with CytoVision 3.6 software (Applied Imaging) on an Axioplan 2 microscope (Zeiss). A total of 50 non-overlapping metaphase spreads were analysed for chromosome-unrepaired breakage. Chromosome aberrations from each metaphase spread were counted blind to hide the identity of the cells analysed. The prevalence of specific chromosomal aberrations between the cells transfected with BRCA2 siRNA, APRIN siRNA and control (siCON) was compared using two-tailed heteroscedastic t-test. P-values <0.05 were considered significant.
Invasive breast cancer TMA
In all, 245 invasive breast carcinomas from patients diagnosed and managed at the Royal Marsden Hospital (RMH) were arrayed in a TMA constructed with replicate 0.6 mm cores. In total, this collection encompassed 27 invasive lobular carcinomas, 25 invasive mixed carcinomas, 185 invasive ductal carcinomas and eight invasive breast carcinomas of other special types. All patients were treated with surgery (69 mastectomy and 156 wide local excision) followed by anthracycline-based chemotherapy. In addition, endocrine therapy (adjuvant tamoxifen) was also prescribed for patients with ER-positive tumours. In each case, tumour histology was reviewed by two qualified pathologists and graded according to a modified Bloom–Richardson scoring system (Elston and Ellis, 1991). Tumour size was categorised according to the TNM staging criteria (Singletary and Connolly, 2006). The study was approved by the RMH Ethics Committee.
In all, 3 μm sections of the TMA were mounted on polylysine-coated slides and then immunostained using and antibodies targeting: APRIN (see below), ER, PR, HER2, EGFR, cytokeratin (Ck)5/6, Ck14, Ck17, KI67, p53, topoisomerase II α, CAV1 and CAV2, E-cadherin and FoxA1 (Arriola et al, 2007; Mahler-Araujo et al, 2008; Tan et al, 2008; Thorat et al, 2008). Positive and negative controls (omission of the primary antibody and IgG-matched serum) were included in every immunohistochemical staining experiment. In addition to antibody staining, TMA sections were also subjected to chromogenic in situ hybridisation (CISH) using SpotLight probes for CCND1, MYC, HER2, TOP2A and the centromere of chromosome 8, as described elsewhere (Arriola et al, 2007; Mahler-Araujo et al, 2008; Tan et al, 2008; Thorat et al, 2008). The immunohistochemical analysis and CISH signals scoring was performed by at least two observers who were blinded to the results of the patients’ outcome. Based on HER2, ER, Ck5/6 and EGFR expression, each tumour was classified according to the scheme described by Nielsen et al (2004), that is, HER2+ (HER2+, ER+/−, Ck5/6 and EGFR+/−), luminal (HER2–, ER+, Ck5/6 and EGFR+/−) and basal-like (ER−, HER2–, Ck5/6 and/or EGFR+). HER2 amplification was also used to define the HER2+ subgroup (Arriola et al, 2007; Tan et al, 2008).
To assess APRIN expression, we used an anti-APRIN antibody from Bethyl Laboratories (cat no. A300-537) at a 1:250 dilution. TMA slides were blocked in 1.5% H2O2 in methanol for 10 min and detection was achieved using the Dual Envision kit (K4061, Dako, Glostrup, Denmark) according to the manufacturer's recommendations. The anti-APRIN antibody was applied for 30 min at 37°C following heat-induced antigen retrieval for 2 min in EDTA buffer pH 8.0 in a pressure cooker. Signal development was performed using chromagen 3,3′-diaminobenzidine (Dako, Glostrup, Denmark). Sections were then counterstained with haematoxylin. Positive controls included formalin fixed pellets of 293T cells, which express high levels of APRIN by western blotting, and samples of normal breast, where APRIN is expressed in stromal cells and in a proportion of breast epithelial cells. In addition, we silenced APRIN expression in 293T cells using a validated siRNA and made formalin-fixed paraffin-embedded pellets, which were also subjected to immunohistochemical analysis using the above protocol. Positive and negative controls consistently displayed the expected staining patterns.
APRIN expression was semi-quantitatively analysed by three individuals blinded to the clinicopathological data and results of qRT–PCR and methylation analysis. Only nuclear expression was considered specific. For the semi-quantitative analysis, first, a proportion score was assigned, which represented the estimated proportion of positive-staining tumour cell nuclei (0, none; 1, <1%; 2, 1–10%; 3, >10–33%; 4, >33–66%; 5, >66–90%; 6, >90%). Next, an intensity score was assigned, which represented the average intensity of positive tumour cell nuclei (0, none; 1, weak, 2, intermediate and 3, strong). The proportion and intensity scores were then added to obtain a total score, which ranged from 0 to 9. Cases with scores 8 and 9 were considered normal for APRIN expression; cases with <8 scores were considered to have a reduction in APRIN expression levels. Representative tumour TMA cores from 180 out of the 245 cohort were interpretable and used in the analysis.
Statistical analysis of clinical data
The Statview version 5 statistical package was used for the statistical analysis of clinical data. Correlations between categorical variables were performed using the χ2 test and Fisher's exact test. Correlations between continuous and categorical variables were performed with the Mann–Whitney U-test. DFS was expressed as the number of months from diagnosis to the occurrence of either local recurrence in the form of ductal carcinoma in situ and/or invasive breast cancer or distant relapse. MFS was expressed as the number of months from diagnosis to the occurrence of distant relapse. BCSS was expressed as the number of months from diagnosis to the occurrence of an event (disease-related death). Cumulative survival probabilities were calculated using the Kaplan–Meier method. Differences between survival rates were tested with the log-rank test. All tests were two-tailed, with a confidence interval of 95%. Multivariate analysis was performed using the Cox multiple hazards model. Cases with missing values were excluded from this analysis. Two multivariate Cox Hazard models were analysed.
Quantitative RT–PCR
Total RNA from cell lines was extracted using Trizol (Invitrogen) according to the manufacturer's instructions. cDNA was synthesised using Omniscript Reverse Transcriptase System for RT–PCR (Qiagen) with oligo dT as per the manufacturer's instructions. Assay-on-Demand primer/probe sets were purchased from Applied Biosystems. Real-Time qPCR was performed on the 790DHT Fast Real-Time PCR System (Applied Biosystems), using GAPDH or 18S as an endogenous control. Standard curves were calculated for all reactions with serial dilutions of control cDNA to calculate reaction efficiency. Gene expression was calculated relative to expression of GAPDH endogenous control, and adjusted relative to expression in control cDNA. Samples were quantified in triplicate.
Bisulphite modification and MSP
Genomic DNA was subjected to modification with sodium bisulphite as described previously (Smith et al, 2007). MSP was performed with multiple primer pairs encompassing different regions of the CpG island located at the 5′ end of the APRIN gene (http://genome.ucsc.edu/). Primer sequences are available upon request.
Supplementary Material
Acknowledgments
We thank Dr N Siaud and Dr M Jasin for providing DR-GFP. This work was funded by Breakthrough Breast Cancer and Cancer Research UK.
Author contributions: RB and IB performed the experiments described in Figures 1, 2, 3, 4, 5 and 6 and the Supplementary Figures. RB, RV, RN and JSRF performed the tumour analyses described in Figure 7. CJL and AA conceived and supervised the project and sourced funding for the work. RB, CJL and AA drafted the manuscript. All authors read and approved the final manuscript.
Footnotes
AA and CJL are inventors named on patents describing the use of PARP inhibitors and may benefit under the ICR ‘Rewards to Investors’ Scheme.
References
- Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P (2010) ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J 29: 3156–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriola E, Rodriguez-Pinilla SM, Lambros MB, Jones RL, James M, Savage K, Smith IE, Dowsett M, Reis-Filho JS (2007) Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat 106: 181–189 [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB (2004) Phosphatases join kinases in DNA-damage response pathways. Trends Cell Biol 14: 339–341 [DOI] [PubMed] [Google Scholar]
- Beckmann MW, Picard F, An HX, van Roeyen CR, Dominik SI, Mosny DS, Schnurch HG, Bender HG, Niederacher D (1996) Clinical impact of detection of loss of heterozygosity of BRCA1 and BRCA2 markers in sporadic breast cancer. Br J Cancer 73: 1220–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y (2008) GammaH2AX and cancer. Nat Rev Cancer 8: 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough R, Wei D, Leulier S, Lord CJ, Rong YS, Ashworth A (2008) Functional analysis of Drosophila melanogaster BRCA2 in DNA repair. DNA Repair (Amst) 7: 10–19 [DOI] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, Hodgson A, George RA, Hoskins RA, Laverty T, Muzny DM, Nelson CR, Pacleb JM, Park S, Pfeiffer BD, Richards S et al. (2002) Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 3: RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R (1998) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2: 317–328 [DOI] [PubMed] [Google Scholar]
- Chen W, Salto-Tellez M, Palanisamy N, Ganesan K, Hou Q, Tan LK, Sii LH, Ito K, Tan B, Wu J, Tay A, Tan KC, Ang E, Tan BK, Tan PH, Ito Y, Tan P (2007) Targets of genome copy number reduction in primary breast cancers identified by integrative genomics. Genes Chromosomes Cancer 46: 288–301 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson MK, Hartmann LC, Cliby WA, DeLacey KA, Keeney GL, Ritland SR, Su JQ, Podratz KC, Jenkins RB (1993) Comparison of loss of heterozygosity patterns in invasive low-grade and high-grade epithelial ovarian carcinomas. Cancer Res 53: 4456–4460 [PubMed] [Google Scholar]
- Dodson H, Morrison CG (2009) Increased sister chromatid cohesion and DNA damage response factor localization at an enzyme-induced DNA double-strand break in vertebrate cells. Nucleic Acids Res 37: 6054–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echalier G, Ohanessian A (1969) [Isolation, in tissue culture, of Drosophila melanogaster cell lines]. C R Acad Sci Hebd Seances Acad Sci D 268: 1771–1773 [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A (2008) Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115 [DOI] [PubMed] [Google Scholar]
- Edwards SM, Dunsmuir WD, Gillett CE, Lakhani SR, Corbishley C, Young M, Kirby RS, Dearnaley DP, Dowe A, Ardern-Jones A, Kelly J, Spurr N, Barnes DM, Eeles RA (1998) Immunohistochemical expression of BRCA2 protein and allelic loss at the BRCA2 locus in prostate cancer. CRC/BPG UK Familial Prostate Cancer Study Collaborators. Int J Cancer 78: 1–7 [DOI] [PubMed] [Google Scholar]
- Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19: 403–410 [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Forget AL, Kowalczykowski SC (2010) Single-molecule imaging brings Rad51 nucleoprotein filaments into focus. Trends Cell Biol 20: 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geck P, Maffini MV, Szelei J, Sonnenschein C, Soto AM (2000) Androgen-induced proliferative quiescence in prostate cancer cells: the role of AS3 as its mediator. Proc Natl Acad Sci USA 97: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geck P, Sonnenschein C, Soto AM (2001) The D13S171 marker, misannotated to BRCA2, links the AS3 gene to various cancers. Am J Hum Genet 69: 461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geck P, Szelei J, Jimenez J, Lin TM, Sonnenschein C, Soto AM (1997) Expression of novel genes linked to the androgen-induced, proliferative shutoff in prostate cancer cells. J Steroid Biochem Mol Biol 63: 211–218 [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A (2004) BRCA2 in meiosis: turning over a new leaf. Trends Cell Biol 14: 401–404 [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A (2006) The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 25: 5864–5874 [DOI] [PubMed] [Google Scholar]
- Hansen LL, Jensen LL, Dimitrakakis C, Michalas S, Gilbert F, Barber HR, Overgaard J, Arzimanoglou (2002) Allelic imbalance in selected chromosomal regions in ovarian cancer. Cancer Genet Cytogenet 139: 1–8 [DOI] [PubMed] [Google Scholar]
- Harada H, Uchida N, Shimada Y, Kumimoto H, Shinoda M, Imamura M, Ishizaki K (2001) Polymorphism and allelic loss at the AS3 locus on 13q12-13 in esophageal squamous cell carcinoma. Int J Oncol 18: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V (2000) Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol 151: 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC (2004) Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64: 9152–9159 [DOI] [PubMed] [Google Scholar]
- Hucl T, Rago C, Gallmeier E, Brody JR, Gorospe M, Kern SE (2008) A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res 68: 5023–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, Ananth S, Davies A, Masson JY, Moses R, West SC, de Winter JP, Ashworth A, Jones NJ, Mathew CG (2004) Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Genet 13: 1241–1248 [DOI] [PubMed] [Google Scholar]
- Hussain S, Witt E, Huber PA, Medhurst AL, Ashworth A, Mathew CG (2003) Direct interaction of the Fanconi anaemia protein FANCG with BRCA2/FANCD1. Hum Mol Genet 12: 2503–2510 [DOI] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23: 787–799 [DOI] [PubMed] [Google Scholar]
- Liu Y, Heyman M, Wang Y, Falkmer U, Hising C, Szekely L, Einhorn S (1994) Molecular analysis of the retinoblastoma gene in primary ovarian cancer cells. Int J Cancer 58: 663–667 [DOI] [PubMed] [Google Scholar]
- Lo T, Pellegrini L, Venkitaraman AR, Blundell TL (2003) Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair (Amst) 2: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T (2005) Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 118 (Part 10): 2133–2141 [DOI] [PubMed] [Google Scholar]
- Mahler-Araujo B, Savage K, Parry S, Reis-Filho JS (2008) Reduction of E-cadherin expression is associated with non-lobular breast carcinomas of basal-like and triple negative phenotype. J Clin Pathol 61: 615–620 [DOI] [PubMed] [Google Scholar]
- Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A (1999) Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol 19: 4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD (2009) FANCD2 hurdles the DNA interstrand crosslink. Cell 139: 1222–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10: 5367–5374 [DOI] [PubMed] [Google Scholar]
- Oyola SO, Bringaud F, Melville SE (2009) A kinetoplastid BRCA2 interacts with DNA replication protein CDC45. Int J Parasitol 39: 59–69 [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J (2008) The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Porteus MH, Yu H (2006) Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J 25: 3377–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Golic KG (2003) The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M (2003) Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol 23: 5706–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singletary SE, Connolly JL (2006) Breast cancer staging: working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J Clin 56: 37–47; quiz 50--51 [DOI] [PubMed] [Google Scholar]
- Sjogren C, Nasmyth K (2001) Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol 11: 991–995 [DOI] [PubMed] [Google Scholar]
- Sjogren C, Strom L (2010) S-phase and DNA damage activated establishment of Sister chromatid cohesion-importance for DNA repair. Exp Cell Res 16: 1445–1453 [DOI] [PubMed] [Google Scholar]
- Slabicki M, Theis M, Krastev DB, Samsonov S, Mundwiller E, Junqueira M, Paszkowski-Rogacz M, Teyra J, Heninger AK, Poser I, Prieur F, Truchetto J, Confavreux C, Marelli C, Durr A, Camdessanche JP, Brice A, Shevchenko A, Pisabarro MT, Stevanin G et al. (2010) A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol 8: e1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Nicholson LJ, Syed N, Payne A, Hiller L, Garrone O, Occelli M, Gasco M, Crook T (2007) Epigenetic inactivation implies independent functions for insulin-like growth factor binding protein (IGFBP)-related protein 1 and the related IGFBPL1 in inhibiting breast cancer phenotypes. Clin Cancer Res 13: 4061–4068 [DOI] [PubMed] [Google Scholar]
- Tan DS, Marchio C, Jones RL, Savage K, Smith IE, Dowsett M, Reis-Filho JS (2008) Triple negative breast cancer: molecular profiling and prognostic impact in adjuvant anthracycline-treated patients. Breast Cancer Res Treat 111: 27–44 [DOI] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM (2009) Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat Res 668: 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorat MA, Marchio C, Morimiya A, Savage K, Nakshatri H, Reis-Filho JS, Badve S (2008) Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J Clin Pathol 61: 327–332 [DOI] [PubMed] [Google Scholar]
- Van C, Yan S, Michael WM, Waga S, Cimprich KA (2010) Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol 189: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL (1997) RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem 272: 31941–31944 [DOI] [PubMed] [Google Scholar]
- Wooster R, Weber BL (2003) Breast and ovarian cancer. N Engl J Med 348: 2339–2347 [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729 [DOI] [PubMed] [Google Scholar]
- Yanagawa S, Lee JS, Ishimoto A (1998) Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem 273: 32353–32359 [DOI] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J (2007) Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development 134: 3191–3201 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang X, Qi J, Yan C, Xu X, Han Y, Wang M (2008) Correlation of genomic and expression alterations of AS3 with esophageal squamous cell carcinoma. J Genet Genomics 35: 267–271 [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu W, Dai Q, Sun L, Tang Q (2000) [The sensitivity to apoptosis is enhanced in U937-ASPI-3K cells which stably express antisense ATM/PI-3K]. Zhonghua Xue Ye Xue Za Zhi 21: 641–643 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.